Abstract
Purpose
In Uygur populations, exfoliation syndrome (XFS) and exfoliation glaucoma (XFG) occurred at a high frequency. In this study, we evaluate the association profiles of the lysyl oxidase-like 1 (LOXL1) gene polymorphisms with XFS in the Uygur population.
Methods
Sixty-four unrelated Uygur patients with XFS and 127 Uygur control subjects were included in this study. Genotypes of the three single nucleotide polymorphisms (SNPs) of LOXL1 (rs1048661, rs2165241, and rs3825942) were analyzed by direct sequencing, and a case-control association study was performed.
Results
The three SNPs were significantly associated with XFS and XFG individually. The G allele of rs1048661 (OR [95%CI]: 1.92 [1.14–3.22]), G of rs3825942 (OR [95%CI]: 4.86 [2.02–11.68]), and T of rs2165241 (OR [95%CI]: 3.98 [2.54–6.25]) were risk alleles for the disorder. The genotypes GG for rs1048661 (OR [95%CI]: 2.13 [1.14–3.97]), GG for rs3825942 (OR [95%CI]: 5.68 [2.28–14.17]), and TT for rs2165241 (OR [95%CI]: 6.13 [2.68–14.01]) were risk genotypes for the disease. The haplotypes G-G for the SNPs rs1048661and rs3825942, G-T for the SNPs rs1048661 and rs2165241, and SNPs rs3825942 and rs2165241 were found to be significantly associated with XFS/G. The haplotypes G-G-T for the three SNPs were determined to be significantly associated with XFS/G. There were significant differences of the allelic and genotypic proportion in different gender/age patients and controls for all three SNPs. T allele of rs2165241 and G of rs3825942 were risk alleles for the disorder in both the male and female groups. G allele of rs1048661 was a risk allele for the disorder in the below 65-year-old group. T of rs2165241 was a risk allele for the disorder in both age groups. G of rs3825942 was a risk allele for the disorder in the over 65-year-old group. The genotypes also showed significant differences in the below 65-year-old group of rs1048661, both groups of rs2165241, and over 65-year-old group of rs3825942.
Conclusions
LOXL1 is a susceptibility gene of XFS/XFG in Uygur populations. The risk alleles of rs1048661, rs3825942 and rs2165241 in Uygur subjects were found to be similar to those of populations in Iceland and the United States and different from Han populations in China. The genotypic and allelic distributions of these SNPs are similar between XFS and XFG.
Introduction
Exfoliation syndrome (XFS) is an age-related, systemic, elastic microfibrillopathy affecting both intraocularand extraocular tissues [1]. The clinical features: white flake-like material on the anterior lens surface, the pupillary border, trabecular meshwork, zonula, ciliary body, and other anterior segment structures etc were initially described by Lindberg in 1917 [2]. Exfoliation glaucoma (XFG) is characterized by rapid progression, high resistance to medical therapy, and poor prognosis. XFS can not only lead to severe chronic open-angle glaucoma but also to acceleration of cataract formation, lens subluxation, angle-closure glaucoma, and severe complications at the time of cataract extraction, such as zonular dialysis, capsular rupture, and vitreous loss [2-5]. Especially in the Uygur populations in Xinjiang, because of lack of medicines and awareness of the disease, XFS/G patients go to the hospital when they have almost lost their visual acuity.
Epidemiological studies have shown that the prevalence of XFS varies greatly among ethnic groups, with a prevalence of 28% in Iceland, 20% in Finland, a little lower in Norway and Switzerland [6], and 1.6~6.3% in north America and western Europe. The highest incidence is in the Scandinavian peninsula [7], while prevalence is 0% in the Greenland Inuit population [8,9]. It can affect 10%–20% of the elderly populations, so the elderly populations and glaucoma patients have a higher incidence [10]. In Asian populations, the prevalence is 3.01%–6.28% in Indians aged over 40 years [11,12], 3.4% in Japanese aged over 50 years [13], 0.4% in Hong Kong Chinese aged over 60 years [14], 0.2% and 0.7% in Singaporean Chinese aged over 40 and over 60 years [15], 3.3% in the Han people in China [16], 5.1% in the Kashi Uygur [17], and 2.2% and 9.5% in the Kuche Uygur aged over 60 and 80 years, respectively [18].
According to studies worldwide, there is evidence showing that genetic factors may play an important role in the pathogenesis of XFS [19]. Recently, Thorleifsson et al. [20] performed a genome-wide association study and identified a strong association of XFG with three single nucleotide polymorphisms (SNPs) in the lysyl oxidase-like 1 gene (LOXL1) on 15q24.1. They identified one intronic SNP (rs2165241) and two nonsynonymous coding SNPs (rs3825942 and rs1048661) with significant disease association in Icelandic and Swedish individuals. This association was recently replicated in the midwestern United States [21], Australian populations [22], Indian populations [23], Japanese populations [24], and Chinese populations [25]. XFS has a high prevalence in Uygur populations [17,18]. Therefore it is logically important to perform a case-control study using another ethnic population like the Uygur minority in China who are different from the Han people, to uncover the pathogenesis of XFS/G in Uygur patients.
Xinjiang lies in northwest China, bordering Gansu and Qinghai provinces to the southeast and the Tibet Autonomous Region to the south, and shares a 5,000-km border with eight countries. It is the largest Chinese administrative division, with a total population of nearly 21.56 million, and it is home to 47 of China's 56 ethnic groups, such as Uygur (47.47%), Han (37.58%), and Kazak (7.3%). We recruited our Uygur case and control subjects from Kuche and Kashi, located in the south of the Tianshan Mountains, where the urban population is mainly Uygur.
Methods
Study subjects
The diagnostic criterion for XFS was the existence of exfoliation material on the anterior lens capsule or on the pupil margin in either eye with dilation of the pupils. Patients with intraocular pressure (IOP) of less than 21 mmHg and no clinical evidence of glaucomatous optic neuropathy were classified as XFS. XFG was diagnosed if the patient had the above characteristics of exfoliation syndrome and all of the following features: (1) IOP ≥22 mmHg in either eye; (2) glaucomatous changes on the optic disc, defined as cup to disc ratio >0.7 in either eye or an asymmetric cup to disc ratio of >0.2 or notching of the disc rim; and (3) characteristic glaucomatous visual field loss [26]. Patients with other causes of secondary glaucoma, such as uveitis, pigment dispersion syndrome, and iridocorneal endothelial syndrome, were excluded. The patients were recruited from Kashi and Kuche Uygur people over 45 years of age. The controls were enrolled by the following criteria: (1) no signs of XFS or XFG, (2) no glaucomatous changes on the optic disc, (3) normal visual field and intraocular pressure, (4) no family history of glaucoma, and (5) no other eye diseases except mild refractive errors. All study subjects were unrelated. All study subjects received comprehensive ophthalmic examinations.
The research protocol was approved by the Ethics Committee for Human Research of the First Affiliated Hospital of Xinjiang Medical University, China. Informed consent was obtained from all participants after explaining the objective and nature of the study. The study was conducted in accordance with the Declaration of Helsinki.
Peripheral blood samples (2–3 ml) were collected from each subject by venipuncture and genomic DNA was extracted from whole blood by using a Genomic DNA Extraction Kit, (The Beijing Genomics Institute, Beijing, China). The three SNPs (rs1048661, rs3825942, and rs2165241) in the LOXL1 gene, in accordance with previous reports, were amplified by PCR and directly sequenced [20,23]. Two sets of primers were used for amplification by PCR (Table 1). The PCR protocol was as follows: For rs2165241: initial denaturation at 94 °C/5 min, followed by 10 cycles (94 °C/30 s, 65–60 °C touchdown for 1 min with 0.5 °C decrement, 72 °C/45 s), 30 cycles (94 °C/30 s, 60 °C/1 min, 72 °C/45 s), and a final extension at 72 °C/1 min. For rs1048661 and rs3825942: initial denaturation at 94 °C for 5 min followed by denaturation at 94 °C for 30 s, annealing at 58–55 °C touchdown for 30 s with 0.5 °C decrement for the first six cycles, extension at 72 °C for 45 s for 35 cycles, and a final extension of 72 °C for 5 min.
Table 1. Primer sequences for PCR for SNPs of LOXL1.
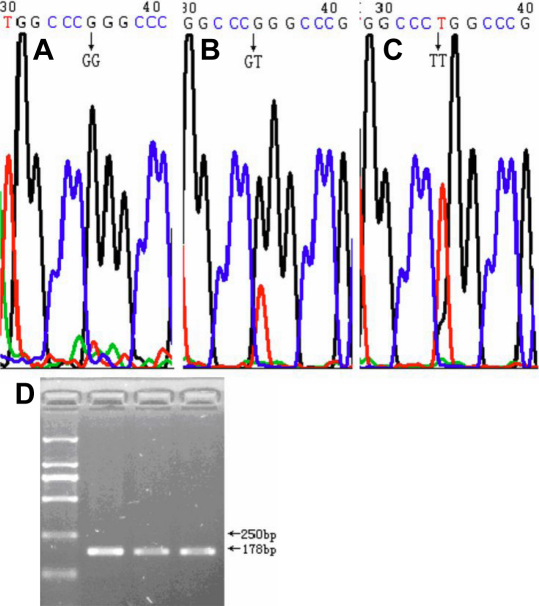
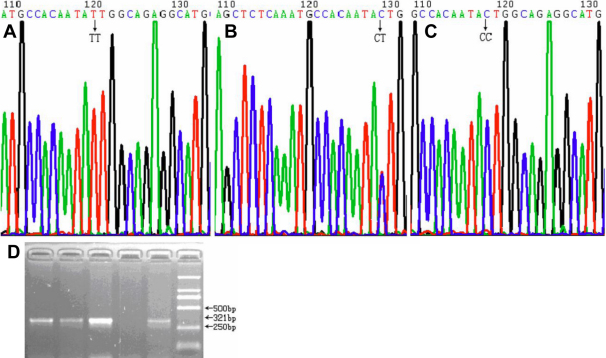
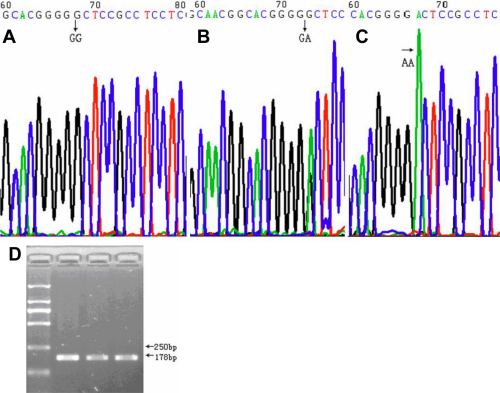
Genotypes of the three LOXL1 SNPs were determined by direct DNA sequencing, using BigDye Terminator v3.1 Kit (Applied Biosystems, Foster City, CA) in a 3730XL capillary sequencer (Applied Biosystems). The sequences were analyzed by sequencing analysis software Chromas (Technelysium Pty Ltd., Queensland, Australia). Representative chromatograms displaying these three SNPs are provided in Figure 1 (rs1048661), Figure 2 (rs2165241), and Figure 3 (rs3825942).
Figure 1.
Representative chromatograms displaying and the PCR electrophoresis image for the SNP rs1048661. A, B, C: The three different genotypes of the SNP rs1048661. D: The PCR electrophoresis image for the SNP rs1048661.
Figure 2.
Representative chromatograms displaying and the PCR electrophoresis image for the SNP rs2165241. A, B, C: The three different genotypes of the SNP rs2165241. D: The PCR electrophoresis image for the SNP rs2165241.
Figure 3.
Representative chromatograms displaying and the PCR electrophoresis image for the SNP rs3825942. A, B, C: The three different genotypes of the SNP rs3825942. D: The PCR electrophoresis image for the SNP rs3825942
Statistical analysis
Statistical analysis was performed using SPSS v13.0 software package (SPSS Inc., Chicago, IL). Hardy–Weinberg equilibrium (HWE) analysis was tested by using the χ2 test in SAS/Genetics v9.1 (SAS Institute Inc., Cary, NC). The comparison of allelic and genotypic frequencies between the patient and control groups as well as haplotype association analysis were performed using a standard χ2 test with a p-value <0.05 considered statistically significant. Relative risk association was estimated by calculating odds ratios (OR) along with 95% confidence intervals (CI).
Results
A total of 64 Uygur patients with exfoliation syndrome, including 7 cases of XFG, 4 cases of XFS with high IOP, and 53 cases of XFS without glaucoma, as well as 127 Uygur control individuals, were recruited into this study. The age at recruitment was 52–95 years (mean age 70.5±9.3 years) in the patient group, and45–84 years (mean age 63.7±10.0 years) in the control group. The gender distribution between the patient and control groups was not significantly different (χ2=0.01, p=0.97), with 41 (64.1%) males and 23 (35.9%) females in the patient group and 81 (63.8%) males and 46 (36.2%) females in the control group. The clinical features of the patients are shown in Table 2. Eleven of the 64 XFG patients had severe glaucoma and high IOP. Only one subject had trabeculectomy and iridotomy, and two were treated with medicine.
Table 2. Clinical features of the patients with XFS/XFG.
|
|
Total (n=64) |
||
|---|---|---|---|
| Clinical features | XFG (n=7) | XFS with high IOP (n=4) | XFS (n=53) |
| Age at recruitment (mean±SD) |
67.00±7.16 |
67.75±10.01 |
71.15±9.53 |
| Range |
58–76 |
60–81 |
52–95 |
| Gender (male/female) |
5/2 |
3/1 |
33/20 |
| History of trabeculectomy |
n=1 |
n=0 |
n=0 |
| History of laser trabeculoplasty |
n=0 |
n=0 |
n=0 |
| History of laser iridotomy |
n=1 |
n=0 |
n=0 |
| Treated with medicine | n=2 | n=0 | n=0 |
A total of 64 patients with exfoliation syndrome were recruited into this study, including 7 cases with XFG and 53 cases with XFS without glaucoma. Only one of the 7 XFG patients had trabeculectomy and laser iridotomy and 2 patients were treated with medicine.
The genotype distribution of SNP rs1048661 (p=0.84 for patients and p=0.68 for controls), rs3825942 (p=0.69 for patients and p=0.12 for controls), and rs2165241 (p=0.37 for patients and p=0.24 for controls) followed the HWE in patients and controls. Allelic association analysis showed that there were significant differences in the allelic distributions between the two groups for the three SNPs (Table 3). The frequency of allele G of rs1048661 was higher in the patient group than in the control group (p=0.013, OR=1.92, 95% CI: 1.14–3.22). The frequency of allele T of rs2165241 was significantly higher in the patient group than in the control group (p=0.000, OR=3.98, 95% CI: 2.54–6.25). The frequency of allele G of rs3825942 was significantly higher in the patient group than in the control group (p=0.000, OR=4.86, 95% CI: 2.02–11.68). The genotypic frequencies for each of the three SNPs were also compared between patients and controls (Table 3). The frequency of genotype GG at SNP rs1048661 was higher in patients than in controls (p=0.016, OR=2.13, 95% CI: 1.14–3.97), and the frequencies of TT genotypes were not significantly different between the two groups. The frequency of genotype TT at SNP rs2165241 was significantly higher in the patient group than in the control group (p=0.000, OR=6.13, 95% CI: 2.68–14.01), and the frequency of CC genotypes was significantly lower in the patients than in the controls. The frequency of genotype GG at SNP rs3825942 was significantly higher in the patient group than in the control group (p=0.000, OR=5.68, 95% CI: 2.28–14.17), and the frequencies of AA genotypes were not significantly different between the two groups.
Table 3. Allele and genotype association analysis for the three SNPs of LOXL1.
| SNP | XFS/XFG | Control | χ2 | p value | OR (95%CI) |
|---|---|---|---|---|---|
|
rs1048661 | |||||
|
Allele | |||||
| G |
104 (0.81) |
176 (0.69) |
6.22 |
0.013 |
1.92 (1.14–3.22) |
| T |
24 (0.19) |
78 (0.31) |
|||
|
Genotype | |||||
| GG |
42 (0.66) |
60 (0.47) |
5.78 |
0.016 |
2.13 (1.14–3.97) |
| GT |
20 (0.31) |
56 (0.44) |
|
|
|
| TT |
2 (0.03) |
11 (0.09) |
2.06 |
0.152 |
0.34 (0.07–1.58) |
| Total |
64 |
127 |
6.37 |
0.041 |
NA |
|
rs2165241 | |||||
|
Allele | |||||
| T |
72 (0.56) |
62 (0.24) |
37.89 |
0.000 |
3.98 (2.54–6.25) |
| C |
56 (0.44) |
192 (0.76) |
|||
|
Genotype | |||||
| TT |
22 (0.34) |
10 (0.08) |
21.43 |
0.000 |
6.13 (2.68–14.01) |
| CT |
28 (0.44) |
42 (0.33) |
|
|
|
| CC |
14 (0.22) |
75 (0.59) |
23.64 |
0.000 |
0.19 (0.10–0.39) |
| Total |
64 |
127 |
31.79 |
0.000 |
NA |
|
rs3825942 | |||||
|
Allele | |||||
| G |
122 (0.95) |
205 (0.81) |
14.73 |
0.000 |
4.86 (2.02–11.68) |
| A |
6 (0.05) |
49 (0.19) |
|||
|
Genotype | |||||
| GG |
58 (0.91) |
80 (0.63) |
16.21 |
0.000 |
5.68 (2.28–14.17) |
| GA |
6 (0.09) |
45 (0.35) |
|
|
|
| AA |
0 (0.00) |
2 (0.02) |
1.02 |
0.313 |
NA |
| Total | 64 | 127 | 16.33 | 0.000 | NA |
There were significant differences for the allelic proportion between the patient and control groups for all three SNPs. G allele of rs1048661, G of rs3825942, and T of rs2165241 were risk alleles for the disorder. The genotypes GG for rs1048661, GG for rs3825942, and TT for rs2165241 were risk genotypes for the disease. Total indicates the general test of association in the 2-by-3 table of disease-by-genotype. NA: not available. The asterisks indicates the OR values and p values derived from comparison of the specific genotype with all the others, i.e., GG versus GT+TT at rs1048661, TT versus CT+CT at rs2165241.
Haplotypes defined by the three SNPs were analyzed (Table 4). For the SNPs rs1048661 and rs3825942, three haplotypes were observed. The haplotype G-G (p=0.006, OR=3.81, 95% CI: 1.40–10.34) was identified to be significantly associated with the patient group. For SNPs rs1048661 and rs2165241, four haplotypes were observed, among which the haplotype G-T was significantly associated with the patient group (p=0.000, OR=3.66, 95% CI: 1.99–6.90). For the SNPs rs3825942 and rs2165241, three haplotypes were observed, with the haplotype G-T showing a significantly higher frequency in the patients than in the controls (p=0.000, OR=3.38, 95% CI: 1.80–6.34). The haplotypes for the three SNPs were also estimated. The haplotype G-G-T had a significantly higher frequency in the patients than in the controls (p=0.000, OR=3.58, 95% CI: 1.90–6.76).
Table 4. Haplotype Association analysis between the LOXL1 SNPs and XFS/XFG.
|
Haplotype |
Proportion |
χ2 |
p value |
OR (95%CI) |
|||
|---|---|---|---|---|---|---|---|
| Case | Control | ||||||
|
rs1048661 |
rs3825942 |
|
|
|
|
|
|
| T |
G |
|
0.047 |
0.181 |
6.52 |
0.011 |
0.22 (0.06–0.77) |
| G |
G |
|
0.922 |
0.756 |
7.66 |
0.006 |
3.81(1.40–10.34) |
| G |
A |
|
0.031 |
0.063 |
0.86 |
0.353 |
0.48 (0.10–2.33) |
| Total |
|
|
|
|
7.90 |
0.019 |
NA |
|
rs1048661 |
rs2165241 |
|
|
|
|
|
|
| T |
C |
|
0.031 |
0.157 |
6.65 |
0.010 |
0.17 (0.04–0.76) |
| G |
T |
|
0.563 |
0.260 |
16.89 |
0.000 |
3.66 (1.99–6.90) |
| T |
T |
|
0.016 |
0.024 |
0.13 |
0.716 |
0.66 (0.07–6.44) |
| G |
C |
|
0.391 |
0.559 |
4.83 |
0.028 |
0.51 (0.27–0.93) |
| Total |
|
|
|
|
19.21 |
0.000 |
NA |
|
rs3825942 |
rs2165241 |
|
|
|
|
|
|
| G |
C |
|
0.406 |
0.660 |
11.35 |
0.001 |
0.35 (0.19–0.65) |
| A |
T |
|
0.016 |
0.008 |
0.25 |
0.619 |
2.00(0.12–32.51) |
| G |
T |
|
0.563 |
0.280 |
15.00 |
0.000 |
3.38 (1.80–6.34) |
| A |
C |
|
0.016 |
0.055 |
1.65 |
0.198 |
0.27 (0.03–2.26) |
| Total |
|
|
|
|
16.06 |
0.001 |
NA |
|
rs1048661 |
rs3825942 |
rs2165241 |
|
|
|
|
|
| T |
G |
C |
0.031 |
0.157 |
6.65 |
0.010 |
0.17 (0.04–0.76) |
| G |
G |
C |
0.375 |
0.504 |
2.85 |
0.092 |
0.59 (0.32–1.09) |
| G |
G |
T |
0.547 |
0.252 |
16.25 |
0.000 |
3.58 (1.90–6.76) |
| T |
G |
T |
0.016 |
0.024 |
0.13 |
0.716 |
0.66 (0.07–6.44) |
| G |
A |
C |
0.016 |
0.055 |
1.65 |
0.198 |
0.27 (0.03–2.26) |
| G |
A |
T |
0.016 |
0.008 |
0.25 |
0.619 |
2.00(0.12–32.51) |
| Total | 19.93 | 0.001 | NA | ||||
The haplotypes G-G for the SNPs rs1048661 and rs3825942, G-T for the SNPs rs1048661 and rs2165241, and G-T for the SNPs rs3825942 and rs2165241 were identified to be significantly associated with XFS/G. The haplotype G-G-T for the three SNPs was identified to be significantly associated with XFS/G. Total indicates omnibus tests, while the others indicate the haplotype-specific tests. The OR values were obtained from comparison of each individual haplotype with all the other haplotypes, i.e., T-G versus G-G + G-A for the SNPs rs1048661 and rs3825942, etc. NA: not available.
Allelic and genotypic association analysis by gender showed that there are significant differences in the allelic and genotypic distributions between the two groups for the three SNPs (Table 5). The G allele of rs1048661 was not significantly different in either the male group or the female group. At rs2165241 frequency of the T allele was higher in male patients than in male controls (p=0.001, OR=2.51, 95% CI: 1.44–4.39) and was significantly higher in female patients than in female controls as well. (p=0.000, OR=9.75, 95% CI: 4.31–22.07). At rs3825942 frequency of the G allele was higher in male patients than in male controls (p=0.005, OR=3.79, 95% CI: 1.42–10.14) and was significantly higher in female patients than in female controls (p=0.007, OR=10.2, 95% CI: 1.31–79.26). The genotypic frequencies for each of the three SNPs were also compared between the patient and control groups by gender (Table 5). The frequency of genotype GG at SNP rs1048661 was not significantly different in either the male group or the female group. The frequencies of genotype TT at SNP rs2165241 and GG at SNP rs3825942 were significantly higher in the patients than in the controls in both gender groups.
Table 5. Allele and genotype association analysis between the LOXL1 SNPs and different gender of XFS/XFG.
|
|
|
|
Genotype count (proportion) |
Allele count (proportion) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | Group | rs1048661 (n) | TT | GT | GG | χ2 | p value | G | T | χ2 | p value | OR (95%CI) |
| Male |
XFS/XFG |
41 |
1 (0.02) |
14 (0.34) |
26 (0.63) |
4.08 |
0.130 |
66 (0.80) |
16 (0.20) |
3.92 |
0.048 |
1.90 (1.00–3.59) |
| |
Control |
81 |
10 (0.12) |
31 (0.38) |
40 (0.49) |
|
|
111 (0.69) |
51 (0.31) |
|
|
|
| Female |
XFS/XFG |
23 |
1 (0.04) |
6 (0.26) |
16 (0.70) |
4.98 |
0.083 |
38 (0.83) |
8 (0.17) |
2.32 |
0.128 |
1.97 (0.82–4.78) |
| |
Control |
46 |
1 (0.02) |
25 (0.54) |
20 (0.43) |
|
|
65 (0.71) |
27 (0.29) |
|
|
|
|
|
|
|
Genotype count (proportion) |
Allele count (proportion) |
||||||||
|
Gender |
Group |
rs2165241 (n) |
CC |
CT |
TT |
χ2 |
p value |
T |
C |
χ2 |
p value |
OR (95%CI) |
| Male |
XFS/XFG |
41 |
12 (0.29) |
19 (0.46) |
10 (0.24) |
9.45 |
0.009 |
39 (0.48) |
43 (0.52) |
10.8 |
0.001 |
2.51 (1.44–4.39) |
| |
Control |
81 |
46 (0.57) |
27 (0.33) |
8 (0.10) |
|
|
43 (0.27) |
119 (0.73) |
|
|
|
| Female |
XFS/XFG |
23 |
2 (0.09) |
9 (0.39) |
12 (0.52) |
27.6 |
0.000 |
33 (0.72) |
13 (0.28) |
34.1 |
0.000 |
9.75 (4.31–22.07) |
| |
Control |
46 |
29 (0.63) |
15 (0.33) |
2 (0.04) |
|
|
19 (0.21) |
73 (0.79) |
|
|
|
|
|
|
|
Genotype count (proportion) |
Allele count (proportion) |
||||||||
|
Gender |
Group |
rs3825942 (n) |
GG |
GA |
AA |
χ2 |
p value |
G |
A |
χ2 |
p value |
OR (95%CI) |
| Male |
XFS/XFG |
41 |
36 (0.88) |
5 (0.12) |
0 (0.00) |
8.41 |
0.015 |
77 (0.94) |
5 (0.06) |
7.89 |
0.005 |
3.79 (1.42–10.14) |
| |
Control |
81 |
51 (0.63) |
28 (0.35) |
2 (0.02) |
|
|
130 (0.80) |
32 (0.20) |
|
|
|
| Female |
XFS/XFG |
23 |
22 (0.96) |
1 (0.04) |
0 (0.00) |
8.46 |
0.004 |
45 (0.98) |
1 (0.02) |
7.19 |
0.007 |
10.2 (1.31–79.26) |
| Control | 46 | 29 (0.63) | 17 (0.37) | 0 (0.00) | 75 (0.82) | 17 (0.18) | ||||||
There were significant differences in allelic and genotypic proportion between the genders in the patient and control groups for the SNPs rs2165241 and rs3825942. T allele of rs2165241 and G of rs3825942 were risk alleles for the disorder for both the male and female groups. Total indicates the general test of association in the 2-by-3 table of disease-by-genotype.
Allelic and genotypic association analysis in different aged groups showed that there were significant differences in the allelic and genotypic distributions between the two groups for the three SNPs (Table 6). At rs1048661 the frequency of the G allele was higher in the patients than in the controls in the below 65-year-old group (p=0.011, OR=3.86, 95% CI: 1.29–11.58). At rs2165241 the frequency of the T allele was significantly higher in the patients than in the controls in both aged groups. (≤65:p=0.000, OR=4.41, 95% CI: 2.09–9.30; >65: p=0.000, OR=3.93, 95% CI: 2.18–7.10). At rs3825942 the frequency of the G allele was significantly higher in the patients than in the controls in the over 65-year-old group. (p=0.000, OR=9.04, 95% CI: 2.66–30.71). The genotypic frequencies for each of the three SNPs were also compared between patients and controls in the different age groups (Table 6). The frequency of genotype GG at SNP rs1048661 was significantly higher in the patients than in the controls in the below 65-year-old age group. The frequency of genotype TT at SNP rs2165241 was significantly higher the patients than in the controls in both aged groups. GG at SNP rs3825942 was significantly higher in patients than in controls in the over 65-year-old group.
Table 6. Allele and genotype association analysis between the LOXL1 SNPs and different aged XFS/XFG.
|
|
|
|
Genotype count (proportion) |
Allele count (proportion) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Years | Group | rs1048661 (n) | TT | GT | GG | χ2 | p value | G | T | χ2 | p value | OR (95%CI) |
| ≤65 |
XFS/XFG |
20 |
0 (0.00) |
4 (0.20) |
16 (0.80) |
7.32 |
0.026 |
36 (0.90) |
4 (0.10) |
6.48 |
0.011 |
3.86 (1.29–11.58) |
| |
Control |
65 |
4 (0.06) |
31(0.48) |
30 (0.46) |
|
|
91 (0.70) |
39 (0.30) |
|
|
|
| >65 |
XFS/XFG |
44 |
2 (0.05) |
16(0.36) |
26 (0.59) |
2.04 |
0.360 |
68 (0.77) |
20 (0.23) |
1.95 |
0.163 |
1.56 (0.83–2.92) |
| |
Control |
62 |
7 (0.11) |
25 (0.40) |
30 (0.48) |
|
|
85 (0.69) |
39 (0.31) |
|
|
|
|
|
|
|
Genotype count (proportion) |
Allele count (proportion) |
||||||||
|
Years |
Group |
rs2165241 (n) |
CC |
CT |
TT |
χ2 |
p value |
T |
C |
χ2 |
p value |
OR (95%CI) |
| ≤65 |
XFS/XFG |
20 |
2 (0.10) |
12 (0.60) |
6 (0.30) |
16.3 |
0.000 |
24 (0.60) |
16 (0.40) |
16.5 |
0.000 |
4.41 (2.09–9.30) |
| |
Control |
65 |
40 (0.62) |
17 (0.26) |
8 (0.12) |
|
|
33 (0.25) |
97 (0.75) |
|
|
|
| >65 |
XFS/XFG |
44 |
12 (0.27) |
16 (0.36) |
16 (0.36) |
21.7 |
0.000 |
48 (0.55) |
40 (0.45) |
21.6 |
0.000 |
3.93 (2.18–7.10) |
| |
Control |
62 |
35 (0.56) |
25 (0.40) |
2 (0.03) |
|
|
29 (0.23) |
95 (0.77) |
|
|
|
|
|
|
|
Genotype count (proportion) |
Allele count (proportion) |
||||||||
|
Years |
Group |
rs3825942 (n) |
GG |
GA |
AA |
χ2 |
p value |
G |
A |
χ2 |
p value |
OR (95%CI) |
| ≤65 |
XFS/XFG |
20 |
17 (0.85) |
3 (0.15) |
0 (0.00) |
1.61 |
0.204 |
37 (0.93) |
3 (0.07) |
1.38 |
0.241 |
2.11 (0.59–7.54) |
| |
Control |
65 |
46 (0.71) |
19 (0.29) |
0 (0.00) |
|
|
111 (0.85) |
19 (0.15) |
|
|
|
| >65 |
XFS/XFG |
44 |
41 (0.93) |
3 (0.07) |
0 (0.00) |
18.4 |
0.000 |
85 (0.97) |
3 (0.03) |
16.9 |
0.000 |
9.04 (2.66–30.71) |
| Control | 62 | 34 (0.55) | 26 (0.42) | 2 (0.03) | 94 (0.76) | 30 (0.24) | ||||||
There were significant differences of the allelic and genotypic proportion in different aged patient and control groups for all three SNPs. G allele of rs1048661 was a risk allele for the disorder in the below 65-year-old group. T of rs2165241 was a risk allele for the disorder in both aged groups. G of rs3825942 was a risk allele for the disorder in the over 65-year-old group. The genotypes also showed significant differences in the below 65-year-old group of rs1048661, both aged groups of rs2165241, and over 65-year-old group of rs3825942. Total indicates the general test of association in the 2-by-3 table of disease-by-genotype.
Discussion
This is the first study to show that the polymorphisms of LOXL1 are associated with Uygur patients with XFS/G. These results are important in understanding the role of LOXL1 polymorphisms in the pathogenesis of XFS/G in Uygur patients. The association between LOXL1 and XFS/XFG in the Han population has rarely been reported except in one recent report from Beijing Tongren Eye Center [25]. In this study, we recruited samples of XFS/XFG patients from Kuche and Kashi which are inhabited mostly by Uygur people. Three major LOXL1 SNPs were found to be significantly associated with XFS/XFG, even though the sample size was small at 64. Hence, the effect sizes of these SNPs in the Uygur population were large, and 64 samples provided a good statistical power to detect significant association. In Xinjiang, it is usually difficult to recruit a large sample of XFS/XFG throughout the autonomous region to ensure frequency matching because of the long distance between cities.
The reason for the higher prevalence in Uygur individuals than in Han individuals is still unknown [16-18]. In this study, we found that all at-risk alleles and genotypes of the LOXL1 SNPs in the Uygur population are different from the Han population but similar to Iceland [20] and the United States [27]. Compared to the frequency of the G allele at rs1048661, the frequency of the T allele at rs2165241 was significantly higher in the patients than in the controls (p=0.000, OR=3.98, 95% CI: 2.54–6.25), and the G allele at rs3825942 was also significantly higher in the patients than in the controls (p=0.000, OR=4.86, 95% CI: 2.02–11.68).
Such a discrepancy might not be fully explained by the ethnic difference in the frequencies of the at-risk alleles of the LOXL1 SNPs (Table 7), and other genetic and/or environmental factors might be involved in the development of the disorder.
Table 7. Risk alleles and MAF for the three SNPs of LOXL1 in different populations.
|
|
rs1048661 (G/T) |
rs3825942 (G/A) |
rs2165241 (T/C) |
|
|||
|---|---|---|---|---|---|---|---|
| Population | Risk allele | MAF | Risk allele | MAF | Risk allele | MAF | Reference |
| Iceland |
G |
0.349(T) |
G |
0.153 (A) |
T |
0.473 (T) |
[20] |
| Sweden |
G |
0.318 (T) |
G |
0.121 (A) |
T |
0.465 (C) |
|
| Austria |
G |
0.329 (T) |
G |
0.183 (A) |
NA |
NA |
[28] |
| United States |
G |
0.335 (T) |
G |
0.156 (A) |
T |
0.487 (T) |
[27] |
| United States |
G |
0.297 (T) |
G |
0.202 (A) |
T |
0.448 (T) |
[29] |
| United States |
G |
0.400 (T) |
G |
0.120 (A) |
NA |
NA |
[30] |
| United States |
G |
0.281 (T) |
G |
0.205 (A) |
T |
0.456 (T) |
[31] |
| Germany and Italy |
G |
0.348 (T) |
G |
0.149 (A) |
T |
0.488 (T) |
[32] |
| Australia |
G |
NA |
G |
NA |
NA |
NA |
[22] |
| refSNP (European) |
NA |
0.040 (T) |
NA |
0.172 (T) |
NA |
0.392 (T) |
NCBI Database |
| India |
* |
0.270 (T) |
G |
0.070 (A) |
NA |
NA |
[23] |
| Japan |
T |
0.450 (G) |
G |
0.147 (A) |
NA |
NA |
[33] |
| Japan |
T |
0.497 (G) |
G |
0.137 (A) |
C |
0.102 (T) |
[34] |
| Japan |
T |
0.460 (G) |
G |
0.143 (A) |
NA |
NA |
[35] |
| Japan |
T |
NA |
G |
NA |
NA |
NA |
[36] |
| Japan |
T |
0.493 (T) |
G |
0.123 (A) |
NA |
NA |
[37] |
| Japan |
T |
0.450 (T) |
G |
0.194 (A) |
C |
0.124 (T) |
[38] |
| Singapore (Chinese) |
* |
0.444 (G) |
G |
0.082 (A) |
NA |
NA |
[39] |
| China (Beijing) |
T |
0.484 (G) |
G |
0.104 (A) |
C |
0.100 (T) |
[25] |
| China (Xinjang Uygur) |
G |
0.188 (T) |
G |
0.047 (A) |
T |
0.244 (T) |
present study |
| China |
|
|
|
|
|
|
[40] |
| (Hong Kong) |
NA |
0.470 (G) |
NA |
0.124 (A) |
NA |
0.102 (T) |
|
| (Beijing) |
NA |
0.497 (G) |
NA |
0.135 (A) |
NA |
0.084 (T) |
|
| Asian (China) |
NA |
NA |
NA |
0.111 (T) |
NA |
0.067 (T) |
|
| Asian (Japan) | NA | 0.438 (G) | NA | 0.125 (T) | NA | 0.167 (T) | |
NA indicates not available. The asterisk indicates that no association was found. MAF indicates minor allele frequency.
In the present study, alleles and genotypes were also significantly associated with XFS/XFG subjects between different gender and age groups. The alleles and genotypes in rs1048661 were not statistically significant between gender groups, as opposed to rs2165241 and rs3825942, which showed higher OR and 95%CI in the female group than in the male group. At rs1048661 the frequency of the G allele was higher in the patients than in the controls in the below 65-year-old group. At rs2165241 the frequency of the T allele was significantly higher in the patients than in the controls in both age groups. At rs3825942 the frequency of the G allele was significantly higher in the patients than in the controls in the over 65-year-old group. Therefore, gender and age might also play an important role in the pathogenesis of XFS/G in Uygur patients.
Although this is our first step in researching the polymorphisms of LOXL1 in Uygur patients, the role of this SNP in pathogenesis of the disorder in Uygur patients needs to be studied further, and further investigations are also needed to determine additional genetic or environmental factors modifying the development of this disorder.
Acknowledgments
We thank all the patients and healthy participants in this study. We also thank the Xinjiang Key Laboratory of Molecular Biology.
References
- 1.Damji KF. Progress in understanding pseudoexfoliation syndrome and pseudoexfoliation-associated glaucoma. Can J Ophthalmol. 2007;42:657–8. doi: 10.3129/i07-158. [DOI] [PubMed] [Google Scholar]
- 2.Ritch R, Schlotzer-Schrehardt U, Konstas AG. Why is glaucoma associated with exfoliation syndrome? Prog Retin Eye Res. 2003;22:253–75. doi: 10.1016/s1350-9462(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 3.Naumann GO, Schlotzer-Schrehardt U, Kuchle M. Pseudoexfoliation syndrome for the comprehensive ophthalmologist. Intraocular and systemic manifestations. Ophthalmology. 1998;105:951–68. doi: 10.1016/S0161-6420(98)96020-1. [DOI] [PubMed] [Google Scholar]
- 4.Vesti E, Kivel AT. Exfoliation syndrome and exfoliation glaucoma. Prog Retin Eye Res. 2000;19:345–68. doi: 10.1016/s1350-9462(99)00019-1. [DOI] [PubMed] [Google Scholar]
- 5.Ritch R, Schlotzer-Schrehardt U. Exfoliation syndrome. Surv Ophthalmol. 2001;45:265–315. doi: 10.1016/s0039-6257(00)00196-x. [DOI] [PubMed] [Google Scholar]
- 6.Forsius H, Forsman E, Fellman J. Exfoliation syndrome: frequency, gender distribution and association with climatically induced alterations of the cornea and conjunctiva. Acta Ophthalmol Scand. 2002;80:478–84. doi: 10.1034/j.1600-0420.2002.800504.x. [DOI] [PubMed] [Google Scholar]
- 7.Schlotzer-Schrehardt U, Naumann GO. Ocular and systemic pseudoexfoliation syndrome. Am J Ophthalmol. 2006;1:921–37. doi: 10.1016/j.ajo.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 8.Challa P. Genetics of pseudoexfoliation syndrome. Curr Opin Ophthalmol. 2009;20:88–91. doi: 10.1097/ICU.0b013e328320d86a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lantukh VV, Piatin MM. Features of ocular pathology among the indigenous inhabitants of Chukotka. Vestn Oftalmol. 1982;(4):18–20. [PubMed] [Google Scholar]
- 10.Ringvold A. Epidemiology of the pseudoexfoliation syndrome. Acta Ophthalmol Scand. 1999;77:371–5. doi: 10.1034/j.1600-0420.1999.770401.x. J. [DOI] [PubMed] [Google Scholar]
- 11.Krishnadas R, Nirmalan PK, Ramakrishnan R, Thulasiraj RD, Katz J, Tielsch JM, Friedman DS, Robin AL. Pseudoexfoliation in a rural population of southern India: the Aravind Comprehensive Eye Survey. Am J Ophthalmol. 2003;135:830–7. doi: 10.1016/s0002-9394(02)02271-7. [DOI] [PubMed] [Google Scholar]
- 12.Thomas R, Nirmalan PK, Krishnaiah S. Pseudoexfoliation in southern India: the Andhra Pradesh Eye Disease Study. Invest Ophthalmol Vis Sci. 2005;46:1170–6. doi: 10.1167/iovs.04-1062. [DOI] [PubMed] [Google Scholar]
- 13.Miyazaki M, Kubota T, Kubo M, Kiyohara Y, Iida M, Nose Y, Ishibashi T. The prevalence of pseudoexfoliation syndrome in a Japanese population: the Hisayama study. J Glaucoma. 2005;14:482–4. doi: 10.1097/01.ijg.0000185436.15675.b3. [DOI] [PubMed] [Google Scholar]
- 14.Young AL, Tang WW, Lam DS. The prevalence of pseudoexfoliation syndrome in Chinese people. Br J Ophthalmol. 2004;88:193–5. doi: 10.1136/bjo.2003.021816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster PJ, Seah SK. The prevalence of pseudoexfoliation syndrome in Chinese people: the Tanjong Pagar Survey. Br J Ophthalmol. 2005;89:239–40. doi: 10.1136/bjo.2004.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Gao Y. Pseudoexfoliation syndrome in the aged people. Chinese Journal of Geriatrics. 1994;13:209–21. [Google Scholar]
- 17.Xiao L, Liu L. Investigation of sight restoring operation of pseudoexfoliation syndrome associated cataract in Kashi area in Xinjiang. Chinese Journal Of Ocular Trauma and Occupational Eye Disease. 2005;28:485–456. [Google Scholar]
- 18.Xie T, Chen X, Mutelli P. Epidemiology of pseudoexfoliation syndrome in aged Uygur farmers in Xinjiang. Chinese Journal of Geriatrics. 2008;27:229–30. [Google Scholar]
- 19.Allingham RR, Loftsdottir M, Gottfredsdottir MS. Pseudoexfoliation syndrome in Icelandic families. Br J Ophthalmol. 2001;85:702–7. doi: 10.1136/bjo.85.6.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorleifsson G, Magnusson KP, Sulem P, Walters GB, Gudbjartsson DF, Stefansson H, Jonsson T, Jonasdottir A, Jonasdottir A, Stefansdottir G, Masson G, Hardarson GA, Petursson H, Arnarsson A, Motallebipour M, Wallerman O, Wadelius C, Gulcher JR, Thorsteinsdottir U, Kong A, Jonasson F, Stefansson K. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317:1397–400. doi: 10.1126/science.1146554. [DOI] [PubMed] [Google Scholar]
- 21.Fingert JH, Alward WL, Kwon YH, Wang K, Streb LM, Sheffield VC, Stone EM. LOXL1 Mutations Are Associated with Exfoliation Syndrome in Patients from the Midwestern United States. Am J Ophthalmol. 2007;144:974–5. doi: 10.1016/j.ajo.2007.09.034. [DOI] [PubMed] [Google Scholar]
- 22.Hewitt AW, Sharma S, Burdon KP, Wang JJ, Baird PN, Dimasi DP, Mackey DA, Mitchell P, Craig JE. Ancestral LOXL1 variants are associated with pseudoexfoliation in Caucasian Australians but with markedly lower penetrance than in Nordic people. Hum Mol Genet. 2008;17:710–6. doi: 10.1093/hmg/ddm342. [DOI] [PubMed] [Google Scholar]
- 23.Ramprasad VL, George R, Soumittra N. Association of non-synonymous single nucleotide polymorphisms in the LOXL1 gene with pseudoexfoliation syndrome in India. Mol Vis. 2008;14:318–22. [PMC free article] [PubMed] [Google Scholar]
- 24.Mabuchi F, Sakurada Y, Kashiwagi K. Lysyl oxidase-like 1 gene polymorphisms in Japanese patients with primary open angle glaucoma and exfoliation syndrome. Mol Vis. 2008;14:1303–8. [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Jia L, Wang N. Evaluation of LOXL1 polymorphisms in exfoliation syndrome in a Chinese population. Mol Vis. 2009;15:2349–57. [PMC free article] [PubMed] [Google Scholar]
- 26.Foster PJ, Buhrmann R, Quigley HA, Johnson GJ. The definition and classification of glaucoma in prevalence surveys. Br J Ophthalmol. 2002;86:238–42. doi: 10.1136/bjo.86.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Challa P, Schmidt S, Liu Y, Qin X, Vann RR, Gonzalez P, Allingham RR, Hauser MA. Analysis of LOXL1 polymorphisms in a United States population with pseudoexfoliation glaucoma. Mol Vis. 2008;14:146–9. [PMC free article] [PubMed] [Google Scholar]
- 28.Mossböck G, Renner W, Faschinger C, Schmut O, Wedrich A, Weger M. Lysyl oxidase-like protein 1 (LOXL1) gene polymorphisms and exfoliation glaucoma in a Central European population. Mol Vis. 2008;14:857–61. [PMC free article] [PubMed] [Google Scholar]
- 29.Aragon-Martin JA, Ritch R, Liebmann J, O'Brien C, Blaaow K, Mercieca F, Spiteri A, Cobb CJ, Damji KF, Tarkkanen A, Rezaie T, Child AH, Sarfarazi M. Evaluation of LOXL1 gene polymorphisms in exfoliation syndrome and exfoliation glaucoma. Mol Vis. 2008;14:533–41. [PMC free article] [PubMed] [Google Scholar]
- 30.Fingert JH, Alward WL, Kwon YH, Wang K, Streb LM, Sheffield VC, Stone EM. LOXL1 mutations are associated with exfoliation syndrome in patients from the midwestern United States. Am J Ophthalmol. 2007;144:974–5. doi: 10.1016/j.ajo.2007.09.034. [DOI] [PubMed] [Google Scholar]
- 31.Fan BJ, Pasquale L, Grosskreutz CL, Rhee D, Chen T, DeAngelis MM, Kim I, del Bono E, Miller JW, Li T, Haines JL, Wiggs JL. DNA sequence variants in the LOXL1 gene are associated with pseudoexfoliation glaucoma in a U.S. clinic-based population with broad ethnic diversity. BMC Med Genet. 2008;9:5–11. doi: 10.1186/1471-2350-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasutto F, Krumbiegel M, Mardin CY, Paoli D, Lammer R, Weber BH, Kruse FE, Schlotzer-Schrehardt U, Reis A. Association of LOXL1 common sequence variants in German and Italian patients with pseudoexfoliation syndrome and pseudoexfoliation glaucoma. Invest Ophthalmol Vis Sci. 2008;49:1459–63. doi: 10.1167/iovs.07-1449. [DOI] [PubMed] [Google Scholar]
- 33.Mabuchi F, Sakurada Y, Kashiwagi K, Yamagata Z, Iijima H, Tsukahara S. Lysyl oxidase-like 1 gene polymorphisms in Japanese patients with primary open angle glaucoma and exfoliation syndrome. Mol Vis. 2008;14:1303–8. [PMC free article] [PubMed] [Google Scholar]
- 34.Ozaki M, Lee KY, Vithana EN, Yong VH, Thalamuthu A, Mizoguchi T, Venkatraman A, Aung T. Association of LOXL1 gene polymorphisms with pseudoexfoliation in the Japanese. Invest Ophthalmol Vis Sci. 2008;49:3976–80. doi: 10.1167/iovs.08-1805. [DOI] [PubMed] [Google Scholar]
- 35.Hayashi H, Gotoh N, Ueda Y, Nakanishi H, Yoshimura N. Lysyl oxidase-like 1 polymorphisms and exfoliation syndrome in the Japanese population. Am J Ophthalmol. 2008;145:582–5. doi: 10.1016/j.ajo.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 36.Mori K, Imai K, Matsuda A, Ikeda Y, Naruse S, Hitora-Takeshita H, Nakano M, Taniguchi T, Omi N, Tashiro K, Kinoshita S. LOXL1 genetic polymorphisms are associated with exfoliation glaucoma in the Japanese population. Mol Vis. 2008;14:1037–40. [PMC free article] [PubMed] [Google Scholar]
- 37.Fuse N, Miyazawa A, Nakazawa T, Mengkegale M, Otomo T, Nishida K. Evaluation of LOXL1 polymorphisms in eyes with exfoliation glaucoma in Japanese. Mol Vis. 2008;14:1338–43. [PMC free article] [PubMed] [Google Scholar]
- 38.Tanito M, Minami M, Akahori M, Kaidzu S, Takai Y, Ohira A, Iwata T. LOXL1 variants in elderly Japanese patients with exfoliation syndrome/glaucoma, primary open-angle glaucoma, normal tension glaucoma, and cataract. Mol Vis. 2008;14:1898–905. [PMC free article] [PubMed] [Google Scholar]
- 39.Lee KY, Ho SL, Thalamuthu A, Venkatraman A, Venkataraman D, Pek DC, Aung T, Vithana EN. Association of LOXL1 polymorphisms with pseudoexfoliation in the Chinese. Mol Vis. 2009;15:1120–6. [PMC free article] [PubMed] [Google Scholar]
- 40.Gong WF, Chiang SW, Chen LJ, Tam PO, Jia LY, Leung DY, Geng YQ, Tham CC, Lam DS, Ritch R, Wang N, Pang CP. Evaluation of LOXL1 polymorphisms in primary open-angle glaucoma in southern and northern Chinese. Mol Vis. 2008;14:2381–9. [PMC free article] [PubMed] [Google Scholar]