Abstract
Recent evidence suggests that particular species of non-coding RNAs can modulate gene transcription in human cells. While such observations were in the past relegated to imprinted genes, it is now becoming apparent that several different genes in differentiated cells may be under some form of RNA based regulatory control. Studies carried out to date have begun to discern the mechanism of action whereby non-coding RNAs modulate gene transcription by the targeted recruitment of epigenetic silencing complexes to homology containing loci in the genome. The results of these studies will be considered in detail as well as the implications that a vast array of non-coding RNA based regulatory networks may be operative in human cells.
Keywords: concordant, discordant, transcriptional silencing, non-coding RNA, epigenetic
Introduction
The paradigm biologists operate in has been constructed upon the seminal work of Darwin and the eventual genesis of the theory of evolution: which surmises that all living organisms are the descendants of a common ancestor. The theory of evolution argues that species undergo “descent with modification”, whereby mutations in the genome are selected over time leading to the evolution of new traits and indeed new species. Such selective pressures would be thought to result in new species acquiring or loosing genes over time, as they adapt to the particular selective pressures which have driven them into the frame work of a new species. Not un-common to theories there appear to be exceptions to the rules. When one assesses the genome of a single celled organism relative to a multicellular organism this does not appear to be the case. When scientists sequenced the entire genome of the ~2000 celled Volvox carteri and the single-celled green alga, they found very few if any genomic differences. In fact the only difference they were able to discern was that the Volvox contained increased repetitive non-coding DNA sequences [1]. One has to wonder; what might this interesting observation be telling us and what exactly are non-coding DNA sequences? How can these non-coding DNAs be functional in differentiating between single celled and multicelled organism?
To begin with what exactly are non-coding DNAs and what role might they be playing in the evolution of multi-cellular organisms from a single celled organismal state. Simply stated, non-coding DNAs are those regions in the genome that do not code for proteins. In eukaryotes actually a large percentage, estimates in humans range from the low 70 to upper 90% of the genome is non-coding (reviewed in [2, 3]. The use of genome-wide analytical tools such as deep sequencing have begun to shed some light on those RNAs emanating from non-coding DNAs in human cells [4]. As these RNAs are transcribed from non-coding DNAs they are referred to as non-coding RNAs.
Evidence has begun to emerge suggesting that non-coding RNAs can functionally alter gene expression in a manner that is both directly targeted as well as long-lasting. The impact of a set of molecules that govern this form of control cannot be overstated as long lasting effects can, by extension, have an impact on the evolution of the cell. Functionally, long non-coding RNAs were first shown to be involved in dosage compensation and X-inactivation in undifferentiated primordial cells (reviewed [5]). These long-non-coding RNAs appeared to function by the targeting the recruitment of different epigenetic regulatory complexes to their intended targets. In the context of X-chromosome inactivation in human cells, one of the X- chromosomes is silenced by the coating of the chromosome with TSIX, which is the antisense non-coding RNA for Xist [6]. The coating of the chromosome results in epigenetic silencing of those genes on one of the X chromosomes. The result, for females, is in essence the expression of only one X chromosome. This form of silencing in essence allows females to control the dosage of genes expressed from the rather large X-chromosome and in essence express the same amount of genomic material as males, whom only have one X chromosome. Beyond the quintessential examples of non-coding RNA regulatory networks found in X-chromosome inactivation and imprinting related genes reside what appears to be a sea of genes being regulated by non-coding RNAs. Recent evidence is emerging which suggests that many more genes than previously envisioned, beyond X-inactivation and imprinting related genes, might actually be regulated by non-coding RNAs. In fact non-coding RNAs might be actively switching on and off genes in an orchestral regulation that governs the fidelity of the cell. Some examples in humans are those genes involved in cell-cycle regulation, such as E-cadherin, p21, and p15 [7–9]. The actual mechanism for how these long non-coding RNAs are functional in regulating their particular candidate gene is only just now becoming appreciated and is precipitated from work carried out initially with de novo derived small interfering RNAs. The first clue came from studies where siRNAs derived to specifically target a gene promoter were found to transcriptionally control the promoter/genes transcription [10].
Small non-coding RNAs
The endogenous form of the small interfering RNA, is the microRNA (miRNA), which functions primarily to stymie protein expression by modulating translation efficiencies. MiRNAs might also have several yet to be discovered functions and even be involved in human cells and the regulation of long non-coding RNAs (discussed in detail in [11]). Certainly, the most well characterized regulatory small non-coding RNAs in human cells have been the ability of the miRNAs to regulate gene expression in a post-transcriptional manner. Functionally, these miRNAs bind their target mRNAs, via homology dependent interactions. The result of this binding is the obstruction of the target mRNAs ability to be translated [12].
Small non-coding RNAs can however have different effects in human cells, beyond those described for miRNAs and their respective post-transcriptional regulation. For instance we now also know that small single stranded RNAs, that are antisense to their compliment, can be implicit in directing the localization of epigenetic silencing complexes to homology containing loci in the genome [13–17]. There is also evidence that small hairpin RNAs (shRNAs) or siRNAs in human cells can recruit and direct epigenetic complexes to their respective homology containing target loci in the context of the chromatin [13]. This is an important distinction, as these observations suggest that non-coding RNAs can in principle play a role in directing proteins involved in epigenetic regulation of gene expression. If for instance the small RNA is targeted to a region in the genome where a gene promoter resides the result can be epigenetic remodeling which can lead to transcriptional silencing of the particular targeted gene. A model has been proposed, based on the observations of several different groups (reviewed in [18, 19]), whereby the small single stranded antisense RNA directs the localization of epigenetic silencing complexes to target loci (Figure 1A). While these observations are powerful with regards to de novo designed approaches to specifically control gene transcription, it has remained unknown, until recently, as to whether there was an active mechanism functional in human cells whereby endogenous non-coding RNAs were actively controlling gene transcription.
Figure 1. Antisense non-coding RNA mediated regulation of gene transcription.
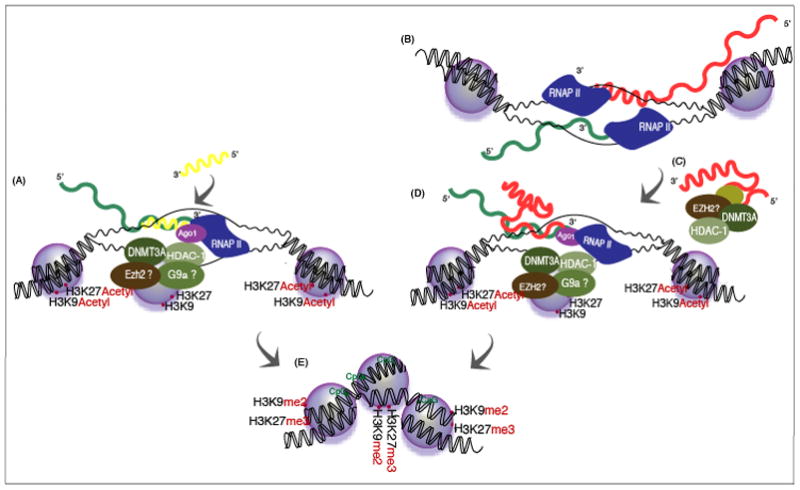
(A) de novo derived small antisense RNAs, targeted to a gene promoter interact with the promoter RNA at the transcribed promoter and facilitate the recruitment of epigenetic remodeling complexes. (B) Endogenous long antisense non-coding RNAs can emanate from bidrectionally transcribed loci. (C) The antisense non-coding RNA might then interact with epigenetic remodeling proteins and (D) guide them to the actively transcribed sense/mRNA transcript. (E) the result of either small or long antisense non-coding RNA directed targeting could be epigenetic silencing of the targeted loci.
Recent work [20] as well as previously published observations [7, 9] have begun to suggest that the endogenous mechanism, which these de novo derived small antisense RNAs are utilizing to control gene transcription, actually involves long antisense non-coding RNAs (Figure 1B–D). This model presumes that the long antisense non-coding RNAs have multiple functions. They can function as both scaffolding for proteins to bind (Figure 1C), as well as a target recognition motif (Figure 1D), whereby the bound proteins can be directly guided to particular loci in the genome. Interestingly, the de novo DNA methyltransferse 3a (DNMT3a) has been shown previously to bind non-coding RNAs [21] as well as to complex with enhancer of zeste (Ezh2) and histone deacetylase 1 (HDAC-1)[22–24]. Moreover, promoter targeted small antisense RNAs have been directly localized at their target loci with DNMT3a and the histone 3 lysine 27 tri-methly mark, imparted on the histone by the action of Ezh2 [17] and HDAC-1[15].
The targeting of these epigenetic complexes to the genome by the action of the de novo derived small or long antisense non-coding RNAs is presumed to be via the interaction between the antisense non-coding RNA and actively transcribed regions at it’s homology containing target. The target could be a promoter-associated RNA [25] whereby the transcription of a gene is controlled (Figure 1), or possibly interactions with complimentary non-coding RNAs that are acting as local chromatin scaffolds for genes [26, 27] or intergenic regions [26, 28]. All of these examples appear to have a direct impact on the accessibility and fidelity of the genome, i.e. access of the DNA to the cellular machinery.
While much has been gleamed from studies with de novo derived small non-coding RNAs, presumably even more striking is the recent escalation of findings suggesting that long antisense non-coding RNAs can functionally regulate a genes transcription in human cells [7, 9, 29–31]. These observations are suggestive of a role for non-coding RNAs in gene regulation that is upstream of the well defined post-transcriptional arena where miRNAs are generally functional. Specifically, these observations suggest a role for long antisense non-coding RNAs in the transcriptional regulation of gene expression. Mechanistically, these particular genes appear to exhibit bidirectional transcription, often times exhibiting a lower copy non-coding RNA, which is antisense to the sense/mRNA compliment (Figure 1B–D). Studies whereby these antisense non-coding RNAs have been suppressed demonstrate appreciable increase’s in the sense counterparts transcriptional fidelity with a concomitant loss of silent state epigenetic marks in the promoter for the sense/mRNA [7]. This form of regulation, termed discordant regulation [32], suggests that the antisense non-coding RNA is directly involved in governing some aspects of the epigenetic regulation of the sense/mRNA gene promoter. An example of discordant regulation is depicted in Figure 2, whereby the antisense non-coding RNA enhances/guides the recruitment of silent state epigenetic modifying complexes to a particular genomic loci. If the targeted loci is a promoter the results can be transcriptional silencing of gene expression (reviewed in [11])(Figure 1B–D). Interestingly, long antisense non-coding RNAs also appear to be involved in concordant regulation, whereby the suppression of the antisense non-coding RNA correlates with a concomitant suppression of the sense/mRNA transcript [32]. What dictates the differences between concordant and discordant regulation remains enigmatic. Recent results however indicate that where the antisense non-coding RNA targets might dictate to what extent the gene is discordantly or concordantly regulated.
Figure 2. Antisense non-coding RNA mediated discordant regulation.
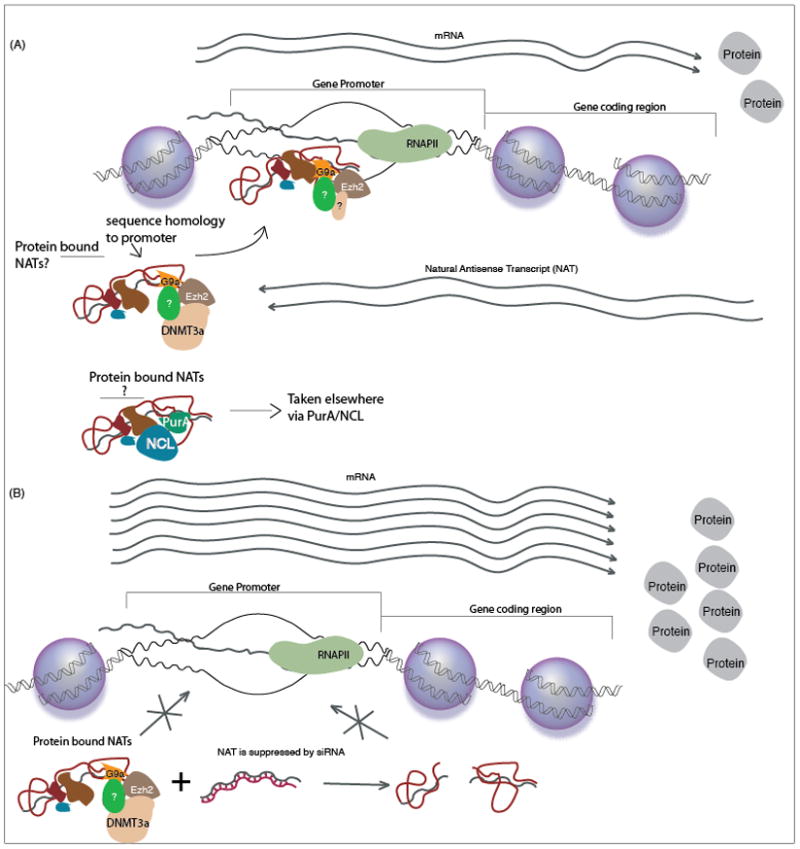
(A) Bidirectionally transcribed genes produce antisense non-coding RNAs which can fold into secondary structures and interact with various proteins as well as target these proteins to homology containing loci in the genome. The antisense RNAs, might operate as a feedback mechanism to apply an epigenetic brake to gene transcription whereby some non-coding RNA complexes are targeted to the chromatin and others are removed from the nucleus. (B) The suppression of the antisense non-coding RNA, by RNAi, can result in a loss of this epigenetic brake and significant increases in gene transcription and eventual protein production.
Non-coding RNA directed discordant vs. concordant regulation
Recent studies with the p21 and p15 genes have indicated that bidirectional transcription is operative in human cells and that the long antisense non-coding RNA, from these particular bidirectional transcribed genes, is functional in epigenetically regulating the sense/mRNA expression (reviewed in [11]. The majority of genes which have been found to be bidirectional, in that they express both sense/mRNAs as well as antisense non-coding RNAs, appear to be involved in cell cycle regulation (reviewed in [33]). More interesting is the observation that those genes exhibiting bidirectional characteristics are often times found to be epigenetically silenced in human cancers [33]. From these limited studies as well as a recent study much has been gleamed regarding bidirectional genes and discordant regulation of gene transcription.
Recent studies with the bidirectionally transcribed Oct-4 gene have shown that an antisense non-coding RNA derived from an Oct-4 psuedogene is actively involved in discordant regulation of Oct-4 transcription [20]. This antisense non-coding RNA does not appear to be poly-adenylated, contains Oct-4 intronic sequences, and was found to specifically span the Oct-4 promoter. The suppression of this Oct-4 antisense non-coding RNA resulted in a loss of H3K27me3, H3K9me2 at the Oct-4 promoter and activation of Oct-4 transcription. The suppression of Ezh2 and G9a also had an impact on Oct-4 transcription, the loss of these factors resulted in increased transcription of Oct-4 [20]. More interesting was the observation that this Oct-4 antisense non-coding RNA, when directly immunoprecipitated and assessed by mass spectrophotometry, was found to associate with several new proteins. When these proteins, namely PurA and Nucleolin were suppressed by RNAi, the Oct-4 mRNA was suppressed even further [20]. These data have provided a greater detailed picture of discordant regulation and suggest that a pathway exists whereby the long antisense non-coding RNAs can be shunted away from their target, most likely depending on whether they are bound to epigenetic remodeling proteins or not (Figure 3). Supporting the observations of antisense non-coding RNA discordant regulation of Oct-4 transcription are recent genome-wide analysis of non-poly adenylated antisense non-coding RNAs, where there appears to be a strong correlation with the abundance of antisense non-coding RNA, the relative amount of DNA methylation at gene promoters, and the suppression of sense/mRNA gene expression from these promoters in human cells [34].
Figure 3. Antisense non-coding RNA mediated Discordant regulation of Oct-4.
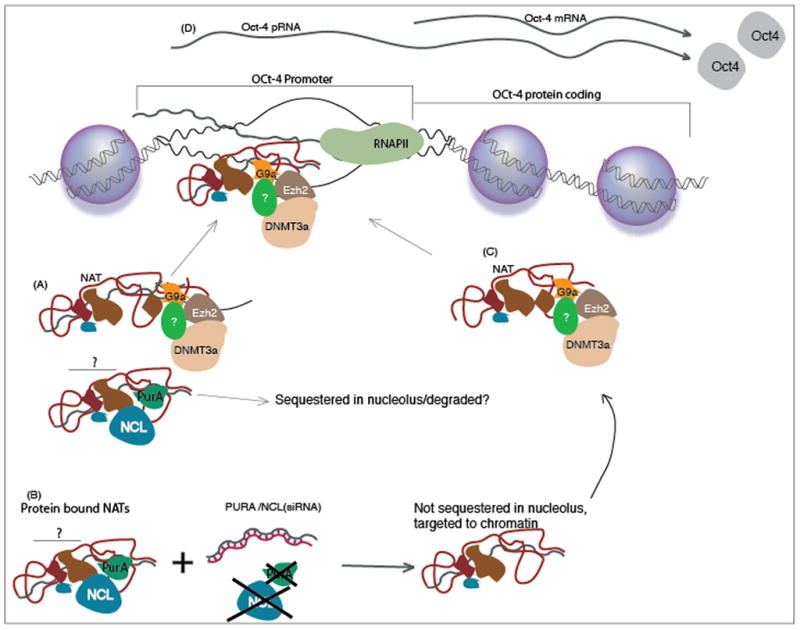
An Oct-4 specific antisense non-coding RNA is expressed capable of targeting epigenetic silencing complexes to the Oct-4 promoter and discordantly regulating transcription. A subset of RNA binding proteins, Nucleolin (NCL) and PurA, were found to directly associate with the Oct-4 regulatory antisense non-coding RNA. When NCL and PurA expression was suppressed by RNAi, an enhanced suppression of Oct-4 was observed, suggesting that one role for NCL and PurA is to sequester the antisense non-coding RNAs away from their intended target.
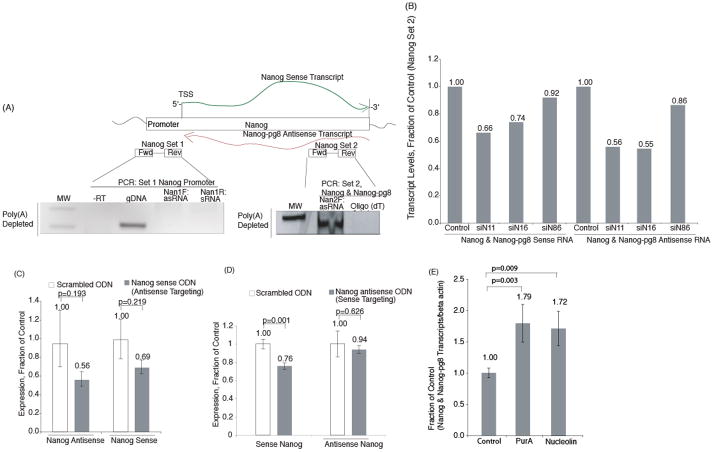
Not all bidirectionally transcribed genes appear to regulate their sense counterpart in a discordant manner. Studies carried out with the gene Nanog, also involved in stem cell genesis, have determined that like Oct-4, there are also Nanog specific psuedogenes, which express long antisense non-coding RNAs (Figure 4A). Interestingly, while these long antisense non-coding RNAs were also not poly-adenylated they don’t appear to span the Nanog promoter (Figure 4A). When these antisense non-coding RNAs were suppressed using shRNAs (Figure 4B) or antisense phosphorothiate oligonucleotides (Figure 4C) the Nanog mRNA expression was also reduced in a concordant manner. Conversely, the suppression of the sense mRNA with phosphorothiate oligonucleotides also resulted in a modest reduction in antisense non-coding RNA (Figure 4D). While the reduction in either Nanog sense or antisense non-coding RNAs with phosphorothiate oligonucleotides tends to be modest, the overall trends appear to be the same, i.e. when the antisense non-coding RNA is suppressed there is a concordant reduction in the sense/mRNA transcript as well. Furthermore, when those proteins, Nucleolin and PurA, found in previous work to be directly associated with the Oct-4 antisense non-coding RNA, were suppressed there appeared to be significant increases in Nanog expression (Figure 4E). These data are the direct opposite of what was observed previously with the discordant regulated Oct-4 [20].
Figure 4. Characterization of an antisense non-coding RNA involved in regulating Nanog.
(A) Detection of a Nanog specific antisense transcript by directional RT and PCR with Nanog specific psuedogene 8 primers from MCF-7 cellular RNA depleted of Poly A (techniques described in [7, 20]). (B) The Nanog specific antisense non-coding RNA was targeted with three different shRNAs relative to the Oct18 scrambled control [20]. (C and D) Single stranded phosphorothioates were generated to target the Nanog 16 site (refer to B) at either (C) the antisense non-coding RNA or (D) sense mRNA, and transfected into MCF-7 cells (100nM, Lipofectamine 2000). The cultures were collected 72hrs later and assessed for Nanog expression. (E) PurA and Nucleolin were suppressed (transfection of 1μg Mission shRNA plasmids/10^6 MCF7 cells (a gift Carol Krieder Sigma)). Nanog expression was determined 72 hrs later. (C-E) The averages of triplicate treated cultures are shown with the standard deviations and p values from a paired T-test.
Taken together the Oct-4 discordant/Nanog concordant model systems might be telling us something important. What is observed here is that both gene systems have endogenous psuedogenes that express antisense non-coding RNAs that affect the transcriptional fidelity of the Oct-4 or Nanog sense counterparts. Both of these systems appear to exhibit non-poly-adenylated antisense non-coding RNAs and both seem to depict opposite effects when the antisense non-coding RNA, PurA, or Nucleolin are suppressed. The only striking difference between these two systems is the observation that the Oct-4 antisense non-coding RNA appears to span the Oct-4 promoter whereas the Nanog antisense non-coding RNA does not appear to span the Nanog promoter. As such the Nanog antisense non-coding RNA would be expected to direct epigenetic silencing marks to the coding region of Nanog (Figure 5). This is a crucial observation and suggestive of separate and distinct roles for antisense non-coding RNAs in gene expression. This observation suggests that the major difference in the role of antisense non-coding RNA in Oct-4 (Figure 3) and Nanog (Figure 5) regulation resides primarily in where the antisense non-coding RNA targets the gene.
Figure 5. Antisense non-coding RNA mediated concordant regulation of Nanog.
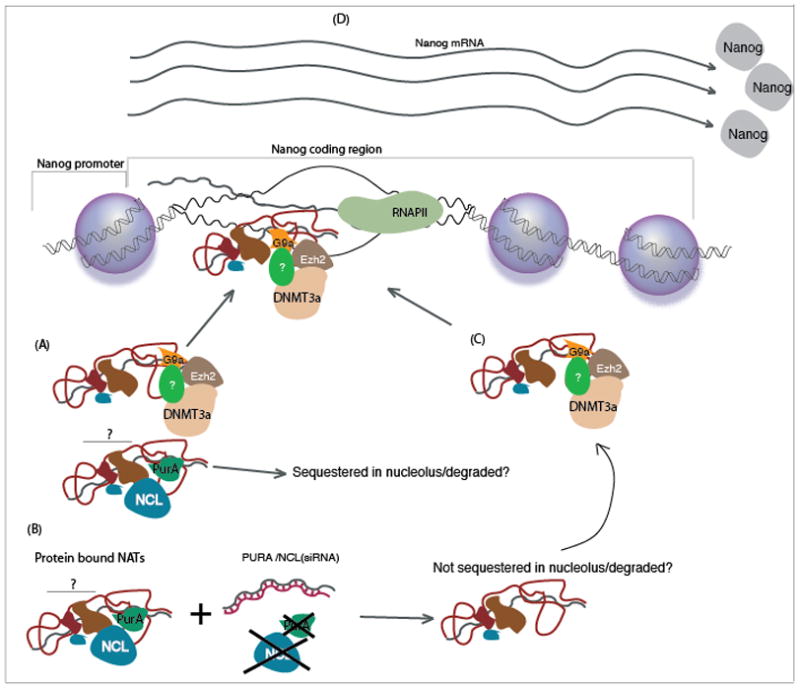
(A) The antisense non-coding RNA expressed from the Nanog psuedogene is able to interact with various protein complexes. (B) Suppression of PurA or Nucleolin (NCL) results in a loss of one protein complex involved in regulating or trafficking the particular antisense non-coding RNA resulting in (C) the enhanced targeting of the antisense non-coding RNA and an epigenetic silencing complex to the homology containing loci in the coding region of Nanog. (D) The result of enhanced epigenetic targeting of the coding region in Nanog is increased mRNA and protein expression.
Based on many previous studies (reviewed in [11]) as well as those carried out with Oct-4 and it’s regulatory antisense non-coding RNA [20], a paradigm is emerging where antisense non-coding RNAs appear to be functional in human cells in targeting the recruitment of epigenetic silencing complexes to homology containing loci in the chromatin. This non-coding RNA based targeting is known in some instances to involve DNMT3a [17] and result in DNA methylation at the targeted site [13, 16]. If the non-coding RNA targets a gene promoter the result is the methylation of both histones and CpG sites in the genomic DNA resulting ultimately in transcriptional gene silencing (reviewed in [19]). Notably, this form of silencing appears to be long-lasting and can be passaged to daughter cells [13]. But what happens when these non-coding RNAs are guiding DNA methylation or epigenetic silencing complexes to the body/coding region of a gene? Recent findings suggest that DNA methylation, within the exons or coding regions of a gene, may in fact facilitate gene expression, splicing, elongation and termination of transcription [35, 36]. Moreover, DNMT3a dependent non-promoter DNA methylation has been directly linked to activation of gene transcription [37]. These observations bolster those observations with Nanog and it’s concordant regulation by antisense non-coding RNAs. One would expect that as the antisense non-coding RNA involved in directing epigenetic changes to the body of Nanog is reduced so too would be the overall expression of Nanog mRNA, i.e. due to the loss of DNA methylation at the Nanog antisense non-coding RNA target loci. Conversely, when Nucleolin or PurA are suppressed there is a gain of Nanog targeted antisense non-coding RNAs, due to an inability of PurA and Nucleolin to properly traffic the antisense non-coding RNA, ultimately resulting in appreciable increases in targeting of the Nanog coding region by the Nanog antisense non-coding RNA. In essence the story of discordant vs. concordant regulation, might simply boil down to the target loci of the respective antisense non-coding RNA; i.e. promoter targeted non-coding RNAs=discordant regulation and coding region targeting RNAs=concordant regulation. However, so little is currently known about concordant regulation that many of these notions are merely speculation at this time.
Conclusion
Over the last decade much hype has been generated around RNA interference (RNAi) and the ability of double stranded RNAs, siRNAs, shRNAs and miRNAs to regulate gene expression in a post-transcriptional manner [38]. The observation that double stranded RNAs can modulate mRNA expression post-transcriptionally is no doubt a paradigm shift. It was indeed observations that double stranded RNAs could also modulate gene transcription in plants and S. Pombe that precipitated earlier work showing siRNAs could also modulate gene transcription in human cells (reviewed in [39]). However, over the last ~2 years it has become increasingly apparent that RNAi is the proverbial “tip of the iceberg” with regards to RNA based regulation in human cells. Studies carried out to discern how siRNAs are able to modulate gene transcription in human cells have led to insights suggesting that antisense non-coding RNAs, be them short or long, can transcriptionally regulate gene expression. But why should these relatively low expressed natural antisense transcripts be of any interest? The answer to this question lies in the fact that antisense non-coding RNAs appear in some instances to regulate gene transcription via the targeting of epigenetic changes to particular loci in the genome. The targeted recruitment of epigenetic changes can have a significant impact as mounting evidence suggest that epigenetic changes are heritable and able to be transmitted across generations [40, 41]. These observations argue that in essence non-coding RNAs possess the ability to instill long term changes in gene expression that are capable of being passed on to daughter cells. Such a notion runs counter to a Darwinian existence and in essence suggest that changes outside of the DNA, epigenetic, can have an impact on generations of daughter cells. Curiously this presumed model for how genes are regulated by non-coding RNAs in fact insinuates that a Mendellian sort of existence might also be operative inside the human cell. In essence changes outside the DNA are affecting the expression of the DNA and these epigenetic changes can be passed to daughter cells and potentially to offspring. Such a notion brings forth the question as to what extent non-coding RNAs are involved in ongoing selective pressures and/or the cell responding to selective pressures. Such a paradigm would be envisioned to operate from a selective pressure resulting in increased expression of a particular non-coding RNA that affects the epigenetic architecture of downstream targets resulting in enhanced cell survival due a particular gene expression profile. However, such a scenario might be a bit forthcoming and maybe the role of miRNAs is to in essence govern the relative ratios of antisense non-coding RNAs to sense mRNAs and fundamentally prohibit such profound changes.
The miRNA mediated targeting of transcripts is a relatively potent suppressor of gene expression, but this suppression is transient, lasting until new transcripts are generated or the mRNA complexed miRNAs are degraded. This is a feature that may lend miRNAs to function more as a regulator of general RNA abundance, allowing for the cell to augment gene expression accordingly based on particular selective pressures [3, 12]. Such a scenario might also allow for transient non-coding RNA based responses to be initiated without permanent epigenetic changes being instilled on the cell. Thus, if favorable, those cells expressing higher proportions of particular non-coding RNAs can outcompete their neighbors and the expression persists long enough to modulate particular epigenetic changes that become instilled in the chromatin of the dominant cells. There is precedence for such regulation when considering the androgen receptor and specific patterns of antisense non-coding RNAs being expressed in response to androgen receptor activation [42]. Supportive of miRNA regulation of non-coding RNAs is the observation that double stranded RNAs can actually target both sense or antisense strands [43], and maybe the reason miRNAs are abundantly expressed and linked to several forms of cancer in human cells is due to their role in regulating bidriectionally transcribed loci (discussed in detail in [3]). As the majority of those bidirectionally transcribed genes detected and/or characterized to date appear to be involved in cell-cycle, the loss of which can result in oncogenic transformation, it is not too far of a stretch to envision a role for miRNAs in regulating antisense non-coding RNAs. In fact one example has been recent described whereby miRNAs have been shown to be involved in the control of psuedogene RNAs for the cell cycle related gene p16 [44].
While there has been much progress gained over the last few years with regards to the realization that a complex paradigm is active in Human cells whereby vast species of non-coding RNAs are active in either regulating one another or sense/mRNA expressing genes, many questions remain. Such as what governs the expression of antisense non-coding RNAs, are miRNAs actually involved in regulating both sense and antisense non-coding RNAs, and to what extent are non-coding RNAs involved in the ability of the cell to either detect and/or respond to selective pressures. Furthermore, what dictates a gene being under either concordant or discordant non-coding RNA based regulation and how pronounced are these forms of regulation in the human cells. These are but only a few of the questions that if addressed might provide an inkling of insight into the role of non-coding RNAs in the fabric of life.
Acknowledgments
This project is funded by NIH R01 HL083473 and NIH R01 AI084406 to KVM. We thank Carol Kreider, of Sigma-Aldrich, for providing shRNA constructs to suppress PURA and NCL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- 1.Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 329:223–6. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–99. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 3.Morris KV. Non-coding RNAs, epigenetic memory, and the passage of information to progeny. RNA Biol. 2009;6 doi: 10.4161/rna.6.3.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 5.Latos PA, Barlow DP. Regulation of imprinted expression by macro non-coding RNAs. RNA Biol. 2009:6. doi: 10.4161/rna.6.2.7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–42. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–92. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 11.Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics. 2009:4. doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–44. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D, Kelleher A. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. Journal of RNAi and Gene Silencing. 2005;1:66–78. [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper DA, et al. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J Biol Chem. 2008 doi: 10.1074/jbc.M709651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner AM, De La Cruz J, Morris KV. Mobilization-competent Lentiviral Vector-mediated Sustained Transcriptional Modulation of HIV-1 Expression. Mol Ther. 2009;17:360–8. doi: 10.1038/mt.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256–62. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malecova B, Morris KV. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Curr Opin Mol Ther. 2010;12:214–22. [PMC free article] [PubMed] [Google Scholar]
- 19.Morris KV. RNA-Directed Transcriptional Gene Silencing and Activation in Human Cells. Oligonucleotides. 2009 doi: 10.1089/oli.2009.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.hawkins PGaKVM. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5 Transcription. 2010 doi: 10.4161/trns.1.3.13332. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffery L, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J Biol Chem. 2004;279:49479–87. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- 22.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. Embo J. 2001;20:2536–44. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–40. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 24.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2005 doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 25.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci U S A. 2007;104:12422–7. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–88. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spicer DB, Sonenshein GE. An antisense promoter of the murine c-myc gene is localized within intron 2. Mol Cell Biol. 1992;12:1324–9. doi: 10.1128/mcb.12.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebralidze AK, Guibal FC, Steidl U, Zhang P, Lee S, Bartholdy B, et al. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev. 2008;22:2085–92. doi: 10.1101/gad.1654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahmoudi S, Henriksson S, Corcoran M, Mendez-Vidal C, Wiman KG, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell. 2009;33:462–71. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 32.Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–8. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 33.Morris KV, Vogt PK. Long antisense non-coding RNAs and their role in transcription and oncogenesis. Cell Cycle. 2010:9. doi: 10.4161/cc.9.13.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe Y, Numata K, Murata S, Osada Y, Saito R, Nakaoka H, et al. Genome-wide analysis of expression modes and DNA methylation status at sense-antisense transcript loci in mouse. Genomics. 2010 doi: 10.1016/j.ygeno.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, et al. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–605. doi: 10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–8. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–29. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 39.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 40.Franklin TB, Russig H, Weiss IC, Graff J, Linder N, Michalon A, et al. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68:408–15. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 41.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Louro R, Nakaya HI, Amaral PP, Festa F, Sogayar MC, da Silva AM, et al. Androgen responsive intronic non-coding RNAs. BMC Biol. 2007;5:4. doi: 10.1186/1741-7007-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei JX, Yang J, Sun JF, Jia LT, Zhang Y, Zhang HZ, et al. Both strands of siRNA have potential to guide posttranscriptional gene silencing in mammalian cells. PLoS One. 2009;4:e5382. doi: 10.1371/journal.pone.0005382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rigoutsos I, Furnari F. Gene-expression forum: Decoy for microRNAs. Nature. 2010;465:1016–7. doi: 10.1038/4651016a. [DOI] [PubMed] [Google Scholar]