Abstract
Studies using dogs provide an ideal solution to the gap in animal models of natural disease and translational medicine. This is evidenced by approximately 400 inherited disorders being characterized in domesticated dogs, most of which are relevant to humans. There are several hundred isolated populations of dogs (breeds) and each has vastly reduced genetic variation compared to humans; this simplifies disease mapping and pharmacogenomics. Dogs age five to eight-fold faster than humans, share environments with their owners, are usually kept until old age, and receive a high level of health care. Farseeing investigators recognized this potential and, over the last decade, developed the necessary tools and infrastructure to utilize this powerful model of human disease, including the sequencing of the dog genome in 2005. Here we review the nascent convergence of genetic and translational canine models of spontaneous disease, focusing on cancer.
Keywords: canine, comparative oncology, dog genome, translational medicine
New Models of Complex Disease Needed
The greatest challenge facing clinical scientists is an incomplete understanding of the genetic basis for complex human diseases [1]. Despite numerous technological advances in genetics, progress has been slow. This is owed, in part, to intricate gene-gene interactions and poorly understood environmental effects [2]. The identification of these interactions and environmental influences is difficult to dissect in humans due to the high level of genetic heterogeneity [3]. Most genome-wide association studies (GWAS) have only identified a small fraction of the genetic basis of complex diseases [4]. And yet disease heritability is critical to understanding disease risk, the effects of environment and lifestyle on disease development, and response to treatment.
Much of the research on human disease genetics relies on animal models. The most frequently used model, the mouse, has several advantages. Mice have short gestation times and are small, making their generation relatively rapid and inexpensive compared to other mammals. Moreover, technologies exist to manipulate the expression of genes in the entire organism or in select cells or tissues [5]. However, mouse models of cancer have limitations. The most notable is that tumors arise spontaneously in humans, but must be induced in most mouse models. Whereas human disease is polygenic, genetic manipulations in mouse models often involve one or a few genes and/or environmental conditions that affect expression of specific genes in an inbred mouse line with undetermined human relevance [3]. Mouse models of cancer in humans are thus missing vast gene networks and interactions that are responsible for, or contribute to, disease in humans. Here we discuss the advantages of tumor-bearing dogs as an alternative model for understanding the genetic basis of human disease [6], highlighting three cancer types as examples.
Advantages of Dog Models
Domesticated dogs (Canis lupus familiaris) are excellent models of human complex diseases for several reasons, including their easy accessibility and prominent status in diverse cultures. For instance, >73 million dogs live in ~40% of households in the USA [7] and 54% of them are considered to be a “family member” [8]. Over 40B (USD) is spent annually on dog health care [8], and is second only to humans in the level of health care received [9]. That, combined with the shared environment of owners and dogs, can be exploited for epidemiological studies of diseases common to dogs and humans.
Next to humans, domesticated dogs have the most phenotypic diversity and known naturally-occurring diseases of all land mammals [10]. For example, the average weight of Chihuahuas and English Mastiffs differs by 65-fold. Dogs share over ~650 Mb of ancestral sequence in common with humans that is absent in mice, and canine DNA and protein sequence is more similar to human than mouse is [11; Fig. 1A]. Analysis of the 13,816 protein coding genes with 1:1:1 orthology in human, mouse and dog showed that the numbers of lineage-specific non-synonymous substitutions (i.e., amino acid changing; KA) are 0.017, 0.038, and 0.021, respectively [11]. Thus, many aspects of human biology are presumably more relevant in dogs than in mice [12]. Approximately 400 inherited diseases similar to those of humans are characterized in dogs, including complex disorders such as cancers, heart disease, and neurological disorders [13, 14]. Indeed, more than 40 naturally occurring canine diseases have mutations in a homologous human gene associated with a similar disease [15]. Additionally, depending on breed size, dogs have a five to eight-fold accelerated aging process compared to humans [http://www.avma.org/animal_health/care_older_pet_faq.asp]. Moreover, dogs are kept as companion animals well into their old age [16, 17]. The most recently available data (2006) shows that ~45% of companion dogs were >6 years old [8], the human equivalent of ~60-95. Thus, dog models hold great promise for accelerating the understanding of genetic and environmental contributions to human disease, particularly those that are chronic or associated with aging.
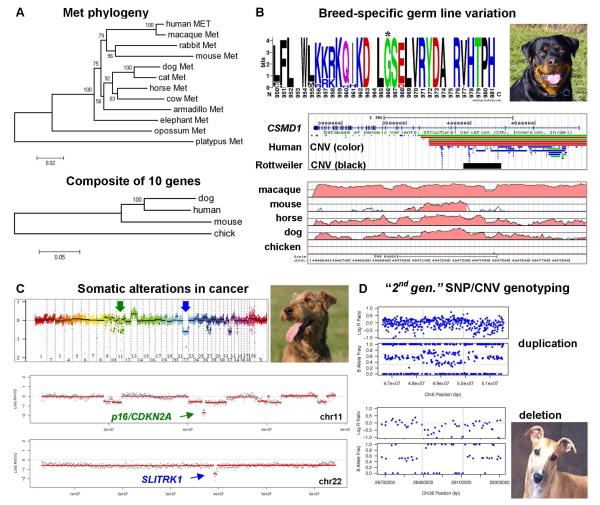
Figure 1. Dog cancer genetics.
(A) Protein sequence conservation in dogs. (i) Phylogenetic tree of the mammalian c-Met receptor. The branching pattern corresponds well with the organismal relationships. For example, the Boreoeutheria clade comprises two sister taxa which include primates, rodents, rabbits and a taxa including carnivorans and most hoofed animals. Although mouse and human c-Met branch together, the branch length of mouse c-Met shows that the protein sequence is more divergent than human and dog (scale bar shows amino acid changes per site). (ii) Dog proteins are more similar to those of humans than are mouse proteins. Phylogenetic treeing analysis of a composite of 10 cancer proteins branches human and dog proteins apart from mouse with a bootstrap value of 100. The following proteins were included: MYC, ERBB2, KIT, ret proto-oncogene (RET), v-raf murine sarcoma viral oncogene homolog B1 (BRAF), PTEN, RB1, CDKN2A, breast cancer 1, early onset (BRCA1), TP53. [Neighbor-Joining trees shown (500-replicate bootstrap values); Maximum Parsimony topology is the same]. (B) Examples of breed-specific germ line variation with potential cancer relevance. (i) Common missense variant in Rottweiler c-Met receptor. WebLogo analysis shows a close-up of the consensus amino acid sequence of c-Met from 23 mammals. Letter height corresponds to the frequency of a given amino acid at each position, with the highest letters signifying complete conservation. 70% of Rottweiler’s have a missense variant at Gly 966, which is located in the extracellular region and could thus affect ligand binding or receptor signaling [71]. (ii) More than 60% of Rottweilers have a 273-kb copy number variant (CNV) in an intron of CSMD1, but it has not been observed in diverse other breeds (UCSC Browser; human gene transcribed right to left) [72]. (iii) Close-up of one of several non-coding conserved elements within the CSMD1 CNV (Vista Browser, conservation with human >60% shown by red coloring). The most conserved region within this area contains three candidate binding sites for the tumor suppressor transcription factor E2A (another conserved element contains TP53 binding sites [72]). The conservation (which is absent in chicken) is reduced in mouse in comparison to more distantly related mammals, horse and dog. (C) Somatic genome alterations in canine cancer. Kisseberth et al. isolated the OSW T cell lymphoma cell line and identified several genomic alterations [73]. A single two copy loss was found, and it affects the CDKN2A tumor suppressor gene. Subsequent analysis of OSW by high resolution tiling ologinucleotide-array CGH revealed many additional alterations, including focal two-copy deletions affecting as few as a single gene [72]. (i) Whole genome display of CGH analysis of OSW [72]. The midline shows a 1:1 DNA ratio to the reference genome of a boxer. Deletion CNVs are segments below the midline, and gains are above the midline (log 2 scale). “Un” denotes unmapped contigs and is highly enriched for repetitive sequences; the Y chromosome is absent from the canFam2 genome assembly. (ii, iii) Close-up of CGH analysis of chromosomes 11 and 22. Both chromosomes have 2-copy microdeletions. One confirms complete deletion of the tumor suppressor, p16/CDKN2A. The other spans a single active gene, SLITRTK1, which was previously implicated in malignant hematopoiesis [74]. This illustrates how dogs can be used as translational models of known human cancer genetics, as well as for discovery of novel genes in the same genetic pathways. (D) Second generation genotyping technology allows the integration of single nucleotide polymorphism and CNV maps. CNVs from two Greyhounds are shown. This 170k oligonucleotide array enables simultaneous SNP genotyping and DNA copy number determination (Illumina CanineHD). For each pair, the top window (i) shows DNA copy number as Log 2 R ratios, with the midline generally corresponding to copy number of 2. The bottom windows (ii) show allele frequencies. A copy number gain is detected as an upward shift on Log R ratio, and as a shift from B allele ratios of 1:1 (left and right segments) to 1:2 and 2:1 allele ratios (center segment). A copy number loss is detected as a downward shift in Log R ratio, and as a shift from allele ratios of 1:1 (left, right segments) to an allele ratio of 1:0 (or loss of heterozygosity; center).
The greatest advantage of dog models is the result of their evolutionary history which involved at least two severe population bottlenecks [14]. The first occurred when dogs were domesticated from wolves ~15,000-40,000 years ago [18]. The second was most pronounced ~200 years ago when most dog breeds were created by selection of morphological and behavioral traits. Today there are ~400 isolated populations or breeds. Breed creation inadvertently selected many “founder” mutations that are associated with specific traits and diseases; this translates into reduced disease and genetic heterogeneity, consistent with the fact that most breeds are predisposed to a distinct set of diseases. Because linkage disequilibrium is up to 100-fold greater in dogs than humans, single breeds are powerful subjects for broad genetic mapping [14]. By contrast, related breeds that share a trait are powerful subjects for fine mapping. This advantage is illustrated by the recent analysis of polyneuropathy with juvenile onset in dogs, which is similar to human Charcot-Marie-Tooth (CMT) syndrome [19]. The comparison of 7 affected and 17 related unaffected control Greyhounds identified a 19.5 Mb region that was homozygous in the affected dogs, and contained a 10 bp deletion in N-myc downstream regulated 1 (NDRG1), orthologous to a known human CMT gene. Pedigree information and the extended homozygosity suggest that the mutation arose in a popular sire in 1968. Now the disease can be eradicated from the breed through selective breeding, and the dog model can be used to better understand and treat human CMT [19]. Additionally, dogs might provide clues about the “missing heritability” of human complex genetics. Recently, a group of 300 investigators [20] performed a meta-analysis of GWAS (an approach using SNP markers across the entire genomes of many people to find genetic variations associated with a particular disease) of 180,000 individuals characterized for height (known to be 80% heritable). They identified 180 loci that together explain 10% of height heritability. Similarly, Boyko et al. studied 57 quantitative morphological traits in 915 dogs that included samples from 80 breeds; traits included body size and external dimensions, and cranial/dental/long bone size and shape [21]. In contrast to human studies, they found that one to three quantitative trait loci explain the majority of phenotypic variation for most of the dog traits examined. The question now is whether canine complex diseases will turn out to have a similarly simplified genetic architecture.
Cancer Development in Dogs
Dogs are exceptional models of cancer because they naturally develop the same cancers as humans [22]. Indeed, dog tumors are histologically similar to human, and respond similarly to conventional therapies [6]. Although disease course is reported to be more aggressive in dogs than humans for some cancer types [6], it is not clear whether dog cancer is generally more aggressive than human. This issue is complicated because dog cancers are not treated as aggressively as human, and therefore result in shorter survival times and faster evaluations of outcomes. Moreover, disease-bearing dogs tend to present for treatment at later stages than humans. Regardless, the significantly shorter duration time of canine clinical trials is a major advantage [6; Fig. 2]. The disease-free time interval in dogs treated for cancer is 18 months, compared to >7 years needed to assess treatment outcomes in humans [6]. Additionally, many histological types of cancer are associated with similar genetic alterations in humans and dogs. For instance, statistical analysis of genomic alterations in human and dog colorectal tumors showed that samples clustered according to stage, origin and instability status across species [23]. Strikingly, cluster analysis of genome regions affected by DNA copy number alterations showed branching together of human and dog tumors according to colorectal cancer subtypes (vs. species) [23]. This suggests that the same genetic pathways are affected in colorectal tumorigenesis in both species. By contrast, species-specific alterations tended to localize to evolutionarily unstable genome regions. These observations thus hint that the alterations common to both species are more likely to cause cancer than those found in only one (i.e., the latter could be irrelevant species-specific mutation hotspots). In summary, dogs are useful in multiple approaches to cancer investigation [24]: breed-specific risk can be used to discover disease pathways; human cancer pathways can be tested for roles, and targeted for treatment, in canine disease; and canine somatic mutations and genome alterations can be used to narrow down human mutations (Fig. 1B-D). Below we provide three examples of canine-human comparative oncology.
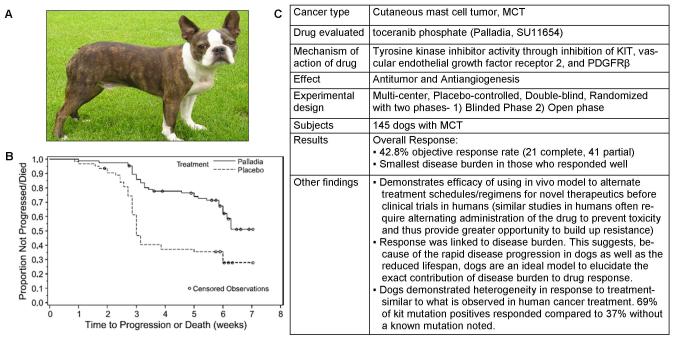
Fig. 2. An example of the clinical relevance of dogs for cancer treatments.
Canines are increasingly being used in clinical cancer drug trials to determine the efficacy of treatment given how closely many of the cancer they develop recapitulate the human cancer. (A) A picture of a Boston terrier, a breed predisposed to the development of Mast cell tumors. (B) London et al. conducted a clinical trial of an oral receptor tyrosine kinase inhibitor, Palladia on dogs with recurrent mast cell tumors. Shown here is a Kaplan-Meier survival analysis demonstrating time to tumor progression in placebo-treated and Palladia-treated dogs with Mast Cell Tumors [75]. (C) A breakdown of the clinical trial of Palladia, including the demonstrated advantages of dogs as models of pharmacologic cancer intervention. Reproduced with Permission, from [75].
Soft-tissue sarcomas
Soft tissue sarcomas (STS) comprise 1% of all newly diagnosed cancer types in humans [25] and represent a heterogeneous group of mesenchymal neoplasms which demonstrate a high degree of variation in clinical presentation and cellular morphology [26]. These genetically complex cancers include angiosarcomas (hemangiosarcomas in dogs), fibrosarcomas, and histiocytomas. Recent advances in immunohistochemistry, cytogenetics, and molecular genetic analysis have allowed a clinically relevant division of STS to improve diagnosis and treatment [27]. Based on clinical and biological variation among these neoplasms, STS can be broadly dichotomized into two groups. One is characterized by specific, balanced chromosomal translocations, whereas the other typically shows more extensive chromosomal rearrangements leading to recurrent, but non-specific, chromosomal gains and losses [27]. Owing to their complex nature, the specific cells from which most of this group of cancers develop remain largely unknown. Although some strains of mice have developed spontaneous STS, rodent models generally require induction of STS [28]. By contrast, dogs are an excellent model of STS because they have similar tumor genetic complexity to humans [29]. For instance, two poorly differentiated fibrosarcomas taken from Labrador Retrievers had large chromosomal rearrangements, amplifications and deletions similar to those observed in human fibrosarcoma [30]. Notably, these fibrosarcomas had loss of heterozygosity affecting cyclin-dependent kinase family 2A and 2B (CDKN2A/CDKN2B). Given that deletions of CDKN2A and CDKN2B have been reported in other cancer types, including STS in humans, this offers a novel target for discovering common pathways and genes affected in both dogs and humans that affects the development or progression of STS [29].
Another advantage of using canines for studying STS is breed predispositions to specific types of STS, including increased incidences in Flat-Coated Retrievers and Rhodesian Ridgebacks [13]. For example, hemangiosarcomas are relatively common in dogs, accounting for ~5-7% of all observed tumors [31]. The dogs at greatest risk for hemangiosarcoma are Golden Retrievers (GR), German Shepherds, and Boxers [32]. One group recently compared gene expression profiles in hemangiosarcoma tumors from multiple dog breeds [22]. They found that the GR was unique in over-expression of vascular endothelial growth factor 1 (VEGF1) compared to other breeds, whereas VEGF2 was more highly expressed in the other breeds compared to the GR. When VEGF2 expression was blocked in hemangiosarcoma-derived tumor cell lines, the rate of cell growth slowed – except in cell lines derived from GR tumors. This finding implies that the unique genetic background of the GR influenced this breed’s susceptibility to the development of hemangiosarcoma, suggesting that canine tumors can be used to understand how genetic background can influence susceptibility of an individual to non-inherited cancers. Clinical trials involving tyrosine kinase inhibitor (TKI) treatment of STS found that the most effective TKIs (e.g. Sorafenib), also targeted all VEGF isoforms [33]. Performing clinical trials on pedigree dogs, such as the GR, could provide novel information regarding genetic background effects on tumor progression. Thus, given the increased incidence of STS in dogs, the diversity of naturally occurring ‘complex’ and ‘simple’ sarcoma similarity in humans and dogs, and the availability of different genetic backgrounds across breeds for clinical therapy testing, the canine model is more relevant than other animal models for direct human STS applications.
Osteosarcoma
In humans, the most commonly diagnosed primary malignant tumor of the bone is osteosarcoma (OSA). It is the third most frequent cause of cancer in adolescents and represents over 56% of all bone tumors. The prognosis for patients with metastatic OSA is poor, with only 20% surviving event-free for 5 years post-diagnosis [34], and > 30% of patients do not respond to chemotherapy [35]. Roughly 10,000 dogs are diagnosed with OSA yearly in the USA [36], compared to 2,650 new cases of human primary bone cancer [including OSA, Ewing sarcoma, malignant fibrous histiocytoma, and chondrosarcoma; http://www.cancer.gov/cancertopics/types/bone/]. Because there is no consistent method for reporting cancer in dogs, we estimate OSA incidence is at least 13.9/100,000 [8, 37], as opposed to the actual incidence of 1.02/100,000 in humans (across all ages) [38]. In both humans and dogs, OSA has a bimodal age distribution and the main cause of death is pulmonary metastasis. It accounts for 85% of malignancies originating in the bone [39] in large and giant dog breeds [40], which have an OSA risk 61 times higher than all breeds [32]. The canine disease is much more aggressive than the human, with surgical treatment alone producing a 5% survival rate [36]. The same treatments for OSA are used in both humans and dogs [41]. Dogs develop OSA at similar sites as humans and both have similar histology and response to treatment [36, 42]. Indeed, dogs have been a valuable model of OSA since they first participated in clinical trials pioneering limb salvage techniques that are now used in humans [43].
In addition to the similarity of tumor biological behavior of human and dog OSA, recent studies identified parallel genetic features [44]. Both human and canine OSA have a 75% aneuploid DNA index, and both share similar genetic alterations [42]. Moreover, many candidate genes implicated in pediatric OSA have also been implicated in the canine disease: phosphatase and tensin homolog (PTEN), retinoblastoma 1 (RB1), ezrin (EZR), met proto-oncogene (hepatocyte growth factor receptor; MET), v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 (ERBB2), and tumor protein 53 (TP53) [45]. The commonly affected p53 tumor suppressor pathway has similar alterations in human and canine OSA [46]. Because human TP53 is more similar to dog than mouse [47], and because mutations occur naturally in dogs, the canine OSA model is presumably more relevant to humans. Additionally, recent work in dogs focused on differential OSA tumor expression of genes associated with short and long term survival [48]. In experiments using cDNA microarrays, investigators found deregulated expression of the following signaling pathways that were previously reported in human OSA: Wnt, chemokine/cytokine, apoptosis signaling, interleukin, and Ras [48]. Co-expression of hepatocyte growth factor (HGF) and the proto-oncogenic receptor c-Met are implicated in growth, invasion, and metastasis in human OSA. Although more frequently over-expressed in human OSA, another study found co-expression of HGF and c-Met in all 59 OSA canine tumors in the study, with over-expression of both present in 24% of cases [49]. Other investigators identified two genes, interleukin 8 (IL8) and solute carrier family member 3 (SLC1A3) that were uniformly expressed in all canine OSA tumors, but not in all human pediatric OSA tumors. However, pediatric patients who did over-express IL8 and SLC1A3 had poorer outcomes then those who did not [50]. Yet another gene expression study of canine OSA tumors identified 10 significantly differentiated pathways between responders to treatment and non-responders [51]. These pathways [including cAMP signaling; chemokines and adhesion; Sonic Hedgehog and Parathyroid Hormone Signaling Pathways in Bone and Cartilage Development] are also disrupted in human cancers. These various findings suggest that alterations of similar pathways occur in human and canine OSA, but that species-specific genetic changes might account for overall disparity in incidence and aggressiveness. Related to that, Phillips et al. used a whole genome linkage approach to map OSA segregating in a four generation pedigree of Scottish Deerhounds [56]. They found evidence of linkage (Zmax=5.766) consistent with a dominant OSA mutation in a 4.5 Mb region of chromosome 34q16.2–q17.1 (syntenic to human 3q26). Because OSA is relatively rare and most cases are sporadic in humans, inherited forms and different risks across dog breeds offer a great opportunity to identify pathogenetic pathways.
OSA tumors in dogs and humans also share DNA structural changes. Analyzing 38 OSA tumors from 29 Rottweilers and 9 Golden Retrievers, a recent study demonstrated that, as with its human counterpart, dog OSA has a tendency toward highly complex and chaotic karyotypes [52]. These are comprised of structural and numerical aberrations, including gene dosage imbalances of known oncogenes and tumor suppressors. The most frequently observed genome alteration was an amplification affecting both the MYC and KIT (c-KIT) oncogenes. This is consistent with observations of genome alterations in human OSA that are predictive of clinical outcome. Notably, KIT was recently proposed as a novel therapeutic target for pediatric OSA [53]. This not only supports the genetic relevance of the canine model, but also the clinical utility of including dogs in OSA clinical trials. Thus, the canine OSA model recapitulates the human cancer and, because OSA occurs 20 times more often in dogs than humans [42], it provides an unparalleled opportunity for identifying key cellular pathways in this cancer [12].
Lymphomas
A group of cancers affecting the lymph tissue are collectively known as lymphomas. Lymphomas represent ~5% of all human cancers in the United States and account yearly for 4.6 Billion USD worth of treatment [http://www.cancer.gov/aboutnci/servingpeople/snapshots/lymphoma]. One specific class, non-Hodgkin’s lymphoma (NHL) occurs in B or T-cells, with >65,000 new cases reported in 2009 [for types of NHL, see http://www.cancer.gov/cancertopics/types/non-hodgkins-lymphoma]. Notably, the incidence of NHL is increasing but the etiology remains obscure [54]. Thus, an alternative model of lymphoma is needed to elucidate the causes and identify clinically meaningful cancer biology. Dogs and humans have similar tumor biology, tumor biological behavior, and genetic aberrations. The incidence of lymphoma in humans and dogs is similar [55]: 15.5-29.9 [56] and 15-30 [57] per 100,000, respectively. The most common type of NHL is the same in both humans and dogs – diffuse large B-cell – and the same chemotherapy agents are used to treat it [55]. An additional advantage of the dog model is the increased prevalence of lymphoma within specific dog breeds. Lymphoma is the most common life-threatening cancer in all dogs, accounting for 24% of all canine cancers [58]. Approximately, 1 in 4 Boxers and 1 in 8 Golden Retrievers develop lymphoma [32]. Additionally, there is a breed-specific distribution of B-cell and T-cell lymphomas [59; Fig. 3]: whereas excess incidence of T-cell lymphomas were noted in 10 breeds, the most striking occurred (in order of observed frequency) in Irish Wolfhounds, Siberian Huskies, and Shih Tzus. By contrast, the breeds with excessive occurrence of B-cell lymphomas were Cocker Spaniels and Basset Hounds. A second study conducted in Norway grouped together all types of lymphomas and also identified excessive occurrence of lymphomas in specific breeds, lending credence to a breed specific risk for lymphoma development [60]. They found the relative risk for lymphoma was highest in the Boxer and Flat-Coated Retriever. More recently, a study examined records from the Veterinary Medical Database and selected cases with a diagnosis of lymphoma type not specified, giant follicular lymphoma, and lymphosarcoma and used controls with any diagnosis other than lymphoma [61]. This study also identified a breed specific risk for lymphoma with the highest breeds including Bullmastiff [odds ratio (OR) 4.83 vs control], Boxer [OR 4.05 vs control] and Bernese Mountain dog [OR 3.64 vs control]. Notably, although the former and latter studies examined different subsets of lymphoma, they included many of the same breeds and had similar findings. For instance, the Irish Wolfhound had the highest rate of T-cell lymphoma in the Modiano et al. study [59], and also had an OR of 3.23 for lymphoma compared to other dogs in the Villamil et al. study [61]. The underlying cytogenetic basis of lymphoma appears to be shared in human and dog. The examination of three canine hematological cancers, including Burkitt lymphoma and small lymphocytic lymphoma [62], showed that these canine cancers shared cytogenetic abnormalities with those characteristic of their human counterparts. This suggests that humans and dogs share common pathways or an ancestrally retained pathogenetic basis for lymphoma [62]. Consequently, by using the dog genome in comparison with the human genome, relevant genetic aberrations can be identified.
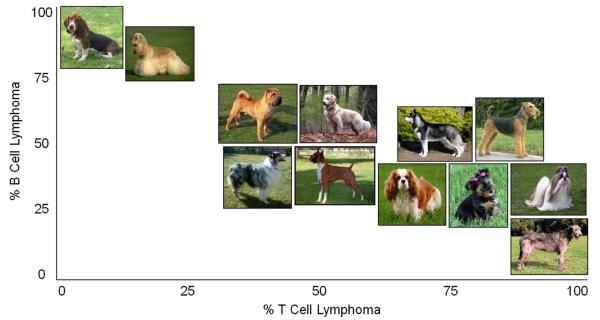
Fig. 3. Prevalence of B and T-cell lymphoma in Dog breeds.
A varying excess of T and B-cell lymphoma, in a breed specific manner, has been noted. Presented here is the observed percentage of T vs B-cell lymphoma by breed: Irish wolfhounds (100:0 Siberian huskies (88.9:11.1), Shih Tzus (81:19), Airedale terriers (80:20), Cavalier King Charles spaniels (80:20), and Yorkshire terriers (80:20). By contrast, the breeds with an excessive occurrence of B-cell compared to T-cell lymphomas were cocker spaniels (93.2:6.8) and basset hounds (94.4:5.6) [59]. Photo sources follow: http://www.dublinirishfestival.org/animals/irishwolfhound.php; http://blogneffy.blogspot.com/2010/06/wanted-shih-tzu-breeders-in-davao-city.html; http://sentinelkennels.com/images/airedale.jpg; http://www.justdogbreeds.com/images/breeds/cavalier-king-charles-spaniel.jpg; http://www.petsflick.com/images/yorkshire-terrier.jpg; http://tidyyourdog.com/wp-content/uploads/2009/04/siberian-husky.jpg; http://www.petside.com/breeds/chinese-shar-pei.php; http://www.fordogtrainers.com/ProductImages/dog-breeds-muzzles/Australian-Shepherd-muzzle-Australian-Shepherd.jpg; http://www.breederretriever.com/photopost/pindex/516/; http://retrieverman.files.wordpress.com/2009/01/white-golden-retriever-wikipedia.jpg; http://www.dogtastic.org/dogtastic/images/BreedPics/cocker%20spaniel.jpg; http://a1.cdnsters.com/static/images/dogster/breeds/basset_hound.jpg.
Finally, the relevance of dogs as a lymphoma model is supported by use in clinical trials. Given that dogs develop spontaneous B-cell NHL and share many characteristics in common with human B-cell NHL [such as diagnostic criteria and response to a chemotherapy based regimen that includes cyclophosphamide, doxorubicin, vincristine, and prednisone (commonly referred to as the CHOP chemotherapy)], dogs were recently enrolled in a clinical trial of a selective and irreversible Bruton tyrosine kinase (Btk) inhibitor, PCI-32765, which blocks B-cell activation [63]. Activation of the B-cell antigen receptor signaling pathway contributes to the initiation and maintenance of B-cell [63]. This clinical trial research began when the same group described the synthesis of a series of Btk inhibitors that bind covalently to a cysteine residue leading to potent and irreversible inhibition of Btk enzymatic activity. In that study, after additional analysis of this agent in both cell lines and mouse models, they initiated a canine clinical trial. Although the clinical trial is ongoing to date, 8 dogs have been treated with 3 demonstrating stable disease and 3 with partial responses including one dog with a 77% decrease in tumor size [this drug is now undergoing human clinical development in patients with B-cell malignancies]. Finally, a recent pilot study used anti-human leukocyte antigen (HLA)-DR monoclonal antibody (mAb) as a treatment for dogs with lymphoma [64]. Preliminary results demonstrated that humanized IgG4 anti-HLA-DR, currently under evaluation pre-clinically for human trials, also bound malignant canine lymphocytes. These findings provide justification for the using dogs with lymphoma in safety and efficacy evaluations of therapy for both veterinary and human purposes [64, Fig. 4].
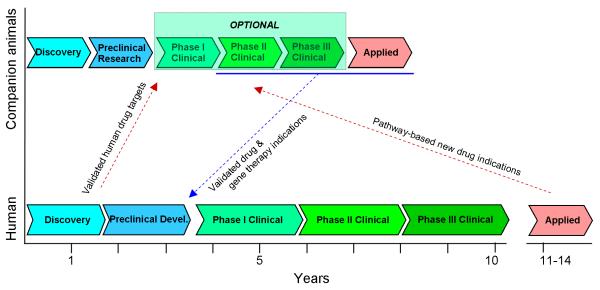
Fig. 4. Translational potential of tumor bearing dogs.
On the bottom is the typical course of human drug research and development. There is no established paradigm for the drug research and development in dogs and other companion animals [6]. Although our schematic mirrors the same process in pets, most drugs used on patient animals are taken from human drug development or are approved human drugs used off-label. Indeed, few regulations exist for Phase I/II/III clinical trials before drugs are used in pets.
Potential utility of dogs in translational medicine
The naturally occurring relevance of the canine model to cancer in humans can be exploited to generate new treatments relatively quickly (Fig. 4). Whereas there are strict Food and Drug Administration (FDA) regulations concerning treatments to be used and commercialized, as well as for clinical trials in humans, there are fewer regulations for Phase I/II/III clinical trials before drugs use in pets [65; http://prsinfo.clinicaltrials.gov]. Rather, it is left to the discretion of the owner, who could approve the use of investigational therapeutics before conventional treatments. There are several trends in drug development that suggest increased use of dogs as translational models. Two of these are the rising proportion of biological vs. chemical compounds, and the growing focus on targeting genetic/biochemical pathways (or disease subtypes) vs. broad diseases or types of cancer. Here we propose that dogs are ideal patients in which to develop novel therapeutics. Several facts indicate using dogs in translational medicine can hugely accelerate drug development: reduced regulatory guidelines, vastly diminished and soon-to-be fully defined genetic variation within breeds (but similar levels of variation occur across all breeds as humans), reduced disease heterogeneity (i.e., breed-specific risks of diseases are often associated with a single founder mutation), and accelerated aging/disease progression compared to humans. These genetic benefits translate into faster progress at every stage – e.g., identifying disease mutations in discovery, identifying biomarkers and endpoints in clinical trials, and using pharmacogenetics from preclinical research to post-approval studies. Indeed, dogs have been instrumental in rapid development of biological and biological-like therapeutics, including gene therapies (e.g., for specific inherited forms of muscular and retinal dystrophies [66]) and antisense morpholino oligonucleotides (e.g., to alter mRNA splicing and avert nonsense-mediated decay of dystrophin [67]). However, we believe dog patients are greatly underutilized in development of therapeutic interventions. Drug development is difficult and risky, with the average drug costing ~800M USD to develop. One of the most challenging go/no go decision points is determination that a therapeutic agent is effective in humans. This is established by a small clinical study of select subjects that will likely respond to therapy. Dog breeds with known disease mutations are ideal lead-ins to such studies. Depending on the disease, such proof-of-concept studies could be done robustly in even fewer than 10 subjects, and at a pace proportional to the accelerated disease progression. Such studies would not only establish efficacy, pharmco-kinetics/dynamics, and toxicity, but also dosing, biomarkers/endpoints, and adverse effects. This could dramatically reduce the failure rate of human proof-of-concept studies, and thus time and costs.
Concluding remarks and future perspectives
Dogs are uniquely suited for use as an animal model of complex human disease due to their phenotypic diversity and naturally occurring disease similarity to human conditions. The evolutionary history of dogs, their position as a family member in many households, and the high level of health care they receive offer tremendous opportunities. That, combined with recently developed genetic resources, makes dogs outstanding models for the study of known genetic pathways, discovery of genetic and environmental contributions to disease, and translational studies in cancer risk, prevention, and treatments [6, 14]. The full utilization of canine models of cancer will require expertise in basic science, translation, and direct clinical relevance. This will necessitate large collaborations across almost all aspects of veterinary and human medicine: including molecular biology and genetics, epidemiology, pharmacology, bioinformatics, statistics, and engineering. Developing these pipelines now will speed potential therapeutic outcomes. Although this review focused on the relevance of the dog as a model for research in cancer genetics, biomedical research has long included canine models of numerous other diseases and their treatments [14]. For example, dogs are also increasingly used in behavioral research, including learning [68], social cognition [69], and the effects of diet and behavior enrichment on executive functioning [70]. Increased appreciation of the unique and comparative value of the dog as a model of diverse human diseases should accelerate research leading to new treatments, and improved health care for both we humans and for our best friend.
Acknowledgments
We thank C.A. London for critically reading the manuscript, and W.C. Kisseberth and C.G. Couto for providing their veterinary clinical expertise. CEA is supported by R210602710 from NIH/NIHGR, and funding from The Research Institute at Nationwide Children’s Hospital. JLR is supported by a fellowship award F31NR011559 from NIH/NINR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265(5181):2037–48. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 2.Strauch K, et al. How to model a complex trait. 1. General considerations and suggestions. Hum Hered. 2003;55(4):202–10. doi: 10.1159/000073204. [DOI] [PubMed] [Google Scholar]
- 3.Karlsson EK, Lindblad-Toh K. Leader of the pack: gene mapping in dogs and other model organisms. Nat Rev Genet. 2008;9(9):713–25. doi: 10.1038/nrg2382. [DOI] [PubMed] [Google Scholar]
- 4.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gondo Y, et al. Next-generation gene targeting in the mouse for functional genomics. BMB Rep. 2009;42(6):315–23. doi: 10.5483/bmbrep.2009.42.6.315. [DOI] [PubMed] [Google Scholar]
- 6.Paoloni M, Khanna C. Translation of new cancer treatments from pet dogs to humans. Nat Rev Cancer. 2008;8(2):147–56. doi: 10.1038/nrc2273. [DOI] [PubMed] [Google Scholar]
- 7.American Pet Products Manufacturers Association (APPMA) Report. 2005 [Google Scholar]
- 8.U.S. pet ownership & demographics sourcebook. 2007 v.:]. Available from: http://www.avma.org/reference/marketstats/sourcebook.asp.
- 9.Patterson DF. Companion animal medicine in the age of medical genetics. J Vet Intern Med. 2000;14(1):1–9. [PubMed] [Google Scholar]
- 10.Starkey MP, et al. Dogs really are man’s best friend--canine genomics has applications in veterinary and human medicine! Brief Funct Genomic Proteomic. 2005;4(2):112–28. doi: 10.1093/bfgp/4.2.112. [DOI] [PubMed] [Google Scholar]
- 11.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438(7069):803–19. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 12.Khanna C, et al. The dog as a cancer model. Nat Biotechnol. 2006;24(9):1065–6. doi: 10.1038/nbt0906-1065b. [DOI] [PubMed] [Google Scholar]
- 13.Sargan DR. IDID: inherited diseases in dogs: web-based information for canine inherited disease genetics. Mamm Genome. 2004;15(6):503–6. doi: 10.1007/s00335-004-3047-z. [DOI] [PubMed] [Google Scholar]
- 14.Parker HG, Shearin AL, Ostrander EA. Man’s best friend becomes biology’s best in show: genome analyses in the domestic dog. Annu Rev Genet. 2010;44:309–36. doi: 10.1146/annurev-genet-102808-115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrander EA, Galibert F, Patterson DF. Canine genetics comes of age. Trends Genet. 2000;16(3):117–24. doi: 10.1016/s0168-9525(99)01958-7. [DOI] [PubMed] [Google Scholar]
- 16.Cummings BJ, et al. The canine as an animal model of human aging and dementia. Neurobiol Aging. 1996;17(2):259–68. doi: 10.1016/0197-4580(95)02060-8. [DOI] [PubMed] [Google Scholar]
- 17.Bonnett BN, Egenvall A. Age patterns of disease and death in insured Swedish dogs, cats and horses. J Comp Pathol. 2010;142(Suppl 1):S33–8. doi: 10.1016/j.jcpa.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Germonpré M, et al. Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: osteometry, ancient DNA and stable isotopes. Journal of Archaeological Science. 2009;36(2):473–490. [Google Scholar]
- 19.Drogemuller C, et al. A deletion in the N-myc downstream regulated gene 1 (NDRG1) gene in Greyhounds with polyneuropathy. PLoS One. 2010;5(6):e11258. doi: 10.1371/journal.pone.0011258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyko AR, et al. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 2010;8(8):e1000451. doi: 10.1371/journal.pbio.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamburini BA, et al. Gene expression profiles of sporadic canine hemangiosarcoma are uniquely associated with breed. PLoS One. 2009;4(5):e5549. doi: 10.1371/journal.pone.0005549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang J, et al. Copy number abnormalities in sporadic canine colorectal cancers. Genome Res. doi: 10.1101/gr.092726.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breen M. Update on genomics in veterinary oncology. Top Companion Anim Med. 2009;24(3):113–21. doi: 10.1053/j.tcam.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krikelis D, Judson I. Role of chemotherapy in the management of soft tissue sarcomas. Expert Rev Anticancer Ther. 10(2):249–60. doi: 10.1586/era.09.176. [DOI] [PubMed] [Google Scholar]
- 26.Guillou L, Aurias A. Soft tissue sarcomas with complex genomic profiles. Virchows Arch. 456(2):201–17. doi: 10.1007/s00428-009-0853-4. [DOI] [PubMed] [Google Scholar]
- 27.Mertens F, Panagopoulos I, Mandahl N. Genomic characteristics of soft tissue sarcomas. Virchows Arch. 2010;456(2):129–39. doi: 10.1007/s00428-009-0736-8. [DOI] [PubMed] [Google Scholar]
- 28.Cohen SM, et al. Hemangiosarcoma in rodents: mode-of-action evaluation and human relevance. Toxicol Sci. 2009;111(1):4–18. doi: 10.1093/toxsci/kfp131. [DOI] [PubMed] [Google Scholar]
- 29.Aguirre-Hernandez J, et al. Disruption of chromosome 11 in canine fibrosarcomas highlights an unusual variability of CDKN2B in dogs. BMC Vet Res. 2009;5:27. doi: 10.1186/1746-6148-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sargan DR, et al. Chromosome rearrangements in canine fibrosarcomas. J Hered. 2005;96(7):766–73. doi: 10.1093/jhered/esi122. [DOI] [PubMed] [Google Scholar]
- 31.Modiano J. Canine Hemangiosarcoma - The Road from Despair to Hope. National Canine Cancer Foundation; 2008. [Google Scholar]
- 32.Shearin AL, Ostrander EA. Leading the way: canine models of genomics and disease. Dis Model Mech. 2010;3(1-2):27–34. doi: 10.1242/dmm.004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chao J, Chow WA, Somlo G. Novel targeted therapies in the treatment of soft-tissue sarcomas. Expert Rev Anticancer Ther. 2010;10(8):1303–11. doi: 10.1586/era.10.100. [DOI] [PubMed] [Google Scholar]
- 34.Mialou V, et al. Metastatic osteosarcoma at diagnosis: prognostic factors and long-term outcome--the French pediatric experience. Cancer. 2005;104(5):1100–9. doi: 10.1002/cncr.21263. [DOI] [PubMed] [Google Scholar]
- 35.Mankin HJ, et al. Survival data for 648 patients with osteosarcoma treated at one institution. Clin Orthop Relat Res. 2004;(429):286–91. doi: 10.1097/01.blo.0000145991.65770.e6. [DOI] [PubMed] [Google Scholar]
- 36.Withrow Stephen J., Vail David M., editors. Withrow & MacEwen’s small animal clinical oncology. 4th ed. Saunders Elsevier; St. Louis, Mo: 2007. p. xvii.p. 846. ed, ed. S.J. Withrow and D.M. Vail. [Google Scholar]
- 37.Withrow SJ. In: Small animal clinical oncology. 3rd ed. Withrow Stephen J., MacEwan E. Gregory., editors. W. B. Saunders; Philadelphia: 2001. p. xvii.p. 718. ed, ed. E.G. MacEwan. [Google Scholar]
- 38.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115(7):1531–43. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harari J. The prevalence of and risk factors for canine appendicular osteosarcoma.(study recaps and comments) Veterinary Medicine. 2008;103(2):79(1). [Google Scholar]
- 40.Ru G, Terracini B, Glickman LT. Host related risk factors for canine osteosarcoma. Vet J. 1998;156(1):31–9. doi: 10.1016/s1090-0233(98)80059-2. [DOI] [PubMed] [Google Scholar]
- 41.Messerschmitt PJ, et al. Osteosarcoma. J Am Acad Orthop Surg. 2009;17(8):515–27. doi: 10.5435/00124635-200908000-00005. [DOI] [PubMed] [Google Scholar]
- 42.Withrow SJ, Wilkins RM. Cross talk from pets to people: translational osteosarcoma treatments. ILAR J. 2010;51(3):208–13. doi: 10.1093/ilar.51.3.208. [DOI] [PubMed] [Google Scholar]
- 43.LaRue SM, et al. Limb-sparing treatment for osteosarcoma in dogs. J Am Vet Med Assoc. 1989;195(12):1734–44. [PubMed] [Google Scholar]
- 44.Mueller F, Fuchs B, Kaser-Hotz B. Comparative biology of human and canine osteosarcoma. Anticancer Res. 2007;27(1A):155–64. [PubMed] [Google Scholar]
- 45.Kirpensteijn J, et al. TP53 gene mutations in canine osteosarcoma. Vet Surg. 2008;37(5):454–60. doi: 10.1111/j.1532-950X.2008.00407.x. [DOI] [PubMed] [Google Scholar]
- 46.Levine RA, Fleischli MA. Inactivation of p53 and retinoblastoma family pathways in canine osteosarcoma cell lines. Vet Pathol. 2000;37(1):54–61. doi: 10.1354/vp.37-1-54. [DOI] [PubMed] [Google Scholar]
- 47.Nasir L, et al. Nucleotide sequence of a highly conserved region of the canine p53 tumour suppressor gene. DNA Seq. 1997;8(1-2):83–6. doi: 10.3109/10425179709020890. [DOI] [PubMed] [Google Scholar]
- 48.Selvarajah GT, et al. Gene expression profiling of canine osteosarcoma reveals genes associated with short and long survival times. Mol Cancer. 2009;8:72. doi: 10.1186/1476-4598-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fieten H, et al. Expression of hepatocyte growth factor and the proto-oncogenic receptor c-Met in canine osteosarcoma. Vet Pathol. 2009;46(5):869–77. doi: 10.1354/vp.08-VP-0155-F-FL. [DOI] [PubMed] [Google Scholar]
- 50.Paoloni M, et al. Canine tumor cross-species genomics uncovers targets linked to osteosarcoma progression. BMC Genomics. 2009;10:625. doi: 10.1186/1471-2164-10-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Donoghue LE, et al. Expression profiling in canine osteosarcoma: identification of biomarkers and pathways associated with outcome. BMC Cancer. 2010;10:506. doi: 10.1186/1471-2407-10-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas R, et al. Influence of genetic background on tumor karyotypes: evidence for breed-associated cytogenetic aberrations in canine appendicular osteosarcoma. Chromosome Res. 2009;17(3):365–77. doi: 10.1007/s10577-009-9028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Entz-Werle N, et al. KIT gene in pediatric osteosarcomas: could it be a new therapeutic target? Int J Cancer. 2007;120(11):2510–6. doi: 10.1002/ijc.22593. [DOI] [PubMed] [Google Scholar]
- 54.Institute, N.C. Cancer Trends Progress Report-2009/2010 Update. 2010 http://progressreport.cancer.gov/doc_detail.asp?pid=1&did=2009&chid=93&coid=920&mid=#measuring.
- 55.Hansen K, Khanna C. Spontaneous and genetically engineered animal models; use in preclinical cancer drug development. Eur J Cancer. 2004;40(6):858–80. doi: 10.1016/j.ejca.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 56.Vail DM, MacEwen EG. Spontaneously occurring tumors of companion animals as models for human cancer. Cancer Invest. 2000;18(8):781–92. doi: 10.3109/07357900009012210. [DOI] [PubMed] [Google Scholar]
- 57.Hahn KA, et al. Naturally occurring tumors in dogs as comparative models for cancer therapy research. In Vivo. 1994;8(1):133–43. [PubMed] [Google Scholar]
- 58.Breen M. The Prognostic Significance of Chromosome Aneuploidy in Canine Lymphoma. 2009 http://www.akcchf.org/pdfs/2009FundingRequest.pdf.
- 59.Modiano JF, et al. Distinct B-cell and T-cell lymphoproliferative disease prevalence among dog breeds indicates heritable risk. Cancer Res. 2005;65(13):5654–61. doi: 10.1158/0008-5472.CAN-04-4613. [DOI] [PubMed] [Google Scholar]
- 60.Gamlem H, Nordstoga K, Glattre E. Canine neoplasia--introductory paper. APMIS Suppl. 2008;(125):5–18. doi: 10.1111/j.1600-0463.2008.125m2.x. [DOI] [PubMed] [Google Scholar]
- 61.Villamil JA, et al. Hormonal and sex impact on the epidemiology of canine lymphoma. J Cancer Epidemiol. 2009;2009:591753. doi: 10.1155/2009/591753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Breen M, Modiano JF. Evolutionarily conserved cytogenetic changes in hematological malignancies of dogs and humans--man and his best friend share more than companionship. Chromosome Res. 2008;16(1):145–54. doi: 10.1007/s10577-007-1212-4. [DOI] [PubMed] [Google Scholar]
- 63.Honigberg LA, et al. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci U S A. 2010;107(29):13075–80. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stein R, et al. Evaluation of anti-human leukocyte antigen-DR monoclonal antibody therapy in spontaneous canine lymphoma. Leuk Lymphoma. 2010 doi: 10.3109/10428194.2010.535182. [DOI] [PubMed] [Google Scholar]
- 65.Khanna C, et al. Guiding the optimal translation of new cancer treatments from canine to human cancer patients. Clin Cancer Res. 2009;15(18):5671–7. doi: 10.1158/1078-0432.CCR-09-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Z, et al. Gene therapy in large animal models of muscular dystrophy. Ilar J. 2009;50(2):187–98. doi: 10.1093/ilar.50.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yokota T, et al. Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann Neurol. 2009;65(6):667–76. doi: 10.1002/ana.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Elgier AM, et al. Communication between domestic dogs (Canis familiaris) and humans: dogs are good learners. Behav Processes. 2009;81(3):402–8. doi: 10.1016/j.beproc.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 69.Morell V. Animal behavior. Going to the dogs. Science. 2009;325(5944):1062–5. doi: 10.1126/science.325_1062. [DOI] [PubMed] [Google Scholar]
- 70.Cotman CW, Head E. The canine (dog) model of human aging and disease: dietary, environmental and immunotherapy approaches. J Alzheimers Dis. 2008;15(4):685–707. doi: 10.3233/jad-2008-15413. [DOI] [PubMed] [Google Scholar]
- 71.Liao AT, McMahon M, London CA. Identification of a novel germline MET mutation in dogs. Anim Genet. 2006;37(3):248–52. doi: 10.1111/j.1365-2052.2006.01415.x. [DOI] [PubMed] [Google Scholar]
- 72.Chen WK, et al. Mapping DNA structural variation in dogs. Genome Res. 2009;19(3):500–9. doi: 10.1101/gr.083741.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kisseberth WC, et al. A novel canine lymphoma cell line: A translational and comparative model for lymphoma research. Leuk Res. 2007 doi: 10.1016/j.leukres.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 74.Milde T, et al. A novel family of slitrk genes is expressed on hematopoietic stem cells and leukemias. Leukemia. 2007;21(4):824–7. doi: 10.1038/sj.leu.2404525. [DOI] [PubMed] [Google Scholar]
- 75.London CA, et al. Multi-center, placebo-controlled, double-blind, randomized study of oral toceranib phosphate (SU11654), a receptor tyrosine kinase inhibitor, for the treatment of dogs with recurrent (either local or distant) mast cell tumor following surgical excision. Clin Cancer Res. 2009;15(11):3856–65. doi: 10.1158/1078-0432.CCR-08-1860. [DOI] [PubMed] [Google Scholar]