Abstract
The genomes of herpesviruses establish latency as a circular episome. However, Human herpesvirus-6 has been shown to specifically integrate into the telomeres of chromosomes during latency and vertically transmit through the germ-line. This review will focus on the telomere integration of HHV-6, the potential viral and cellular genes that mediate integration, and the clinical impact on the host.
Keywords: HHV-6, gliHHV-6, CIHHV-6, Telomere Integration, Chromosome Integration, Viral Latency, Chromosomally Integrated HHV-6
1. Introduction
Human herpesvirus-6 (HHV-6) was initially identified as human B-lymphotropic virus (HBLV) by Salahuddin et al. in 1986, given that the virus was isolated primarily from B-lymphocytes of patients infected with HIV, HTLV, and lymphoproliferative disorders [1]. However, through further molecular and clinical studies, and sequencing of the viral genome, the virus was found to include two subgroups; HHV-6A and HHV-6B [2–4]. Furthermore in 1988, Yamanishi et al. establish that HHV-6B primary infection in young children is the etiological agent of exanthema subitum (roseola infantum), which is characterized by high fever, diarrhea, and a mild skin rash along the trunk, neck, and face [4]. Serologic studies have found that by the age of two, 90 % of children have acquired a primary HHV-6B infection [5]. As for HHV-6A, the virus has been associated with several adult diseases; including cofactor in AIDS progression, and various neurological disorders including encephalitis, ataxia, seizures, and chronic fatigue syndrome, however the causal link between human disease and virus infection remains to be fully elucidated [6–10].
Following primary infection, the genome of herpesviruses establishes latency as a nuclear circular episome [11–13]. The subsequent decrease in lytic viral genes expression allows the stabile replication of the viral genome in the absence of newly formed infectious virions. However in 1993, Luppi et al. identified the HHV-6 genome linked to high molecular weight cellular DNA in peripheral blood mononuclear (PBMCs) of three patients [14]. The initial observation of HHV-6 integration into the human genome was confirmed by other laboratories [15–18]; however the significance of this event and whether the integrated viral genome reactivates from its integrated state has not been fully characterized. Recent findings by Arbuckle et al. demonstrated the in vivo and in vitro integration of HHV-6 into the telomeres of human chromosomes during latency [19]. Moreover, viral reactivation from its latent integrated state was demonstrated in patient PBMCs and in vitro integrated cell lines. This review details current knowledge of HHV-6 genomic structure during latent infections, the viral and cellular genes that could perhaps mediate integration, and discuss the cellular as well as the clinical impact of viral integration into the telomeres.
2. Genome structure
Herpesviruses are characterized by their large double stranded DNA genomes of 90–220 kb in length that are packaged within an icosahedral capsid [11]. The viral capsid is surrounded by a proteinaceous tegument often consisting of proteins necessary for initiation of genome replication. Following the capsid layer the virus is encased in a viral envelope that is obtained from the host cell’s plasma membrane during the budding process. In particular, the viral genome of HHV-6B is 162 kb long, containing 119 unique open reading frames (ORF), while HHV-6A genome contains 119 unique ORF and genome is 159 kb long [2, 3]. The genomic architecture of HHV-6 is organized into two major regions (Fig. 1a). The unique long (UL) region of the genome which is ~143 kb contains seven major core gene blocks that are conserved amongst herpesviruses. These conserved genes are responsible for replication of the viral genome, cleavage, and packaging of the viral genome into the mature virion. Additionally, these viral genes include: DNA binding protein, DNA polymerase, capsid, tegument, glycoproteins, and proteinase. Also, the UL region contains blocks of CMV related genes known as US22 family that are conserved amongst betaherpesviruses.
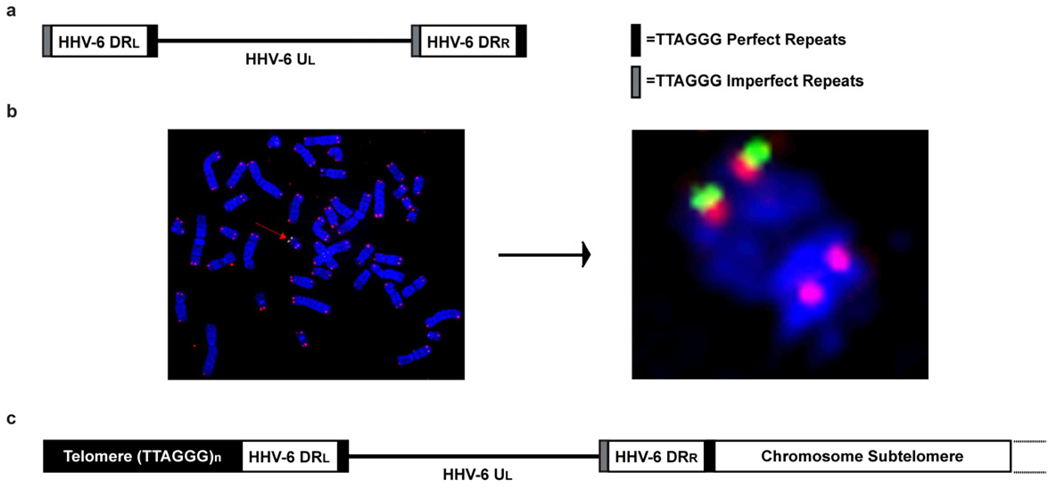
Fig. 1.
Schematic comparing the genome structure of HHV-6 to germ-line-integrated HHV-6. (gliHHV-6). (a) The linear HHV-6 genome contains a unique long (UL) region of 13–160 kb with left and right direct repeats (DR) of 7–8 kb at the termini. Perfect and imperfect telomere sequence (TTAGGG) positioned at end of DR play role HHV-6 chromosome integration into the telomere. (b) Fluorescence in situ hybridization (FISH) of PBMCs from a patient with gliHHV-6A integrated into the telomere of chromosome 22q. Metaphase chromosomes counterstained with DAPI (blue), cy5-PNA telomere probe (red), and FITC-conjugated HHV-6 cosmid probe (green). (c) Genome structure of gliHHV-6 in which HHV-6 DRR is fused with the telomere repeats near the chromosome subtelomere and the DRL fused with the remaining telomere repeat array (TTAGGG)n [19].
The second major genomic region of HHV-6 is the ~8 kb left and right direct repeats (DR) located at the termini of the UL region [2, 3]. Within the right end direct repeat (DRR) and the left end direct repeat (DRL) the HHV-6 genome encodes a perfect TTAGGG telomere repeat array and an imperfect TTAGGG repeat array. Arbuckle et al. and others have shown that viral integration into the telomere of chromosomes is mediated by the telomere repeats encoded within the DR of HHV-6 [16, 19]. Details about this phenomenon will be further discussed in subsequent sections.
The distinction between HHV-6A and HHV-6B subtypes can not only be classified by their differences in clinical disease association, but also differences in their restriction fragment length polymorphism and the nucleotide sequence of their genomes [2, 3, 19]. The nucleotide sequence identity between HHV-6A and HHV-6B is 90 %. The variation in nucleotide sequence is not only observed between HHV-6A and HHV-6B subtypes, but also between clinical isolates and laboratory strains of the virus. Variability in the number of telomere repeats within the DR of clinical HHV-6 isolates is observed in the range of 15 to 180 repeats [19, 20]. Furthermore, preliminary experiments suggest that ORF U2 through U5 [21], as well as DR1 through the first exon of DR6 [22] are dispensable for in vitro viral replication. These examples demonstrate the plasticity of the HHV-6 genome as well as variability in the sequences of HHV-6A and HHV-6B which warrant consideration for these viruses to be classified as different herpesvirus species rather than merely a subtype distinction between “A” and “B” [23].
3. Latency and chromosome integration
Human herpesviruses can be further subdivided into three major subfamilies characterized by their genome sequence and structure, as well as host cell range during lytic and latent infection [11]. The alphaherpesviruses include HSV-1, HSV-2, and VZV infect and establish latency within neurons, while gamaherpesviruses include EBV and KSHV infect monocytes and undergo oncogenic transformation during latent infection. Betaherpesviruses, HHV-6 as well as HHV-7, and CMV are characterized based upon their ability to establish a latent infection in monocytes and retain similarities in their genes and genomic structure amongst these viruses.
During the course of lytic infection, the viral envelope of herpesviruses fuses with the cell’s plasma membrane and the capsid releases the linear viral genome into the nucleus [11]. The genome then circularizes and rolling-circle replication through the help of viral DNA polymerase leads to the formation of viral concatemers [12, 13]. The concatemers consist of linear viral genomes linked in an end-to-end conformation that are subsequently cleaved to give rise to single copy viral genomes that are packaged into the capsid during virion maturation. However during latency, the genome of herpesviruses remains as a nuclear circular episome, while the cellular DNA polymerase and latency-associated viral genes promote replication of the viral genome without production of infectious virus. Therefore, the central dogma of herpesvirus replication is that the viral genome establishes latency as a nuclear circular episome and reactivation occurs through rolling-circle DNA replication [11–13]. However findings over the last two decades suggest this may not be the case as pertaining to HHV-6 latency.
Primary infection of HHV-6 leads to an increase in viral copies present in peripheral blood of children and subsequent decrease in viral copies during latency [4, 5]. However, in a select number of patients including adults and children, there are high copies of viral genome consistently detected in peripheral blood measuring greater than 1 million per ml or 1 copy of HHV-6 per cell [17, 19, 24, 25]. In 1993 Luppi et al. identified a unique high molecular weight viral fragment present in PBMCs of three patients with elevated copies of HHV-6 [14]. This high molecular weight fragment resolved by pulse field gel electrophoresis demonstrated the earliest observation of viral integration into the genome. Subsequently, several other labs have shown integration of HHV-6 into human chromosomes through fluorescent in situ hybridization (FISH) and PCR amplification allowed for the detection of HHV-6 sequences in hair follicles [15–17].
Despite previous publications, the uncertainty of HHV-6 chromosome integration still prevailed and whether the integrated viral genome represented a latent infection remained in question. However, a publication by Arbuckle et al. illustrated that HHV-6 is unique among human herpesviruses: it specifically and efficiently integrates into telomeres of chromosomes during latency [19]. Experiments performed in this study evaluated HHV-6 integration resulting from in vitro infection of cell lines and PBMCs of six different families. These families originated from different geographical locations across the globe. FISH identified that HHV-6 probe co-hybridized specifically with the telomere of one chromosome in each patient (Fig. 1b). However, to confirm the integration of HHV-6 into the telomere of chromosomes, amplification of the virus-chromosome integration site was completed using two different primer sets. One primer was designed to specifically anneal to the unique chromosome ends and the second set of primers were designed to anneal adjacent to the telomere repeats of HHV-6 DRL and DRR. Sequencing identified that integration of HHV-6 occurs through recombination of the perfect telomere repeats encoded in the DRR juxtaposed to the end of the chromosome and the tandem array of telomere repeats [(TTAGGG)n] at the end of the chromosome extend beyond the DRL (Fig. 1c).
The next step was to determine whether the integrated virus within patients’ PBMCs represents a lytic or latent infection [19]. To do so, the vertical agarose gel technique described in Gardella et al. was performed [26]. Briefly, one million cells were placed in a well overlaid with SDS and pronase to gently lyse the cells. Next and an electrical current was applied across the wells to separate the high molecular weight human genomic DNA present in the loading well from the circular latent viral episome in the middle of the gel, and replicating linear viral DNA at the bottom of the gel. Through Southern hybridization with HHV-6 cosmid probes, the viral genome co-migrated with the human genomic DNA and the assay did not detect the presence of replicating linear viral DNA which indicates that the patient cells are latently integrated [19]. In addition, the presence of a circular episome was undetectable. Furthermore these findings were confirmed using PCR amplification of HHV-6 sequences from circular fractions of CsCl/Ethidium bromide density gradients and again this assay showed no proof of circular episome. Therefore, the genome of HHV-6 forms a covalent linkage with the telomeres of chromosomes during latency rather than forming a circular episome like other human herpesviruses.
4. Transmission of germ-line integrated (gliHHV-6)
Following primary infection, HHV-6 establishes a latent infection primarily within monocytes/macrophages and within the salivary gland [7, 27]. It is believed that the predominant means of acquiring HHV-6 is a result of HHV-6 horizontal transmission through viral shedding from the saliva of infected individuals to uninfected infants and children. However, vertical transmission of integrated HHV-6 through the germ-line (gliHHV-6) has proven to be a novel mechanism by which the virus integrates during latency in all cells. Daibata et al. described the first reported case of gliHHV-6 vertical transmission through the germ-line of parent to their child [28]. This initial observation was confirmed by several other laboratories [18, 19, 29]. In each of these reports, both parents and children with gliHHV-6 contained identical sites of chromosome integration. Furthermore, the same virus strain of HHV-6 was inherited through the germ-line between parent and child. This was evident because gliHHV-6 integrated in each family member contained similar restriction fragment polymorphism profiles and similar sequence of select viral genes [19]. Furthermore, germ-line transmission results in viral integration within the chromosome of every cell that resides in the individual. This statement coincides with the information previously describing the viral load of patients consistently at or above 1 copy per cell [18, 19, 29].
In patients receiving hematopoietic stem cell transplants, HHV-6 may reactivate and induce graft rejection, bone marrow suppression, and encephalitis [7, 9, 11, 30]. Moreover, even after ablation of the recipient’s bone marrow in preparation for transplantation, it has been shown that the donor stem cells may transfer gliHHV-6 to the receiving donor [31–33]. This results in the subsequent rise of HHV-6 viral copies within the recipient peripheral blood as well as well as reintroduction of gliHHV-6 into donor stem cells such as circulating white blood cells. The initial studies on the transmission of integrated HHV-6 into chromosome of hematopoietic stem cells during transplantation has not been studied sufficiently, and additional studies must be performed to understand the long-term effects on the patients who received stem cell transplants with gliHHV-6. Investigations are also necessary to understand how immunosuppressive agents used to prevent graft rejection affect infected patients. Will clinicians have to consider screening for gli-HHHV-6 during blood donations? There are many questions that remain unanswered and need resolving to avoid possible widespread reactivation and subsequent secondary diseases in immune-compromised individuals.
5. Frequency of HHV-6 telomere integration
During latency, the episomes of herpesviruses associate with chromosomes through the expression of viral DNA binding proteins such as EBV nuclear antigen-1 (EBNA-1) and KSHV’s latency-associated nuclear antigen (LANA) [34, 35]. The binding of EBNA1 and LANA to chromosomes is not chromosome-specific, but this association of herpesvirus episomes with chromosomes ensures each infected cell obtains at least one copy of the viral episome during cell division. However, in rare cases as shown by EBV the entire genome infrequently integrate into random chromosome sites [36, 37]. The integration EBV into chromosomes has been demonstrated during long-term passage of infected cell lines and the isolation of virus from these cell lines have yet to be established.
Similar to the integration of HHV-6A and HHV-6B into telomeres of human chromosomes during latency [19], the gammaherpesvirus Marek’s disease virus (MDV) frequently integrates into the telomere of chickens through telomeric repeats (TTAGGG)n encoded in the internal repeat short (IRS) of the viral genome [38–40]. MDV is acquired from inhalation of virions released from the dander of infected chickens. During latency, the linear ~180 kb dsDNA genome integrates into multiple telomeres of chickens in each cell. The expression of viral oncogene Meq and viral telomerase RNA (vTR) induce transformation of T-cells and the rapid formation of solid visceral tumors and T-cell lymphomas within two to six weeks. Furthermore, the sequence specific integration of the retrotransposable element TRAS1 (telomeric repeat-associated sequence) was found to frequently integrate within the (TTAGG)n telomere repeat of the silkworm, Bombyx mori [41]. The process of integration occurs through cleavage of the telomere TTAGG sequence by the TRAS1 endonculease and the fusion with the TTAGG repeats encoded in TRAS1. In summary, the specific integration of HHV-6 into telomeres during latency is a novel process for a human herpesvirus. However, the commonality of telomere integration seems to be evident amongst other species and viral strains such as the avian herpesrirus MDV and the silkworm retrotransposon TRAS1.
To appreciate the significance of latent HHV-6 integration into the telomere of chromosomes, several studies investigated the prevalence of HHV-6 integration amongst normal blood donors and hospitalized patients with high viral loads (106–107 copies per ml of blood) [24, 25]. In normal healthy blood donors it was found that 0.8 % (4/500) [24] and 1.5 % (10/653) [25] had high viral loads most likely attributed to HHV-6 integration in the germ-line. In contrast, prevalence of integrated HHV-6 in hospitalized patient was higher, 2.9 % (13/449) [24] and 3.3 % (6/184) [25]. In conclusion, the transmission of HHV-6 through the germ-line and its prevalence in the population implies the necessity for research to characterize the significance and possible pathologies of HHV-6 integration. Moreover, it is essential that researchers determine the overall impact integrated virus may have on hospitalized patients and understand disease progression associated with gliHHV-6.
6. Telomere biology and HHV-6 integration
The integration of HHV-6 into telomeres is a newly identified form of human herpesvirus latency and the knowledge attained from the virus telomere biology may play a critical role in understanding the process of virus replication and integration. The length of telomeres in somatic cells is 5–15 kb in length and after every cell division 250–300 bp are lost from the end of telomeres due to the challenges encountered with the process of end replication [42]. Cells reach the Hayflick limit once their telomeres reach a critical length, and undergo replicative senescence or apoptosis.
Alternatively, telomeres can be lengthened by telomerase reverse transcriptase (TERT) through the addition of the TTAGGG repeat [42]. Telomerase mediated telomere lengthening has been detected in germ-line cells, cancer cells, and stem cells. The repetitive telomere sequence and the complex of six telomere associated proteins protect the ends of chromosomes from double stranded breaks, chromosome fusions, and inhibits telomerase mediated lengthening [43, 44]. The complex of telomere associate proteins known as the shelterin complex contains: TRF1 (TTAGGG Repeat Factor 1), TRF2 (TTAGGG Repeat Factor 2), and Pot1 (Protection of Telomeres 1) which directly binds to the telomeric TTAGGG repeat. The complex protein interaction of the shelterin complex is maintained through TIN2 (TRF1-Interacting Nuclear Factor 2), TPP1 (TINT1, PIP1, PYOP1), and RAP1 (Repressor Activator Protein 1). Until recently, telomeres were believed to be transcriptionally silent; however mammalian telomeres were shown to transcribe telomere-repeat-encoding RNA (TERRA) [45]. TERRA stabilizes the shelterin complex and facilitates telomere heterochromatin formation through the direct interaction with TRF1 and TRF2 [46].
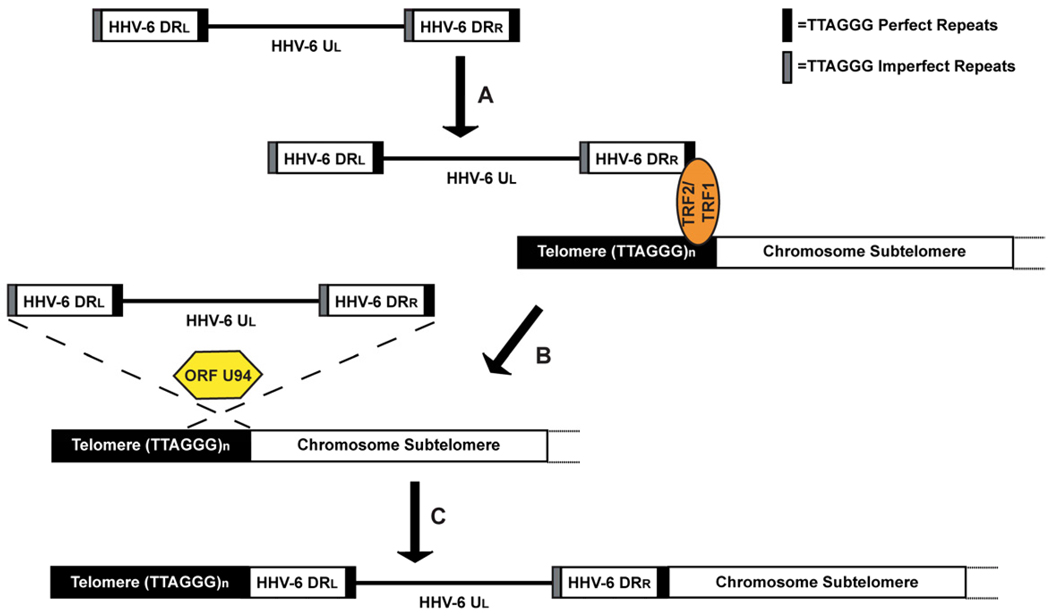
From the six shelterin proteins, TRF2 was shown to bind to the three nonamer EBV encoded TTAGGGTTA repeat (imperfect TTAGGG repeat) within the origin of plasmid replication (OriP) in cooperation with viral latency gene EBNA-1 [47]. TRF2 and EBNA-1 binding of OriP stabilizes the EBV episome during latency and enables non-covalent attachment of the viral genome to metaphase chromosomes [35]. This mechanism ensures division of the EBV genome amongst daughter cells during mitosis. Moreover, Lieberman et al. also found that EBNA-1 directly interacted with TERRA, however the principle of this interaction has not been fully elucidated [46]. Similarly, we hypothesize that the binding of telomere repeats encoded in the DR of HHV-6 by TRF2/TRF1 plays a role in telomere mediated integration by facilitating the localization of the viral genome to the telomere of chromosomes (Fig. 2). This in turn promotes homologous recombination of the viral telomere repeats and that of the chromosome telomere.
Fig. 2.
Proposed model of HHV-6 integration into chromosome telomeres. (a) We hypothesizes that localization of the linear HHV-6 genome to chromosome telomeres occurs through TRF2 and TRF1 bidding of chromosome and HHV-6 DR encoded telomere repeat. (b) The viral rep gene ORF U94 then promotes homologous recombination between viral and chromosome telomere repeats, (c) which results in the integration of HHV-6 into chromosome telomeres.
The integration of HHV-6 into the same chromosome telomeres of family members lends itself to vertical transmission through the germ-line [18, 19, 29]. Moreover, studies have shown that HHV-6 preferentially integrates into telomeres of chromosomes 9q34.4, 17p13.3, 18q23, 19q13.4, and 22q13.3 [15, 16, 18, 19, 31]. These results may be explained by two different mechanisms. First, a number of chromosome ends may be more accessible for homologous recombination between the viral and chromosomal TTAGGG repeats due to heterochromatin structure of the chromosome telomere. Alternatively, HHV-6 may integrate randomly into all 92 telomere regions at the end of chromosomes, but the integrated genome may be unstable in some telomeres [19]. After a series of cell divisions, it’s possible that cells with unstable chromosomes would be “negatively” selected over those with stable chromosomes.
Results thus far have shown that between in vivo integrated gliHHV-6 amongst patients and in vitro integrated cell lines, the viral DRR of the genome is integrated within 5–41 telomere repeats from the end of the chromosome (Fig. 1) [19]. The proximity of the ~160 kb HHV-6 genome integrated between the end of the chromosome and tandem array of telomere repeats suggests that the virus has a minimal impact on the telomere/shelterin complex. However, if the HHV-6 genome was to integrate near the end of the telomere, one would expect progressive shortening of the telomere leading to loss of viral genome sequences. Subsequently, this loss may lead to chromosome instability as the result of chromosome fusion, replicative senescence, or the inability of the shelterin complex to bind the non-telomere coding sequence of the virus to form the cap at the end of telomere.
Furthermore, the physiological effect on the length of the telomere after the integration of HHV-6 remains unknown. This is of interest since telomere shortening leads to cell senescence and/or telomerase activation and chromosome translocation in somatic cells, which is a hallmark of malignant transformation and chromosome instability [44].
7. Clinical impact of HHV-6 and gliHHV-6
HHV-6 infection occurs by means of attachment and entry into cells through the complement regulator receptor CD46 [48]. The diverse cell tropism and multiple disease associations with HHV-6 are due, in part, to the expression of CD46 present on the surface of all nucleated cells. In particular, HHV-6B primary infection of young children is the etiological agent of exanthema subitum (roseola), which is characterized by high fever, diarrhea, and a mild skin rash along the trunk, neck, and face [4]. There is no classic textbook clinical disease caused by HHV-6A at this time. However, HHV-6A has been suggested to act as a co-factor in AIDS progression by contributing to the killing of CD-4 lymphocytes and enhancement of HIV replication through activation of the long terminal repeats (LTR) [7, 8, 49]. Following primary infection, reactivation of HHV-6 in immune compromised patients has led to graft rejection in solid organ and bone marrow transplants, seizures, and encephalitis [7, 9, 10, 30].
The current understanding of HHV-6 chromosome integration and direct clinical link to disease is in its initial stages of development, nevertheless over the last 20 years significant progress has been accomplished. Considering the clinical implications that gliHHV-6 has when manifested into a patients’ DNA, it’s crucial to determine whether reactivation of virus occurs from its latent integrated state. The first experiment conducted by Daibata et al. induced lytic genes and capsid mRNA expression from chromosomally integrated HHV-6 Burkitt’s lymphoma cell line [15]. The induction of lytic mRNA expression from chromosomally integrated HHV-6 was achieved by culturing of cell lines with the phorbol ester TPA (12-O-tetradecanoyl-phorbol-13-acetate). Furthermore, Arbuckle et al. demonstrated reactivation of gliHHV-6 from patient T-cells as well as viral integration amongst HEK-293 cells and T-cell lines in vitro [19]. Reactivation was achieved by means of co-culturing patient T-cells with the HDAC inhibitor trichostatin-A (TSA) or with TPA plus hydrocortisone. The reactivation of the integrated virus induced cytopathic effects which were visibly evident by cell lysis and syncytium formation in a naïve T-cell line. Moreover, partial sequencing of the viral genome confirmed the gliHHV-6 reactivation indeed occurs from its integrated state. This evidence further demonstrates that chromosome integration of HHV-6 into telomeres is an alternative mechanism for a human herpesvirus to establish latency, reactivates, and induce disease.
Patients with gliHHV-6 present a wide range of symptoms and various diseases associated with the integrated viral genome, while other patients show no signs of clinical disease [14–19, 24, 31]. It remains unknown why some patients manifest signs (such as acute viral infection) of the disease and others don’t. However in vitro gliHHV-6 reactivation of patient T-cells [15, 19], suggests that the virus may be in a latent integrated state in patients showing no signs of disease, while patients presenting symptoms of disease may have HHV-6 reactivated from its integrated state. Furthermore, when considering the transmission of HHV-6 through the germ-line, in every cell of the patient there is at least one integrated copy of the viral genome [18, 19, 29]. Reactivation can occur in any number of tissues within the body of the patient, which can lead to cell death, changes in cell function attributed to viral gene expression, and subsequent spread to neighboring CD46+ cells. Therefore, the ability of HHV-6 to infect the majority of nucleated cells through CD46 and the presences of gliHHV-6 integrated in every cell sheds light on the various symptoms and diseases associated with this virus.
The neurotropic nature of HHV-6 has been linked to chronic fatigue syndrome, ataxia, hypersomnia, mild dementia, encephalitis, seizures, and multiple sclerosis (MS) [6, 7, 10, 50–54]. MS is a debilitating autoimmune disease of the central nervous system (CNS) that is induced by autoreactive CD4+ T-cells toward the myelin sheath [55]. Inflammation of the central nervous system through the release of cytokines, results in the recruitment of macrophages and the development of plaques as well as the degradation of the myelin sheath. Studies have suggested that HHV-6 can act as a co-factor in the progression of MS due to the neurotropic nature of the virus [6, 7, 10, 50–54]. Several investigators reported an increase in anti-HHV-6 IgG and IgM antibody titers as well as HHV-6 DNA and RNA copies while monitoring clinical relapse of MS in patients compared to control subjects [50, 53, 54]. HHV-6 DNA was also detected in CSF, brain tissues, and plaques of patients with MS at higher levels than control subjects [51, 52]. However pertaining to gliHHV-6, the direct clinical link between neurological disease and active viral replication is still underway. It remains uncertain if CNS dysfunction is due to reactivation of gli-HHV6 or if the virus is merely an opportunistic infection within the brain.
As previously stated, primary infection of young children with HHV-6B are subjected to contracting exanthema subitum (roseolla), and the disease ultimately leads to a persistent latent state for the life of the host [4]. However as frequently seen in hematopoietic stem cell transplant patients undergoing immunosuppressive therapy, can lead to reactivation of HHV-6 and has cause graft rejection in solid organ and bone transplants, encephalitis, ataxia, and seizures [7, 10, 30]. Similarly, studies have come across patients with gliHHV-6 presenting neurological symptoms and various lymphomas, which include Hodgkin’s lymphoma, Burkitt’s lymphoma, acute lympoblastic leukaemia (ALL), chronic myelogenous leukemia (CML), and acute myeloid leukemia (AML) [14, 15, 28, 56–58]. However, the prevalence of gliHHV-6 in children with ALL and AML does not differ significantly from healthy populations [58]. Therefore the direct clinical link between cancer and gliHHV-6 is not completely conclusive, but reactivation of the virus can be linked to graft rejection in hematopoietic stem cell transplant patients treated with immunosuppressive drugs [7, 30].
HHV-6 may not play a direct role in cancer development, but the virus may play a role in cancer progression by complementing oncogenic viruses in an environment that condones cancer development. Similarly, HHV-6 has the ability to increase HIV replication through activation of the long terminal repeats (LTR) and enhance AIDS progression in macaques [7, 8, 49]. Also, HHV-6 can increase RNA expression of human papillomavirus virus (HPV) oncogenes E6 and E7 in cervical epithelial cells [59]. HHV-6 and HPV co-infection of cervical epithelial cells increased production of E6 and E7 mRNA and accelerated tumor growth within mice by 3 weeks as compared to the 6 week time period normally expected with HPV infection alone.
During the course of human evolution, human endogenous retroviruses (HERV) integrated into the germ-line roughly 10 million years ago [60]. HERVs no longer produce mature virus due to mutations and deletions in their sequence, however in a study by zur Hausen et al., demonstrated that HHV-6 induced transcription of gag, pol, and env genes from HERV found in normal PBMCs and in cell lines [61]. Moreover, HERV env protein has also been found to be expressed in various cancer cells. Duelli et al. hypothesized that cell fusion caused by endogenous viruses and syncytium-forming viruses like HHV-6 fuse cells leading to aneuploidy, subsequently induce chromosome instability and promote malignant transformation of cells [62]. Therefore the ability of HHV-6 and HERV to induce cell fusion, and increase expression of HERV env protein by HHV-6, emphasizes the need of further clinical studies to determine the potential impact of HHV-6 and gliHHV-6 on cancer as well as their effect on MS, CFS, DRESS, and bone marrow suppression in transplants.
8. HHV-6 putative latency gene ORF U94
Latency genes expressed by human herpesviruses promote maintenance and replication of the viral episome while inhibiting the expression of lytic genes [7]. Examples of herpesviruses latency genes include EBV nuclear antigen-1 (EBNA-1) and KSHV latency associated nuclear antigen (LANA) [34, 35]. The Epstein-Barr virus latency gene EBNA-1 was shown to bind to the origin of plasmid replication (OriP) of the EBV encoded TTAGGGTTA repeat (imperfect telomere repeat) in cooperation with telomere binding protein TRF2 binding [46, 47]. TRF2 and EBNA-1 binding of OriP stabilizes the EBV episome during latency and enables the non-covalent attachment of the EBV genome to metaphase chromosomes [35]. Similarly, LANA binds within the KSHV terminal repeats (TR) to form the episome [63]. Within the TR of KSHV, the promoter of transmembrane glycoprotein K1 is responsible for activating the signaling pathways for lytic infection [64]. As a result of LANA binding to the TR, the transcription of K1 is inhibited, therefore preventing the initiation of lytic infection. A second target for LANA to establish latency is through inhibition of Rta, which is an IE gene responsible for promoting lytic gene replication [65]. Lan et al. established that LANA has the ability to directly repress Rta’s promoter activity, and inhibit the auto-activation of viral transcription. Furthermore, non-homologous association of KSHV episome is achieved by binding LANA’s C-terminus to the TR of KSHV and the N–terminus to histones H2A–H2B [34, 66].
Currently, there are several lines of evidence suggesting that the unique gene of ORF U94 encoded by HHV-6 may have a role in the establishment and maintenance of latency. First, the expression of ORF U94 transcripts were observed in latently infected PBMCs from healthy donors, while lytic transcripts were not expressed [67]. Second, lymphoid cell lines stably expressing ORF U94, were permissive to HHV-6A infection, however no cytopathic effect was observed. There was a steady decline in copies of viral DNA and transcripts detected by 22 days post-infection. Furthermore, ORF U94 expression resulted in decrease in viral replication of betaherpesviruses CMV and HHV-7 [68]. Moreover, ORF U94 has ssDNA binding activities [69] that have an impact on viral replication perhaps through transcriptional regulation. This hypothesis is supported by studies that have shown ORF U94 to inhibit H-ras mediated transformation in NIH3T3 cells and also inhibits HIV-1 LTR expression cells at the level of the promoter [70]. The transcriptional regulation by ORF U94 was further verified by in vivo and in vitro immunoprecipitation assay; these assays found that ORF U94 binds to the human TATA-Binding Protein (hTBP) [71]. hTBP functions as a subunit of the transcription factor TFIID, and binds to the TATA box. This binding activity is a critical step for the formation of the pre-initiation complex. Therefore, the transcriptional regulation of HHV-6 lytic genes could possibly occur through the interaction of ORF U94 with hTBP and thus creates a means by which HHV-6 can shift from a lytic to a latent infection.
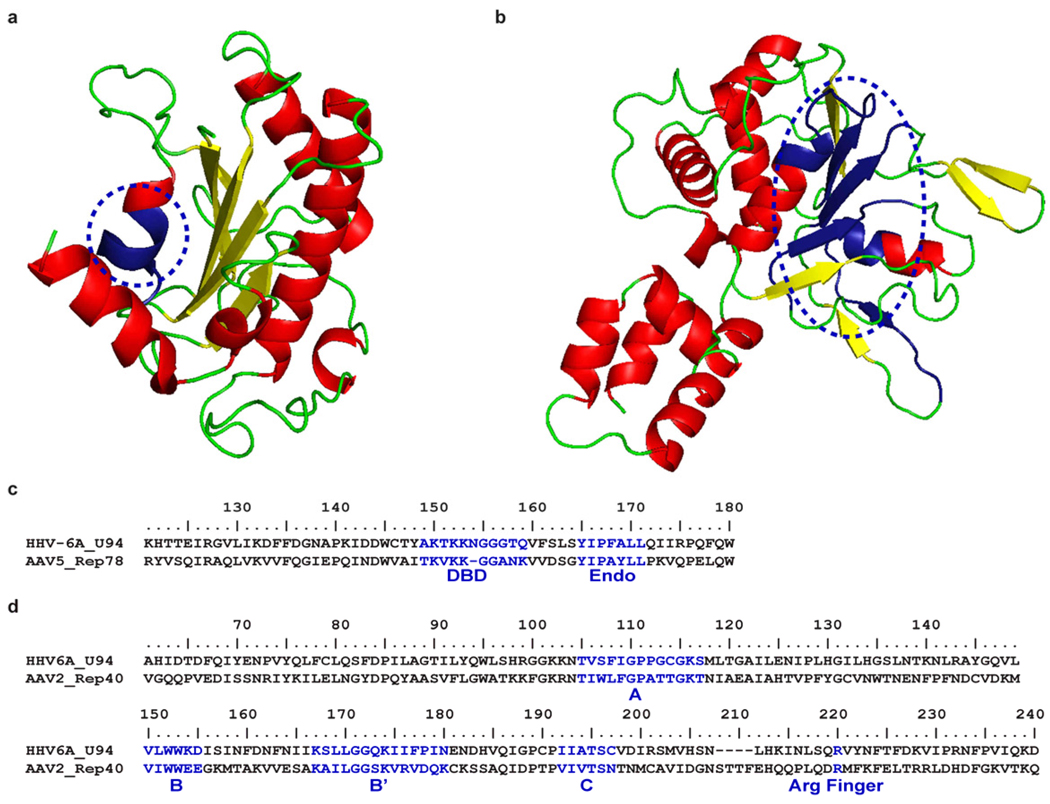
Interestingly, ORF U94 shares 24 % sequence identity with latency gene rep68/78, derived from human adeno-associated virus type 2 (AAV-2) [72]. Rep68/78 regulates viral gene expression and directs the site specific integration of AAV-2 into the AAVS1 site in chromosome 19q13.4 [73–76]. The functions of rep68/78 are through DNA binding, endonuclease and helicase activities. Furthermore, in a rep68/78 deficient AAV-2 model, ORF U94 was able to complement the activity of the mutant by allowing the virus to replicate and integrate [77]. Therefore, this suggests that ORF U94 may function as a latency gene and possibly facilitate HHV-6 specific integration into telomeres similar to its AAV-2 encoded rep68/78 counterpart.
Despite the fact that the 490 amino acids of U94 shares only 24 % sequence identity with rep68/78 [72], there are structural features that exhibit similar biological functions (Fig. 3). The ORF U94 C-terminus (residues 229–490 aa) corresponds to the helicase and ATPase domain of rep68/78. Specifically, the sequence codes for Walker A and B motifs form the core of the class SF3 helicase active site and an arginine finger which is critical player in ATP hydrolysis [78]. Furthermore, the N-terminus (15–206 aa) of ORF U94 corresponds to the DNA binding and endonuclease domain of rep68/78 [76]. It has been found that the N-terminus of ORF U94 has ssDNA-binding activity [69] and the endonuclease domain of rep68/78 is required for the site specific cleavage of the AAVS1 site which is shared by both the inverted terminal repeats of AAV-2 and the site in chromosome 19q13.4 [76]. In conclusion, the structural features shared between ORF U94 and rep68/78, and the act of promoting transcriptional regulation emphasizes the necessity to understand the function of this protein and implications it may have on viral replication, integration, and latency.
Fig. 3.
Structural SWISS-MODEL and consensus sequence of HHV-6A ORF U94 N and C-terminus. (a) The predicted ribbon diagrams of the U94 N-terminus (15–206 aa) corresponds to the AAV rep nuclease domain [Protein Data Bank (PDB) accession number 1uut [76]) and the (b) C-terminus (229–490 aa) of U94 corresponds to the AAV rep helicase domain [PDB accession number 1s9h [78]] circled in blue. (c) Consensus protein sequence of AAV-2 Rep78 illustrates the DNA-binding domain (DBD) and endonuclease (Endo) domain for the N-terminus of HHV-6A ORF U94. (d) The consensus protein sequence of AAV-2 Rep40 helicase domain and C-terminus of HHV-6A U94. The Walker (A, B, B’, and C) motifs form the core of the SF3 helicase and the arginine (Arg) finger catalyzes ATP hydrolysis when the protein forms a hexamer structure.
9. Proposed model for HHV-6 telomere integration and reactivation
As previously discussed, integration of HHV-6 into the telomeres of chromosomes is unique and alterative means for a human herpesvirus to establish latency [19]. Comparable, the chicken hepresvirus MDV also integrates into the telomeres of chromosomes during latency [38–40]. Furthermore, both HHV-6 and MDV reactivate from a latent, integrated state to produce a lytic infection of naïve T-cells through chemicals inducers [15, 19, 40]. However, the process of viral HHV-6 and MDV integration and reactivation is unclear and remains to be fully elucidated.
In this next section, we propose a model (Fig. 2) to explain how integration of HHV-6 into chromosome telomeres occurs during latency. The process of integration emerges through homologous recombination between the viral and host telomere repeats. During this process, we believe that the TRF1 and TRF2 binding to the viral encoded TTAGGG repeats (similar to the binding of EBV Orip TTAGGGTTA repeat [46, 47]). This allows the linear HHV-6 genome to sit proximally to the chromosome telomere and permit homologous recombination. Second, during the process of latency, ORF U94 may play a role in transcriptional repression of lytic gene expression and aid in the specific integration of HHV-6 into to the telomeres. Similar actions were observed in the rep68/78 mediated integration of AAV-2 into chromosome 19q13.4 [73–75]. Third, reactivation of HHV-6 from its linear integrated state serves as a template for the replication of a linear dsDNA genome. The treatment of HHV-6 and MDV with HDAC inhibitors supports the notion that reactivation precedes chromatin decondensation [15, 19, 40]. This is followed by circularization of the genome and subsequent rolling-circle replication leads to the concatemer of viral genomes linked in an end to end conformation. Finally, cleavage of concatamers to unit-length viral genomes takes place followed by encapsidation of the viral DNA (Arbuckle et al. unpublished).
10. Conclusions
The objective of this review was to present the current and preceding knowledge of gliHHV-6 and introduce new areas of research. Investigators have established that HHV-6B is the etiological agent of roseolla and HHV-6A is identified as a cofactor in AIDS progression [4, 5, 8, 9]. HHV-6 is perhaps associated with MS, while both subtypes are linked to graft rejection in transplants, and various neurological disorders [7, 9, 10, 30, 50–55]. Furthermore, rather than establishing latency as a circular episome like other herpesviruses, HHV-6A and HHV-6B specifically integrate into the telomeres of human chromosomes through the viral encoded TTAGGG repeats within the direct repeat (DR) of the viral genome [16, 19]. The HHV-6 genome was also found to be vertically transmitted through the germ-line [18, 19, 28, 29]. Therefore parents and children inherit the same strain of HHV-6 integrated within the same chromosome. Moreover, the integration of HHV-6 into chromosomes is not a dead end pathway; the virus is shown to reactivate from its latent integrated state and leads to infection of naïve cells resulting in cell death [15, 19]. We hypothesize that during latency, HHV-6 integrates into the telomeres of human chromosomes through recombination with the TTAGGG viral repeats, while latency is sustained through the expression of viral encoded ORF U94 and cellular encoded telomere proteins TRF1 and TRF2.
Despite the current advancement in the knowledge of HHV-6 latency and integration, there are many questions left unanswered. We have proposed that integration of HHV-6 into the telomere of chromosomes occurs through homologous recombination. However it remains unknown whether the viral latency gene ORF U94 and telomere binding proteins TRF1 and TRF2 do in fact play a role during the process of integration. Furthermore, the physiological effect and impact on the stability of gliHHV-6 has on the telomere remains unknown. These questions are of interest to investigators since telomere shortening leads to cell senescence and/or telomerase activation, which is a hallmark of malignant transformation and chromosome instability [44]. However, there is a greater need to determine the relationship between HHV-6 infection and MS, CFS, cancer, AIDS progression, and various neurological disorders.
The use of BAC vectors to clone the entire genome of herpesviruses are instrumental in understanding the means by which viruses replicate, while the creation of knockout-mutants characterize viral proteins. Initial attempts to generate an infectious HHV-6 BAC cloned virus were unsuccessful [21, 79]. However, Arbuckle et al. were the first to successfully produce an infectious HHV-6A virus that expresses GFP and a selectable marker [19]. The reproducible cloning system was then verified by Tang et al. [80]. The knowledge gained from HHV-6 integration and adaption of recombinant HHV-6 viruses focuses future studies to comprehend the balance that viral infection and latency/integration has on the host.
Over 90 % of the population has acquired a primary HHV-6 infection by three years of age and the virus remains latent for the life of the host [4, 5]. Moreover, the integration of HHV-6 in chromosomes resulting in high viral load (106–107 copies per ml of blood) has been described [24, 25]. The prevalence of high viral load in normal blood donors are 0.8 % [24] and 1.5 % [25]. In contrast, prevalence of gliHHV-6 in hospitalized patient cohorts are higher, 2.9 % [24] and 3.3 % [25]. Therefore, this review concludes that the current knowledge of HHV-6 integration/latency supports a concerted effort to elucidate the mechanism by which HHV-6 integrates into chromosome telomeres, to understand the mediation of integration through viral and cellular genes, as well as the physiological and biological impact that reactivation of gliHHV-6 has on patients.
ACKNOWLDEGMENTS
This work was supported by grants from the HHV-6 Foundation and the National Institutes of Health (5R01CA111196).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Salahuddin SZ, Ablashi DV, Markham PD, Josephs SF, Sturzenegger S, Kaplan M, Halligan G, Biberfeld P, Wong-Staal F, Kramarsky B, et al. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986;234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 2.Dominguez G, Dambaugh TR, Stamey FR, Dewhurst S, Inoue N, Pellett PE. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 1999;73:8040–8052. doi: 10.1128/jvi.73.10.8040-8052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gompels UA, Nicholas J, Lawrence G, Jones M, Thomson BJ, Martin ME, Efstathiou S, Craxton M, Macaulay HA. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 4.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;1:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- 5.Okuno T, Takahashi K, Balachandra K, Shiraki K, Yamanishi K, Takahashi M, Baba K. Seroepidemiology of human herpesvirus 6 infection in normal children and adults. J. Clin. Microbiol. 1989;27:651–653. doi: 10.1128/jcm.27.4.651-653.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron B, Flamand L, Juwana H, Middeldorp J, Naing Z, Rawlinson W, Ablashi D, Lloyd A. Serological and virological investigation of the role of the herpesviruses EBV, CMV and HHV-6 in post-infective fatigue syndrome. J. Med. Virol. 2010;82:1684–1688. doi: 10.1002/jmv.21873. [DOI] [PubMed] [Google Scholar]
- 7.De Bolle L, Naesens L, De Clercq E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin. Microbiol. Rev. 2005;18:217–245. doi: 10.1128/CMR.18.1.217-245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lusso P, Crowley RW, Malnati MS, Di Serio C, Ponzoni M, Biancotto A, Markham PD, Gallo RC. Human herpesvirus 6A accelerates AIDS progression in macaques. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5067–5072. doi: 10.1073/pnas.0700929104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ablashi DV, Devin CL, Yoshikawa T, Lautenschlager I, Luppi M, Kuhl U, Komaroff AL. Review Part 3: Human herpesvirus-6 in multiple non-neurological diseases. J. Med. Virol. 2010;82:1903–1910. doi: 10.1002/jmv.21860. [DOI] [PubMed] [Google Scholar]
- 10.Yao K, Crawford JR, Komaroff AL, Ablashi DV, Jacobson S. Review part 2: Human herpesvirus-6 in central nervous system diseases. J. Med. Virol. 2010;82:1669–1678. doi: 10.1002/jmv.21861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields BN, Knipe DM, Howley PM, editors. Fields virology. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 12.Jacob RJ, Morse LS, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J. Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poffenberger KL, Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J. Virol. 1985;53:587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luppi M, Marasca R, Barozzi P, Ferrari S, Ceccherini-Nelli L, Batoni G, Merelli E, Torelli G. Three cases of human herpesvirus-6 latent infection: integration of viral genome in peripheral blood mononuclear cell DNA. J. Med. Virol. 1993;40:44–52. doi: 10.1002/jmv.1890400110. [DOI] [PubMed] [Google Scholar]
- 15.Daibata M, Taguchi T, Taguchi H, Miyoshi I. Integration of human herpesvirus 6 in a Burkitt's lymphoma cell line. Br. J. Haematol. 1998;102:1307–1313. doi: 10.1046/j.1365-2141.1998.00903.x. [DOI] [PubMed] [Google Scholar]
- 16.Nacheva EP, Ward KN, Brazma D, Virgili A, Howard J, Leong HN, Clark DA. Human herpesvirus 6 integrates within telomeric regions as evidenced by five different chromosomal sites. J. Med. Virol. 2008;80:1952–1958. doi: 10.1002/jmv.21299. [DOI] [PubMed] [Google Scholar]
- 17.Ward KN, Leong HN, Nacheva EP, Howard J, Atkinson CE, Davies NW, Griffiths PD, Clark DA. Human herpesvirus 6 chromosomal integration in immunocompetent patients results in high levels of viral DNA in blood, sera, and hair follicles. J. Clin. Microbiol. 2006;44:1571–1574. doi: 10.1128/JCM.44.4.1571-1574.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka-Taya K, Sashihara J, Kurahashi H, Amo K, Miyagawa H, Kondo K, Okada S, Yamanishi K. Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J. Med. Virol. 2004;73:465–473. doi: 10.1002/jmv.20113. [DOI] [PubMed] [Google Scholar]
- 19.Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, Lund TC, Tolar J, De Meirleir K, Montoya JG, Komaroff AL, Ambros PF, Medveczky PG. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5563–5568. doi: 10.1073/pnas.0913586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Achour A, Malet I, Deback C, Bonnafous P, Boutolleau D, Gautheret-Dejean A, Agut H. Length variability of telomeric repeat sequences of human herpesvirus 6 DNA. J. Virol. Methods. 2009;159:127–130. doi: 10.1016/j.jviromet.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Kondo K, Nozaki H, Shimada K, Yamanishi K. Detection of a gene cluster that is dispensable for human herpesvirus 6 replication and latency. J. Virol. 2003;77:10719–10724. doi: 10.1128/JVI.77.19.10719-10724.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borenstein R, Zeigerman H, Frenkel N. The DR1 and DR6 first exons of human herpesvirus 6A are not required for virus replication in culture and are deleted in virus stocks that replicate well in T-cell lines. J. Virol. 2010;84:2648–2656. doi: 10.1128/JVI.01951-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ablashi D. Human herpesvirus-6 strain groups: a nomenclature. Arch. Virol. 1993;129:363–366. doi: 10.1007/BF01316913. [DOI] [PubMed] [Google Scholar]
- 24.Leong HN, Tuke PW, Tedder RS, Khanom AB, Eglin RP, Atkinson CE, Ward KN, Griffiths PD, Clark DA. The prevalence of chromosomally integrated human herpesvirus 6 genomes in the blood of UK blood donors. J. Med. Virol. 2007;79:45–51. doi: 10.1002/jmv.20760. [DOI] [PubMed] [Google Scholar]
- 25.Ward KN, Thiruchelvam AD, Couto-Parada X. Unexpected occasional persistence of high levels of HHV-6 DNA in sera: detection of variants A and B. J. Med. Virol. 2005;76:563–570. doi: 10.1002/jmv.20399. [DOI] [PubMed] [Google Scholar]
- 26.Gardella T, Medveczky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J. Virol. 1984;50:248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo K, Kondo T, Shimada K, Amo K, Miyagawa H, Yamanishi K. Strong interaction between human herpesvirus 6 and peripheral blood monocytes/macrophages during acute infection. J. Med. Virol. 2002;67:364–369. doi: 10.1002/jmv.10082. [DOI] [PubMed] [Google Scholar]
- 28.Daibata M, Taguchi T, Nemoto Y, Taguchi H, Miyoshi I. Inheritance of chromosomally integrated human herpesvirus 6 DNA. Blood. 1999;94:1545–1549. [PubMed] [Google Scholar]
- 29.Mori T, Tanaka-Taya K, Satoh H, Aisa Y, Yamazaki R, Kato J, Ikeda Y, Okamoto S. Transmission of chromosomally integrated human herpesvirsus 6 (HHV-6) variant A from a parent to children leading to misdiagnosis of active HHV-6 infection. Transpl. Infect. Dis. 2009;11:503–506. doi: 10.1111/j.1399-3062.2009.00430.x. [DOI] [PubMed] [Google Scholar]
- 30.Dockrell DH, Paya CV. Human herpesvirus-6 and -7 in transplantation. Rev. Med. Virol. 2001;11:23–36. doi: 10.1002/rmv.299. [DOI] [PubMed] [Google Scholar]
- 31.Clark DA, Nacheva EP, Leong HN, Brazma D, Li YT, Tsao EH, Buyck HC, Atkinson CE, Lawson HM, Potter MN, Griffiths PD. Transmission of integrated human herpesvirus 6 through stem cell transplantation: implications for laboratory diagnosis. J. Infect. Dis. 2006;193:912–916. doi: 10.1086/500838. [DOI] [PubMed] [Google Scholar]
- 32.Jeulin H, Salmon A, Gautheret-Dejean A, Agut H, Bordigoni P, Fortier B, Venard V. Contribution of human herpesvirus 6 (HHV-6) viral load in whole blood and serum to investigate integrated HHV-6 transmission after haematopoietic stem cell transplantation. J. Clin. Virol. 2009;45:43–46. doi: 10.1016/j.jcv.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Kamble RT, Clark DA, Leong HN, Heslop HE, Brenner MK, Carrum G. Transmission of integrated human herpesvirus-6 in allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 2007;40:563–566. doi: 10.1038/sj.bmt.1705780. [DOI] [PubMed] [Google Scholar]
- 34.Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 35.Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas JC. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 1999;73:4385–4392. doi: 10.1128/jvi.73.5.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurley EA, Agger S, McNeil JA, Lawrence JB, Calendar A, Lenoir G, Thorley-Lawson DA. When Epstein-Barr virus persistently infects B-cell lines, it frequently integrates. J. Virol. 1991;65:1245–1254. doi: 10.1128/jvi.65.3.1245-1254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, Luo X, Tang K, Li X, Li G. Epstein-Barr virus integrates frequently into chromosome 4q, 2q, 1q and 7q of Burkitt's lymphoma cell line (Raji) J. Virol. Methods. 2006;136:193–199. doi: 10.1016/j.jviromet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 38.Delecluse HJ, Hammerschmidt W. Status of Marek's disease virus in established lymphoma cell lines: herpesvirus integration is common. J. Virol. 1993;67:82–92. doi: 10.1128/jvi.67.1.82-92.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S. Marek's disease virus: from miasma to model. Nat. Rev. Microbiol. 2006;4:283–294. doi: 10.1038/nrmicro1382. [DOI] [PubMed] [Google Scholar]
- 40.Delecluse HJ, Schuller S, Hammerschmidt W. Latent Marek's disease virus can be activated from its chromosomally integrated state in herpesvirus-transformed lymphoma cells. EMBO J. 1993;12:3277–3286. doi: 10.1002/j.1460-2075.1993.tb05997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anzai T, Takahashi H, Fujiwara H. Sequence-specific recognition and cleavage of telomeric repeat (TTAGG)(n) by endonuclease of non-long terminal repeat retrotransposon TRAS1. Mol. Cell Biol. 2001;21:100–108. doi: 10.1128/MCB.21.1.100-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riethman H. Human telomere structure and biology. Annu. Rev. Genomics Hum. Genet. 2008;9:1–19. doi: 10.1146/annurev.genom.8.021506.172017. [DOI] [PubMed] [Google Scholar]
- 43.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 44.Raynaud CM, Sabatier L, Philipot O, Olaussen KA, Soria JC. Telomere length, telomeric proteins and genomic instability during the multistep carcinogenic process. Crit. Rev. Oncol. Hematol. 2008;66:99–117. doi: 10.1016/j.critrevonc.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 45.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 46.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng Z, Lezina L, Chen CJ, Shtivelband S, So W, Lieberman PM. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell. 2002;9:493–503. doi: 10.1016/s1097-2765(02)00476-8. [DOI] [PubMed] [Google Scholar]
- 48.Santoro F, Kennedy PE, Locatelli G, Malnati MS, Berger EA, Lusso P. CD46 is a cellular receptor for human herpesvirus 6. Cell. 1999;99:817–827. doi: 10.1016/s0092-8674(00)81678-5. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi K, Sonoda S, Higashi K, Kondo T, Takahashi H, Takahashi M, Yamanishi K. Predominant CD4 T-lymphocyte tropism of human herpesvirus 6-related virus. J. Virol. 1989;63:3161–3163. doi: 10.1128/jvi.63.7.3161-3163.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caselli E, Boni M, Bracci A, Rotola A, Cermelli C, Castellazzi M, Di Luca D, Cassai E. Detection of antibodies directed against human herpesvirus 6 U94/REP in sera of patients affected by multiple sclerosis. J. Clin. Microbiol. 2002;40:4131–4137. doi: 10.1128/JCM.40.11.4131-4137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cermelli C, Berti R, Soldan SS, Mayne M, D'Ambrosia J M, Ludwin SK, Jacobson S. High frequency of human herpesvirus 6 DNA in multiple sclerosis plaques isolated by laser microdissection. J. Infect. Dis. 2003;187:1377–1387. doi: 10.1086/368166. [DOI] [PubMed] [Google Scholar]
- 52.Rotola A, Merlotti I, Caniatti L, Caselli E, Granieri E, Tola MR, Di Luca D, Cassai E. Human herpesvirus 6 infects the central nervous system of multiple sclerosis patients in the early stages of the disease. Mult. Scler. 2004;10:348–354. doi: 10.1191/1352458504ms1045oa. [DOI] [PubMed] [Google Scholar]
- 53.Soldan SS, Leist TP, Juhng KN, McFarland HF, Jacobson S. Increased lymphoproliferative response to human herpesvirus type 6A variant in multiple sclerosis patients. Ann. Neurol. 2000;47:306–313. [PubMed] [Google Scholar]
- 54.Yao K, Gagnon S, Akhyani N, Williams E, Fotheringham J, Frohman E, Stuve O, Monson N, Racke MK, Jacobson S. Reactivation of human herpesvirus-6 in natalizumab treated multiple sclerosis patients. PLoS One. 2008;3:e2028. doi: 10.1371/journal.pone.0002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hafler DA, Slavik JM, Anderson DE, O'Connor KC, De Jager P, Baecher-Allan C. Multiple sclerosis. Immunol. Rev. 2005;204:208–231. doi: 10.1111/j.0105-2896.2005.00240.x. [DOI] [PubMed] [Google Scholar]
- 56.Daibata M, Taguchi T, Sawada T, Taguchi H, Miyoshi I. Chromosomal transmission of human herpesvirus 6 DNA in acute lymphoblastic leukaemia. Lancet. 1998;352:543–544. doi: 10.1016/S0140-6736(05)79251-5. [DOI] [PubMed] [Google Scholar]
- 57.Yasukawa M, Ohminami H, Sada E, Yakushijin Y, Kaneko M, Yanagisawa K, Kohno H, Bando S, Fujita S. Latent infection and reactivation of human herpesvirus 6 in two novel myeloid cell lines. Blood. 1999;93:991–999. [PubMed] [Google Scholar]
- 58.Hubacek P, Muzikova K, Hrdlickova A, Cinek O, Hyncicova K, Hrstkova H, Sedlacek P, Stary J. Prevalence of HHV-6 integrated chromosomally among children treated for acute lymphoblastic or myeloid leukemia in the Czech Republic. J. Med. Virol. 2009;81:258–263. doi: 10.1002/jmv.21371. [DOI] [PubMed] [Google Scholar]
- 59.Chen M, Popescu N, Woodworth C, Berneman Z, Corbellino M, Lusso P, Ablashi DV, DiPaolo JA. Human herpesvirus 6 infects cervical epithelial cells and transactivates human papillomavirus gene expression. J. Virol. 1994;68:1173–1178. doi: 10.1128/jvi.68.2.1173-1178.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. U.S.A. 2004;101 Suppl 2:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prusty BK, zur Hausen H, Schmidt R, Kimmel R, de Villiers EM. Transcription of HERV-E and HERV-E-related sequences in malignant and non-malignant human haematopoietic cells. Virology. 2008;382:37–45. doi: 10.1016/j.virol.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 62.Duelli D, Lazebnik Y. Cell-to-cell fusion as a link between viruses and cancer. Nat. Rev. Cancer. 2007;7:968–976. doi: 10.1038/nrc2272. [DOI] [PubMed] [Google Scholar]
- 63.Verma SC, Choudhuri T, Kaul R, Robertson ES. Latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus interacts with origin recognition complexes at the LANA binding sequence within the terminal repeats. J. Virol. 2006;80:2243–2256. doi: 10.1128/JVI.80.5.2243-2256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verma SC, Lan K, Choudhuri T, Robertson ES. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen modulates K1 expression through its cis-acting elements within the terminal repeats. J. Virol. 2006;80:3445–3458. doi: 10.1128/JVI.80.7.3445-3458.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lan K, Kuppers DA, Verma SC, Robertson ES. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 2004;78:6585–6594. doi: 10.1128/JVI.78.12.6585-6594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 67.Rotola A, Ravaioli T, Gonelli A, Dewhurst S, Cassai E, Di Luca D. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13911–13916. doi: 10.1073/pnas.95.23.13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caselli E, Bracci A, Galvan M, Boni M, Rotola A, Bergamini C, Cermelli C, Dal Monte P, Gompels UA, Cassai E, Di Luca D. Human herpesvirus 6 (HHV-6) U94/REP protein inhibits betaherpesvirus replication. Virology. 2006;346:402–414. doi: 10.1016/j.virol.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 69.Dhepakson P, Mori Y, Jiang YB, Huang HL, Akkapaiboon P, Okuno T, Yamanishi K. Human herpesvirus-6 rep/U94 gene product has single-stranded DNA-binding activity. J. Gen. Virol. 2002;83:847–854. doi: 10.1099/0022-1317-83-4-847. [DOI] [PubMed] [Google Scholar]
- 70.Araujo JC, Doniger J, Kashanchi F, Hermonat PL, Thompson J, Rosenthal LJ. Human herpesvirus 6A ts suppresses both transformation by H-ras and transcription by the H-ras and human immunodeficiency virus type 1 promoters. J. Virol. 1995;69:4933–4940. doi: 10.1128/jvi.69.8.4933-4940.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mori Y, Dhepakson P, Shimamoto T, Ueda K, Gomi Y, Tani H, Matsuura Y, Yamanishi K. Expression of human herpesvirus 6B rep within infected cells and binding of its gene product to the TATA-binding protein in vitro and in vivo. J. Virol. 2000;74:6096–6104. doi: 10.1128/jvi.74.13.6096-6104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomson BJ, Efstathiou S, Honess RW. Acquisition of the human adeno-associated virus type-2 rep gene by human herpesvirus type-6. Nature. 1991;351:78–80. doi: 10.1038/351078a0. [DOI] [PubMed] [Google Scholar]
- 73.Im DS, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 74.Henckaerts E, Dutheil N, Zeltner N, Kattman S, Kohlbrenner E, Ward P, Clement N, Rebollo P, Kennedy M, Keller GM, Linden RM. Site-specific integration of adeno-associated virus involves partial duplication of the target locus. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7571–7576. doi: 10.1073/pnas.0806821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Samulski RJ, Zhu X, Xiao X, Brook JD, Housman DE, Epstein N, Hunter LA. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hickman AB, Ronning DR, Perez ZN, Kotin RM, Dyda F. The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol. Cell. 2004;13:403–414. doi: 10.1016/s1097-2765(04)00023-1. [DOI] [PubMed] [Google Scholar]
- 77.Thomson BJ, Weindler FW, Gray D, Schwaab V, Heilbronn R. Human herpesvirus 6 (HHV-6) is a helper virus for adeno-associated virus type 2 (AAV-2) and the AAV-2 rep gene homologue in HHV-6 can mediate AAV-2 DNA replication and regulate gene expression. Virology. 1994;204:304–311. doi: 10.1006/viro.1994.1535. [DOI] [PubMed] [Google Scholar]
- 78.James JA, Escalante CR, Yoon-Robarts M, Edwards TA, Linden RM, Aggarwal AK. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure. 2003;11:1025–1035. doi: 10.1016/s0969-2126(03)00152-7. [DOI] [PubMed] [Google Scholar]
- 79.Borenstein R, Frenkel N. Cloning human herpes virus 6A genome into bacterial artificial chromosomes and study of DNA replication intermediates. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19138–19143. doi: 10.1073/pnas.0908504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang H, Kawabata A, Yoshida M, Oyaizu H, Maeki T, Yamanishi K, Mori Y. Human herpesvirus 6 encoded glycoprotein Q1 gene is essential for virus growth. Virology. 2010;407:360–367. doi: 10.1016/j.virol.2010.08.018. [DOI] [PubMed] [Google Scholar]