Abstract
Genes influencing body weight and composition and serum concentrations of leptin, insulin, and insulin-like growth factor I (IGF-I) in nonfasting animals were mapped in an intercross of the extreme high-growth mouse line DU6i and the inbred line DBA/2. Significant loci with major effects (F > 7.07) for body weight, obesity, and muscle weight were found on chromosomes 1, 4, 5, 7, 11, 12, 13, and 17, for leptin on chromosome 14, for insulin on chromosome 4, and for IGF-I on chromosome 10 at the Igf1 gene locus itself and on chromosome 18. Significant interaction between different quantitative trait loci (QTL) positions was observed (P < 0.01). Evidence was found that loci having small direct effect on growth or obesity contribute to the obese phenotype by gene–gene interaction. The effects of QTLs, epistasis, and pleiotropy account for 64% and 63% of the phenotypic variance of body weight and fat accumulation and for over 32% of muscle weight and serum concentrations of leptin, and IGF-I in the F2 population of DU6i x DBA/2 mice.
[The quantitative trait loci described in this paper have been submitted to the Mouse Genome Database.]
Growth is a complex trait that is genetically determined by gene variants controlling nutrient turnover and energy balance. Genetic studies based on natural pedigrees in humans and on crossbred populations in mouse, rat, and livestock have led to the identification of genetic components responsible for body weight, obesity, and diabetes (for review, see Chagnon et al. 2000). Additionally, quantitative trait loci (QTLs) related to energy balance have been identified in human (Norman et al. 1998) and mice (Moody et al. 1999). Furthermore, statistical methods have been developed for the analysis of epistatic effects between loci controlling the trait (Cheverud and Routman 1995; Mitchell et al. 1997; Wagner et al. 1998). For example, evidence for increased susceptibility to diabetes caused by interaction between genes has been demonstrated in Mexican Americans (Cox et al. 1999).
In order to identify genetic components underlying high body weight and obesity, we have explored the genetically unique, extremely large and fat mouse line DU6, which has twice the body weight and three times the fat content of unselected control mice (Bünger et al. 1990). Muscle hypertrophy with increased muscle diameter was found for high body weight in line DU6 after 40 generations of selection (Rehfeld and Bünger 1990). Recently, we have detected nine QTLs for body weight and eight QTLs for fat accumulation in a cross between line DU6 and the unselected control, DUKs. These QTLs control about one-third of the phenotypic variance of the traits within the F2 population (Brockmann et al. 1998). The QTLs have been designated as Bw4–Bw12 and Afw1–Afw8, respectively (Mouse Genome Database [MGD]). Line DU6 has been selected for 78 generations for high body weight at the age of six weeks. The detected QTLs are responsible for weight and obesity control at this age, which corresponds to the end of the juvenile phase, when animals are fertile. This age is interesting for human researchers because obesity or extreme size during youth is critical for later onset of severe health impairment, and it is of importance for animal breeders because it coincides with the period of slaughtering in meat production.
The extreme phenotype of DU6 mice is accompanied by hyperleptinemia, hyperinsulinemia, significantly elevated insulin-like growth factor I (IGF-I) serum concentrations, and low growth hormone serum levels (Timtchenko et al. 1999). Leptin, insulin, and IGF-I are well known for their growth stimulating effects on metabolism and their influence upon differentiation. Leptin is a peripheral signal modulating the appetite and food consumption in interaction with the central nerve system. Although leptin is essential for avoidance of obesity, the increased leptin concentration in DU6 animals is in concordance with the observation of leptin-resistance in most obese human and rodent models (Maffei et al. 1995). Candidate genes responsible for the extremely high concentrations of leptin, insulin and IGF-I in DU6 mice include Lep, Ins, or Igf1 genes themselves, as well as their receptors and binding proteins or other mediating factors. The two mouse models of obesity, ob and db, are recessively inherited mutations of the leptin encoding gene (Lep) and its receptor (Lepr), respectively. The Lepr gene has been identified as a candidate gene in our previous study (Brockmann et al. 1998). At present, little is known about additional genetic determination of the serum concentrations of leptin, insulin, and IGF-I. Recently, a locus contributing to plasma leptin concentrations but not obesity was identified on chromosome 4, near the leptin receptor gene in a cross between inbred mouse strains C57BL/6J and CAST/Ei (Mehrabian et al. 1998). Evidence for linkage to insulin was found for mouse chromosome 2 and the human homologous chromosome 20 (Lembertas et al. 1997). Genetic loci influencing insulin have been demonstrated in diabetic rats (Galli et al. 1999). To our knowledge, IGF-I concentration has never been analyzed in QTL studies. The control of IGF-I by Igf1 gene in our DU6 mice has been shown by gene expression analyses (Brockmann et al. 1996). To date, our analyses of body weight and obesity have not found evidence of linkage to other strong candidates such as Cpe, Lep, Tub, Ay, Ucp, and Sim1. Thus, we assume that growth and obesity, including serum proteins, are regulated by additional genes that might interact with these genes to influence body weight and obesity.
This study of an intercross between the high-body-weight selected line DU6i and the commercial inbred line DBA/2 is aimed at the discovery of QTLs underlying body weight and obesity and genetic links between loci controlling growth-related physiological parameters and QTLs responsible for body weight and development of obesity. For the convenience of the genetic research (fixation of microsatellite marker alleles), the outbred selected line DU6 has been inbred for four generations to generate the partially inbred line DU6i, which we used in this study.
We examined simultaneously the inheritance of QTLs influencing body weight, fat accumulation, and muscle development with the genetic control of serum parameters leptin, insulin, and IGF-I. This experiment permits the (1) analysis of the genetic correlation between the different growth measures and physiological parameters, (2) the mapping of single quantitative trait loci contributing to the complexity of body weight and fat accumulation, as well as the pattern of genetic determinants of the physiological action of leptin, insulin, and IGF-I, and (3) the search for the genetic interaction between QTLs responsible for individual phenotype differences.
RESULTS
Line and Pedigree Characteristics
Data on the body composition characteristics and serum concentrations of leptin, insulin, and IGF-I of males from the lines DU6i and DBA/2 and the F2 are presented in Table 1. Male mice of the selected line DU6i show a mean body weight of 68.6 g at 42 d, which is 274% greater than DBA/2 animals. The extreme body weight coincides with high mean weights of abdominal fat tissue and muscle. Line DU6i differs from the inbred line DBA/2 by 12.7 S.D. in body weight, 2.8 S.D. in abdominal fat weight, and 9.0 S.D. in muscle weight. Leptin, insulin, and IGF-I serum concentrations were elevated 12-fold, 63-fold, and 3.2-fold in line DU6i as compared with line DBA/2, respectively.
Table 1.
Characteristics of the Parental Mouse Lines
| DU6i Mean ± s.d. (17 males) | DBA/2 Mean ± s.d. (20 males) | F2 Population Mean ± s.d. (411 animals)a | |
|---|---|---|---|
| Body composition | |||
| Body weight (g) | 68.6 ± 3.96 | 18.3 ± 1.6 | 31.05 ± 3.83 |
| Abdominal fat weight (mg) | 1801 ± 560 | 241 ± 58 | 486 ± 197 |
| Abdominal fat percentage (%) | 2.62 ± 0.74 | 1.31 ± 0.28 | 1.49 ± 0.50 |
| Muscle weight (mg) | 617 ± 42 | 240 ± 28 | 339 ± 52 |
| Liver weight (mg) | 4077 ± 520 | 930 ± 166 | 1745 ± 266 |
| Kidney weight (mg) | 882 ± 78 | 240 ± 28 | 386 ± 73 |
| Spleen weight (mg) | 275 ± 67 | 78 ± 13 | 156 ± 51 |
| Serum parameters | |||
| Leptin (ng/ml) | 21.16 ± 11.52 | 1.72 ± 0.56 | 5.55 ± 3.66 |
| Insulin (pmol/L) | 1651 ± 982 | <26 | 248 ± 139 |
| IGF-I (ng/ml) | 750 ± 151 | 238 ± 39 | 616 ± 121 |
Individual values were adjusted for the fixed effects of sex, number of repeated parity, subfamily, and pupsize.
For linkage analysis, a pedigree of F2 intercross design by crossing the DU6i selected inbred line and the DBA/2 inbred line has been generated (Table 2). Pearson's correlation coefficients (r) between recorded traits in mice of the F2 population are presented in Table 3. Body weight, abdominal fat weight, and muscle weight were highly correlated (r > 0.61). A high correlation was also evident between abdominal fat weight and the serum concentrations of leptin (r = 0.69), as well as insulin (r = 0.49). The correlation between muscle weight and leptin, and muscle weight and insulin were r = 0.34 and r = 0.49, respectively. A low correlation was found between IGF-I and body composition (r < 0.31). Sex differences were found for the correlation between serum proteins and the weights of abdominal fat and muscle. For the serum concentrations, we found a high correlation between leptin and insulin (0.46), a low correlation between leptin and IGF-I (0.17), and no correlation between IGF-I and insulin.
Table 2.
F2 Pedigree Structure of the Initial Mating between DBA/2 (Male) and DU6i (Female)
| F2 Subfamily | Number of offspring (male; female) | Number of offspring with serum parameters (male; female) |
|---|---|---|
| 1 (F1♂5 × F1♀17) | 32 (17; 15) | 32 (17; 15) |
| 2 (F1♂6 × F1♀18) | 34 (19; 15) | 33 (19; 14) |
| 3 (F1♂7 × F1♀19) | 26 (13; 13) | 25 (13; 12) |
| 4 (F1♂8 × F1♀20) | 45 (24; 21) | 43 (23; 20) |
| 5 (F1♂9 × F1♀21) | 25 (17; 8) | 24 (17; 7) |
| 6 (F1♂10 × F1♀22) | 28 (17; 11) | 14 (8; 6) |
| 7 (F1♂11 × F1♀23) | 20 (10; 10) | 20 (10; 10) |
| 8 (F1♂12 × F1♀24) | 51 (31; 20) | 32 (18; 14) |
| 9 (F1♂13 × F1♀25) | 38 (21; 17) | 37 (20; 17) |
| 10 (F1♂14 × F1♀26) | 50 (23; 27) | 49 (23; 26) |
| 11 (F1♂15 × F1♀27) | 37 (26; 11) | 37 (26; 11) |
| 12 (F1♂16 × F1♀28) | 25 (15; 10) | 24 (14; 10) |
| Total | 411 (233; 178) | 370 (208; 162) |
Table 3.
Pearson's Correlation Coefficients between Measures of Body Composition and Serum Parameters
| Sex | BW | AFW | AFP | MW | LW | KW | SW | Leptin | Insulin | IGF-I | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BW | male | 1 | 0.75 | 0.57 | 0.84 | 0.88 | 0.77 | 0.16** | 0.52 | 0.57 | 0.31 |
| female | 1 | ||||||||||
| AFW | male | 0.76 | 1 | 0.95 | 0.61 | 0.57 | 0.50 | (−0.02) | 0.69 | 0.49 | 0.28 |
| female | 0.74 | 1 | |||||||||
| AFP | male | 0.64 | 0.96 | 1 | 0.49 | 0.39 | 0.35 | (−0.08) | 0.67 | 0.39 | 0.23 |
| female | 0.58 | 0.97 | 1 | ||||||||
| MW | male | 0.80 | 0.58 | 0.53 | 1 | 0.72 | 0.74 | (0.00) | 0.34 | 0.49 | 0.19 |
| female | 0.81 | 0.60 | 0.48 | 1 | |||||||
| LW | male | 0.88 | 0.62 | 0.50 | 0.69 | 1 | 0.75 | 0.22 | 0.37 | 0.58 | 0.27 |
| female | 0.74 | 0.42 | 0.28 | 0.50 | 1 | ||||||
| KW | male | 0.69 | 0.47 | 0.40 | 0.69 | 0.64 | 1 | 0.11* | 0.26 | 0.51 | 0.24 |
| female | 0.81 | 0.55 | 0.42 | 0.70 | 0.72 | 1 | |||||
| SW | male | 0.18** | (−0.08) | −0.15* | (−0.06) | 0.26 | (0.08) | 1 | (0.03) | 0.11* | (0.02) |
| female | (0.03) | (−0.03) | (−0.04) | (−0.10) | (0.08) | (−0.02) | 1 | ||||
| Leptin | male | 0.53 | 0.69 | 0.69 | 0.28 | 0.39 | 0.18** | (0.00) | 1 | 0.46 | 0.17 |
| female | 0.49 | 0.67 | 0.66 | 0.36 | 0.27 | 0.33 | (−0.05) | 1 | |||
| Insulin | male | 0.47 | 0.51 | 0.50 | 0.36 | 0.46 | 0.32 | (−0.05) | 0.53 | 1 | (0.06) |
| female | 0.37 | 0.33 | 0.28*** | 0.34 | 0.28*** | 0.38 | (0.07) | 0.28 | 1 | ||
| IGF-I | male | 0.35 | 0.31 | 0.27 | 0.21** | 0.33 | 0.28 | (0.01) | 0.17* | (0.03) | 1 |
| female | 0.26*** | 0.21** | 0.17* | 0.15* | 0.21** | 0.24** | (0.01) | (0.12) | (0.08) | 1 |
BW, body weight; AFW, abdominal fat weight; AFP, abdominal fat percentage; MW, weight of quadriceps (musculus rectus femulus, m. vastus intermedius, m. vastus lateralis, m. vastus medialis); LW, liver weight; KW, kidney weight; SW, spleen weight. Values above the diagonal are for correlation over both sexes together; values below the diagonal are for males and females separately. Bold values are significant at P < 0.0001. Asterisks mark different levels of significance: *P < 0.05, **P < 0.01, ***P < 0.001. Values in parentheses are not significant.
Genome-Wide QTL Mapping
The F2 pedigree was genotyped for 93 loci. The markers and their chromosomal locations are listed in Table 4. The experiment–specific, genome-wide significance levels were determined by permutation analysis. The empirical F-value threshold was 9.86 for genome-wide highly significant (P < 0.01) and 7.07 for genome-wide significant (P < 0.05) linkage. The F-value thresholds for the genome-wide suggestive levels of linkage, equivalent to the F-values at the chromosome-wise 5% levels, are shown in Table 5. They vary between chromosomes depending on their length and the markers they contain.
Table 4.
Markers Used for Linkage Analysis
| Marker | Location (cM) | Marker | Location (cM) | Marker | Location (cM) |
|---|---|---|---|---|---|
| D1Mit68 | 9.0 | D7Mit21 | 0.5 | D13Mit16 | 10.0 |
| D1Mit236 | 25.7 | D7Mit25 | 16.0 | D13Mit139 | 32.0 |
| D1Mit214 | 32.1 | D7Mit26 | 24.0 | D13Mit186 | 36.0 |
| D1Mit46 | 43.1 | D7Mit250 | 37.0 | D13Mit130 | 61.0 |
| D1Mit217 | 63.1 | D7Mit253a | 52.8 | D13Mit78 | 75.0 |
| D1Mit33 | 81.6 | D7Mit259a | 72.4 | ||
| D1Mit16 | 87.2 | D14Mit11 | 3.0 | ||
| D1Mit36 | 92.8 | D8Mit64 | 16.0 | D14Mit183 | 19.5 |
| D1Mit293 | 109.6 | D8Mit249 | 37.0 | D14Mit37 | 27.5 |
| D8Mit245 | 72.0 | D14Mit87 | 28.5 | ||
| D2Mit6a | 9.0 | D14Mit162 | 44.0 | ||
| D2Mit81b | 13.0 | D8Mit112c | 21.0 | D14Mit165 | 52.0 |
| D2Mit92 | 40.0 | D9Mit229 | 28.0 | ||
| D2Mit447 | 60.0 | D9Mit136 | 54.0 | D15Mit12 | 6.4 |
| D2Mit106 | 75.0 | D9Mit55a | 61.0 | D15Mit46 | 26.4 |
| D2Mit266 | 109.0 | D15Mit37 | 48.5 | ||
| D10Mit16 | 16.0 | D15Mit193 | 57.9 | ||
| D3Mit264 | 2.4 | D10Mit186 | 40.0 | ||
| D3Mit94 | 22.0 | D10Mit73 | 62.0 | D16Mit146 | 16.9 |
| D3Mit25 | 29.5 | D10Mit102 | 69.0 | D16Mit5 | 38.0 |
| D3Mit73 | 39.7 | ||||
| D3Mit42 | 58.8 | D11Mit71 | 1.0 | D17Mit113 | 6.5 |
| D3Mit59 | 84.1 | D11Mit19 | 14.0 | D17Mit49 | 23.2 |
| D11Mit310 | 25.0 | D17Mit72 | 47.4 | ||
| D4Mit94 | 10.6 | D11Mit5 | 37.0 | D17Mit123 | 56.7 |
| D4Mit196 | 12.1 | D11Mit30 | 39.8 | ||
| D4Mit205 | 45.2 | D11Mit120 | 47.5 | D18Mit60 | 16.0 |
| D4Mit37 | 56.5 | D11Mit326 | 49.0 | D18Mit7 | 50.0 |
| D4Mit54 | 66.0 | D11Mit67 | 58.0 | ||
| D11Mit100 | 68.0 | D19Mit30 | 20.0 | ||
| D5Mit66 | 17.0 | D11Mit104 | 79.0 | D19Mit10 | 47.0 |
| D5Mit58 | 41.0 | ||||
| D5Mit24a | 60.0 | D12Mit46 | 16.0 | DXMit192 | 16.0 |
| D5Mit95 | 68.0 | D12Mit36 | 28.0 | DXMit170 | 44.0 |
| D5Mit221 | 81.0 | D12Mit141 | 55.0 | DXMit131 | 59.0 |
| D6Mit139 | 2.5 | ||||
| D6Mit17 | 30.5 | ||||
| D6Mit149 | 46.3 | ||||
| D6Mit14 | 74.0 |
Heterozygous DU6i parent.
Heterozygous DBA/2 parent.
This marker has been reassigned to chromosome 9.
Table 5.
Most Likely Positions and Effects of QTLs
| Chromosome | F-ratio thresholds 1%; 5%; 10%a | QTL position (cM)b | Trait | F-ratio 1 vs. 0 QTLc | Additive effect | Dominance effect | % F2 Symbol Variancegd | ||
|---|---|---|---|---|---|---|---|---|---|
| a | s.e. | d | s.e. | ||||||
| 1 | 6.6; 5.1; 4.3 | 35 (11; 97) | MW (mg) | 6.10 | 13 | 4 | −9 | 7 | 3.18 |
| 36 (25; 51) | BW (g) | 10.44 | 1.3 | 0.30 | −0.94 | 0.49 | 5.36 Bw5 | ||
| 39 (10; 59) | LW (mg) | 5.71 | 74 | 22 | −3 | 33 | 2.99 | ||
| 86 (65; 103) | KW (mg) | 4.99 | 11 | 6 | −22 | 8 | 2.61 | ||
| 100 (80; 109) | Leptin | 4.94 | −0.036 | 0.022 | 0.103 | 0.038 | 2.89 | ||
| 2 | 7.2; 5.1; 4.3 | 26 (11; 39) | KW (mg) | 7.59 | 27 | 7 | −7 | 12 | 3.93 Kwq6 |
| 88 (57; 100) | IGF-I | 4.46 | 0.013 | 0.009 | 0.041 | 0.016 | 2.58 | ||
| 3 | 7.4; 5.0; 4.2 | 28 (22; 35) | Leptin | 5.47 | −0.042 | 0.021 | −0.078 | 0.031 | 3.19 |
| 29 (23; 37) | AFP (%) | 6.17 | −0.094 | 0.040 | −0.150 | 0.060 | 3.24 Afpq1 | ||
| 30 (23; 36) | AFW (mg) | 4.23 | −32 | 15 | −41 | 22 | 2.27 Afw1 | ||
| 4 | 7.0; 4.6; 3.8 | 33 (21; 52) | LW (mg) | 7.69 | 95 | 27 | 89 | 48 | 3.98 Lwq1 |
| 59 (34; 72) | BW (g) | 5.34 | 0.88 | 0.33 | −0.85 | 0.48 | 2.81 Bw7 | ||
| 61 (49; 72) | Insulin (pmol/l) | 7.18 | 52.76 | 14.1 | 16.4 | 20.9 | 4.19 Insq1 | ||
| 66 (58; 74) | AFW (mg) | 7.61 | 74 | 19 | 17 | 32 | 4.00 Afw2 | ||
| 66 (60; 72) | AFP (%) | 5.72 | 0.160 | 0.047 | 0.001 | 0.077 | 3.01 Afpq2 | ||
| 66 (59; 72) | Leptin | 6.39 | 0.091 | 0.026 | 0.015 | 0.043 | 3.71 | ||
| 5 | 5.9; 4.6; 3.7 | 18 (0; 39) | IGF-I | 4.28 | 0.020 | 0.008 | −0.018 | 0.012 | 2.47 |
| 25 (0; 64) | Leptin | 3.75 | 0.058 | 0.025 | −0.061 | 0.039 | 2.21 | ||
| 80 (69; 91) | AFW (mg) | 5.67 | 54 | 16 | 11 | 23 | 3.01 Afw3 | ||
| 81 (73; 89) | BW (g) | 11.69 | 1.45 | 0.30 | 0.25 | 0.42 | 5.96 Bw13 | ||
| 81 (77; 85) | LW (mg) | 9.34 | 77 | 21 | 68 | 30 | 4.79 Lwq2 | ||
| 7 | 7.0; 5.1; 4.2 | 22 (13; 27) | AFP (%) | 18.50 | 0.249 | 0.043 | −0.121 | 0.063 | 9.11 Afpq9 |
| 23 (16; 28) | AFW (mg) | 24.93 | 112 | 16 | −47 | 24 | 12.02 Afw9 | ||
| 28 (23; 33) | BW (g) | 25.91 | 2.34 | 0.33 | −0.94 | 0.51 | 12.31 Bw14 | ||
| 28 (21; 35) | KW (mg) | 14.50 | 35 | 6 | −3 | 10 | 7.25 Kwq7 | ||
| 29 (20; 34) | MW (mg) | 19.49 | 27 | 4 | −11 | 7 | 9.55 Mwq1 | ||
| 32 (27; 44) | LW (mg) | 28.76 | 173 | 23 | −50 | 35 | 13.42 Lwq6 | ||
| 32 (23; 56) | Insulin (pmol/l) | 6.20 | 42.4 | 13.8 | −42.2 | 20.8 | 3.64 | ||
| 9 | 6.8; 4.6; 3.8 | 27 (21; 35) | LW (mg) | 9.41 | 92 | 23 | 54 | 31 | 4.83 Lwq7 |
| 37 (22; 51) | BW (g) | 6.23 | 1.28 | 0.36 | −0.18 | 0.56 | 3.27 | ||
| 10 | 5.9; 4.7; 3.8 | 38 (32; 45) | IGF-I | 29.70 | 0.056 | 0.007 | −0.011 | 0.010 | 14.98 Igf1q1 |
| 64 (47; 80) | SW (mg) | 10.93 | −18 | 4 | 0 | 6 | 5.50 Swq5 | ||
| e−8 | 59 | −11 | 10 | ||||||
| f−29 | 7 | 42 | 12 | ||||||
| 11 | 7.9; 5.4; 4.6 | 4 (0; 12) | Leptin | 5.08 | 0.057 | 0.022 | −0.058 | 0.032 | 2.97 |
| 9 (0; 19) | AFP (%)* | 5.86 | 0.116 | 0.043 | −0.139 | 0.064 | 3.08 Afpq5 | ||
| e0.079 | 0.054 | 0.004 | 0.083 | ||||||
| f0.146 | 0.069 | −0.314 | 0.096 | ||||||
| 10 (0; 18) | MW (mg)* | 5.96 | 11 | 4 | −14 | 6 | 3.13 Mwq2 | ||
| 12 (2; 19) | AFW (mg)* | 6.48 | 49 | 16 | −47 | 23 | 3.42 Afw5 | ||
| e27 | 21 | 4 | 32 | ||||||
| f74 | 27 | −117 | 38 | ||||||
| 14 (6; 17) | BW (g)* | 7.52 | 1.06 | 0.31 | −0.81 | 0.43 | 3.92 Bw16 | ||
| 39 (23; 42) | SW (mg) | 8.13 | 14 | 4 | 10 | 5 | 4.16 Swq1 | ||
| 55 (36; 65) | BW (g)* | 9.52 | 1.32 | 0.31 | −0.28 | 0.45 | 4.91 Bw4 | ||
| 59 (53; 69) | AFP (%)* | 4.26 | 0.083 | 0.040 | 0.123 | 0.059 | 2.26 | ||
| 59 (50; 65) | MW (mg)* | 11.50 | 17 | 4 | −13 | 6 | 5.86 Mwq3 | ||
| 61 (53; 71) | AFW (mg)* | 4.79 | 41 | 16 | −44 | 24 | 2.55 | ||
| 12 | 6.4; 4.3; 3.5 | 16 (8; 24) | Leptin | 6.02 | 0.058 | 0.020 | −0.053 | 0.029 | 3.50 |
| 17 (0; 50) | BW (g) | 3.87 | 0.77 | 0.29 | −0.35 | 0.43 | 2.05 Bw9 | ||
| 18 (10; 26) | AFP (%) | 7.48 | 0.125 | 0.041 | −0.140 | 0.061 | 3.89 Afpq10 | ||
| 20 (14; 26) | MW (mg) | 12.25 | 19 | 4 | −9 | 6 | 6.23 Mwq4 | ||
| 21 (0; 30) | SW (mg) | 8.90 | −15 | 4 | 13 | 6 | 4.52 Swq2 | ||
| 21 (15; 27) | AFW (mg)* | 8.56 | 59 | 21 | −82 | 30 | 4.50 Afw10 | ||
| 43 (26; 60) | AFW (mg)* | 4.37 | 5 | 23 | 111 | 37 | 2.34 | ||
| 46 (31; 61) | LW (mg) | 4.02 | 33 | 24 | 105 | 42 | 2.12 | ||
| 13 | 5.7; 4.5; 3.8 | 10 (3; 16) | BW (g)* | 7.36 | −1.23 | 0.39 | −0.90 | 0.50 | 3.84 Bw15 |
| 10 (0; 21) | MW (mg)* | 4.42 | −140 | 5 | −6 | 7 | 2.32 | ||
| 11 (4; 18) | AFW (mg) | 6.03 | −39 | 17 | −54 | 22 | 3.20 Afw6 | ||
| 13 (0; 46) | AFP (%) | 4.25 | −0.105 | 0.046 | −0.101 | 0.065 | 2.25 Afpq4 | ||
| 13 (0; 21) | Leptin | 4.32 | 0.013 | 0.023 | −0.098 | 0.033 | 2.54 | ||
| 43 (33; 52) | MW (mg)* | 9.75 | 26 | 6 | −9 | 8 | 5.00 Mwq5 | ||
| 47 (33; 61) | BW (g)* | 6.39 | 1.42 | 0.40 | −0.48 | 0.63 | 3.35 Bw10 | ||
| 14 | 6.6; 4.7; 3.9 | 3 (0; 13) | SW (mg) | 4.66 | 5 | 4 | −15 | 5 | 2.43 Swq3 |
| 18 (6; 29) | AFW (mg) | 5.58 | 53 | 16 | 12 | 22 | 2.97 | ||
| 28 (21; 41) | Leptin | 7.58 | 0.061 | 0.021 | 0.075 | 0.030 | 4.36 Lepq1 | ||
| 43 (16; 70) | KW (mg) | 4.02 | 16 | 6 | 4 | 8 | 2.12 | ||
| 15 | 6.2; 4.5; 3.7 | 16 (0; 38) | Insulin (pmol/l) | 3.90 | 31.3 | 14.3 | −43.6 | 24.1 | 2.32 |
| 50 (33; 56) | LW (mg) | 6.31 | 77 | 22 | −27 | 32 | 3.29 Lwq5 | ||
| 57 (49; 65) | SW (mg) | 3.71 | 10 | 4 | −6 | 5 | 1.94 | ||
| 56 (47; 65) | IGF-I | 4.29 | 0.019 | 0.007 | −0.008 | 0.009 | 2.48 | ||
| 17 | 6.5; 4.7; 3.8 | 36 (27; 51) | AFP (%) | 8.92 | −0.205 | 0.048 | −0.007 | 0.083 | 4.61 Afpq6 |
| 39 (30; 52) | AFW (mg) | 9.00 | −77 | 18 | 8 | 30 | 4.70 Afw7 | ||
| 18 | 5.9; 3.9; 3.2 | 16 (0; 32) | LW (mg) | 5.07 | 53 | 21 | −53 | 29 | 2.66 |
| 19 (0; 35) | KW (mg) | 3.83 | 9 | 6 | −21 | 9 | 2.02 | ||
| 39 (26; 54) | IGF-I | 9.51 | 0.034 | 0.008 | 0.002 | 0.013 | 5.34 Igf1q2 | ||
| 19 | 5.6; 3.6; 3.0 | 47 (32; 55) | Leptin | 6.22 | 0.079 | 0.022 | −0.005 | 0.032 | 3.61 |
| X | 4.5; 3.0; 2.2 | 16 (0; 32) | AFP (%) | 2.61 | e0.053 | 0.037 | 1.39 | ||
| f−0.077 | 0.042 | ||||||||
| 17 (0; 39) | AFW (mg) | 4.78 | e43 | 15 | 2.54 Afw11 | ||||
| f−23 | 17 | ||||||||
| 44 (32; 58) | LW (mg) | 5.85 | e−68 | 20 | 3.02 Lwq8 | ||||
| f−18 | 22 | ||||||||
Trait acronyms are given below Table 3. For leptin and IFG-I, logarithmic transformed data were used for the analysis. Thus, the estimates are given as logarithmic units as well.
The chromosome specific 1%, 5%, and 10% F-value thresholds estimated via permutation analyses.
The most likely locations are given as distance from the centromere.
At loci in which QTL effects were sex specific (P < 0.05), below the joint estimate over both sexes estimates are given separately for males and females.
F values for traits in which two QTLs were found in a linkage group (P < 0.05); the presented F values result from one QTL analysis after fitting the second QTL on the chromosome as the background genetic effect.
The QTL effects are given as reduction of the residual sum of squares fitting 1 vs. 0 QTLs.
Males.
Females.
Symbols were given for QTLs of the genome-wide 5% significant level at F > 7.07 for loci in which two QTLs were found in a linkage group at P < 0.05 and both single QTLs have chromosome-wide significant level at P < 0.05 and for suggestive QTLs, which have been confirmed from our previous experiment (Brockmann et al. 1998). Bold printed names indicate QTL symbols that have been assigned in this study.
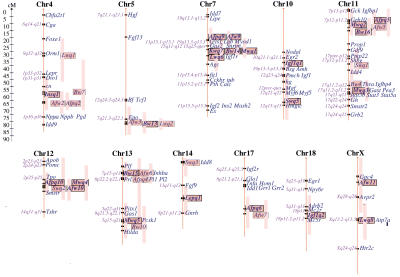
The estimates for the most likely positions and effects of QTLs suggestive at the chromosome-wise level of significance detected in the pedigree are presented for every chromosome in Table 5. Highly significant QTLs (F > 9.86) were mapped for body weight on chromosomes 1, 5, and 7; for abdominal fat weight and liver and kidney weights on chromosome 7; for muscle weight on chromosomes 7, 11, and 12; for spleen weight on chromosome 10. Highly significant influence on the serum concentrations of IGF-I was found for a QTL on chromosome 10. Positive estimates of genetic effects indicate that alleles from the selected line (DU6i) increase the trait. As expected, the great majority of estimated genetic effects are in this direction. However, DU6i QTL alleles with decreasing effects compared to the DBA/2 allele were also detected. Figure 1 displays the chromosomal positions of significant QTLs relative to map positions of candidate genes for the QTL effects.
Figure 1.
Depiction of chromosomes harboring QTLs newly identified in this study. QTL symbols of newly identified QTLs were labeled in bold text in the last column of Table 5. Shown are the chromosomal positions and confidence intervals of significant QTLs for body weight (Bw*), abdominal fat weight (Afw*), abdominal fat percentage (Afpq*), muscle weight (Mwq*), liver weight (Lwq*), kidney weight (Kwq*), spleen weight (Swq*), leptin (Lepq*), insulin (Insq*), and IGF-I (Igf1q*) relative to map positions of candidate genes for the QTL effects. Candidate genes were taken from the Mouse Genome Database (MGD), section endocrine defects, hormones, growth, and obesity (MGD), and from the Human Obesity Gene Map (Chagnon et al. 2000). The human homology is shown on the left side of every chromosome.
The highest effect on body weight has been estimated at a peak F-value of 25.91 at 28 cM from the centromere on chromosome 7 (Bw14). The one-LOD confidence interval of this QTL comprises a region of 10 cM with additional effect on fat accumulation (F = 24.93 at 23 cM) and muscle development (F = 19.49 at 29 cM), liver and kidney weight, and the serum concentration of insulin (F = 6.2 at 32 cM). The effect of the body weight QTL accounts for 12.3% of the phenotypic variance within the F2 population. The DU6i allele had an additive effect of 2.34 g on body weight. The highly significant QTLs on chromosomes 1 (Bw5) and 5 (Bw13) account for 5.4 and 6%, respectively, of the phenotypic F2 variance. The test for multiple QTLs influencing body weight was significant for the highly significant effect of chromosome 11 (P < 0.01) and the significant QTL on chromosome 13 (P < 0.05). At least two QTLs reside on chromosome 11 at 14 cM (Bw16) and 55 cM (Bw4) and on chromosome 13 at 10 cM (Bw15) and 47 cM (Bw10). Most QTLs influencing body weight were additive with an increasing effect of the DU6i allele. The only exception was Bw15 at chromosome 13 near the centromere, where the DU6i allele caused a lower body weight of 1.23 g compared to the DBA/2 allele. QTLs at the genome-wide suggestive level were mapped to chromosomes 4 (Bw7 which has been confirmed in our previous study) and 9. The net effect of all detected QTLs for body weight explained 34.9 % of the phenotypic variance in the F2 population.
Because total body weight is a reflection of fat accumulation and muscle development, these two traits were analyzed separately for their contribution to body weight. The analyzed F2 population gave highly significant evidence for QTLs responsible for muscle weight on chromosomes 7, 11, and 12 and significant evidence on chromosomes 1 and 13, in chromosomal regions influencing body weight. Chromosome 7 was harboring the QTL with the largest effect; this was at 23 cM and accounted for 9.6% of the phenotypic F2 variance (Mwq1). The analyses identified two QTLs responsible for muscle weight (P < 0.05) on chromosome 11 at 10 cM (Mwq2) and 59 cM (Mwq3) and on chromosome 13 at10 cM and 43 cM (Mwq5). Corresponding to the development of body weight, the additive genetic effects of QTL alleles inherited from line DU6i increased muscle development at all loci, except the DU6i allele at 10 cM on chromosome 13, which reduced the muscle weight by 0.14 g. Together, muscle weight QTLs accounted for 27.9% of the phenotypic variance of muscle weight in the F2 population.
The inclusion of body weight as a covariate in the analyses indicated that all of the QTLs influencing body weight have an effect on fat and muscle weight proportional to the overall phenotypic association between body weight, fatness, and muscle weight. Thus, inclusion of body weight as a covariate in the analyses removed all evidence of all QTLs identified for muscle weight.
Estimated positions of QTLs responsible for abdominal fat weight coincide with body-weight QTLs on chromosomes 4, 5, 7, 11, 12, and 13. The largest effect on abdominal fat weight was mapped at a peak F-value of 24.9 on chromosome 7 (Afw9), and this accounted for 12% of the phenotypic F2 variance. Although three peaks were visible in the F-value curve pertaining to chromosome 11, only two QTLs at 12 (Afw5) and 61 cM were found at P < 0.05 with influence on abdominal fat weight. Evidence for two fat QTLs was found on chromosome 12 at 21 (Afw10) and 43 cM (P < 0.05). Additionally, QTLs for abdominal fat weight were detected on chromosomes 3 (Afw1), 14 , 17 (Afw7), and X (Afw11) these might be specific for fat accumulation in the abdominal fat tissue, because their effect on total body weight was not significant. The identified fat QTLs differ in their effects. They were mainly positive for the DU6i allele. However, the DU6i allele caused less abdominal fat weight by an additive genotype effect at chromosomes 3 (Afw1), 13 (Afw6), and 17 (Afw7). Negative dominance components of the QTL effect were present for DU6i alleles at Afw5 on chromosome 11 and at Afw10 on chromosome 12. Among the loci responsible for abdominal fat accumulation, the QTL on the proximal part of chromosome 11 (Afw5) acts significantly differently (P < 0.05) in males and females. It mainly influences fat accumulation in females and shows negative heterosis (d = −117 mg, a = 74 mg), so that heterozygous animals showed a higher mean reduction of abdominal fat weight than half the difference between the homozygous genotypes. For males, an additive effect of 27 mg was found for the DU6i allele. The net effect of all detected QTLs influencing abdominal fat weight explained 34.1% of the phenotypic variance in the F2 population.
Because the DU6i parent of the pedigree comes from an outbred line following four generations of inbreeding, five of the 93 markers (D2Mit6, D5Mit24, D7Mit253, D7Mit259, D9Mit55) still were heterozygous and, hence, a linked QTL might be heterozygous as well. Therefore, we have analyzed the interactions between a QTL and subfamily for QTLs at the proximal region of chromosome 2, the distal part of chromosomes 5, 7, and 9. There was significant evidence (P < 0.001) that QTL genotypes differed between subfamilies for the QTL influencing abdominal fat weight at 81 cM at the distal part of chromosome 5 (Afw3). The inclusion of body weight as covariate in the analyses removed QTLs responsible for abdominal fat weight on chromosomes 5, 7, 11, 12, 13, and 14 but not those on chromosomes 1, 4, 17 and X.
The most likely positions of QTLs influencing abdominal fat percentage coincided with the chromosomal intervals identified for QTLs for abdominal fat weight on chromosomes 3, 4, 7, 11, 12, 13, 17, and X. No QTLs for abdominal fat percentage were found on chromosomes 5 and 14 by which there were mapped QTLs influencing abdominal fat weight. Corresponding to abdominal fat weight, DU6i QTL alleles for fat percentage on chromosomes 3, 13, and 17 and the female-specific effect on chromosome 11 at 12 cM act by negative additive or dominant effect, while the other DU6i QTL alleles had positive additive effects on abdominal fat percentage.
QTLs for liver weight coincided with QTL positions for abdominal fat weight on chromosomes 5, 7, and 12 and for body weight QTLs on chromosomes 1, 5, 7, and 9. Additionally, loci on chromosomes 4 (outside the most likely QTL interval for body weight and fat), 15, and 18 were responsible for liver weight. The genetic effects of the identified loci were predominantly additive. The joint effect of all QTLs accounted for 31.3% of the F2 variance.
Significant QTLs affecting kidney weight were mapped on chromosomes 2 and 7 at F-values of 7.59 and 14.5, respectively. Additional QTLs at the chromosome-wise suggestive level of significance were mapped at the distal part of chromosomes 1, 14, and 18. The identified QTLs accounted for 17.3% of the phenotypic F2 variation.
The genetic control of spleen weight differs from the regulation of the other traits characterizing body composition. A highly significant effect on spleen weight was found for the QTL on chromosome 10, with a peak F-value of 10.9 at 64 cM (Swq5). Genome-wide significant loci with influence on spleen weight were found on chromosomes 11 and 12. Chromosome-wise suggestive QTLs were located on chromosomes 14 and 15. The DU6i alleles of the spleen QTLs on chromosomes 10 and 12 reduced spleen weight relative to the DBA/2 allele, while the other DU6i QTL alleles had an increasing effect.
Significant evidence for the genetic regulation of leptin concentration in serum, with a peak F-value of 7.58, was found on chromosome 14 at 28 cM (Lepq1). The DU6i allele of this locus had an increasing additive effect accounting for 4.4% of the phenotypic F2 variance of leptin, which was accompanied by increased fat accumulation. Other genome-wide suggestive loci changing the leptin concentration were identified on chromosome 3 at 28 cM (F = 5.47), on the distal part of chromosome 4 at 66 cM (F = 6.39), on chromosome 12 at 16 cM (F = 6.02), and on chromosome 19 at 47 cM (F = 6.22). The loci on chromosomes 3 and 12 mapped to regions that also effected fat accumulation. The leptin QTL on chromosome 4 coincided with QTLs affecting fat accumulation, body weight, and insulin. However, no influence of the leptin QTL on chromosome 19 was found on abdominal fat weight. Additionally, QTLs with lesser effects on the serum concentration of leptin were detected on chromosomes 1, 5, 11, and 13. An increase of the serum leptin level by heterosis was caused by the DU6i alleles on chromosome 1. The chromosomal region on chromosome 13 that causes a reduction of the serum leptin level by heterosis in DU6i also has a reducing effect on fat accumulation. The joint effect of all loci influencing leptin was 24.0% of the phenotypic F2 variance.
For insulin concentration in serum, there was one QTL on chromosome 4 at 61 cM (Insq1) that reached the genome-wide significance threshold at F = 7.09. This insulin locus coincided with a locus that effects leptin concentration and Afw2 increasing fat accumulation in DU6i. A genome-wide suggestive locus affecting the serum insulin concentrations was detected on chromosome 7, and a chromosome-wise suggestive locus was mapped to chromosome 15. The QTL at 32 cM on chromosome 7 mapped together with QTLs influencing fat accumulation (Afw9), body weight (Bw14), muscle weight (Mwq1), and liver weight (Lwq6) in a 10 cM interval. The estimated effect of all QTLs in this interval was additive, with increasing effects of the DU6i alleles. The insulin QTL on chromosome 15 did not have any apparent effect on other traits. The joint effect of the detected loci affecting serum insulin concentration was 9.1% of the phenotypic variance within the F2 population.
IGF-I was expected to influence body weight mainly by promoting protein accretion and muscle development. A major QTL (Igf1q1) responsible for differences in the serum IGF-I concentration on chromosome 10 with a peak F-value of 29.7 at 38 cM from the centromere, with a one LOD support interval of 13 cM contributed most of the genetic variation detected. The strong additive effect (0.056 logarithmic units, equivalent to 81 ng/ml) accounted for 15.0% of the phenotypic variance of the trait in the F2 population. A second highly significant QTL (Igf1q2) having additive increasing effect of the DU6i allele was found on chromosome 18 with a peak F-value of 9.51 at 39 cM. The regions harboring the Igf1q1 and Igf1q2 QTLs did not show any linkage to body, muscle, or fat weight. Additionally, QTLs influencing the IGF-I concentration were found on chromosomes 2, 5, and 15. On chromosome 5, the loci influencing the serum concentration of IGF-I and leptin map within a 5-cM region. The identified IGF-I QTLs accounted for 22.5% of the phenotypic variance within the F2 population.
Interaction between QTLs
The effect of interaction between loci was tested for body weight, abdominal fat weight, muscle weight, and serum concentrations of leptin, insulin, and IGF-I. The analysis of a specific trait (e.g., body weight) included the test of interaction between markers linked to QTLs influencing this specific trait (e.g., QTLs for body weight) and, additionally, the test of interaction between markers linked to QTLs responsible for differences of the other analyzed traits (e.g., QTLs for fat, leptin, etc.). We chose this strategy because of the high correlation between the body-composition traits and the concentration of serum proteins and because of our observation of colocated, but often small, subsignificant effects of QTLs identified for one trait on other related traits.
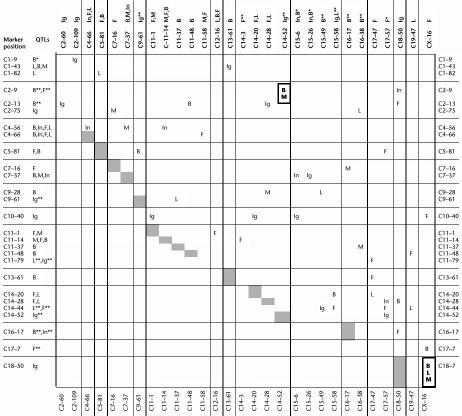
Here we report results that proved significant interaction effects at P < 0.01. Five out of 40 (insulin) to 12 out of 264 (abdominal fat weight ) tests of epistasis were significant at this level of significance for the different traits (Table 6). As we wanted to report all potentially biologically important interactions, we chose a relatively relaxed threshold, but in this study we focused our discussion on the most significant results. The direct effects of markers and the effects of locus*locus interaction are given for every trait in Table 6. Highly significant epistatic effects at P < 0.001 were evident for the serum concentrations of leptin (C9-61*C11-37), insulin (C4-56*C11-14) and IGF-I (C1-43*C13-61, C7-37*C15-26, C14-44*C15-49).
Table 6.
Account of Interaction to the Phenotypic F2 Variance for Every Trait
| BW Position | Position effect (%) | C9–61 0.5 | C11–48 6.1 | C14–52 0.3 | C15–58 0.9 | C17–47 0.3 | C18–50 0.3 | CX–16 0.5 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C1–82 | 2.1 | 4.8 | |||||||||
| C2–9 | 1.3 | 3.7 | |||||||||
| C2–13 | 0.9 | 3.9 | |||||||||
| C5–81 | 5.8 | 4.2 | |||||||||
| C11–14 | 3.6 | 4.6 | |||||||||
| C14–20 | 0.7 | 3.8 | |||||||||
| C14–28 | 1.0 | 4.1 | |||||||||
| C17–7 | 0.2 | 5.1 | |||||||||
| C18–50 | 0.5 | 4.5 | |||||||||
| AFW Position | Position effect (%) | C11–58 3.0 | C12–16 2.9 | C14–3 0.3 | C15–58 0.2 | C17–47 4.7 | C17–57 4.5 | C18–50 1.0 | C19–47 0.6 | CX–16 1.4 | |
| C2–13 | 1.1 | 3.8 | |||||||||
| C4–66 | 0.5 | 3.7 | |||||||||
| C5–81 | 3.5 | 5.2 | |||||||||
| C10–40 | 0.0 | 3.8 | |||||||||
| C11–1 | 1.0 | 4.4 | |||||||||
| C11–14 | 1.7 | 3.9 | |||||||||
| C11–48 | 1.8 | 3.6 | |||||||||
| C11–79 | 0.5 | 4.2 | |||||||||
| C13–61 | 0.0 | 4.4 | |||||||||
| C14–44 | 0.6 | 4.3 | 4.1 | ||||||||
| C16–17 | 0.2 | 4.0 | |||||||||
| MW Position | Position effect (%) | C7–16 9.4 | C7–37 9.4 | C14–28 0.2 | C14–52 0.2 | C16–17 0.2 | C16–38 0.5 | CX–16 0.8 | |||
| C2–9 | 1.1 | 4.3 | |||||||||
| C2–75 | 0.0 | 4.2 | |||||||||
| C4–56 | 0.8 | 4.3 | |||||||||
| C7–16 | 9.4 | 4.0 | |||||||||
| C9–28 | 1.8 | 3.7 | |||||||||
| C11–37 | 4.9 | 3.8 | |||||||||
| C18–50 | 0.2 | 4.1 | |||||||||
| Leptin Position | Position effect (%) | C5–81 2.7 | C11–37 0.9 | C15–49 0.1 | C16–38 0.3 | C17–47 0.6 | C19–47 3.2 | CX–16 0.1 | |||
| C1–82 | 2.0 | 4.9 | |||||||||
| C2–75 | 0.6 | 5.0 | |||||||||
| C9–28 | 0.3 | 4.3 | |||||||||
| C9–61 | 0.2 | 8.9 | |||||||||
| C14–20 | 2.1 | 4.6 | |||||||||
| C14–44 | 1.5 | 4.5 | |||||||||
| C18–50 | 1.3 | 4.4 | |||||||||
| Insulin Position | Position effect (%) | C4–66 3.3 | C11–14 0.5 | C15–6 1.5 | C17–57 1.3 | C18–50 0.2 | |||||
| C2–9 | 0.9 | 4.5 | |||||||||
| C4–56 | 2.8 | 4.9 | 6.7 | ||||||||
| C7–37 | 2.4 | 4.3 | |||||||||
| C14–28 | 0.9 | 4.6 | |||||||||
| IGF-I Position | Positive effect (%) | C2–60 1.6 | C2–109 0.3 | C11–1 0.2 | C13–61 0.1 | C14–20 0.2 | C14–28 0.5 | C15–6 0.6 | C15–26 0.9 | C15–49 0.8 | C17–57 1.8 |
| C1–9 | 2.7 | 4.8 | |||||||||
| C1–43 | 0.3 | 6.3 | |||||||||
| C2–13 | 0.3 | 4.5 | 3.9 | ||||||||
| C7–37 | 0.7 | 6.0 | |||||||||
| C10–40 | 14.3 | 4.6 | 5.4 | 4.2 | |||||||
| C14–44 | 0.2 | 8.8 | |||||||||
| C14–52 | 0.9 | 4.6 |
The contingency tables for body weight (BW), abdominal fat weight (AFW), muscle weight (MW), and the serum levels of leptin, insulin, and IGF-I present significant effects (P < 0.01) of the locus*locus interaction on every trait. The direct effect of a marker on a trait is given below the marker in the heading and behind the marker in the left column. Significant direct effects at 0.01 are marked bold. The direct marker effects on the F2 variance of a trait are given as reduction of the residual sum of squares with and without direct marker effect in the model. The effect of the locus*locus interaction was estimated as the reduction of the residual sum of squares with and without interaction effects in the model. Empty columns and rows were deleted from the contingency tables. Interaction effects exceeding P < 0.001 significance are framed.
Among the interactions affecting body weight, the loci C18-50 and CX-16 interacted with two or more different loci. For abdominal fat weight C14-44, C17-47 (Afw7), C17-57 (Afw7), and C18-50 were found to interact with two other loci, for muscle weight, C7-16 had interaction effects to two loci, for insulin, C4-56 (Insq1), and for IGF-I the loci C2-13 and C10-40 (Igf1q1), and C14-20 interacted with several other loci.
The interaction analysis provides evidence for coordinated regulation of body and muscle weight by the interaction of two pairs of loci (C2-9*C14-52 and C18-50*CX-16) (Table 7). Additionally, the interaction between C18-50*CX-16 controls the serum concentration of leptin. These interactions might contribute to the high genetic correlation between body and muscle weight.
Table 7.
Traits Affected by Interaction between Loci that Directly Influence One of the Traits
This is a condensed contingency table of 49 QTL-linked markers that were included in the interaction analysis. It shows affected traits by pairwise locus interaction. Empty columns and rows resulting from markers that did not contribute any significant interaction effect were omitted from the complete contingency table. Grey cells are on the diagonal of the full contingency table. Marker positions are given as chromosome number and (cM) position (e.g., C1–9 for D1Mit68). Behind the marker positions, the traits are given that have direct effect at the marker position (see Table 5). * indicates traits with nonsignificant effect in this study but which were significant in the cross DU6xDUKs (Brockmann et al. 1998). ** indicates traits that suggest effects but did not reach the 0.1 significance threshold in this study. All listed markers have at least interaction effect together with one other marker (i.e., QTL-linked markers were omitted entirely if they were not involved in any interactions). Locus*locus interactions that influence two or more traits are marked bold.
B, body weight; F, abdominal fat weight; M, muscle weight; L, leptin; In, insulin; Ig, IGF-I.
The coincidence of interaction effects influencing different traits by same marker loci is presented in Table 7. The interaction effect of one marker locus on different traits might indicate potential pleiotropic effects. The epistatic effect on the IGF-I serum concentration that was found for the highly significant Igf1q1 QTL on chromosome 10 (C10-40) is of interest in this respect. Although Igf1q1 had no direct effect on any other traits and also interaction with the marker alleles on chromosomes 11 (C11-1), 14 (C14-20), and 15 (C15-6) had significant influence on IGF-I concentration only, the loci interacting with Igf1q1 had potentially pleiotropic effect on fat accumulation (C11-1), on body weight, and leptin (C14-20), and on insulin (C15-6) when interacting with other loci. A coordinate effect on obesity was also found for genotypes of Igf1q2 at C18-50, which affected body weight in a joint action with C14-28 and CX-16 and affected abdominal fat weight in interaction with chromosomes 2 and 16. Furthermore, the leptin-influencing locus C19-47 had no effect on obesity but influenced fat accumulation when interacting with C11-48 (Bw4). Manifold interaction on growth and obesity was also found for chromosome 14 harboring Lepq1. Many effects of interaction between marker loci could be attributed to chromosome 17 (preferentially at C17-57, which is close to Afw7) and mainly affected obesity traits but did not influence muscle weight). The markers covering chromosome 11 interacted with several different loci on other chromosomes to influence specifically either protein, fat accumulation, or serum proteins.
Although the loci contributing to epistatic or pleiotropic effects often individually explained little trait variation, the coordinate effect of these loci on two or even more traits when interacting with other loci indicate a major role of the interacting chromosomal regions in the genetic determination of growth and obesity. Table 8 gives an overview of total effects on the phenotypic F2 population explained by direct QTL effects, interaction effects, and QTL and interaction effects together. The net effects of all interactions found at the 0.01 significance level contributed 33%, 36%, and 21% to the F2 variance of body, fat, and muscle weight, respectively. The interaction effects for the serum concentration of proteins accounted for 30%, 20%, and 33% of the phenotypic F2 variance of leptin, insulin, and IGF-I, respectively. The estimates provide evidence that direct QTL and interaction effects together account for 64% and 63% of the phenotypic variance of body weight and abdominal fat weight, respectively. QTL and interaction effects contribute 32% of the phenotypic F2 variance of muscle weight and 45% of the F2 variance of leptin, 21% of insulin, and 42% of IGF-I.
Table 8.
Joint Effect of the Identified QTLs and Locus*Locus Interactions
| Trait | Joint net effect of QTLs (%)a | Joint net effect of interaction (%)b | Joint net effect of QTLs and interaction together (%)b |
|---|---|---|---|
| BW | 34.9 | 32.9 | 64.4 |
| AFW | 34.1 | 35.5 | 62.8 |
| AFP | 26.2 | n.d. | n.d. |
| MW | 27.9 | 20.7 | 31.8 |
| LW | 31.3 | n.d. | n.d. |
| KW | 16.9 | n.d. | n.d. |
| SW | 17.3 | n.d. | n.d. |
| Leptin | 24.0 | 29.9 | 44.5 |
| Insulin | 9.1 | 20.3 | 21.2 |
| IGF-I | 22.5 | 32.8 | 41.6 |
The net effect is given as reduction of the residual sum of squares with and without inclusion of all QTLs, all interactions, or all QTLs and interactions together in the analysis.
The estimates for the joined net effect of all QTLs result from multipoint linkage analysis using the most likely QTL position.
The estimates for the joined net effect of interaction and of QTLs and interaction together result from the analysis using the marker positions next to the most likely QTL positions found via linkage analysis.
DISCUSSION
We have analyzed a single pedigree from a cross of the inbred high-growth selected mouse line DU6i with the inbred line DBA/2. In this pedigree, we found high correlations between the individual data on body composition and leptin and insulin serum concentrations. The evidence of a considerable correlation between leptin and muscle weight suggests that leptin may have direct effects on fat and muscle tissues. Insulin had similar correlations with both fat and muscle weight. The observed interdependence between leptin and insulin concentrations confirms findings in model animal studies (Sivitz et al. 1997). The inhibition of insulin by leptin in normal rodents (Poitout et al. 1998) seems to be distorted in DU6i mice. The correlation between IGF-I and body composition was much smaller than that for leptin and insulin.
Linkage analysis of the F2 pedigree indicated ten chromosomal regions harboring genes with influence on body weight, 12 regions affecting fat accumulation, and seven regions responsible for muscle weight. Most of the QTLs identified with effects significant at the suggestive level on body weight, abdominal fat weight, and liver weight were coincident to the QTL positions mapped in our previous study (Brockmann et al. 1998). Additionally, we have identified QTLs influencing body weight on chromosomes 7 and 9, but no longer see QTLs on chromosomes 15 and X. For abdominal fat weight, additional QTLs were detected on chromosomes 7, 12, 14 and X, and two previously reported on chromosomes 9 and 19 were not found. The X-chromosome-linked obesity QTL coincided with recently reported QTLs in KK (Taylor et al. 1999), AKR mouse lines (York et al. 1997), and in divergent selected mice (Horvat et al. 2000). In general, loci responsible for growth also effected liver and kidney weight, while spleen weight seemed to be regulated by completely different pathways. Differences in QTL detection between the two crosses (DU6i x DBA/2 versus the previous cross DU6 x DUKs) result primarily from the different genetic contrast but also from different random genotypic samples and changes in environmental conditions.
In this study, the biggest effects on body weight and fatness were detected on chromosomes 7 and 11. Although nonsuggestive linkage results have not been shown here, the data on QTLs that were not seen in one of the crosses have F-values for linkage, which suggests an effect, as they were often just below the threshold for suggestive linkage (data not shown). This is one of the reasons that we have included all these loci into the analysis of interaction between pairs of markers linked with the analyzed traits. Additional support for the existence of QTLs on growth and obesity detected in this linkage analysis is provided by results from other QTL mapping studies for several different loci (West et al. 1994; Warden et al. 1995; Cheverud et al. 1996; Keightley et al. 1996; Lembertas et al. 1996, 1997; Taylor and Phillips 1996, 1997; Rance et al. 1997; Kirkpatrick et al. 1998; Mehrabian et al. 1998; Moody et al. 1999; Morris et al. 1999; Horvat et al. 2000).
The genetic effects of the identified QTLs are complex. Most DU6i QTL alleles had additive increasing effect. However, despite the high selection response of DU6i, there were gene variants with smaller effects than those of the inbred line DBA/2. In some cases, heterosis was found for the action of the QTL alleles with highest increase or reduction of the trait in heterozygous animals. The observed sex-dependent influence on abdominal fat weight of the QTLs on chromosome 11 is likely to be due to the different fat pads measured in the two sexes, in males the testicular fat and in females the perimetrial fat, which may be regulated in a sex-specific manner. The finding of an influence of the subfamilies on the QTL on chromosome 5 affecting accumulation of abdominal fat suggests that even after long-term selection and four generations of inbreeding, not all alleles contributing to the heavy body weight were fixed in line DU6i.
So far, muscle weight has not been included in any linkage analyses. Our results indicate that the contribution of all seven QTLs to the development of the quadriceps is proportional to overall body-weight development. The most obvious candidate genes within the identified region are for chromosome 1, the myostatin gene (Mstn) and genes encoding IGF-binding proteins 2 and 5 (Igfbp-2, Igfbp-5); for chromosome 7, the insulin-like growth factor 1 receptor gene (Igf1r); for chromosome 11, the genes encoding signal transducing factors 5 and 3 (Stat3, Stat5a,b), and growth hormone (Gh); and for chromosome 12 the somatostatin receptor 1 gene (Smstr1). No strong candidate gene for muscle development is known for the two regions on chromosome 13. Except the QTL on chromosome 1, the loci influencing muscle weight on chromosomes 7, 11, 12, and 13 coincide with loci having effect on fat accumulation. This might be an indication either on the pleiotropic effect of the underlying genes or a hint on multiple genes having different effects in the identified linkage groups.
Concerning the inheritance of high or low serum concentrations of leptin, insulin, and IGF-I, we have identified novel QTLs responsible for the extreme difference in all serum parameters between DU6i and DBA/2 under nonfasting conditions. Although serum concentrations have been mostly analyzed under fasting conditions, the proteins are regulated by hunger, as well as by repletion. For our selected line, DU6i, a higher gross energetic efficiency (body energy gain/gross energy intake) has been found in comparison with controls, probably due to their increased capacity for feed consumption resulting in a greater amount of energy available for gain (Klein et al. 1999). Thus, the physiological condition of nonfasting reflects the most likely stage of rapidly-growing DU6i animals at ad libitum food supply. As the serum concentration of the measured proteins is influenced by the individual food consumption at the time of slaughtering, we expected and observed a high residual variance in our linkage analyses, causing less power to detect the genetic determinants for physiological differences. Thus, the joint effects of the identified QTLs influencing serum proteins were smaller compared to joint QTL effects identified for body weight, fat accumulation, and muscle weight. Nevertheless, the identified QTLs for serum parameters and their interaction to other loci contribute to search for nodal points of growth and obesity regulation.
For the regulation of serum leptin concentration, a significant influence of chromosome 14 most likely at 28.9 cM (Lepq1) was discovered. This leptin-regulating locus has not been reported in either humans or in any model animal. So far, no candidate gene is known for the strong effect of this QTL, which might influence the leptin concentration. The identified murine region corresponds to human chromosome 14q11.2, of which there has not been found human linkage to fat distribution (Chagnon et al. 2000).
A locus with suggestive effect on leptin concentration was identified at 66 cM on chromosome 4. The map position coincides with the recently identified QTL in an intercross between C57BL/J6 and CAST/Ei (Mehrabian et al. 1998). The most likely location of the leptin QTL on chromosome 4 is distal to the Lepr gene. Comparative sequencing of the Lepr gene in DU6i and DBA/2 mice revealed an amino acid exchange in the protein at amino acid position 359 (U. Reichart and G. Brockmann, pers. comm.). This position is inside the leptin-binding domain that has been localized to residues 323 to 640 (Fong et al. 1998). Thus, the identified polymorphism might contribute to the identified QTL effect of chromosome 4. Obviously, there is another gene distal to Lepr responsible for the QTL effect on serum leptin, fat accumulation, and body weight that might interact with the Lepr locus. Our analysis indicated a cosegregation of the locus influencing leptin concentration with Insq1. This finding coincides with the mapping of insulin-susceptible loci in NOD mice (Idd9) (Rodrigues et al. 1994) and in NZO mice (Igel et al. 1997). To the same chromosomal region, an effect on heat loss was added (Moody et al. 1999).
Two loci on chromosomes 10 (Igf1q1) and 18 (Igf1q2) had major effect on IGF-I concentration. The Igf1q1 QTL mapped directly at the Igf1 gene locus, which most likely is the QTL underlying gene, as different IGF-I transcript amounts have been shown between selected DU6 and unselected mice (Timtchenko et al. 1999). For Igf1q2 on chromosome 18 no candidate gene is known. The Igf1q1 and Igf1q2 loci itself did not indicate effect on body, fat or muscle weight.
The analysis of interaction between loci contributed to the explanation of phenotypic variance in the F2 population in a different manner: (1) The interaction analysis showed loci that did not attribute to the trait variance by itself but by interaction with other loci. Examples for this kind of conclusion are loci responsible for leptin (Lepq1) or IGF-I (Igf1q1, Igf1q2), which contributed to the phenotypic F2 variance of fat and/or muscle weight only by interaction. (2) The interaction analysis gave evidence for a pair of loci with coordinate effect on correlated traits (e.g., the influence of C18-50*CX-16 on body and muscle weight and leptin concentration). (3) The interaction analysis proved loci potentially playing a central role in the genetic regulation of complex phenotypes related to growth and obesity by various types of regulation contributed by a single marker locus. Two of these loci are the distal part of chromosome 17 (C17-57) and the middle part of chromosome 14 (C14-20) with influence on insulin and IGF-I, which might be an indication of major regulatory factors. (4) The interaction of several different loci on one chromosome affecting diverse traits (fat, muscle, or serum proteins) helps distinguish multiple loci in the identified QTL linkage group contributing by small effects to the variance, rather than having one single gene responsible for the estimated QTL effect. This situation might be assumed for chromosome 11, for example. For chromosome 11, two QTLs have been found for body, fat, and muscle weight by test statistics. However, the genome-wide interaction of different loci on chromosome 11 with loci on other chromosomes allows us to expect at least a third QTL near C11-37. The loci contributing to the phenotypic variance by small effects only are difficult to isolate, even by constructing congenic lines. The identification of interacting chromosomal segments may be used for the targeted construction of bicongenic lines, which combine the additive and dominant effects of every locus and additionally the effect of interaction.
The contribution of interaction effects to the phenotypic F2 variance was estimated at about one-third. Together, the results of the QTL analysis and the locus* locus interaction analysis contributed about two-thirds of the phenotypic variance of body weight and abdominal fat weight in the F2 population. The direct QTL and interaction effects together considerably increase the explained proportion of phenotypic F2 variance of muscle weight and the serum concentrations of leptin, insulin, and IGF-I. The estimates might be biased due to the simplified model we used for the estimation of interaction effect. Our tests are likely conservative in the estimation of effects contributing to epistasis. This results mainly from recombination between the QTL and the marker position, which reduce the estimates for additive and dominant genetic effects and, thus, most likely, as well the genetic interaction. Hence, for a more detailed analysis of the interaction effects, the statistical tests of multipoint linkage analysis have to be combined with interaction analysis of most likely estimates of QTL locations as suggested by Haley and Knott (1992) and Routman and Cheverud (1997). Additionally, the evaluation of quantitative changes of combined additive and dominant effects of epistasis and pleiotropy is necessary, as compensatory up- and down-regulation of correlated traits by gene interaction is an important factor of genetic control of body weight homeostasis (Gibson 1996). Nevertheless, the identified QTL positions and interaction effects from our cross may help to identify genes from the relevant chromosomal region that underlay the complex net of gene actions responsible for growth and obesity regulation.
METHODS
Mouse Lines
This study was carried out on the mouse line DU6i, which has been inbred for four generations from the high-body-weight selected line DU6 and the commercial inbred mouse line DBA/2 (Harlan Nederland, Horst). The outbred line DU6 had been selected for 78 generations for high body weight at the age of 42 d (Bünger et al. 1990). Line DU6 descends from original crosses of four base (NMRI orig., Han: NMRI, CFW, CF1) and four inbred (CBA/Bln, AB/Bln, C57BL/Bln, XVII/Bln) populations at the Research Institute for the Biology of Farm Animals, Dummerstorf, Germany (Schüler 1985). Animals were fed ad libitum with a breeding diet containing 12.5 MJ/kg metabolic energy with an average content of 22.5% crude protein, 5.0% crude fat, 4.5% crude fiber, 6.5% crude ash, 13.5% water, 48.0% N-free extract, vitamins, trace elements, amino acids, and minerals (Altromin diet 1314, Germany). Seventeen and 20 nonfasting males were analyzed for DU6i and DBA/2, respectively, for the characterization of the two mouse lines at 42 d.
Pedigree Design
QTL analysis was performed using a pedigree of F2 intercross design. These were established by crossing one female of the high-growth inbred line DU6i to one male of the contrast line DBA/2. A large pedigree with a total of 411 F2 offspring was generated by repeated mating of the parents and subsequently of repeated mating within subfamilies of 12 pairs of F1 offspring. Mating was initially at the age of 10 wk and repeated after 6 wk. The pedigree structure is shown in Table 2.
Phenotypic Measures
The phenotypic data were recorded at 42 d of age, the age of selection in all generations. The quantitative measurements used in the QTL analysis were the weights of the body (BW), abdominal fat (AFW), muscle (MW), liver (LW), kidney (KW), and spleen (SW), as well as the serum concentrations of leptin, insulin, and IGF-I. Nonfasting F2 animals were slaughtered between 0900 and 1200. Animals were decapitated to obtain the largest possible blood serum volumes for the analysis of physiological parameters. The abdominal fat measured in males was the testicular fat and in females the perimetrial fat. The ratio of abdominal fat to body weight was defined as abdominal fat percentage (AFP). The weight of the quadriceps comprising musculus rectus femulus, musculus vastus intermedicus, musculus vastus lateralis, and musculus vastus medialis was recorded as representative of muscle development (MW).
Leptin was measured by the ‘Quantikine Murine’ enzyme-linked immunosorbert assay (ELISA) (R&D Systems, Wiesbaden, Germany). The determination of insulin in mouse sera was performed by an ultrasensitive rat insulin ELISA (Diagnostic Research Group, Marburg, Germany), which showed a 100% cross reactivity with mouse insulin. IGF-I was determined by ELISA in serum after acid ethanol extraction as described previously (Kratzsch et al. 1993).
Markers
Markers were chosen for their high variability between appropriate inbred lines from the mouse genome database (MGD). Mouse MapPair primers were purchased from Research Genetics. Over 630 microsatellite markers were tested for informative parental alleles, before the parents, and F2 were genotyped for 93 loci covering all chromosomes at an average spacing of 14.1 cM. The marker map is shown in Table 4. DNA preparation and genotyping were performed using standard methods, as described by Das et al. (1996). All genotyping results were scored twice and runs were repeated when there were discrepancies.
Statistical Analysis
For linkage analysis, a pedigree specific marker map initially was generated with the program CRIMAP (Lander and Green 1987). As the map was consistent with the published map—marker D8Mit112 was reassigned to chromosome 9 (Jackson et al. 1999)—we used the marker distances of the consensus map of the mouse genome (Dietrich et al. 1996) for the mapping of QTLs. This allowed the direct incorporation of identified QTL positions to the consensus map in MGD. The differences between the use of the consensus map and the pedigree-specific map slightly changed F-values and estimates but did not change the results in principle.
For the analysis of the pedigree, the influences of sex, parity (five classes), subfamily (i.e., F2 animals from the same pair of F1 parents: 12 classes), and pupsize (number of offspring per litter: five classes) initially were estimated for every trait via variance analysis (SAS 1990). All factors were found to be significant and, thus, were included as fixed effects in the linkage analysis. The test for normality of the distribution after adjusting the raw data for the fixed effects of sex, parity, family, and pupsize (procedure capability, SAS) showed that body weight, abdominal fat weight and percentage, muscle weight, and the weights of the inner organs were normally distributed. Leptin and IGF-I were normally distributed after logarithmic transformation of the original serum concentration data. Therefore, logarithmic transformed data were used throughout all analyses (correlation, linkage, interaction) of leptin and IGF-I.
QTL analyses were performed by multiple regression (Haley et al. 1994). Initially, the standard interval mapping model was used with a single QTL on the linkage group. Once a single QTL on a chromosome was identified, the presence of a second QTL was investigated by performing a grid search at 2 cM intervals. The two-QTLs model was accepted if there was a significant improvement over the best possible one-QTL model at the nominal P < 0.05 using a variance ratio (F) test with 3 degrees of freedom (for the additional additive and dominant effect and position estimated for the second QTL). The obtained estimates were revised by fitting QTLs as background genetic effects on other linkage groups, as suggested by Zeng (1993) and Jansen (1993). Background genetic effects were included as cofactors for all loci showing effects at the suggestive level of significance. During that procedure, a background effect was dropped from the analysis when analyzing its own position. The sex chromosome was analyzed as a pseudo-autosomal chromosome, as all markers were located in that region. In additional analyses, body weight was introduced as a covariate in the analysis of abdominal fat weight and muscle weight to examine the dependence between these traits. The joint effect of all identified QTLs for a trait (i.e., variance explained of the F2) was estimated as reduction of the residual mean square in the one QTL analysis fitting all QTLs in comparison with no QTL fitted. Where a QTL was identified, the interaction of the QTL effects with intrapedigree subfamily and sex were tested and were accepted as significant at the nominal level of P < 0.05.
The experiment-specific empirical threshold values of the test statistics from the regression analysis were estimated with the permutation test proposed by Churchill and Doerge (1994). One thousand replicates were performed with a 1-cM step size. Previous experience has shown that thresholds derived by permutation analysis for different traits such as those analyzed here are very similar. Therefore, we have used a common set of thresholds derived from permutation analysis of body weight for all traits. Levels for genome-wide highly significant (α = 0.01) and significant (α = 0.05) linkage were used (Lander and Kruglyak 1995). The chromosome-wise 0.05 significance levels were taken as genome-wide thresholds for suggestive linkage. This is reasonable because using a 0.05 chromosomal threshold with each of 20 chromosomes (19 autosomes and the pseudo-autosomal region), we expect to see one type-I error on average in the scan on all chromosomes. We used the one-LOD drop to provide an indicative support interval for all genome-wide suggestive QTLs. Gene symbols were assigned to QTLs exceeding the genome-wide significant thresholds, to loci where two QTLs were found in a linkage group (at P < 0.05) and both single QTLs were genome-wide suggestive (chromosome-wide significant at P < 0.05) and to genome-wide suggestive QTLs that have been confirmed from our previous study (Brockmann et al. 1998).
To examine the interaction between QTLs, we used the general linear model of variance analysis (SAS). For a specific QTL detected in the analyses with a single or two-linked QTLs, the nearest marker was taken to analyze the interaction effect between QTLs. The model of variance analysis included the effects of sex, subfamily, parity, pupsize, the single effects of the QTL identified for the specific trait at two selected loci, and the interaction between these two specific loci. In the interaction analysis, two degrees of freedom were used for each marker fitted and four degrees of freedom for the interaction between two markers. Forty-nine out of 93 markers were included in the analyses. Because independent multiple tests were carried out, we used P < 0.001 as a stringent threshold for our test statistics, and P < 0.01 as a relatively relaxed threshold for the acceptance of interaction. We estimated the effect of an interaction on the phenotypic F2 variance as reduction of residual sum of squares with and without inclusion of the marker–marker interaction in the regression model. The effects of single-marker loci on the trait were estimated similarly. These may deviate from the estimated effect of a linked QTL from the linkage analysis if the QTL position does not exactly coincide with the marker position. For easier tracking of interaction results, we used shortcuts for markers that contained the chromosome number and the cM position of the marker (e.g., C1-9 for D1Mit68 on chromosome 1 at 9 cM).
Acknowledgments
Excellent technical assistance was provided by Hannelore Tychsen for DNA preparation and genotyping. This work was supported by the German Research Foundation, Grant No. BR 1285/4 and the H. Wilhelm Schaumann Stiftung. J.K. was supported by the Interdisziplinäres Zentrum für Klinische Forschung and Bundesministerium für Bildung und Forschung (Grant No. B15). C.S.H. acknowledges support from the Biotechnology and Biological Sciences Research Council and Ministry of Agriculture, Fisheries and Food.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Gudrun.Brockmann@fbn-dummerstorf.de; FAX 049 38208 68702.
Article published online before print: Genome Res., 10.1101/gr.149900.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.149900.
REFERENCES
- Brockmann G, Timtchenko D, Das P, Renne U, Freyer G, Kuhla S, Teuscher F, Wolf J, Kühn C, Schwerin M. Detection of QTL for body weight and body fat content in mice using genetic markers. J Anim Breed Genet. 1996;113:373–379. [Google Scholar]
- Brockmann GA, Haley CS, Renne U, Knott SA, Schwerin M. QTLs affecting body weight and fatness from a mouse line selected for high growth. Genetics. 1998;150:369–381. doi: 10.1093/genetics/150.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bünger L, Herrendörfer G, Renne U. Results of long term selection for growth traits in laboratory mice. Proceedings of the 4th World Congress on Genetics Applied to Livestock Production, Univ Edinburgh, Edinburgh, Scotland. 1990;13:321–324. [Google Scholar]
- Chagnon YC, Perusse L, Weisnagel SJ, Rankinen T, Bouchard C. The human obesity gene map: The 1999 update. Obes Res. 2000;8:89–117. doi: 10.1038/oby.2000.12. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Routman EJ. Epistasis and its contribution to genetic variance components. Genetics. 1995;130:1455–1461. doi: 10.1093/genetics/139.3.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Routman EJ, Duarte FAM, van Swinderen B, Cothran K, Perel C. Quantitative trait loci for murine growth. Genetics. 1996;142:1305–1319. doi: 10.1093/genetics/142.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox NJ, Frigge M, Nicolae DL, Concannon P, Hannis CL, Bell GI, Kong A. Loci on chromosomes 2 (NIDDM1) and 15 interact to decrease susceptibility to diabetes in Mexican Americans. Nat Genet. 1999;2:213–215. doi: 10.1038/6002. [DOI] [PubMed] [Google Scholar]
- Das P, Brockmann G, Meyer L, Renne U, Freyer G, Schwerin M. The effect of a restricted region of chromosome 11 on body weight in mice under special consideration of the growth hormone gene locus. Arch Tierz. 1996;39:185–194. [Google Scholar]
- Dietrich WF, Miller J, Steen R, Merchant MA, Damron-Boles D, Husain Z, Dredge R, Daly MJ, Ingalls KA, O'Connor TJ, et al. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- Fong TM, Huang RR, Tota MR, Mao C, Smith T, Varnerin J, Karpitskiy VV, Krause JE, van der Ploeg LH. Localization of leptin binding domain in the leptin receptor. Mol Pharmacol. 1998;53:234–240. doi: 10.1124/mol.53.2.234. [DOI] [PubMed] [Google Scholar]
- Galli J, Li LS, Fakhrai-Rad H, Kamel A, Marcus C, Norgren S, Luthman H. Pathophysiological and genetic characterization of the major diabetes locus in GK rats. Diabetes. 1999;48:2463–2470. doi: 10.2337/diabetes.48.12.2463. [DOI] [PubMed] [Google Scholar]
- Gibson G. Epistasis and pleiotropy as natural properties of transcriptional regulation. Theor Popul Biol. 1996;49:58–89. doi: 10.1006/tpbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Haley CS, Knott SA. A simple method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Haley CS, Knott SA, Elsen LM. Mapping quantitative trait loci in crosses between outbred lines using least squares. Genetics. 1994;136:1195–1207. doi: 10.1093/genetics/136.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvat S, Bunger L, Falconer VM, Mackay P, Law A, Bulfield G, Keightley PD. Mapping of obesity QTLs in a cross between mouse lines divergently selected on fat content. Mamm Genome. 2000;11:2–7. doi: 10.1007/s003350010002. [DOI] [PubMed] [Google Scholar]
- Igel M, Becker W, Herberg L, Joost HG. Hyperleptinemia, leptin-resistance, and polymorphic leptin receptor in the New Zealand obese mouse. Endocrinology. 1997;138:4234–4239. doi: 10.1210/endo.138.10.5428. [DOI] [PubMed] [Google Scholar]
- Jackson AU, Fornes A, Galecki A, Miller RA, Burke DT. Multiple-trait quantitative trait loci analysis using a large mouse sibship. Genetics. 1999;151:785–795. doi: 10.1093/genetics/151.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC. Interval mapping of multiple quantitative trait loci. Genetics. 1993;135:205–211. doi: 10.1093/genetics/135.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Hardge T, May L, Bulfield G. A genetic map of quantitative trait loci for body weight in the mouse. Genetics. 1996;142:227–235. doi: 10.1093/genetics/142.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick BW, Mengelt A, Schulman N, Martin ICA. Identification of quantitative trait loci for prolifacacy and growth in mice. Mamm Genome. 1998;9:97–102. doi: 10.1007/s003359900696. [DOI] [PubMed] [Google Scholar]
- Klein M, Schadereit R, Scholze H, Renne U. Studies on energy metabolism in lines of mice selected for different growth parameters. J Anim Physiol A Anim Nutr. 1999;81:75–89. [Google Scholar]
- Kratzsch J, Blum WF, Schenker E, Keller E, Jahreis G, Haustein B, Ventz M, Rotzsch W. Measurement of insulin-like growth factor I (IGF-I) in normal adults, patients with liver cirrhosis and acromegaly: Experience with a new enzyme immunoassay. Exp Clin Endocrinol. 1993;101:144–149. doi: 10.1055/s-0029-1211221. [DOI] [PubMed] [Google Scholar]
- Lander E, Green P. Construction of multilocus genetic linkage maps in humans. Proc Nat Acad Sci. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lembertas AV, Fisler JS, Warden CH, Wen P-Z, Xia Y-R, Lusis AJ. A locus on the X chromosome is linked to body length in mice. Mamm Genome. 1996;7:171–173. doi: 10.1007/s003359900048. [DOI] [PubMed] [Google Scholar]
- Lembertas AV, Pérusse L, Chagnon YC, Fisler JS, Warden CH, Purcell-Huynh DA, Dionne FT, Gagnon J, Nadeau A, Lusis AJ, et al. Identification of an obesity quantitative trait locus on mouse chromosome 2 and evidence of linkage to body fat and insulin on the human homologous region 20q. J Clin Invest. 1997;100:1240–1247. doi: 10.1172/JCI119637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei M, Halaas J, Ravussin E, Pratley RE, Lee GH, Zhang Y, Fei H, Kim S, Lallone R, Ranganathan S, et al. Leptin levels in human and rodent: Measurement of plasma leptin and ob RNA in obese and weight-reduced subjects. Nat Med. 1995;1:1155–1161. doi: 10.1038/nm1195-1155. [DOI] [PubMed] [Google Scholar]
- Mehrabian M, Wen PZ, Fisler J, Lusis AJ. Genetic loci controlling fat, lipoprotein metabolism, and insulin levels in a multifactoial mouse model. J Clin Invest. 1998;101:2485–2496. doi: 10.1172/JCI1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MGD, Mouse Genome Database, Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, Maine. URL ( http://www.informatics.jax.org/) June 1998.
- Mitchell BD, Gosh S, Schneider JL, Birznieks G, Blangero J. Power of variance component linkage analysis to detect epistasis. Genet Epidemiol. 1997;14:1017–1022. doi: 10.1002/(SICI)1098-2272(1997)14:6<1017::AID-GEPI76>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Moody DE, Pomp D, Nielsen MK, Van Vleck LD. Identification of quantitative trait loci influencing traits related to energy balance in selection and inbred lines of mice. Genetics. 1999;152:699–711. doi: 10.1093/genetics/152.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KH, Ishikawa A, Keightley PD. Quantitative trait loci for growth traits in C57BL/6J x DBA/2J mice. Mamm Genome. 1999;10:225–228. doi: 10.1007/s003359900977. [DOI] [PubMed] [Google Scholar]
- Norman RA, Tataranni PA, Pratley R, Thompson DB, Hanson RL, Prochazka M, Baier L, Ehm MG, Sakul H, Foroud T, et al. Autosomal genome scan for loci linked to obesity and energy metabolism in Pima Indians. Am J Hum Genet. 1998;62:659–668. doi: 10.1086/301758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitout V, Rouault C, Guerre-Millo M, Briand I, Reach G. Inhibition of insulin secretion by leptin in normal rodent islets of Langerhans. Endocrinology. 1998;139:822–826. doi: 10.1210/endo.139.3.5812. [DOI] [PubMed] [Google Scholar]
- Rance KA, Hill WG, Keightley PD. Mapping quantitative trait loci for body weight on the X-chromosome in mice. I Analysis of a reciprocal F2 population. Genetical Research. 1997;70:117–124. doi: 10.1017/s0016672397002917. [DOI] [PubMed] [Google Scholar]
- Rehfeld C, Bünger L. Effects of long-term selection of laboratory mice on parameters of muscle growth and muscle structure. Arch Tierz. 1990;33:507–516. [Google Scholar]
- Rodrigues NR, Cornall RJ, Chandler P, Simpson E, Wicker LS, Peterson LB, Todd JA. Mapping of an insulin-dependent diabetes locus, Idd9, in NOD mice to chromosome 4. Mamm Genome. 1994;5:167–170. doi: 10.1007/BF00352349. [DOI] [PubMed] [Google Scholar]
- Routman EJ, Cheverud JM. Gene effects on a quantitative trait: Two-locus epistatic effects measured at microsatellite markers and at estimated QTL. Evolution. 1997;51:1654–1662. doi: 10.1111/j.1558-5646.1997.tb01488.x. [DOI] [PubMed] [Google Scholar]
- SAS . SAS User's guide: Statistics. Cary, North Carolina: SAS Institute; 1990. [Google Scholar]
- Schüler L. Mouse strain Fzt:Du and its use as model in animal breeding research. Arch Tierz. 1985;28:357–363. [Google Scholar]
- Sivitz WI, Walsh SA, Morgan DA, Thomas MJ, Haynes WG. Effects of leptin on insulin sensitivity in normal rats. Endocrinology. 1997;138:3395–3401. doi: 10.1210/endo.138.8.5327. [DOI] [PubMed] [Google Scholar]
- Taylor BA, Phillips SJ. Obesity QTLs on mouse chromosomes 1 and 7 by selective DNA polling. Genomics. 1996;34:389–398. doi: 10.1006/geno.1996.0302. [DOI] [PubMed] [Google Scholar]
- ————— Detection of obesity QTLs on mouse chromosomes 2 and 17. Genomics. 1997;43:249–247. doi: 10.1006/geno.1997.4835. [DOI] [PubMed] [Google Scholar]
- Taylor BA, Tarantino LM, Phillips SJ. Gender-influenced obesity QTLs identified in a cross involving the KK type II diabetes-prone mouse strain. Mamm Genome. 1999;10:963–968. doi: 10.1007/s003359901141. [DOI] [PubMed] [Google Scholar]
- Timtchenko D, Kratzsch J, Sauerwein H, Wegner J, Souffrant WB, Schwerin M, Brockmann GA. Fat storage capacity in growth-selected and control mouse lines is associated with line-specific gene expression and plasma hormone levels. Int J Obes Relat Metab Disord. 1999;23:586–594. doi: 10.1038/sj.ijo.0800872. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Laubichler MD, Bagheri-Chaichian H. Genetic measurement theory of epistatic effects. Genetica. 1998;102/103:569–580. [PubMed] [Google Scholar]
- Warden CH, Fisler JS, Shoemaker SM, Wen PZ, Svenson KL, Pace MJ, Lusis AJ. Identification of four chromosomal loci determining obesity in a multifactorial mouse model. J Clin Invest. 1995;95:1545–1552. doi: 10.1172/JCI117827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West DB, Waguespack J, York B, Goudey-Lefevre J, Price RA. Genetics of dietary obesity in AKR/J x SWR/J mice: Segregation of the trait and identification of a linked locus on chromosome 4. Mamm Genome. 1994;5:546–552. doi: 10.1007/BF00354928. [DOI] [PubMed] [Google Scholar]
- York B, Lei K, West DB. Inherited non-autosomal effects on body fat in F2 mice derived from a AKR/J X SWR/J cross. Mamm Genome. 1997;8:726–730. doi: 10.1007/s003359900554. [DOI] [PubMed] [Google Scholar]
- Zeng Z-B. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci. 1993;90:10972–10976. doi: 10.1073/pnas.90.23.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]