Abstract
We have determined the solution structure of the complex between an arginine-glycine-rich RGG peptide from the fragile X mental retardation protein (FMRP) and an in vitro-selected guanine-rich sc1 RNA. The bound RNA forms a novel G-quadruplex separated from the flanking duplex stem by a mixed junctional tetrad. The RGG peptide is positioned along the major groove of the RNA duplex, with the G-quadruplex forcing a sharp turn of R10GGGGR15 at the duplex-quadruplex junction. Arginines R10 and R15 form cross-strand specificity-determining intermolecular hydrogen-bonds with the major-groove edges of guanines of adjacent Watson-Crick G•C pairs. Filter binding assays on RNA and peptide mutations identify and validate contributions of peptide-RNA intermolecular contacts and shape complementarity to molecular recognition. These findings on FMRP RGG domain recognition by a combination of G-quadruplex and surrounding RNA sequences have implications for recognition of other genomic G-rich RNAs.
Keywords: FMRP, RGG box, RNA recognition, RNA duplex-quadruplex junction
The regulation of gene expression by interactions between nucleic acid-binding proteins and G-quadruplexes is an area of intense interest. Bioinformatic analyses have predicted that there are potentially over 350,000 G-quadruplex-forming sequences in the human genome1,2, and recent studies of RNA G-quadruplexes in the transcriptome predict that they may be even more extensive than previously appreciated3,4. Functionally, G-quadruplex structures in RNA have been implicated in almost all aspects of pre-mRNA and mRNA metabolism including mRNA stability, IRES-dependent translation initiation5, translational repression3,6,7, alternative splicing8,9 and alternative polyadenylation/3´ end formation10,11 suggesting that the G-quadruplex may be an important regulatory motif in many aspects of gene expression.
The Fragile X mental retardation protein (FMRP) is a regulatory RNA binding protein that binds with high affinity to guanine-rich RNAs capable of G-quadruplex formation12,13. Loss of FMRP function leads to the Fragile X Syndrome, the most common form of inherited mental retardation, afflicting 1 in 2500 males, and is the leading single-gene cause of autism14. The Fragile X Syndrome almost always results from a triplet repeat expansion in the 5’-UTR, leading to abnormal methylation of the gene, repression of transcription, and hence complete loss of FMRP expression. However, one severely affected patient harbors a missense mutation in one of the RNA binding domains15, and mice harboring this mutation are indistinguishable from FMRP null mice in most behavioral and electrophysiologic assays and macroorchidism16, suggesting that loss of RNA binding activity may underlie the synaptic dysfunction observed in the disease.
Recent evidence suggests that FMRP functions to repress mRNA translation in neurons, leading to the widely held view that Fragile X Syndrome is a disease of “runaway translation” resulting in inappropriate gene expression with dire consequences for synaptic plasticity. This occurs despite the presence of two autosomal paralogs, FXR1P and FXR2P, which are also expressed in the brain. While all three proteins share a great deal of homology in their N-termini, KH domains and nuclear export signal, the C-termini containing the RGG box have diverged considerably. Consequently, the KH domains share RNA binding specificity but binding to G-rich sequences capable of forming G-quadruplexes is specific to FMRP17. These observations, together with interest in identifying the in vivo RNA ligands of FMRP suggest that understanding the role of the RGG box - G-rich RNA interaction will be important in understanding the human disease.
Binding of FMRP to G-rich RNAs requires only the RGG box RNA binding domain of FMRP, which is rich in arginines and glycines. In vitro selection identified guanine-rich RNA motifs that can bind tightly to FMRP, such as the 36-nt r(GCUGCGGUGUGGAAGGAGUGGUCGGGUUGCGCAGCG) sequence named sc112. Binding of FMRP to sc1 RNA was shown to depend on G-quadruplex formation based on the following observations: (i) the binding affinity was significantly increased in the presence of K+ as compared to Li+, and (ii) mutations of guanines within G-tracts abolished binding.
To understand how G-rich RNAs could be recognized by the RGG domain of FMRP, we used NMR to characterize the 1:1 complex between the 36-nt sc1 RNA sequence with a 28-aa peptide from the RGG domain of FMRP (RGG peptide) (Fig. 1a). We prepared complexes with various RNA and peptide constructs, including unlabeled, uniformly 13C,15N-labeled, as well as residue-type-specific and site-specific 13C,15N-labeled sc1 RNA and RGG peptide molecules. The structure of the complex revealed a number of new motifs and recognition principles between the FMRP RGG peptide and the sc1 RNA duplex-quadruplex junction. Subsequently, the peptide-RNA intermolecular contacts and the contributions of shape complementarity to molecular recognition were validated following analysis of filter binding assays on structure-guided RNA and peptide mutations.
Figure 1.
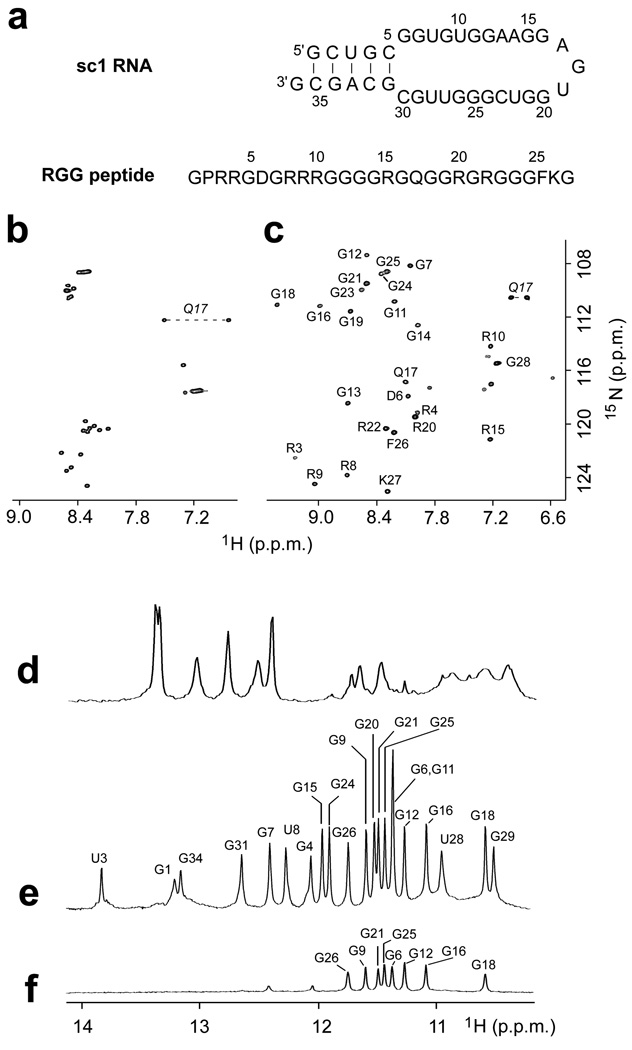
Sequence and NMR spectra of sc1 RNA and RGG peptide. (a) Sequence of 36-mer sc1 stem-loop RNA and 28-mer FMRP RGG peptide. (b,c) 1H,15N HSQC spectra of RGG peptide in the (b) free and (c) sc1 RNA-bound states. Backbone amide resonance assignments are included in panel c. (d–f) Imino proton spectra (10.0 to 14.0 ppm) of (d) free sc1 RNA, (e) sc1 RNA bound to the RGG peptide, and (f) RGG peptide - sc1 RNA complex after 16 hr in D2O. Spectra were recorded in 50 mM K-acetate, pH 6.8 at 25 °C.
RESULTS
Binding of FMRP RGG peptide to sc1 RNA
NMR spectra of both peptide and RNA in 50 mM K-acetate, pH 6.8 at 25 °C, clearly indicated that the RGG peptide binds to sc1 RNA to form a stable complex. From the peptide side, peak dispersion in the fingerprint region of the 1H-15N HSQC spectra of the free peptide (Fig. 1b) and RNA-bound peptide (Fig. 1c) revealed a transition from a ‘random coil’ to a well-ordered conformation of the peptide upon RNA binding. From the RNA side, the imino proton spectrum of the RNA in the free state (Fig. 1d) suggested that besides formation of a Watson-Crick duplex stem (indicated by six imino proton peaks between 12 to 14 ppm), the remainder of the molecule either is essentially not well-structured or adopts multiple conformations, as indicated by broad imino protons between 10 and 12 ppm. Upon binding to the RGG peptide, sc1 RNA becomes well-structured as indicated by a sharp and well-resolved imino proton spectrum containing well-dispersed additional resonances (Fig. 1e). Imino proton peaks from 12 to 14 ppm are characteristic of Watson-Crick base pairs, while those from 10 to 12 ppm are characteristic of G-tetrad formation, with eight imino protons between 10–12 ppm exchanging slowly and observed even 16 hours after transfer to D2O solution (Fig. 1f).
NMR assignments of sc1 RNA in the complex
The imino protons of the FMRP RGG peptide-sc1 RNA complex in K+-containing H2O buffer solution are exceptionally well-dispersed between 10.5 and 14.0 ppm (Fig. 1e; see also 1H-15N HSQC spectrum in Supplementary Fig. 1a). We have unambiguously assigned the guanine imino protons following site-specific incorporation of 2% 15N-labeled deoxyguanine at individual guanine positions in the sc1 RNA sequence18. An example of a 15N-edited NMR spectrum following incorporation of the label at the G31 position is shown below the control spectrum (Fig. 2a) and allows unambiguous assignment of the G31 imino proton resonance. The complete guanine imino proton assignments in the complex using this approach are listed over the spectrum in Fig. 1e. The imino (H1) protons were next correlated to H8 protons within individual guanines by through-bond connectivities via their intervening C5 carbons19. The well-resolved H1-C5 and H8-C5 cross-peaks in the 1H-13C HMBC spectrum allowed unambiguous assignment of all guanine H8 protons based on the known assignments of the corresponding guanine imino protons (Fig. 2b). Aromatic protons including guanine and adenine H8 protons and uracil and cytosine H6 protons were also independently assigned or confirmed with site-specific deoxyribose-for-ribose substitutions20.The aromatic protons were then correlated by through-bond connectivities with the sugar ring protons21. Ribose sugar protons were connected using COSY, TOCSY, and 3D HCCH-COSY and HCCH-TOCSY experiments22.
Figure 2.
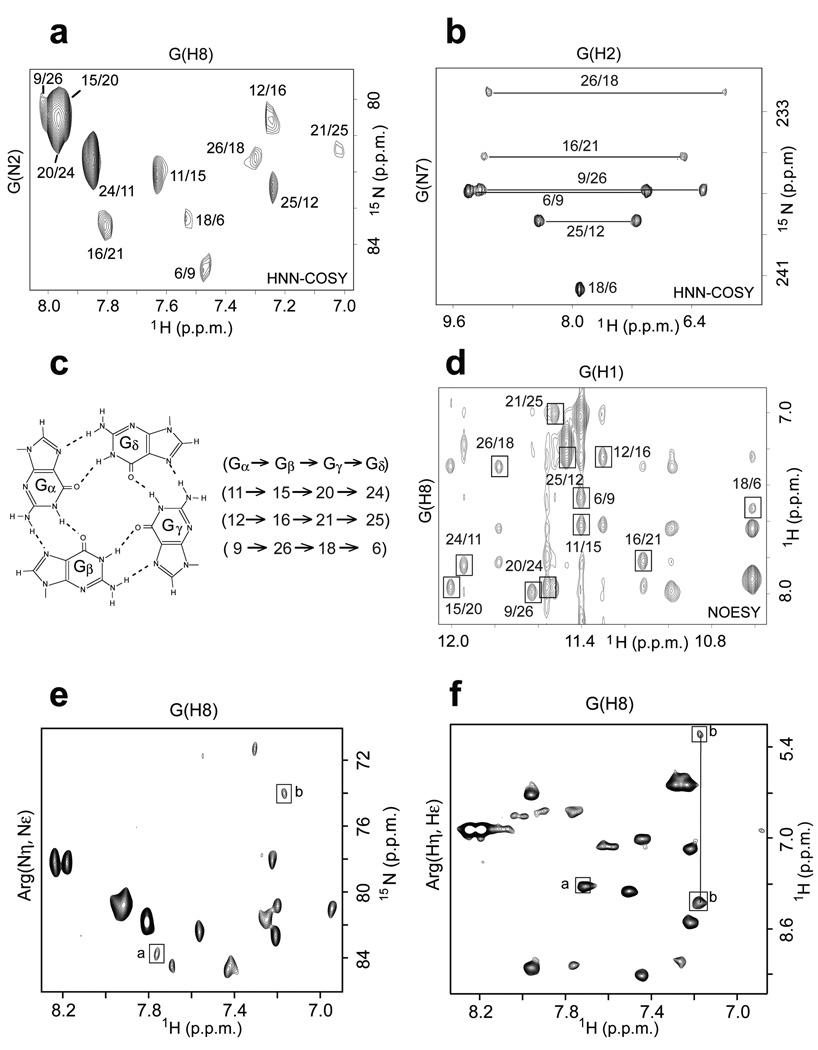
RNA resonance assignments in the RGG peptide - sc1 RNA complex. (a) Example of imino proton assignment: the reference imino proton NMR spectrum of the complex is shown on the top and the corresponding filtered spectrum of the complex following site-specific incorporation of 2% 15N-labeled deoxyguanine at the G31 position in the RNA is shown at the bottom. (b) Guanine H8 protons were assigned by through-bond connectivities to the guanine imino protons via 13C5 by HMBC experiment.
To overcome spectral crowding in the sugar proton region of the spectrum, we prepared sc1 RNA that was selectively 13C,15N-labeled solely at guanines, at adenines and at cytosines, and recorded 13C-edited and/or 15N-edited spectra of these selectively-labeled complexes (Supplementary Fig. 2). The base and sugar proton chemical shifts of sc1 RNA in the RGG peptide-bound state are tabulated in Supplementary Table 1.
Pairing alignments within duplex and quadruplex segments
We have applied through-bond 1H-15N HNN-COSY correlation experiments23,24 to monitor G•C (guanine imino proton to cytosine N3 nitrogen) and A•U (uracil imino proton to adenine N1 nitrogen) Watson-Crick pair formation (Supplementary Fig. 1b) within the sc1 RNA on complex formation with the FMRP RGG peptide. The data support formation of a five-base-pair duplex containing one A•U and four G•C base pairs as shown schematically in Fig. 1a, but also establish formation of an additional G7•C30 Watson-Crick pair, thereby unexpectedly extending the duplex by one base pair.
We have applied through-bond and through-space two-dimensional experiments to monitor G•G•G•G tetrad formation within the guanine-rich sc1 RNA sequence on complex formation with the FMRP RGG peptide (Fig. 3). We have used unambiguous through-bond HNN-COSY experiments to monitor hydrogen-bonding patterns between adjacent guanines around a G-tetrad, with the correlations between guanine N2 nitrogen and guanine H8 proton around individual G-tetrads25 plotted in Fig. 3a and between guanine amino protons and guanine N7 nitrogen around individual G-tetrads26 plotted in Fig. 3b. As an example, we observe connectivities 9/26, 26/18, 18/6 and 6/9 (Figs. 3a,b) identifying G9•G26•G18•G6 as one of the three G-tetrads (Fig. 3c) forming the G-quadruplex. Related through-bond connectivity patterns identify formation of additional G11•G15•G20•G24 and G12•G16•G21•G25 G-tetrads on complex formation (Fig. 3c).
Figure 3.
Identification of G-tetrad alignments and assignment of through-bond correlations in the RGG peptide - sc1 RNA complex. (a) HNN-COSY contour plot showing through-bond connectivities between amino nitrogens and H8 protons around the G-tetrad. The labeling represents through-bond amino/H8 correlations between adjacent guanines within a G-tetrad plane. (b) HNN-COSY contour plot showing through-bond connectivities between amino protons and N7 nitrogens around the G-tetrad. The labeling represents through-bond amino/N7 correlations between adjacent guanines within a G-tetrad plane. (c) Hydrogen bonding alignment around the G-tetrad and data analysis supporting formation of three G-tetrads. (d) NOE connectivities between guanine imino and H8 protons around the G-tetrad. The labeling represents through-space imino/H8 correlations between adjacent guanines within a G-tetrad plane. (e) Identification of intermolecular 1H-15N through-bond correlations in HNN-COSY spectra between guanine H8 protons of RNA and arginine guanidinium group nitrogens in the complex. Other correlations present in the spectra are intramolecular through-bond connectivities. Peak labeled a is between the H8 proton of G31 and Nε nitrogen of R10, while peak labeled b is between the H8 proton of G7 and the Nη nitrogen of R15. (f) Identification of intermolecular 1H-1H through bond correlations in HNN-COSY spectra between guanine H8 protons of RNA and arginine guanidinium group protons in the complex. Peak labeled a is between the H8 proton of G31 and NεH proton of R10, while peaks labeled b are between the H8 proton of G7 and the NηH amino protons of R15. Other correlations present in the spectra are intramolecular through-bond connectivities.
We have also used through-space NOE connectivities between imino and H8 protons (involving spin diffusion via the amino protons) on adjacent guanines around individual G-tetrads, and these data in Fig. 3d provide independent support for formation of the same three G-tetrads listed in the previous paragraph on complex formation.
NMR assignments of FMRP RGG peptide in the complex
Backbone and side-chain protons in the FMRP RGG peptide bound to sc1 RNA were assigned using standard triple-resonance experiments27,28 (see Online Methods section) on a sample of the complex containing uniformly 13C,15N-labeled peptide and unlabeled sc1 RNA. The RGG proton peptide amide and side-chain proton chemical shifts in the sc1 RNA bound state are tabulated in Supplementary Table 2a.
Intermolecular peptide-RNA constraints in the complex
We observe a finite set of intermolecular NOEs between RGG peptide and sc1 RNA in the complex. Many of these involve arginine side-chain protons, particularly between peptide side chains of Arg10 and Arg15 and RNA guanine imino protons (Supplementary Fig 3, left panel) and between peptide side chains of Arg4, Arg8 and Arg15 and RNA base protons (Supplementary Fig. 3, right panel). The complete set of intermolecular NOEs used as inputs in the structure determination computations are graphically listed in Supplementary Table 3.
We have used through-bond connectivities in HNN-COSY spectra to monitor intermolecular hydrogen bonding alignments within arginine•guanine pairs29 using samples containing uniform 13C,15N-labeling of both RGG peptide and sc1 RNA in the complex25,30. Using this approach, we have identified intermolecular through-hydrogen-bond correlations between the H8 proton of G31 from sc1 RNA and the Nε nitrogen (peak a, Fig. 3e) and NεH proton (peak a, Fig. 3f) of R10 from RGG peptide, associated with R10•G31 intermolecular pairing, and between the H8 proton of G7 from sc1 RNA and the Nη nitrogen (peak b, Fig. 3e) and NH2η protons (peak b, Fig. 3f) of R15 from RGG peptide, associated with R15•G7 intermolecular pairing in the complex.
Structure calculations
Structure calculations were guided by 58 intramolecular peptide, 232 intramolecular RNA and 90 intermolecular peptide-RNA restraints. Further, six base pairs within the duplex segment and three G-tetrads within the G-quadruplex segment were aligned using hydrogen-bonding restraints consistent with experimental data. In addition, the side chains of R10 and R15 were aligned with the major groove edges of G31 and G7 respectively, based on observed through-bond connectivities. The molecular dynamics protocols, initially in the absence and subsequently in a water shell containing K+ cations that guided the docking and convergence of the structures of the complex are outlined in the Online Methods and Supplementary sections. The experimental restraints and structural statistics are listed in Table 1.
Table 1.
Statistics of NMR restraints guided computations of the FMRP RGG peptide-RNA complex.
| Peptide | RNA | |
|---|---|---|
| NMR distance and dihedral constraints | ||
| Distance restraints | ||
| Total NOE | 58 | 232 |
| Inter-residue | ||
| Sequential (|i – j| = 1) | 40 | 126 |
| Non-sequential (|i – j| > 1 ) | 18 | 106 |
| Hydrogen bonds | 8 | 67 |
| Peptide–RNA intermolecular | 90 | |
| Total dihedral angle restraints | 9 | 226 |
| Protein | ||
| φ | 10 a | |
| ψ | ||
| RNA | ||
| Sugar pucker | 35 b | |
| Backbone | 71 c | |
| Based on A-form geometry | 120 d | |
| Structure statistics | ||
| Violations (mean and s.d.) | 1.0±0.0 | 1.0±0.0 |
| Max. distance constraint violation (Å) | 0.324 | 0.201 |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.01±0.00 | 0.01±0.00 |
| Bond angles (°) | 0.75±0.01 | 0.75±0.01 |
| Impropers (°) | 0.49±0.01 | 0.49±0.01 |
| Average pairwise r.m.s. deviation (Å) | ||
| Pairwise r.m.s. deviation was calculated among 10 refined structures | ||
| Peptide | ||
| All backbone heavy atoms | 1.32±0.34 | |
| Backbone heavy atoms, residues 7–17 | 0.64±0.12 | |
| RNA | ||
| All RNA heavy except residues 10,13,14,19,22,23,27 | 1.01±0.11 | |
| Complex | ||
| Peptide residues 7–17 & all RNA except residues 10,13,14,19,22,23,27 | 1.14±0.12 | |
The φ values for peptide residues D6, R8, R9, R10, R15, Q17, R20, R22, F26 and K27 were restrained in the −30° > φ> −75 °range, based on the observed HN-Hα vicinal J-couplings (< 5 Hz).
Pseudoration angles for RNA residues 6, 9, 10–13, 15–24, 26–28 were restrained to the C2´-endo domain; those for the residues 1–5, 7–8, 11, 14, 25, 29–36 were restrained to the C3´-endo domain, identified experimentally.
Nucleotide residues were restrained to χ values in the 195 (±50)° range, characteristic of anti glycosidic RNA torsion values, and ε were restrained to the stereochemically allowed range of 200 (±50)°.
α, β, γ, ζ angles of the duplex part of RNA were restrained to values characteristic for A-RNA: 298 (±70)°, 180 (±70)° 315(±15)°, 285 (±70)° correspondingly.
A-form RNA restraints for sugar-phosphate backbone of standard RNA duplex (residues 1–5 and 30–35) were switched off at the final minimization stages.
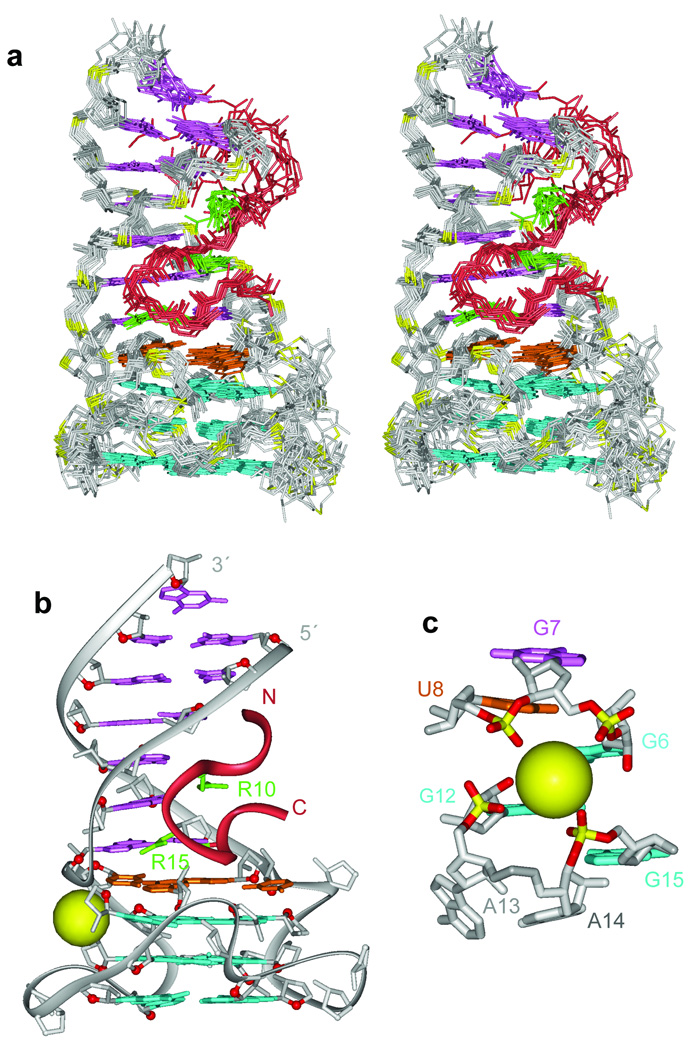
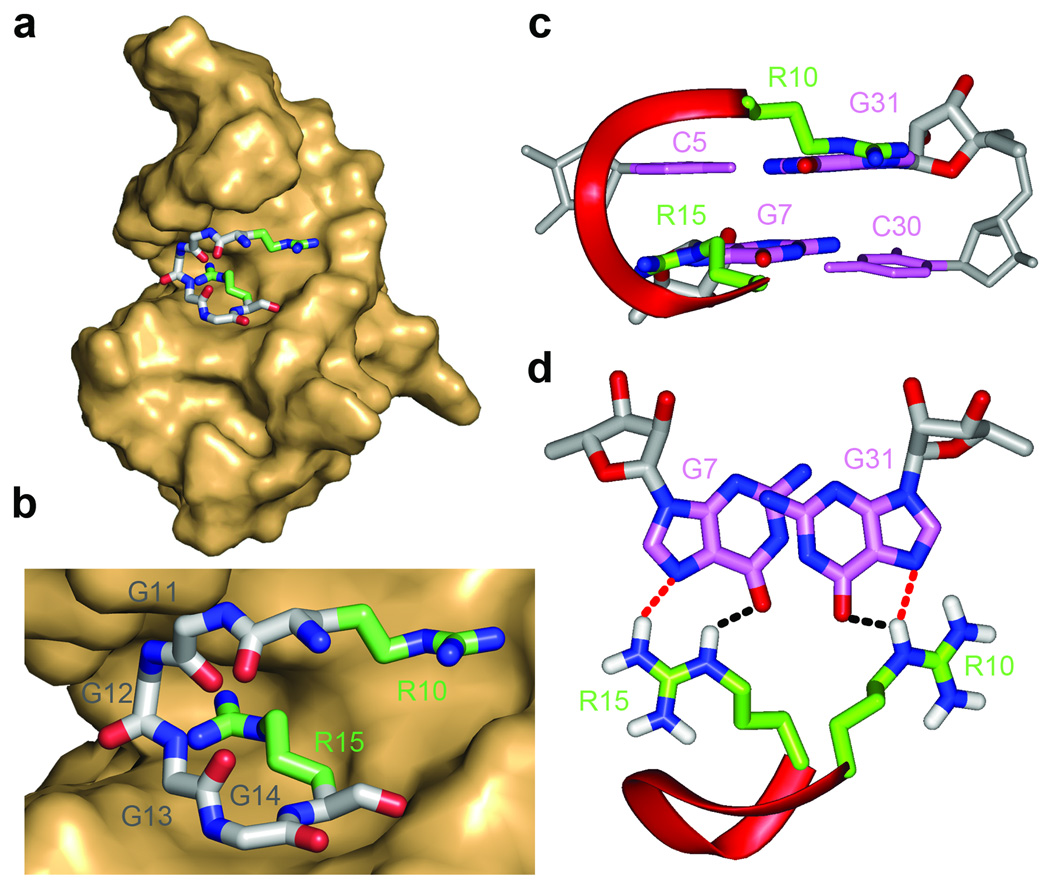
A family of 10 superpositioned structures is shown in stereo in Fig. 4a, with a representative structure shown in Fig. 4b. The peptide is colored in red, while RNA bases within the duplex, quadruplex and junctional segments are colored in purple, cyan and orange, respectively. The RNA sugar-phosphate backbone is colored in silver, while backbone phosphorus atoms are in yellow. The side chains of R8, R10 and R15 are colored in green. A K+ cation in the vicinity of the duplex-quadruplex junction, anchored in place through interaction with oxygens from four phosphate groups, is shown as a yellow sphere in Figs. 4b,c.
Figure 4.
Solution structure of the RGG peptide - sc1 RNA complex and the architecture of the G-quadruplex and duplex-quadruplex junction. (a) Stereo views of 10 superpositioned refined structures of the RGG peptide - sc1 RNA complex. The bound peptide is colored in red, while the duplex, junction and quadruplex segments are colored in magenta, orange and cyan, respectively. The sugar-phosphate backbone of the RNA is in silver, with phosphorus atoms in yellow. Key arginine (R10 and R15) side chains are colored in green. (b) A representative refined structure of the RGG peptide - sc1 RNA complex with the same color coding as in panel a. (c) Location of a potential K+ binding site in the vicinity of the duplex-quadruplex junction following dynamics computations in a water bath containing K+ cations. Note that this K+ cation is anchored through interaction with oxygens from four phosphate groups.
Topology of bound sc1 RNA and RGG peptide in the complex
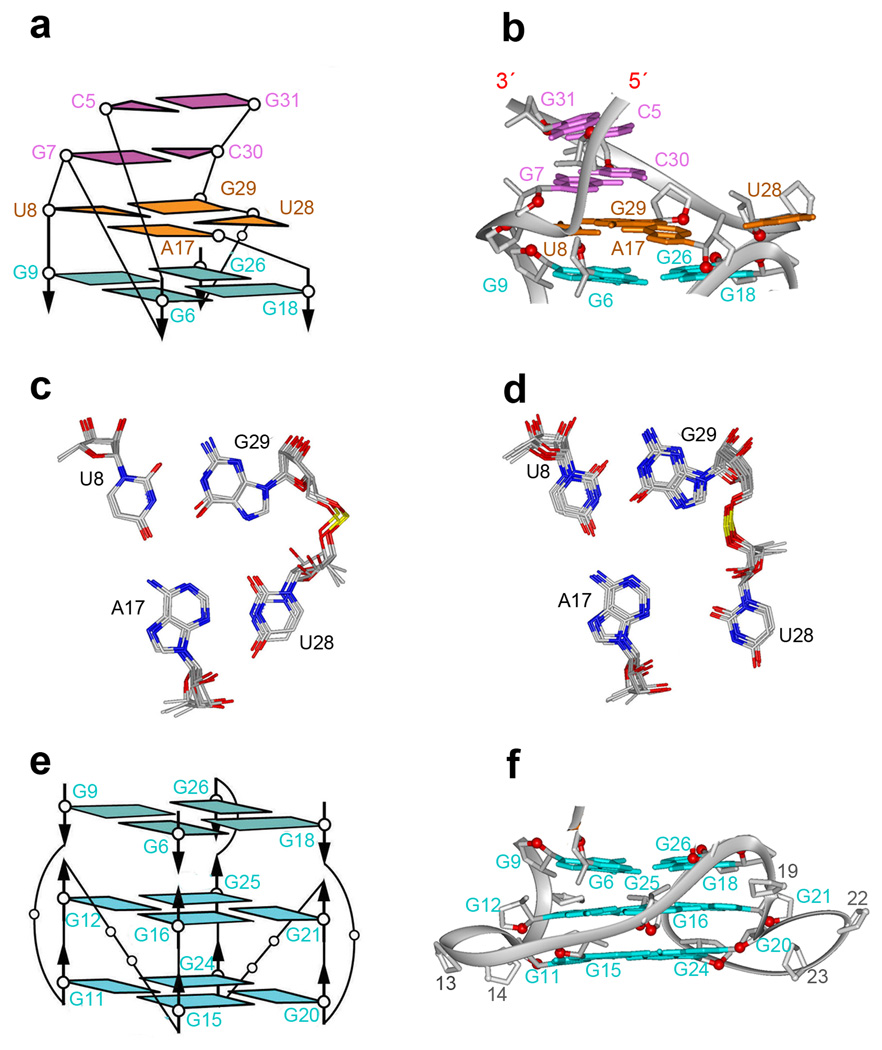
The bound sc1 RNA contains an anti-parallel duplex composed of six base pairs and a G-quadruplex composed of three stacked G-tetrad layers, with these folded elements separated by a junctional mixed tetrad (Fig. 4b). The Watson-Crick pairs (Fig. 1a) of the duplex segment are extended through stacking with Watson-Crick G7•C30 and wobble U8•G29 base pairs (Figs. 5a,b), with the latter pair being part of the junctional U8•A17•U28•G29 tetrad (two alternate alignments between refined structures are shown in Figs. 5c,d and hydrogen bonding alignments are shown in Supplementary Fig. 4a).
Figure 5.
Architecture of the G-quadruplex and duplex-quadruplex junction. (a) Schematic of the pairing alignments and strand connectivities at the duplex (magenta)-quadruplex (cyan) junction mediated by a mixed tetrad (orange). (b) Ribbon representation of schematic in panel a. (c and d). Base alignments around the junctional U8•A17•U28•G29 tetrad in the solution structure of the complex. 4 refined structures showed the pairing alignment in panel c, while 6 others showed the pairing alignment in panel d. (e) Schematic of pairing alignments and strand connectivities within the G-quadruplex. (f) Ribbon representation of schematic in panel e.
The topology of the G-quadruplex scaffold (Figs. 5e,f) is unprecedented: all twelve guanines are anti; the bottom two G-tetrad layers (G11•G15•G20•G24 positioned below G12•G16•G21•G25) are similar to other parallel-stranded G-quadruplexes, with a anti-clockwise hydrogen-bond directionality31–33. The third G-tetrad layer (G9•G26•G18•G6), positioned on top of the other two G-tetrads (Figs. 5e,f) flips over with resulting clockwise hydrogen-bond directionality; three out of four guanines (G6, G9 and G18) from the latter G-tetrad are isolated (not directly connected in the strands that support the G-quadruplex core).
A range of novel loop topologies connect opposingly oriented guanines between the top inverted G-tetrad (G9•G26•G18•G6) layer and the bottom two G-tetrads (G12•G16•G21•G25 and G11•G15•G20•G24) that are part of the all-parallel-stranded G-quadruplex (Figs. 5e,f). Amongst these, single-residue loops connect G9 to G11 and G18 to G20, each of which are separated by a G-tetrad plane, while a single-residue loop connects G16 to G18 on adjacent G-tetrad planes (Supplementary Fig. 4b–d). Strikingly, no nucleotide connects G25 and G26 on adjacent G-tetrads, given that these guanines adopt opposing local strand orientations (Figs. 5e,f). Given the novelty of these loops and the presence of left-handed dinucleotide steps (Supplementary Fig. 5), these topologies are considered further in the Discussion and Supplementary sections.
All residues in the duplex stem adopted the C3’-endo sugar pucker conformation, as usually expected for RNA. By contrast, all residues in the G-tetrad core and loops adopted the C2’-endo sugar pucker conformation, except for G11, G25 and A14, which adopted the C3’-endo conformation. The sugar puckers in the refined structures were consistent with pucker geometries estimated from the magnitude of the COSY peaks between H1’ and H2’ sugar protons in the complex.
The conformation of the bound peptide is best defined for the R10GGGGR15 segment, whose positioning at the duplex-quadruplex junction (Fig. 6a), is defined by the largest number of intermolecular NOEs in the complex (Supplementary Table 3). This R10GGGGR15 segment adopts a turn conformation, centered opposite the duplex-quadruplex junction, with the side chains of R10 and R15 directed in opposite orientations (Fig. 6b). The backbone NH of G13 and CO of G11 point towards each other with the resultant hydrogen bond stabilizing the turn.
Figure 6.
Details of intermolecular peptide-RNA interactions in the solution structure of the RGG peptide - sc1 RNA complex. (a) Positioning of the R10 to R15 segment of the RGG peptide (stick representation) within the major groove of the duplex segment of the sc1 RNA in the complex (surface representation). (b) A blow up emphasizing the intermolecular interaction shown in panel a. The peptide residues are labeled from R10 to R15. (c, d) Two views highlighting the intermolecular hydrogen bonding interactions between the guanidinium groups of R10 and R15 with the major groove edges of G31 and G7, respectively.
RGG peptide-sc1 RNA interface in the complex
The turn conformation of the R10GGGGR15 segment of the RGG peptide recognizes the duplex-quadruplex junction through a combination of shape complementarity (Figs. 6a,b) and intermolecular hydrogen-bonding interactions. Intermolecular steric constraints mandate an absolute requirement for Gly residues at positions 12 and 13, and at best, suggest that only small side chains could be tolerated at positions 11 and 14. The key intermolecular contacts in the complex are between Arg10 and Arg15 of the RGG peptide and the major groove of the Watson-Crick G•C base pairs of the duplex segment adjacent to the duplex-quadruplex junction. Thus, the side chains of Arg10 and Arg15 are directed in opposite directions (Fig. 6b), such that they target adjacent guanines G31 and G7 respectively, on partner strands of the duplex (Figs. 6c,d). Further, the structure of the complex revealed that the two arginines, Arg15 and Arg10, recognize O6 and N7 of guanines along the major-groove edges of G7•C30 and C5•G31 base pairs, respectively (Fig. 6d). In addition, the side chain of Arg8 is positioned in the vicinity of U3 5´-phosphate (Fig. 4a) providing important stabilizing interaction for the complex.
Unexpectedly, no intermolecular NOEs were observed between the RGG peptide and the G-quadruplex segment of the sc1 RNA in the complex.
Minimal RGG peptide and sc1 RNA elements in the complex
The NMR-based structural studies reported above were undertaken on a 1:1 complex of 28-aa RGG peptide and 36-nt sc1 RNA sequence. Given that the R10GGGGR15 segment of the peptide and the duplex-quadruplex junction of the RNA were key to intermolecular recognition, we investigated into the extent to which both components could be truncated and still retain complex formation. The observed dispersion pattern of imino proton resonances in the spectrum of the complex provides a useful set of markers for testing the integrity of complex formation (Fig. 1e). Thus, the length of the peptide could be reduced stepwise from a 28-mer to a 10-mer, with the resulting Arg8 to Gln17 segment still forming a robust complex (Supplementary Fig. 6a–e). Conversely, the RNA could be reduced stepwise in length from a 36-mer to a 31-mer by truncating two terminal base pairs of the duplex, with the resulting U4-to-A33 segment still forming a robust complex (Supplementary Fig. 6f–h).
Impact of RGG peptide mutants on complex formation
Based on the structure of the RGG peptide-sc1 RNA complex (Fig. 4), we investigated how mutations within the peptide sequence affect its structure and binding properties. These mutations were made in the context that the Arg8-to-Gln17 RGG peptide segment targeted the sc1 RNA at the duplex adjacent to the duplex-quadruplex junction.
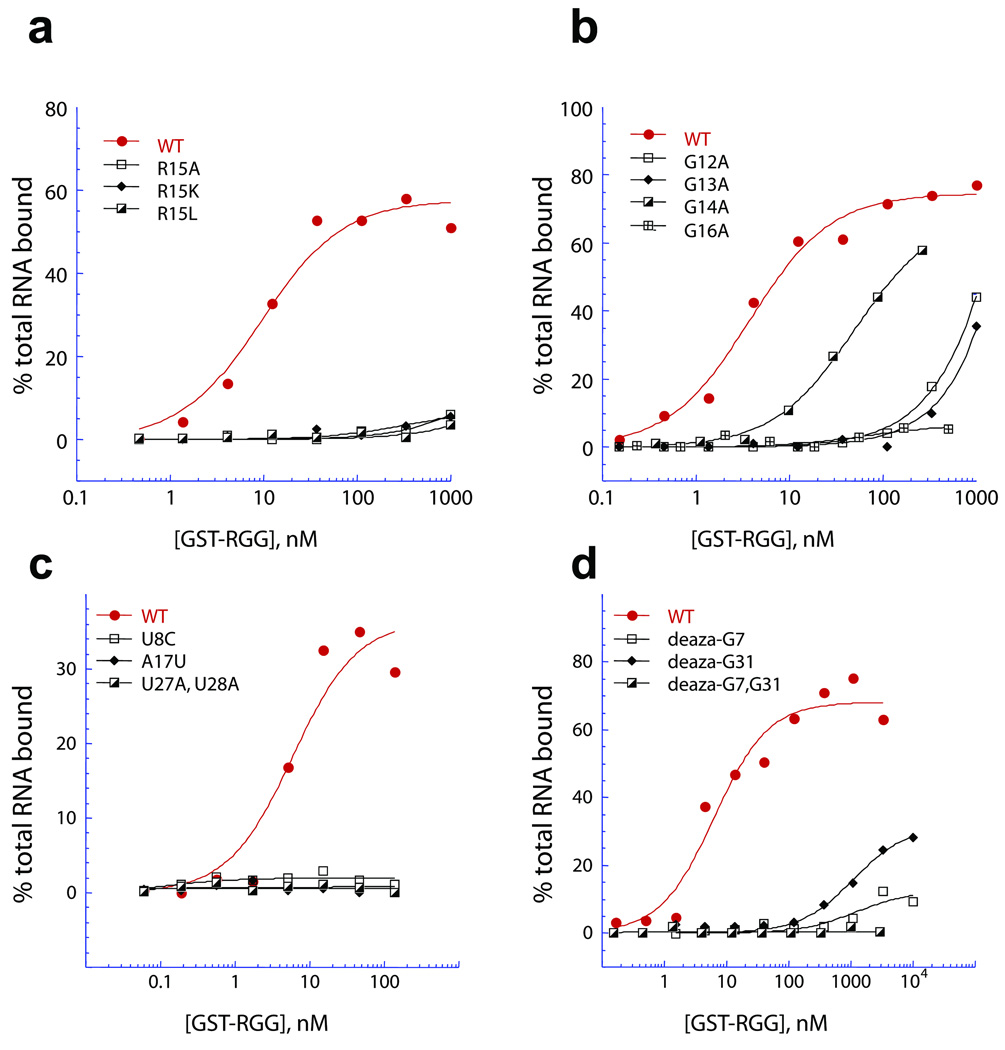
Given that the guanidinium groups of both Arg10 and Arg15 form intermolecular hydrogen bonds with guanine major groove base edges in the complex (Fig. 6d), it was not surprising to observe that mutating Arg10 to either Lys or Leu resulted in complete loss in binding in filter binding assays (Supplementary Fig. 7a), as did mutating Arg15 to either Lys, Leu or Ala (Fig. 7a). Similarly, mutating Gly11 to Asp, but not Ala, resulted in complete loss in binding (Supplementary Fig. 7b), while replacement of Gly12, Gly13 and Gly16 to Ala (Fig. 7b) or Asp (Supplementary Fig. 7c) resulted in either complete or significant loss in binding. Replacement of Gly14 within the R10GGGGR15 segment by Ala (Fig. 7b) or Leu (Supplementary Fig. 7d) results in a 13-fold and 16-fold loss in binding affinity, respectively.
Figure 7.
Assessment of the molecular determinants of the peptide and of the RNA for the FMRP RGG peptide - sc1 RNA interaction by filter binding assay. The affinity of interaction of the FMRP RGG box with 35-mer sc1 RNA (red) and mutations therein was determined by filter binding assay using the indicated concentrations of purified GST-tagged RGG box and 32P end-labeled commercially synthesized RNA. (a) Mutation of Arg15 to Ala, Lys or Leu abolished sc1 RNA binding by the peptide. (b) Mutation of the glycines surrounding these crucial arginines. (WT RGG (red circles, Kd = 3.8 nM), Gly12Ala (open squares, Kd = ND), Gly13Ala (diamonds, Kd = ND), Gly14Ala (half closed squares, Kd = 49.5 nM), Gly16Ala (crossed squares, Kd = ND). (c) Mutation of nucleotides involved in the functional tetrads including U8C, A17U and U27A, U28A resulted in affinities too low to be measured. (d) The affinity of the RGG box for synthetic RNAs incorporating deaza-guanine either singly or in combination in positions G7 and G31 in sc1 RNA was measured to assess the importance of the N7 nitrogen of these guanines. Mutation of either N-7 (G7, open black squares, G31, diamonds) decreased binding by more than two orders of magnitude relative to the WT RNA (red circles) and the double mutation (half-closed black squares) decreased binding to an undetectable level.
We have also mutated Arg residues 3, 4 and 8 to Lys, and observe a modest 2.5-fold loss in binding affinity for the Arg3Lys and Arg4Lys mutants and a 9-fold loss for the Arg8Lys mutant (Supplementary Fig. 7e). Further, replacement of Gln17 by Asn results in a 3.5-fold loss in binding affinity (Supplementary Fig. 7f).
The effect of Arg10 and Arg15 RGG peptide mutations were also tested with various other in vitro-selected RNA sequences (sc2 to sc6 RNAs) identified based on their ability to bind FMRP12. The Arg15 to Lys mutation was more perturbing than the Arg10 to Lys mutation for binding to sc2 to sc6 RNA sequences (Supplementary Figs. 8a–f).
Impact of sc1 RNA mutants on complex formation
We have identified a pair of one-nucleotide loops and a pair of two-nucleotide loops connecting guanines involved in G-quadruplex formation in the complex. Thus, U10 connects G9 to G11 and U19 connects G18 to G20 (Figs. 5e,f), while A13–A14 connects G12 to G15 and C22–U23 connects G21 to G24 (Figs. 5e,f). Replacement of such loop bases U10, U19, C22 and U23 by adenine (Supplementary Fig. 9a) has little impact on binding affinity (Supplementary Fig. 9b). Further, converting two-nucleotide loops into one-nucleotide loops, as for ΔC21 and ΔA14 combined with mutation A13U (Supplementary Fig. 9a) also has little impact on binding affinity (Supplementary Fig. 9b). Finally, U27, which connects G26 of the G-tetrad with U28 of the mixed junctional tetrad, can be replaced by adenine without impacting on binding affinity (Supplementary Fig. 9b). Overall, these results imply that connecting loops can be as small as a single nucleotide, and their base composition is not critical for retention of complex formation.
The integrity of the mixed U8•A17•U28•G29 junctional tetrad (Figs. 5c,d; Supplementary Fig. 4a) is important for complex formation, since single U8C and A17U mutants and the dual U27A/U28A mutants, result in complete loss in binding affinity (Fig. 7c).
We also note that the composition of the G7•C30 and C5•G31 pairs that form intermolecular hydrogen bonds with the side chains of R15 and R10, respectively (Fig. 6d) are also critical, since replacement for instance, of C5•G31 by U5•A31 (designated “long stem” sequence, Supplementary Fig. 9a), for instance resulted in complete loss in binding activity (Supplementary Fig. 9c), while RNA integrity was confirmed by denaturing gel analysis.
Given that the Hoogsteen edges of G7 and G31 pair with the guanidinium groups of R15 and R10 respectively, in the complex (Fig. 6d), we investigated the binding properties of sc1 RNA containing single deaza-G7 and deaza-G31 mutants, as well as the dual deaza-G7/deaza-G31 mutant. These deaza-G7 and deaza-G31 mutants result in complete to pronounced loss in binding affinity (Fig. 7d).
DISCUSSION
Despite considerable interest in the binding of FMRP RGG domain to G-rich sequences capable of G-quadruplex formation, little structural information is available on the molecular events underlying this recognition12,3,17,34–36. Our NMR-based solution structure of the FMRP RGG peptide – sc1 RNA complex has defined how the R10GGGGR15 segment of the RGG peptide targets the duplex-quadruplex junction of the RNA. The structure of the complex explains the role of Gly and Arg residues in mediating shape complementarity and base-specific recognition, as well as key features of the duplex-quadruplex junction topology in shaping the binding pocket and dictating the trajectory of the bound peptide. The RNA quadruplex composed of three stacked G-tetrads adopts an unprecedented fold, with the structure of the bound RNA also defining how a mixed junctional tetrad mediates the facile connection between duplex and quadruplex folds.
Bridging RNA duplex and quadruplex elements in the complex
Both the FMRP RGG peptide (Fig. 1b) and the hairpin loop segment of sc1 RNA (Fig. 1d) are essentially unstructured in the free state, but become structured (Figs. 1c,e) on complex formation, with spectral features characteristic of a single conformational species in solution. We have identified a novel RNA G-quadruplex and an unanticipated duplex-quadruplex junction in the structure of the FMRP RGG peptide - sc1 RNA complex (Figs. 5a,b). The key challenge centered on how to connect an anti-parallel five-base-pair duplex (G1 to C5 paired with G31 to C35, Fig. 1a) to an all-parallel-stranded G-quadruplex composed of two stacked G-tetrads (G11•G15•G20•G24 and G12•G16•G21•G25, Figs. 5e,f). Remarkably, this has been achieved by interjecting three layers at the junction between these two motifs (Figs. 5a,b), that include a base pair, a mixed tetrad and an additional inverted G-tetrad, thereby allowing a smooth transition between the anti-parallel duplex and the all-parallel-stranded quadruplex motifs.
In the first layer, the duplex is extended through formation of a G7•C30 Watson-Crick pair, which is stacked on the terminal C5•G31 Watson-Crick pair of the duplex (Figs. 5a,b). Though G30 and C31 are adjacent to each other in sequence, C5 and G7 are not, but they still retain a pseudo-continuous backbone (Figs. 5a,b).
In the second layer, the duplex is further extended by formation of a U8•G29 wobble pair, which is stacked on the G7•C30 pair mentioned above, and forms part of a U8•A17•U28•G29 mixed tetrad, (Figs. 5a,b,c,d and Supplementary Fig. 4a). Formation of this mixed tetrad, with its U28-G29 platform37, is important since it contributes a bridging function linking the duplex and quadruplex folds.
The third layer is composed of an inverted G9•G26•G18•G6 tetrad, stacked between the mixed U8•A17•U28•G29 and G12•G16•G21•G25 tetrads (Figs. 5a,b and 5e,f). Strikingly, both the strand and hydrogen bond directionalities for the G9•G26•G18•G6 tetrad are the reverse of that observed for the G12•G16•G21•G25 and G11•G15•G20•G24 tetrads (Figs. 5e,f), thereby defining a completely new folding topology between three stacked G-tetrads within a G-quadruplex.
Novel conformational features within junctional elements
The G9•G26•G18•G6 tetrad is connected to the two-G-tetrad-layered all-parallel-stranded G-quadruplex by a novel set of connecting loops, the majority of which have not been observed previously33. Thus, single-residue linkers, U10 and U19, form a new type of loop, which is observed here for the first time, connecting guanines with opposing strand orientations and separated by a G-tetrad, but belonging to the same column of the G-quadruplex (Figs. 5e,f and Supplementary Figs. 4b,c). Single-residue linker A17 forms a second type of new loop connecting guanines with opposing strand orientations on two adjacent G-tetrad layers, while belonging to different columns of the G-quadruplex (Figs. 5e,f and Supplementary Fig. 4d). Linker G7–U8 forms the third type of new loop that connects two guanines G6 and G9 with similar strand orientations that occupy adjacent positions within a G-tetrad (Figs. 5e,f and Supplementary Fig. 4e).
Additional novel conformational features within junctional elements, including left-handed backbone connections (Supplementary Fig. 5), are outlined in the Supplementary section.
R10GGGGR15 adopts a turn fold at duplex-quadruplex junction
Our results establish that the R10GGGGR15 peptide adopts a turn conformation, with the side chains of R10 and R15 pointed in opposite directions (Figs. 6a,b), allowing for interactions with the major groove of flanking G•C pairs of the duplex segment adjacent to the duplex-quadruplex junction (Figs. 6c,d). Unexpectedly, the RGG peptide does not interact directly with the quadruplex, but this scaffold enforces the turn adopted by the R10GGGGR15 peptide, and facilitates its positioning within the binding pocket on the RNA in the complex.
In the case of sc1 RNA, the major groove of the duplex widens somewhat38–40 and opens adjacent to the duplex-quadruplex junction on complex formation as a consequence of three structural factors. First, these include widening of the groove itself as measured by the distance between phosphate groups of constitutive strands, bending of its helical axis, and specific structural nucleotide arrangements. Two structural factors leading to groove widening and generation of a defined ligand-binding pocket are the G29•U8 wobble pair that is shifted towards the major groove, as compared to a standard G•C base pair, and the presence of an extra residue G6 at the pseudo-continuous C5---G7 step, with resulting widening of the major groove at the junction with the quadruplex.
The role of shape complementarity and arginine-guanine pairing to peptide-RNA recognition is outlined in greater detail in the Supplementary section.
Role of G-quadruplex binding in RNA metabolism and disease
Regulation of gene expression through post-transcriptional mechanisms such as splicing, 3’-end formation, translational control and RNA turnover adds multiple layers of refinement to the metabolism of nascent transcripts. The widespread occurrence of G-quadruplex motifs near regulatory sites, and the identification of a number of RNA-binding proteins (RNABPs) which bind to, stabilize or unwind these structures, suggests that greater understanding of these interactions will shed light on these key regulatory steps. Moreover, as loss of similar interactions between DNA binding proteins such as WRN or telomere end-binding proteins and G-quadruplexes in DNA have been linked to human diseases such as Bloom’s and Werner’s syndromes, it is likely that understanding the subtleties of RNABP-RNA G-quadruplex interactions will contribute to the growing list of human diseases resulting from loss-of-function of RNABPs41,42.
Guanine-rich sequences are found in a number of RNAs, other than sc1 RNA, such as the 5’-UTR of certain oncogenes3, that are involved in distinct biological processes. A systematic bioinformatics search found the occurrence of the exact RGGGGR motif in several genes of the H. sapiens genome43–48. The RGG domain of nucleolin, an abundant protein in the nucleolus, has been reported to bind with high affinity to G-rich rDNA and other G-rich sequences49. Thus, the recognition principles reported in this paper for the FMRP RGG peptide - sc1 RNA complex may be applicable to other RGG-containing protein-RNA complexes.
We also discuss in some detail the significance of binding to the duplex-quadruplex junction in determination of FMRP target RNAs in some detail in the Supplementary text section.
A crucial role of RGG box, Arg10 and Arg15 in FMRP function
In the current study we find that arginine interactions play a key role in this recognition. In particular, Arg15 is important for recognition of all six G-quadruplexes identified by RNA selection (sc1–6), while Arg10 is only important for a subset (Supplementary Fig. 8). While these observations suggest that there are multiple ways for the FMRP RGG box to bind G-quadruplexes with high affinity, the observation that Arg15 has at least a 10-fold effect on the binding to each G-quadruplex suggests that a common feature of RGG-box-RNA target recognition may lie in interaction with sequence-specific elements in the stem. Such interactions will be important in further defining RNA targets that are biologically relevant by virtue of their specific interaction with the FMRP RGG-box.
The RGG box is required for several functions of FMRP critical to its role in controlling local mRNA translation to effect activity-dependent synaptic plasticity in neurons, providing a critical link between RNA binding by this domain and protein function in vivo. Deletion of the RGG box resulted in failure of FMRP to nucleate stress granules, thought to be one of the important mechanisms through which FMRP regulates translation in cells50. A finer analysis of the determinants required for FMRP to associate with polyribosomes revealed that the two arginines we find to be directly involved in RNA recognition, Arg10 and Arg15 (R533 and R538 of murine FMRP) are necessary, directly implicating these arginines in proper translational control by FMRP36. Moreover, it has been reported that mono- and dimethylarginine modifications of the RGG box decrease binding to RNA, which may suggest a regulatory role for this modification in vivo. Intriguingly, Arg10 and Arg15 were two of the four arginines found to be modified in vivo by the arginine methyltransferase PRMT151. Finally, it has recently been shown that the RGG box is required for FMRP to mediate MEF2-dependent synapse elimination in response to neuronal activity in mice52. Loss-of-function of FMRP in this regard may cause the increases in spine density observed in both mouse models and human Fragile X patients which may be indicative of a deficit in excitatory spine elimination, and directly contributing to observed defects at the levels of both synapse and circuit function52.
METHOD
Methods and any associated references are available in the Online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yuying Gosser and Stephen Pitt for their participation at the early stage of the project, Olga Mostovetsky, Elizabeth F. Stone, and Ka Ying Sharon Hung for technical assistance, and Alex Lash for bioinformatics of RG4R motif in H. sapiens genome. This research was supported by NIH grant CA049982 to DJP, NIH grant R01 HD040647 to JCD, and Singapore BMRC grant 07/1/22/19/542 to ATP. DJP is a member of the New York Structural Biology Center supported by NIH grant GM66354.
Footnotes
Accession codes
The coordinates for the structure of the RGG-sc1 RNA complex have been deposited in the Protein Data Bank (accession code 2la5).
Note: Supplementary information is available on the Nature Structural & Molecular Biology web site.
AUTHOR CONTRIBUTIONS
ATP, AM and SI were responsible for NMR studies, VK undertook computations, while AS, TR and AP prepared labeled NMR samples, all under the supervision of DJP. CC, DC and JCD undertook filter-binding assays under the joint supervision of JCD and RBD. The paper was written jointly by DJP, ATP, VK, JCD and RBD.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interest.
Reprints and permission information is available online at http://nature.com/reprintsandpermissions/.
REFERENCES
- 1.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5' UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxena S, Miyoshi D, Sugimoto N. Sole and stable RNA duplexes of G-rich sequences located in the 5'-untranslated region of protooncogenes. Biochemistry. 2010;49:7190–7201. doi: 10.1021/bi101093a. [DOI] [PubMed] [Google Scholar]
- 5.Bonnal S, et al. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliver AW, Bogdarina I, Schroeder E, Taylor IA, Kneale GG. Preferential binding of fd gene 5 protein to tetraplex nucleic acid structures. J. Mol. Biol. 2000;301:575–584. doi: 10.1006/jmbi.2000.3991. [DOI] [PubMed] [Google Scholar]
- 7.Arora A, et al. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bensaid M, et al. FRAXE-associated mental retardation protein (FMR2) is an RNA-binding protein with high affinity for G-quartet RNA forming structure. Nucleic Acids Res. 2009;37:1269–1279. doi: 10.1093/nar/gkn1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Didiot MC, et al. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008;36:4902–4912. doi: 10.1093/nar/gkn472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagga PS, Ford LP, Chen F, Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3' end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darnell JC, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 13.Schaeffer C, et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. Embo J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Boulle K, et al. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 16.Zang JB, et al. A mouse model of the human Fragile X syndrome I304N mutation. PLoS Genet. 2009;5:e1000758. doi: 10.1371/journal.pgen.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darnell JC, Fraser CE, Mostovetsky O, Darnell RB. Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs. Hum. Mol. Genet. 2009;18:3164–3177. doi: 10.1093/hmg/ddp255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phan AT, Patel DJ. A site-specific low-enrichment (15)N,(13)C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002;124:1160–1611. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- 19.Phan AT. Long-range imino proton-13C J-couplings and the through-bond correlation of imino and non-exchangeable protons in unlabeled DNA. J. Biomol. NMR. 2000;16:175–178. doi: 10.1023/a:1008355231085. [DOI] [PubMed] [Google Scholar]
- 20.Martadinata H, Phan AT. Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution. J. Am. Chem. Soc. 2009;131:2570–2578. doi: 10.1021/ja806592z. [DOI] [PubMed] [Google Scholar]
- 21.Fiala R, Jiang F, Sklenar V. Sensitivity optimized HCN and HCNCH experiments for 13C/15N labeled oligonucleotides. J. Biomol. NMR. 1998;12:373–383. [Google Scholar]
- 22.Nikonowicz EP, Pardi A. An efficient procedure for assignment of the proton, carbon and nitrogen resonances in 13C/15N labeled nucleic acids. J. Mol. Biol. 1993;232:1141–1156. doi: 10.1006/jmbi.1993.1466. [DOI] [PubMed] [Google Scholar]
- 23.Dingley JC, Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 24.Pervushin K, et al. NMR scalar couplings across Watson-Crick base pair hydrogen bonds in DNA observed by transverse relaxation-optimized spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14147–14151. doi: 10.1073/pnas.95.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majumdar A, Kettani A, Skripkin E, Patel D. Observation of internucleotide NH…N hydrogen bonds in the absence of directly detectable protons. J. Biomol. NMR. 1999;15:207–211. doi: 10.1023/a:1008357304708. [DOI] [PubMed] [Google Scholar]
- 26.Majumdar A, Kettani A, Skripkin E. Observation and measurement of internucleotide 2JNN coupling constants between 15N nuclei with widely separated chemical shifts. J. Biomol. NMR. 1999;14:67–70. doi: 10.1023/a:1008335502416. [DOI] [PubMed] [Google Scholar]
- 27.Muhandiram R, Kay LE. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. 1994;B103:203–216. [Google Scholar]
- 28.Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- 29.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell. Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majumdar A, Gosser Y, Patel DJ. 1H-1H correlations across N-H‥N hydrogen bonds in nucleic acids. J. Biomol. NMR. 2001;21:289–296. doi: 10.1023/a:1013340227140. [DOI] [PubMed] [Google Scholar]
- 31.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 32.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5'-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramos A, Hollingworth D, Pastore A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003;9:1198–1207. doi: 10.1261/rna.5960503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanotti KJ, Lackey PE, Evans GL, Mihailescu MR. Thermodynamics of the fragile X mental retardation protein RGG box interactions with G quartet forming RNA. Biochemistry. 2006;45:8319–8330. doi: 10.1021/bi060209a. [DOI] [PubMed] [Google Scholar]
- 36.Blackwell E, Zhang X, Ceman S. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum. Mol. Genet. 2010;19:1314–1323. doi: 10.1093/hmg/ddq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cate JH, et al. RNA tertiary structure mediation by adenosine platforms. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 38.Puglisi JD, Chen L, Blanchard S, Frankel AD. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science. 1995;270:1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- 39.Ye X, Kumar RA, Patel DJ. Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex. Chem. Biol. 1995;2:827–840. doi: 10.1016/1074-5521(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 40.Davidson A, et al. Simultaneous recognition of HIV-1 TAR RNA bulge and loop sequences by cyclic peptide mimics of Tat protein. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11931–11936. doi: 10.1073/pnas.0900629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 42.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ota T, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 44.Vaccari T, et al. The human gene coding for HCN2, a pacemaker channel of the heart. Biochim. Biophys. Acta. 1999;1446:419–425. doi: 10.1016/s0167-4781(99)00092-5. [DOI] [PubMed] [Google Scholar]
- 45.Wang C, Meier UT. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanhalst K, Kools P, Staes K, van Roy F, Redies C. delta-Protocadherins: a gene family expressed differentially in the mouse brain. Cell. Mol. Life Sci. 2005;62:1247–1259. doi: 10.1007/s00018-005-5021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerdes D, Wehling M, Leube B, Falkenstein E. Cloning and tissue expression of two putative steroid membrane receptors. Biol. Chem. 1998;379:907–911. doi: 10.1515/bchm.1998.379.7.907. [DOI] [PubMed] [Google Scholar]
- 48.Katoh K, et al. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J. Biol. Chem. 2003;278:39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- 49.Hanakahi LA, Sun H, Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999;274:15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 50.Mazroui R, et al. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 51.Stetler A, et al. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum. Mol. Genet. 2006;15:87–96. doi: 10.1093/hmg/ddi429. [DOI] [PubMed] [Google Scholar]
- 52.Pfeiffer BE, et al. Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron. 2010;66:191–197. doi: 10.1016/j.neuron.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Batey RT, Battiste JL, Williamson JR. Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol. 1995;261:300–322. doi: 10.1016/s0076-6879(95)61015-4. [DOI] [PubMed] [Google Scholar]
- 54.Pikovskaya O, Serganov AA, Polonskaia A, Serganov A, Patel DJ. Preparation and crystallization of riboswitch-ligand complexes. Methods Mol. Biol. 2009;540:115–128. doi: 10.1007/978-1-59745-558-9_9. [DOI] [PubMed] [Google Scholar]
- 55.Kuryavyi V, Patel DJ. Solution structure of a unique G-quadruplex scaffold adopted by a guanosine-rich human intronic sequence. Structure. 2010;18:73–82. doi: 10.1016/j.str.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 57.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 58.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 59.Tereshko V, Skripkin E, Patel DJ. Encapsulating streptomycin within a small 40-mer RNA. Chem. Biol. 2003;10:175–187. doi: 10.1016/s1074-5521(03)00024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Placido D, Brown BA, 2nd, Lowenhaupt K, Rich A, Athanasiadis A. A left-handed RNA double helix bound by the Z alpha domain of the RNA-editing enzyme ADAR1. Structure. 2007;15:395–404. doi: 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.