Abstract
Background
Linkage studies have implicated chromosome 1q24 as a putative locus for hereditary prostate cancer. The RNASEL gene maps to 1q24 and has been associated with prostate cancer risk in multiple family-based linkage studies. The RNASEL gene product combats viral infection by degrading viral RNA and inducing apoptosis of infected cells. Few studies have evaluated the role of RNASEL variants in unselected or sporadic prostate cancer, or have considered the potential interaction between RNASEL variants and patient characteristics associated with past viral infection.
Methods
Ten SNPs in the RNASEL gene were genotyped in 1,308 prostate cancer cases and 1,267 age-matched controls from prior population-based, case-control studies. The association between each SNP and haplotype with prostate cancer risk was calculated using logistic regression. Associations stratified by Gleason score were evaluated using polytomous regression. The likelihood ratio test was used to investigate effect modification by history of prostatitis.
Results
Two RNASEL SNPs were associated with overall increases in prostate cancer risk (OR=1.13 for each variant allele of rs12723593; OR=1.88 for any variant allele of rs56250729). Risk estimates did not vary substantially by Gleason score, but there was effect modification for the variant allele of rs635261 by history of prostatitis (p=0.02).
Conclusions
This study identified three RNASEL variants that are associated with risk for prostate cancer. Further research is required to confirm these results and to better understand the potential role RNASEL variants may play in the etiology of sporadic prostate cancer.
Keywords: Prostate cancer, RNASEL, Gleason grade, prostatitis
Introduction
Viruses are known to play a causal role in several types of cancer, including cervical, liver, and head and neck cancers. The association between viral infection and cancer incidence may be due to a virus’ ability to alter host tumor suppressor gene activity by inducing hypermethylation of genomic DNA[1] or to instigate a state of chronic inflammation that leads to carcinogenesis[2]. Studies regarding the role of viral infections in prostate cancer have yielded mixed results. The strongest evidence for an association between viral infection and prostate cancer comes from studies of xenotropic murine leukemia virus-related virus (XMRV), which was first discovered in malignant prostate tumor tissue in 2006[3]. Most recently, a study of prostate tissue collected at prostatectomy (cases) or transurethral resection of the prostate (TURP controls) reported that infection with XMRV was more frequent in tumor tissue from cases than controls, and the frequency of viral infection was highest in cases with higher grade tumors[4].
If viruses play a role in prostate cancer, it is possible that polymorphisms in genes that are activated by a specific viral infection could be associated with prostate cancer risk. The RNASEL gene is one such gene that has several assigned functions for combating viral infection, including causing targeted degradation of viral RNA[5] and inducing apoptosis of infected cells[6]. In particular, RNASEL’s ability to mediate apoptosis may be important in prostate carcinogenesis. In addition, the effect of RNASEL gene variants on prostate cancer risk may vary depending on host characteristics related to immunity and history of exposure to various infectious agents.
Several epidemiologic studies have investigated the association between RNASEL variants and prostate cancer risk, with conflicting results. Most prior studies focused on hereditary prostate cancer (HPC), as this gene was first implicated in a family-based linkage study of HPC where affected men in two of 140 such families demonstrated mutations in RNASEL that segregated in affected men[7]. However, subsequent studies failed to substantiate a role for RNASEL in HPC [8–11]. Few studies have evaluated this gene in relation to sporadic prostate cancer, which constitutes the majority of cases. In this analysis, the association between RNASEL gene variants and prostate cancer was evaluated using data from two population-based, case-control studies. Single nucleotide polymorphisms (SNPs) across the gene region were selected for genotyping. The main effects of RNASEL variants on prostate cancer risk were evaluated, followed by an investigation of whether a history of prostatitis modified the risk estimate associated with each gene variant.
Materials and Methods
Population
Study subjects were participants in one of two previously described population-based case-control studies of prostate cancer in Caucasian and African-American residents of King County, Washington (Study I and Study II) [12,13]. Study I cases were diagnosed at ages 40 to 64 and between January 1, 1993 and December 31, 1996, and Study II cases were diagnosed at ages 35 to 74 and between January 1, 2002 and December 31, 2005. All cases were ascertained through the Seattle-Puget Sound SEER cancer registry. In both studies combined, 2,244 eligible prostate cancer cases were identified, 1,457 (64.9%) of which were interviewed and provided blood for genotyping. Controls without a self-reported physician’s diagnosis of prostate cancer were identified and recruited via random digit dialing. Controls were frequency matched to cases in 5-year age groups and were recruited concurrently with cases. A total of 2,448 eligible controls was identified, 1,352 (55.2%) of which were interviewed and provided sufficient blood for genotyping. The analyses described here were restricted to Caucasian participants (1,308 cases and 1,267 controls).
SNP genotyping
Twelve SNPs in RNASEL were selected for genotyping using data from the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/), the Genome Variation Server (http://gvs.gs.washington.edu/GVS/), and prior publications that evaluated variation in this gene. Six SNPs were nonsynonymous: Gly59Ser (no rs#), Ile97Leu (rs56250729), E265X (no rs#), S406F (no rs#), Arg462Gln (rs486907), and Asp541Glu (rs627928). The remaining SNPs included one within intron 5 (rs11807829), four in the 3′ untranslated region (UTR) (rs627839, rs635261, rs12723593, and rs12729828) and one in the 5′ promoter region (rs682585). All SNPs were genotyped using the Applied Biosystems SNPlex Genotyping System. Genotypes were called using the Applied Biosystems 3730xl DNA Analyzer with GeneMapper Software (www.appliedbiosystems.com). Blind duplicates for 144 participants were evenly distributed throughout the samples for quality control purposes. Cases and controls were evenly distributed across genotyping batches, and laboratory personnel were blinded to the case or control status of samples. Duplicate concordance was 100% for eleven SNPs, and 99% for one SNP (rs627928). Genotyping failed for less than 1.5% of participants for all SNPs. One SNP was found to be monomorphic (E265X) in our population, and genotyping failed for one SNP (S406F).
Statistical analyses
All SNP genotypes were tested for departure from Hardy-Weinberg equilibrium using data from controls. Logistic regression analyses were conducted to calculate odds ratios (ORs) and 95% confidence intervals (CIs) for the odds of prostate cancer in men with various RNASEL SNP genotypes. Two logistic regression models were constructed for each SNP in order to estimate ORs for dominant and recessive genetic effects, and a third model was constructed to estimate allele dosage effect (0, 1, or 2 minor alleles). We considered all three genetic models for each SNP, and selected the model with the most statistically significant odds ratio for presentation. In addition, multinomial logistic regression was used to estimate the odds of lower grade (Gleason score ≤ 3 + 4) and higher grade (Gleason score ≥ 4 + 3) prostate cancer associated with SNP genotypes compared to controls. Multinomial models were also used to compare risk of prostate cancer by stage (local vs. regional/distant) and by a composite measure of aggressiveness (less vs. more). Comparatively more aggressive cases were defined as having: Gleason score ≥7 (4+3), or regional or distant stage, or PSA ≥20 ng/mL at diagnosis [14]. A χ2 test was used to test for significant differences between risk estimates for lower versus higher grade prostate cancer associated with each SNP genotype. All regression models were adjusted for 5-year age groups, to account for the age matching strategy used to select controls.
Family history of prostate cancer in a first-degree relative(s) and prostate cancer screening within the five years prior to reference date (never, digital rectal examination only, or prostate-specific antigen test) were considered as potential confounders. Results from age-adjusted models were compared with models containing these potential confounders, which did not change the risk estimates associated with each SNP by 10% or more. Thus, the final logistic models were not adjusted for additional covariates.
To determine whether the effect of RNASEL polymorphisms on risk differed based on host characteristics, logistic regression models including interaction terms for SNP genotype by family history of prostate cancer, self-reported history of a sexually transmitted disease (STD), and self-reported history of prostatitis were evaluated. Each characteristic was treated as a binary variable, i.e., any first-degree family history of prostate cancer vs. none, history of STD vs. none, and history of prostatitis vs. none. The likelihood ratio test was used to compare the full (i.e., including the interaction term) vs. reduced models, and stratified logistic regression models were constructed for SNPs with significant interactions with family history or prostatitis.
For haplotype analyses, HPlus version 2.5 (http://cdsweb01.fhcrc.org/HPlus/default.aspx) was used to construct haplotypes with the individual SNPs found to have at least a borderline significant association with prostate cancer. In addition, Haploview version 4.1 was used to construct haplotype blocks using all 10 SNPs [15]. Haplotype blocks were assembled in Haploview using the 95% CI method described by Gabriel et al. [16]. The logistic regression analysis function within HPlus was used to estimate age-adjusted ORs and 95% CIs for the association between each haplotype and prostate cancer risk.
A permutation procedure was used to account for multiple testing[17]. Each subject’s case-control status was linked with age and then permuted to approximate the distribution of age-adjusted p-values under the null hypothesis. Maintaining the link between case-control status and age preserved the association between age and prostate cancer risk, and ensured that the p-values calculated for the permutation analysis were comparable with the original age-adjusted p-values. Permutation p-values were compared to nominal p-values to determine if each association was statistically significant after adjustment for multiple comparisons, as described previously [18].
Results
Ninety-one percent of controls and 88% of cases had no missing genotype data, and 3% of controls and 6% of cases were missing data for more than one SNP. Less than 1% of cases and controls were missing data for all SNPs. The median Gleason grade was 6, with 15.5% (N = 202) of cases having a Gleason grade > 7 (4+3). All cases and controls were included in the analyses involving SNPs for which they had genotyping data available, thus, the number of cases and controls varies slightly for each set of analyses. None of the 10 SNPs was found to deviate from Hardy-Weinberg equilibrium in controls (all p-values > 0.05).
Odds ratios for prostate cancer associated with each RNASEL SNP genotype are shown in Table 1. One SNP (rs12723593) was associated with a borderline increase and one SNP (rs56250729) with a significant increase in prostate cancer risk. For rs56250729, there was an 88% (95% CI: 1.05–3.34) elevation in the relative risk for prostate cancer in men carrying the variant allele compared to those homozygous for the common allele. After accounting for the effects of multiple testing, neither of these associations were statistically significant.
Table 1.
Association between ten SNPs in RNASEL and prostate cancer risk
| SNP rs# | Major/minor alleles | Location/effect on RNASEL protein | Position relative to initiation codon | Best-fitting model | Minor allele frequency | OR (95% CI) | Nominal p-value | |
|---|---|---|---|---|---|---|---|---|
| Cases N = 1308 | Controls N = 1267 | |||||||
| rs12723593 | C/G | 3′ | 17,360 | Trend | 0.32 | 0.29 | 1.13 (1.00 – 1.27) | .04 |
| rs635261 | G/C | 3′ | 16,901 | Trend | 0.36 | 0.38 | 0.92 (0.82 – 1.03) | .15 |
| rs12729828 | C/T | 3′ | 15,824 | Dominant | 0.15 | 0.14 | 1.07 (0.89 – 1.27) | .47 |
| rs627839 | G/T | 3′ | 13,861 | Dominant | 0.48 | 0.45 | 1.16 (0.98 – 1.39) | .09 |
| rs11807829 | T/C | Intron 5 | 11,127 | Dominant | 0.32 | 0.34 | 0.90 (0.77 – 1.05) | .18 |
| rs627928 | G/T | Exon 4/Asp541Glu | 4,605 | Recessive | 0.45 | 0.44 | 1.17 (0.97 – 1.40) | .10 |
| rs486907 | G/A | Exon 2/Arg462Gln | 1,385 | Dominant | 0.36 | 0.37 | 0.88 (0.75 – 1.04) | .13 |
| rs56250729 | A/C | Exon 2/Ile97Leu | 289 | Dominant | 0.013 | 0.007 | 1.88 (1.05 – 3.34) | .03 |
| none | G/A | Exon 2/Gly59Ser | 175 | Dominant | 0.004 | 0.008 | 0.57 (0.27 – 1.20) | .14 |
| rs682585 | G/A | 5′ promoter | − 3,568 | Dominant | 0.39 | 0.38 | 1.07 (0.91 – 1.26) | .42 |
OR: Odds ratio adjusted for 5-year age groups; CI: Confidence Interval
When considering clinical features of disease, the rs486907 (R462Q) polymorphism was associated with a significantly decreased relative risk for higher grade prostate cancer. As shown in Table 2, men carrying at least one copy of the variant allele for rs486907 had a 28% decreased risk of higher grade prostate cancer (OR = 0.72, 95% CI: 0.53 – 0.98) compared to controls, but this association was no longer significant after accounting for multiple testing. No association was observed between this SNP genotype and lower grade prostate cancer. The associations observed between the two SNPs associated with overall prostate cancer (rs12723593 and rs56250729) maintained borderline statistical significance for men with lower grade prostate cancers only. None of the ORs, however, was significantly different between lower vs. higher grade tumors. In addition, the results of these analyses did not vary when a composite measure of prostate cancer aggressiveness was used as the outcome variable (data not shown).
Table 2.
Association between ten SNPs in RNASEL and prostate cancer, stratified by Gleason score
| SNP rs# | Best-fitting model | Minor allele frequency, controls N = 1267 | Minor allele frequency, cases with Gleason score ≤ (3 +4) N = 1102 | OR (95% CI) | Minor allele frequency, cases with Gleason score ≥ (4 +3) N = 202 | OR (95% CI) | Nominal p-value† |
|---|---|---|---|---|---|---|---|
| rs12723593 | Trend | 0.29 | 0.32 | 1.14 (1.00 – 1.28) | 0.31 | 1.10 (0.88 – 1.39) | 0.82 |
| rs635261 | Trend | 0.38 | 0.36 | 0.92 (0.81 – 1.03) | 0.37 | 0.96 (0.77 – 1.19) | 0.70 |
| rs12729828 | Dominant | 0.14 | 0.16 | 1.10 (0.91 – 1.32) | 0.14 | 0.93 (0.65 – 1.31) | 0.34 |
| rs627839 | Dominant | 0.45 | 0.48 | 1.15 (0.96 – 1.38) | 0.49 | 1.19 (0.85 – 1.68) | 0.84 |
| rs11807829 | Dominant | 0.34 | 0.32 | 0.91 (0.77 – 1.07) | 0.32 | 0.85 (0.63 – 1.15) | 0.67 |
| rs627928 | Recessive | 0.44 | 0.45 | 1.19 (0.99 – 1.44) | 0.47 | 1.06 (0.75 – 1.51) | 0.53 |
| rs486907 | Dominant | 0.37 | 0.36 | 0.91 (0.77 – 1.08) | 0.33 | 0.72 (0.53 – 0.98) | 0.13 |
| rs56250729 | Dominant | 0.007 | 0.01 | 1.82 (1.00 – 3.31) | 0.01 | 1.86 (0.68 – 5.07) | 0.97 |
| Gly59Ser (no rs#) | Dominant | 0.008 | < 0.01 | 0.62 (0.29 – 1.33) | < 0.01 | 0.32 (0.04 – 2.44) | 0.54 |
| rs682585 | Dominant | 0.38 | 0.39 | 1.07 (0.91 – 1.27) | 0.41 | 1.08 (0.79 – 1.47) | 0.98 |
Note: Four cases for whom Gleason score is unknown were excluded from stratified analyses.
OR: Odds ratio adjusted for 5-year age groups; CI: Confidence Interval;
p-value is Chi square test with 1 degree of freedom, testing the difference between risk estimates for lower- vs. higher-Gleason score groups.
There was no significant interaction between any RNASEL SNP and family history of prostate cancer (data not shown). However, a significant interaction was found between rs635261 and a history of prostatitis (p = 0.02). For this SNP, the minor allele (C) was associated with a significant decrease in the relative risk of prostate cancer in men with a history of prostatitis (OR = 0.51, 95% CI: 0.29 – 0.88), but was not associated with prostate cancer in men with no history of prostatitis (OR = 0.98, 95% CI: 0.83 – 1.16). As was the case for the results described above, this association was no longer significant after accounting for multiple testing.
Four common haplotypes were identified using the two SNPs found to be associated with overall prostate cancer risk (rs56250729 and rs12723593). One haplotype, consisting of the major allele of rs56250729 and the minor allele of rs12723593 was found to be associated with an increased risk of prostate cancer (OR = 1.12, 95% CI: 1.01 – 1.25, p = 0.03). This result does not differ substantially from what was observed in single SNP analyses. Haploview identified three haplotype blocks within the 10 RNASEL SNPs included in this analysis (Figure 1). Block 1 consisted of rs12723593 and rs635261, Block 2 consisted of rs12729828 and rs627839, and Block 3 consisted of rs627928 and rs486907. Only Blocks 1 and 2, both located in the 3′ UTR of RNASEL, were significantly associated with prostate cancer risk. While the association between the GG haplotype (i.e., minor allele of rs12723593 and major allele of rs635261) of Block 1 and prostate cancer did not substantially differ from the single SNP analyses of the constituent SNPs (OR = 1.15, 95% CI: 1.01 – 1.32, p = 0.04), the association between Block 2 and prostate cancer is unexpected, given the null single SNP results. A haplotype consisting of the major alleles of rs12729828 and rs627839 (C and G, respectively) was associated with a modestly reduced relative risk of prostate cancer (OR = 0.86, 95% CI: 0.76 – 0.97, p = 0.02). While this association is consistent with what was observed in the single SNP analyses, it is only statistically significant in the haplotype analysis.
Figure 1.
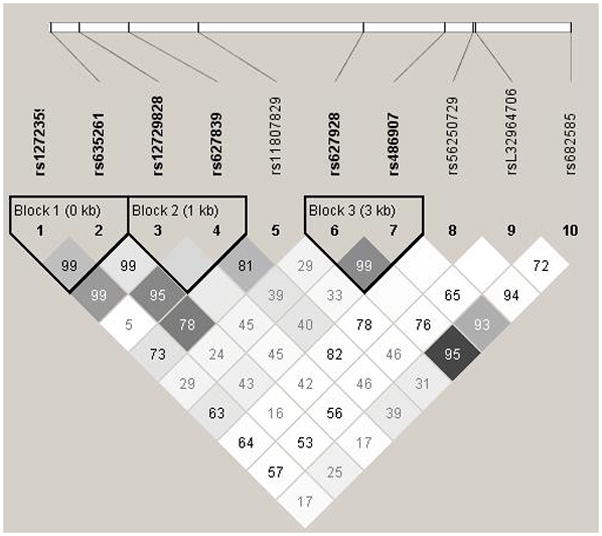
Haplotype blocks in RNASEL, created in Haploview using the 95% CI method described by Gabriel et al.[16] Numbers indicate pairwise D′, with blank boxes indicating D′ = 1. Darker shading indicates higher degrees of pairwise r2 correlation, such that r2 = 1 is shown as black, and r2 = 0 is shown as white.
Discussion
These analyses of ten SNPs in RNASEL in relation to prostate cancer revealed two SNPs that were associated with overall prostate cancer risk. However none of the risk estimates varied substantially by Gleason score. In addition, an interaction was observed between one SNP genotype and self-reported history of prostatitis.
Of the two SNPs associated with prostate cancer in this analysis, rs56250729 was associated with an 88% increase in prostate cancer risk, and rs12723593 was associated with a 13% increase in risk. Table 3 places these results (and those discussed above) in context of prior publications. Rs56250729 has been investigated in two previous studies, neither of which found an association with either familial or unselected prostate cancer [9,11]. This SNP is a coding, non-synonymous polymorphism located in the third ankyrin repeat of the RNASEL protein in a region known to be important in coordinating the conformational changes required for RNASEL dimerization and enzymatic activity [19,20]. Although its placement in the protein suggests that rs56250729 could be functionally important, an in vitro study demonstrated that the variant does not alter RNASEL pro-apoptotic activity[6]. To our knowledge, no previous studies have investigated the association between rs12723593 and risk of prostate cancer.
Table 3.
Association results from previously published studies of RNASEL variants and sporadic and familial prostate cancer
| RNASEL rs# | Author (Year) | Reference number | Family history subgroup* | OR (95% CI) |
|---|---|---|---|---|
| rs12723593 | Fesinmeyer et al. (2011) | all | 1.13 (1.00 – 1.27) | |
| rs635261 | Fesinmeyer et al. (2011) | all | 0.92 (0.82 – 1.03) | |
| Meyer et al. (2010) | 25 | all | 0.90 (0.71 – 1.13) | |
| rs12729828 | Fesinmeyer et al. (2011) | all | 1.07 (0.89 – 1.27) | |
| Meyer et al. (2010) | 25 | all | 1.09 (0.90 – 1.33) | |
| rs627839 | Fesinmeyer et al. (2011) | all | 1.16 (0.98 – 1.39) | |
| rs11087829 | Fesinmeyer et al. (2011) | all | 0.90 (0.77 – 1.05) | |
| Meyer et al. (2010) | 25 | all | 0.90 (0.69 – 1.17) | |
| rs627928 (Asp541Glu) | Fesinmeyer et al. (2011) | all | 1.17 (0.97 – 1.40) | |
| Maier et al. (2005) | 11 | all | 0.83† (0.51 – 1.37) | |
| Meyer et al. (2010) | 25 | all | 0.97† (0.78 – 1.22) | |
| Wiklund et al. (2004) | 8 | sporadic only | 0.77 (0.59 – 1.00) | |
| Rokman et al. (2002) | 10 | sporadic only | NA (no association) | |
| Rokman et al. (2002) | 10 | familial only | NA (no association) | |
| Nakazato et al. (2003) | 30 | familial only, Japanese cases | 6.94 (3.98 – 12.10) | |
| Breyer et al. (2009) | 22 | familial only | 0.64 (p = 0.018) | |
| rs486907 (Arg462Gln) | Fesinmeyer et al. (2011) | all | 0.88 (0.75 – 1.04) | |
| Fesinmeyer et al. (2011) | all Gleason ≥ 7 | 0.72 (0.53 – 0.98) | ||
| Maier et al. (2005) | 11 | all | 0.88 (0.53 – 1.44) | |
| Shook et al. (2007) | 24 | all | 1.04 (0.80 – 1.36) | |
| Meyer et al. (2010) | 25 | all | 0.96 (0.75 – 1.24) | |
| Wang et al. (2002) | 9 | sporadic only | p = 0.92 (OR NA) | |
| Rennert et al. (2005) | 21 | sporadic only | 1.40 (0.99 – 1.90) | |
| Rennert et al. (2005) | 21 | sporadic only Gleason < 7 | 1.50 (1.04 – 2.20) | |
| Rokman et al. (2002) | 10 | sporadic only | 1.12 (0.60 – 2.07) | |
| Casey et al. (2002) | 23 | familial only | 1.51 (1.12 – 2.05) | |
| Wang et al. (2002) | 9 | familial only | 0.55 (0.33 – 0.93) | |
| Breyer et al. (2009) | 22 | familial only | 1.34 (1.03 – 1.76) | |
| Breyer et al. (2009) | 22 | familial only Gleason ≥ 7 | 1.26 (0.85 – 1.88) | |
| Rennert et al. (2005) | 21 | familial only | 0.48 (0.24 – 0.95) | |
| Rennert et al. (2005) | 21 | familial only Gleason <7 | 0.43 (0.21 – 0.88) | |
| Rokman et al. (2002) | 10 | familial only | 1.96 (0.95 – 4.03) | |
| rs56250729 (Ile97Leu) | Fesinmeyer et al. (2011) | all | 1.88 (1.05 – 3.34) | |
| Maier et al. (2005) | 11 | all | 2.32 (0.49 – 11.0) | |
| Wang et al. (2002) | 9 | familial only | p = 0.23 (OR NA) | |
| Gly59Ser | Fesinmeyer et al. (2011) | all | 0.57 (0.27 – 1.20) | |
| Rokman et al. (2002) | 10 | sporadic only | NA (no association) | |
| rs682585 | Fesinmeyer et al. (2011) | all | 1.07 (0.91 – 1.26) | |
| Meyer et al. (2010) | 25 | all | 0.91 (0.73– 1.15) |
OR: Odds Ratio; CI: Confidence Interval; NA: not available (estimates not provided in manuscript text).
Family history subgroups defined as follows: sporadic only = cases with no family history of prostate cancer; familial only = cases with some family history of prostate cancer; all = cases not selected on basis of family history.
inverse of published OR and 95% CI for opposite allele.
When considering clinical features of disease, the minor allele (A) of rs486907 was found to be associated with a 28% decreased relative risk for higher Gleason score cancer, although the test for the difference between the risk estimates for lower vs. higher grade prostate cancer was not significant. A prior case-control study conducted by Rennert et al. also investigated rs486907 in the context of prostate cancer grade, and found that the A allele of rs486907 was associated with an increased risk of lower Gleason score (i.e., Gleason < 7) prostate cancer among sporadic cases, and a decreased risk of lower Gleason score cancer among familial cases [21]. We found no evidence for interaction between any RNASEL SNPs and family history, and thus did not perform a similar stratified analysis. Breyer and colleagues also investigated rs486907 as a risk factor for familial prostate cancer, and found that the A allele was associated with risk for all grades of prostate cancer, but not specifically with higher grade tumors [22]. Although it is difficult to identify a theme in these disparate results, there is a suggestion of some association between rs486907, family history, and prostate cancer grade that remains to be elucidated.
In addition to the two studies cited above [21,22], the rs486907 SNP has been studied in several other studies of familial or hereditary prostate cancer (Table 3). Casey and colleagues found an increased risk for familial prostate cancer associated with the A allele [23], while Wang et al. reported the opposite effect[9]. Rokman et al. found a non-significant association, but with a point estimate indicating increased risk for familial prostate cancer associated with the A allele (OR = 1.96, 95% CI: 0.95 – 4.03)[10]. In contrast to prior research, the present analysis found no association between rs486907 and a first-degree family history of prostate cancer, which was reported by 22% of cases and 11% of controls.
There is more agreement among published studies concerning the association between rs486907 and sporadic or unselected prostate cancer. A case-control study conducted in an ethnically diverse population reported a significant association between rs486907 and unselected prostate cancer in Hispanics and African-Americans, but this association did not extend to Caucasians[24]. Studies of this SNP and sporadic prostate cancer conducted in Finland[10] and the United States[9] found no significant association. Studies of unselected prostate cancer cases conducted in Germany[11] and within a United States cohort study[25] also found no association with rs486907. Finally, our study found no association between this SNP and unselected prostate cancer. Taken together, these results indicate that the biological effect of the rs486907 polymorphism differs between sporadic and familial cases, and perhaps between ethnic groups.
A previous study of the biochemical properties of missense mutations in RNASEL determined that the rs486907 variant strongly affects the pro-apoptotic activity of RNASEL [6]. This SNP corresponds to an amino acid change in the kinase domain of RNASEL that inhibits the ability of RNASEL to dimerize into its active form. Consequently, the rare Arg462Gln substitution produces a protein with approximately one-third the apoptotic activity of wild-type RNASEL protein [6]. Although several other non-synonymous polymorphisms in RNASEL have been identified, none has been shown to substantially alter RNASEL enzymatic activity. As such, additional laboratory studies are needed to determine whether RNASEL enzyme activity is relevant to viral-mediated prostate carcinogenesis, and to the development of higher-grade tumors.
Variation in RNASEL has also been studied in conjunction with XMRV virus infection. The first relevant study was performed in 86 prostate tumor specimens, and reported that viral infection was present in 40% of prostate cancer cases homozygous for the rare allele of rs486907, compared to only 1.6% of cases with one or more copies of the common allele [3]. The overall frequency of the rare allele was 31%. This study, conducted by Urisman and colleagues, did not compare demographic or clinical features of subjects with and without evidence of XMRV infection, therefore it is difficult to evaluate the results of the present study in the context of that prior investigation. A subsequent study conducted by Schlaberg et al. built on these findings by investigating both cancer-free control tissues obtained from TURP procedures and prostate cancer specimens. That study found no association between RNASEL genotype and either prostate cancer risk or the prevalence of XMRV infection. [4]. However, prostate tumor tissue was more likely than TURP control tissue to show evidence of prior XMRV infection. The frequency of the rare allele in prostate cancer cases was higher in the Schlaberg study (44%) than in the Urisman et al. study (31%) or in our study (36%). Neither prior study states whether deviation from Hardy-Weinberg equilibrium was observed for this SNP. In the present study, we observed that the rare allele, which is associated with reduced apoptotic activity, is associated with a decreased relative risk of higher grade prostate cancer. Although Schlaberg et al. reported that patients infected with XMRV were more likely to have higher Gleason grade tumors, they did not investigate whether RNASEL variant status was associated with Gleason grade.
Finally, an interaction between rs635261 genotype and prostate cancer was found in men with a history of prostatitis. A prior study found no association between this SNP and unselected prostate cancer [25]. Prostatitis and prostate inflammation in general has been associated with an increased prostate cancer risk in several epidemiologic studies, although most did not account for potential confounding effects of PSA screening [26]. A prospective study of men with negative initial prostate biopsy results found that 20% of men with chronic prostate inflammation at the time of the initial biopsy had prostate cancer diagnosed within the next five years, compared to 6% of men with no evidence of prostate inflammation at time of initial biopsy[27]. A recent prospective study of over 68,000 men reported that a history of prostatitis was associated with a 30% increased risk of prostate cancer over 3–4 years of follow-up, compared to men with no history of prostate inflammation[28]. In the study presented here, information on prostatitis was limited to self-reported history of prostatitis collected at study enrollment. We were unable to evaluate age of onset or duration of prostatitis, both of which may be important to consider. Further, the majority of prostatitis cases are of unknown etiology, and may or may not be associated with viral infection[29]. Thus, while our observation of statistical interaction between an RNASEL SNP and a history of prostatitis is not surprising, it is impossible to draw conclusions from these data about the role of RNASEL variants in inflammation-related prostate carcinogenesis.
Another RNASEL variant, rs627928, has been inconsistently associated with prostate cancer risk in prior studies, but was not found to be associated with prostate cancer in this analysis. A study conducted in Sweden found a borderline significant association between rs627928 and sporadic prostate cancer [8], and a U.S. study found that this SNP was associated with an OR of 0.64 (p = 0.018) for familial prostate cancer [22]. In an earlier Swedish study of sporadic and familial prostate cancer, no association was found between rs627928 genotype and prostate cancer risk [10]. And, two studies conducted within the Physicians’ Health Study[25] and in Germany[11] found no association between this SNP and unselected prostate cancer in Caucasians[25]. However, a study conducted in a Japanese population reported a significant 7-fold increase in risk for familial prostate cancer in men homozygous for the variant allele of rs627928 [30]. These inconsistencies may be attributable to differences in study design, or in the underlying genetic characteristics of the populations studied.
These analyses did not find any association between any subset of prostate cancer and the remaining three genotyped SNPs: rs11087829, Gly59Ser, and rs682585. This negative finding is consistent with the case-control study conducted within the Physicians’ Health Study, which also found no association between rs11087829 or rs682585 and prostate cancer risk[25], and a Finnish case-control study that found no association between Gly59Ser and prostate cancer[10].
A 2-SNP haplotype in the 3′ UTR of RNASEL was found to be associated with prostate cancer risk. This result did not differ substantially from the single-SNP analyses. However, the 3′ UTR of RNASEL is known to control stability of RNASEL mRNA, [31] and further investigation of variation in this region may be warranted.
A major strength of this study is its population-based, case-control design, and the availability of clinical and family history data on participants. While several other studies have analyzed RNASEL variants as risk factors for sporadic prostate cancer, ours is the first population-based study of unselected prostate cancer conducted in the United States. Other prior studies enrolled subjects from ongoing cohorts (e.g., The Physician’s Health Study in Meyer et al., 2010) or via physician referral (e.g. Wang et al., 2002). Two other population-based studies of RNASEL and non-familial prostate cancer have been conducted in Sweden[8] and Finland[10]. However, these European studies may not be generalizable to the United States due to substantial differences in screening and early detection for prostate cancer in Europe [32], as well as potentially different allele frequencies in Finland’s relatively isolated population[33].
In contrast with previous studies that concentrated on main effects of RNASEL variants, the availability of detailed information on family history and history of prostatitis allowed investigation of effect modification by characteristics potentially related to RNASEL function in vivo. However, these patient characteristics were self-reported, and thus may be susceptible to information bias. This study also contributes to the literature on genetic risk factors for sporadic prostate cancer, which may be etiologically distinct from familial/hereditary prostate cancer. The associations described in this study are modest, and do not persist after adjustment for multiple comparisons. However, this study was motivated by an a priori hypothesis generated by previously published studies, and therefore a lenient approach towards multiple comparisons may be warranted in this context [34]. This study was also limited to a racially homogeneous population, and thus the results may only be applicable to men of European ancestry.
Conclusions
This study identified several RNASEL variants that warrant further investigation for their potential role in prostate cancer etiology. In addition, there is some evidence for effect modification by a history of prostatitis. While several of these associations align with previous studies of RNASEL variants and risk of prostate cancer, they implicate several previously unstudied RNASEL SNPs, and are the first to indicate potential effect modification by prostatitis history. Thus, additional larger studies are required to confirm these observations, particularly the potential for interaction in relation to prostate cancer and genetic variation across the RNASEL gene.
Acknowledgments
This work was funded by NIH Grants RO1 CA056678, RO1 CA092579, and P50 CA097186; with additional support from the Fred Hutchinson Cancer Research Center and the Intramural Program of the National Human Genome Research Institute.
We thank the men who generously participated in these studies.
References
- 1.Li HP, Leu YW, Chang YS. Epigenetic changes in virus-associated human cancers. Cell Res. 2005;15(4):262–271. doi: 10.1038/sj.cr.7290295. [DOI] [PubMed] [Google Scholar]
- 2.Klein EA, Silverman R. Inflammation, infection, and prostate cancer. Curr Opin Urol. 2008;18(3):315–319. doi: 10.1097/MOU.0b013e3282f9b3b7. [DOI] [PubMed] [Google Scholar]
- 3.Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Malathi K, Magi-Galluzzi C, Tubbs RR, Ganem D, Silverman RH, DeRisi JL. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2(3):e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Schlaberg R, Choe DJ, Brown KR, Thaker HM, Singh IR. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc Natl Acad Sci U S A. 2009;106(38):16351–16356. doi: 10.1073/pnas.0906922106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Bisbal C, Silverman RH. Diverse functions of RNase L and implications in pathology. Biochimie. 2007;89(6–7):789–798. doi: 10.1016/j.biochi.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiang Y, Wang Z, Murakami J, Plummer S, Klein EA, Carpten JD, Trent JM, Isaacs WB, Casey G, Silverman RH. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2′,5′-oligoadenylates. Cancer Res. 2003;63(20):6795–6801. [PubMed] [Google Scholar]
- 7.Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, Faruque M, Moses T, Ewing C, Gillanders E, Hu P, Bujnovszky P, Makalowska I, Baffoe-Bonnie A, Faith D, Smith J, Stephan D, Wiley K, Brownstein M, Gildea D, Kelly B, Jenkins R, Hostetter G, Matikainen M, Schleutker J, Klinger K, Connors T, Xiang Y, Wang Z, De Marzo A, Papadopoulos N, Kallioniemi OP, Burk R, Meyers D, Gronberg H, Meltzer P, Silverman R, Bailey-Wilson J, Walsh P, Isaacs W, Trent J. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet. 2002;30(2):181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 8.Wiklund F, Jonsson BA, Brookes AJ, Stromqvist L, Adolfsson J, Emanuelsson M, Adami HO, Augustsson-Balter K, Gronberg H. Genetic analysis of the RNASEL gene in hereditary, familial, and sporadic prostate cancer. Clin Cancer Res. 2004;10(21):7150–7156. doi: 10.1158/1078-0432.CCR-04-0982. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, McDonnell SK, Elkins DA, Slager SL, Christensen E, Marks AF, Cunningham JM, Peterson BJ, Jacobsen SJ, Cerhan JR, Blute ML, Schaid DJ, Thibodeau SN. Analysis of the RNASEL gene in familial and sporadic prostate cancer. Am J Hum Genet. 2002;71(1):116–123. doi: 10.1086/341281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rokman A, Ikonen T, Seppala EH, Nupponen N, Autio V, Mononen N, Bailey-Wilson J, Trent J, Carpten J, Matikainen MP, Koivisto PA, Tammela TL, Kallioniemi OP, Schleutker J. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am J Hum Genet. 2002;70(5):1299–1304. doi: 10.1086/340450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maier C, Haeusler J, Herkommer K, Vesovic Z, Hoegel J, Vogel W, Paiss T. Mutation screening and association study of RNASEL as a prostate cancer susceptibility gene. Br J Cancer. 2005;92(6):1159–1164. doi: 10.1038/sj.bjc.6602401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanford JL, Wicklund KG, McKnight B, Daling JR, Brawer MK. Vasectomy and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 1999;8(10):881–886. [PubMed] [Google Scholar]
- 13.Agalliu I, Salinas CA, Hansten PD, Ostrander EA, Stanford JL. Statin use and risk of prostate cancer: results from a population-based epidemiologic study. Am J Epidemiol. 2008;168(3):250–260. doi: 10.1093/aje/kwn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langeberg WJ, Kwon EM, Koopmeiners JS, Ostrander EA, Stanford JL. Population-based study of the association of variants in mismatch repair genes with prostate cancer risk and outcomes. Cancer Epidemiol Biomarkers Prev. 2010;19(1):258–264. doi: 10.1158/1055-9965.EPI-09-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 16.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 17.Edgington E. Randomization Tests. 3. New York: Marcel Dekker, Inc; 1995. [Google Scholar]
- 18.Fitzgerald LM, Kwon EM, Koopmeiners JS, Salinas CA, Stanford JL, Ostrander EA. Analysis of recently identified prostate cancer susceptibility loci in a population-based study: associations with family history and clinical features. Clin Cancer Res. 2009;15(9):3231–3237. doi: 10.1158/1078-0432.CCR-08-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong B, Niwa M, Walter P, Silverman RH. Basis for regulated RNA cleavage by functional analysis of RNase L and Ire1p. RNA. 2001;7(3):361–373. doi: 10.1017/s1355838201002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka N, Nakanishi M, Kusakabe Y, Goto Y, Kitade Y, Nakamura KT. Structural basis for recognition of 2′,5′-linked oligoadenylates by human ribonuclease L. EMBO J. 2004;23(20):3929–3938. doi: 10.1038/sj.emboj.7600420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rennert H, Zeigler-Johnson CM, Addya K, Finley MJ, Walker AH, Spangler E, Leonard DG, Wein A, Malkowicz SB, Rebbeck TR. Association of susceptibility alleles in ELAC2/HPC2, RNASEL/HPC1, and MSR1 with prostate cancer severity in European American and African American men. Cancer Epidemiol Biomarkers Prev. 2005;14(4):949–957. doi: 10.1158/1055-9965.EPI-04-0637. [DOI] [PubMed] [Google Scholar]
- 22.Breyer JP, McReynolds KM, Yaspan BL, Bradley KM, Dupont WD, Smith JR. Genetic variants and prostate cancer risk: candidate replication and exploration of viral restriction genes. Cancer Epidemiol Biomarkers Prev. 2009;18(7):2137–2144. doi: 10.1158/1055-9965.EPI-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casey G, Neville PJ, Plummer SJ, Xiang Y, Krumroy LM, Klein EA, Catalona WJ, Nupponen N, Carpten JD, Trent JM, Silverman RH, Witte JS. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat Genet. 2002;32(4):581–583. doi: 10.1038/ng1021. [DOI] [PubMed] [Google Scholar]
- 24.Shook SJ, Beuten J, Torkko KC, Johnson-Pais TL, Troyer DA, Thompson IM, Leach RJ. Association of RNASEL variants with prostate cancer risk in Hispanic Caucasians and African Americans. Clin Cancer Res. 2007;13(19):5959–5964. doi: 10.1158/1078-0432.CCR-07-0702. [DOI] [PubMed] [Google Scholar]
- 25.Meyer MS, Penney KL, Stark JR, Schumacher FR, Sesso HD, Loda M, Fiorentino M, Finn S, Flavin RJ, Kurth T, Price AL, Giovannucci EL, Fall K, Stampfer MJ, Ma J, Mucci LA. Genetic variation in RNASEL associated with prostate cancer risk and progression. Carcinogenesis. 2010;31(9):1597–1603. doi: 10.1093/carcin/bgq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennis LK, Lynch CF, Torner JC. Epidemiologic association between prostatitis and prostate cancer. Urology. 2002;60(1):78–83. doi: 10.1016/s0090-4295(02)01637-0. [DOI] [PubMed] [Google Scholar]
- 27.MacLennan GT, Eisenberg R, Fleshman RL, Taylor JM, Fu P, Resnick MI, Gupta S. The influence of chronic inflammation in prostatic carcinogenesis: a 5-year followup study. J Urol. 2006;176(3):1012–1016. doi: 10.1016/j.juro.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 28.Cheng I, Witte JS, Jacobsen SJ, Haque R, Quinn VP, Quesenberry CP, Caan BJ, Van Den Eeden SK. Prostatitis, sexually transmitted diseases, and prostate cancer: the California Men’s Health Study. PLoS One. 2010;5(1):e8736. doi: 10.1371/journal.pone.0008736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Habermacher GM, Chason JT, Schaeffer AJ. Prostatitis/chronic pelvic pain syndrome. Annu Rev Med. 2006;57:195–206. doi: 10.1146/annurev.med.57.011205.135654. [DOI] [PubMed] [Google Scholar]
- 30.Nakazato H, Suzuki K, Matsui H, Ohtake N, Nakata S, Yamanaka H. Role of genetic polymorphisms of the RNASEL gene on familial prostate cancer risk in a Japanese population. Br J Cancer. 2003;89(4):691–696. doi: 10.1038/sj.bjc.6601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XL, Andersen JB, Ezelle HJ, Wilson GM, Hassel BA. Post-transcriptional regulation of RNase-L expression is mediated by the 3′-untranslated region of its mRNA. J Biol Chem. 2007;282(11):7950–7960. doi: 10.1074/jbc.M607939200. [DOI] [PubMed] [Google Scholar]
- 32.Bray F, Lortet-Tieulent J, Ferlay J, Forman D, Auvinen A. Prostate cancer incidence and mortality trends in 37 European countries: an overview. Eur J Cancer. 2010;46(17):3040–3052. doi: 10.1016/j.ejca.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 33.Salmela E, Lappalainen T, Fransson I, Andersen PM, Dahlman-Wright K, Fiebig A, Sistonen P, Savontaus ML, Schreiber S, Kere J, Lahermo P. Genome-wide analysis of single nucleotide polymorphisms uncovers population structure in Northern Europe. PLoS One. 2008;3(10):e3519. doi: 10.1371/journal.pone.0003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice TK, Schork NJ, Rao DC. Methods for handling multiple testing. Adv Genet. 2008;60:293–308. doi: 10.1016/S0065-2660(07)00412-9. [DOI] [PubMed] [Google Scholar]


