Abstract
B cells sense microenvironments through the B cell receptor (BCR) and Toll-like receptors (TLRs). While signals from BCR and TLRs synergize to distinguish self from nonself, inappropriate regulation can result in development of autoimmune disease. Here we show that CD22, an inhibitory co-receptor of BCR, also negatively regulates TLR signaling in B cells. CD22-deficient (Cd22–/–) B cells exhibit hyperactivation in response to ligands of TLRs 3, 4 and 9. Evidence suggests that this results from impaired induction of suppressors of cytokine signaling 1 and 3, well-known suppressors of TLR signaling. Antibody-mediated sequestration of CD22 on wild-type (WT) B cells augments proliferation by TLR ligands. Conversely, expression of CD22 in a Cd22–/– B cell line blunts responses to TLR ligands. We also show that lipopolysaccharide-induced transcription by nuclear factor-κB is inhibited by ectopic expression of CD22 in a TLR4 reporter cell line. Taken together, these results suggest that negative regulation of TLR signaling is an intrinsic property of CD22. Since TLRs and BCR activate B cells through different signaling pathways, and are differentially localized in B cells, CD22 exhibits a broader regulation of receptors that mediate adaptive and innate immune responses of B cells than previously recognized.
Key Words: CD22, Sialic acid binding immunoglobulin-like lectin, Toll-like receptors, B cells, Self nonself recognition
Introduction
CD22 is a sialic acid binding immunoglobulin-like lectin (Siglec) expressed on B cells, which is known for its role in regulation of B cell receptor (BCR) [1,2]. The ligand recognized by CD22 is a sialic acid containing glycan found on mammalian cells that is absent on most pathogens [1]. Simultaneous presentation of antigen and CD22 ligand results in coligation of CD22 with the BCR and inhibition of BCR signaling [2,3,4,5]. Thus, CD22 has been proposed to aid B cells in distinguishing self from nonself [3,4,5].
Recent studies demonstrate that in B cells, Toll-like receptors (TLRs) play an important role in modulation of their function in an immune response [6]. Although not required for induction of humoral immunity, TLR signals independent of BCR ligation induce and enhance proliferation, antigen presentation and immunoglobulin production in B cells [6,7,8,9,10]. However, it has also been suggested that the synergy between BCR and TLR7 and 9 in response to nucleotide-containing self-antigens might result in the aberrant activation of autoreactive B cells [6,11].
Here we report that Cd22–/– B cells exhibit hyperactivation in response to ligands of several TLRs including TLR3, 4 and 9 and that in wild-type (WT) B cells, antibody-mediated sequestration of CD22 from TLRs augments the proliferation by TLR ligands. We also demonstrate that in Cd22–/– B cells, induction of suppressors of cytokine signaling (SOCS) 1 and 3, well-documented suppressors of TLR signaling [12,13], is impaired and that lipopolysaccharide (LPS) induced activation of transcription by nuclear factor-κB (NF-κB), which is a hallmark of TLR signaling, is inhibited by the expression of CD22 in a TLR4 reporter cell line. These observations document a broader role for CD22 in regulation of receptors that mediate B cell signaling that distinguish self and nonself.
Materials and Methods
Mice
C57BL/6J and Cd22–/– mice (a generous gift from Dr. Lars Nitschke, University of Erlangen, Germany) were maintained in pathogen-free conditions at The Scripps Research Institute breeding facility and were used in accordance to the guidelines of the Institutional Animal Care Committee at the National Institutes of Health. Myd88–/– mice were kindly provided by Dr. Bruce Beutler (The Scripps Research Institute, La Jolla, Calif., USA).
Retrovirus Vector and Cell Lines
The retrovirus vector pMXs-IRES-EGFP (pMXs-IG) encoding the cytoplasmic domain of murine CD3ζ and the retrovirus-packaging cell line Plat-E were provided by Dr. Kazuo Yamamoto (University of Tokyo, Tokyo, Japan) by permission of Dr. Toshio Kitamura (University of Tokyo, Tokyo, Japan). Plat-E cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, Calif., USA) supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM glutamine, 50 μM 2-mercaptoethanol, 1.0 μg/ml of puromycin, 10 μg/ml of blasticidin S and 20 mM HEPES. Cd22–/– murine B cell line J2-44 cells were kindly provided by Dr. Henry H. Wortis (Tufts University, Boston, Mass., USA) and maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% heat-inactivated FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM glutamine and 50 μM 2-mercaptoethanol. HEK293 cells expressing mTLR4/MD2-CD14 were obtained from Invivogen (San Diego, Calif., USA) and maintained in the same medium with Plat-E with 50 μg/ml of hygromycin B instead of puromycin.
Isolation of Splenic B Cells
Splenic B cells were isolated from mouse spleens by negative selection using MACS mouse B cell isolation kit (Miltenyi Biotech, Auburn, Calif., USA) or EasySep (StemCell Technologies, Vancouver, B.C., Canada). The purity of isolated B cells was assessed by the staining of isolated cells with anti-mouse CD19 mAb (1D3; BD Biosciences, San Jose, Calif., USA) and was more than 94% throughout this study.
B Cell Proliferation Assay
Purified B cells were suspended at 1.0 × 107 cells/ml in Hank's balanced salt solution (Invitrogen) containing 3% heat-inactivated FCS and 4–6 μM of carboxyfluorescein diacetate succinimidyl ester (Invitrogen) for 7 min at 25°C. Carboxyfluorescein succinimidyl ester (CFSE)-labeled B cells were added in 96-well flat-bottom plate at 2.0 × 105 cells/well in the 200 μl of RPMI medium (Invitrogen) supplemented with 10% heat-inactivated FCS, 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM nonessential amino acid, 1 mM sodium pyruvate and 50 μM 2-melcaptoethanol. For the BCR stimulation, goat F(ab′)2 anti-mouse IgM (Jackson Immunoresearch, West Grove, Pa., USA) was added to each well. For TLR stimulation, poly(I:C), LPS and CpG1826 (Invivogen) were added. All culture was done with triplicate samples. The cells were allowed to proliferate for 2–3 days. Cells were then harvested and analyzed by flow cytometry with the exclusion of dead cells using 1 μg/ml of propidium iodide staining. More than 2.5 × 104 total counts were acquired in FACS Caliber (BD Biosciences) and analyzed with Flowjo (Tree Star, San Carlos, Calif., USA). For the experiment using immobilized antibodies, 96-well ELISA plate was first coated with 20 μg/ml of neutravidin (Pierce, Rockford, Ill., USA), then washed with phosphate-buffered saline (PBS) and added with biotinylated anti-CD22 (Cy34.1; BD Biosciences) or mouse IgG1 isotype-matched control antibody (Biolegend, San Diego, Calif., USA). Then wells were washed with PBS and added with B cells in the presence of LPS, CpG or anti-IgM. To count the living cell number, 1.0 × 104 of Calibrite beads (BD Biosciences) was added to each well before harvesting cells. The living cell number was calculated by the formula: living cells number in the well = the number of living cells acquired × the number of beads acquired /1.0 × 104.
Activation Marker Expression Analysis
Purified B cells or J2-44 cells were cultured for 2 days with or without stimuli as described above. Cells were then harvested, washed with Hank's balanced salt solution containing 0.1% BSA, 1 mM MgSO4, 1.3 mM CaCl2 and 0.1% NaN3 (FACS buffer). Cells were then blocked with anti-mouse CD16/32 (2.4G2; BD Biosciences), and stained with anti-CD86 (GL1; eBiosicence, San Diego, Calif., USA) and MHC class II I-Ab (AF6–120.1; BD Biosciences). The stained cells were washed with FACS buffer and analyzed by flow cytometry as described above.
TLR9 Expression Analysis
Purified B cells were fixed with 3% parafolmaldehyde in PBS for 10 min at 25°C. Then cells were permeabilized with PBS containing 0.1% saponin (Sigma-Aldrich, St. Louis, Mo., USA), 1% BSA and 0.05% NaN3 (PBS-S). Fixed and permeabilized cells were blocked with 1% normal rat serum in PBS-S and stained with anti-TLR9 Ab (IMG-431; Imgenex, San Diego, Calif., USA) followed by phycoerythrin-labeled goat F(ab′)2 anti-rabbit IgG (Southern Biotech, Birmingham, Ala., USA). The stained cells were washed with PBS-S and analyzed by flow cytometry.
Real-Time PCR
Total RNA was isolated from WT and Cd22–/– B cells with TRIzol reagent (Invitrogen). Random hexamer-primed first-strand cDNA was synthesized from DNase-treated total RNA by reverse transcription using Superscript III (Invitrogen). SYBR Green-based real-time PCR was performed using the DyNAmo SYBR Green qPCR Kit (Finnzymes Oy, Espoo, Finland). Signals were normalized to β-actin and normalized data were used to quantitate relative levels of TLR3 and 4 by using ΔΔCt analysis. Primers used were below: 5′-atgatacagggattgcaccc-3′ and 5′-atagggacaaaagtccccca-3′ for TLR3, 5′-ttcacctctgccttcactaca-3′ and 5′-gggacttctcaaccttctcaa-3′ for TLR4, 5′-ccgtgggtcgcgagaac-3′ and 5′-aactcaggtagtcacggagtaccg-3′ for SOCS1, 5′-tcccatgccgctcacag-3′ and 5′-acaggaccagttccaggtaattg for SOCS3 [14], and 5′-tgggacgacatggagaagatctggc-3′ and 5′-tacgaccagaggcatacagggacagc-3′ for β-actin.
Transduction of the J2-44 Cell Line with Murine CD22
cDNA for murine CD22 was subcloned into the pMXs-IG vector. The CD22-pMXs-IG vector and the translation-incompatible CD3ζ-pMXs-IG vector were used for transduction of the J2-44 cell line to establish CD22/J2-44 cells and Mock/J2-44 cells, respectively. Briefly, the retrovirus packaging cell line Plat-E were transfected with the pMXs-IG vectors by Lipofectamin2000 (Invitrogen). After 2 days, the culture supernatant was harvested and added to J2-44 cells with 8 μg/ml of Polybrane (Millipore, Bedford, Mass., USA). J2-44 cells expressing EGFP were sorted to more than 95% by the FACSVantage SE (BD Biosciences).
Western Blot
Purified B cells (2.2 × 107 cells) were rested for 2 h before stimulation in 1 ml of the medium. Cells were stimulated with 10 μg/ml anti-mouse IgM and 1 μM CpG for the indicated time period. Stimulation was stopped by fast spin down and direct lysis in 80 μl of ice-cold lysis buffer containing 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% Triton X-100, protease inhibitors set III (Calbiochem, San Diego, Calif., USA), and phosphatase inhibitor set II (Calbiochem) for 30 min on ice. After cell debris was removed by spin down at 13,000 g at 4°C for 15 min, proteins in the supernatant were separated by polyacrylamide gel electrophoresis using 4–12% 1-mm NuPAGE gels (5 × 106 cells/lane) (Invitrogen) and transferred to nitrocellulose membrane (Pall Corporation, Pensacola, Fla., USA). The membrane was blocked with 5% skim milk (LabScientific, Livingstone, N.J., USA) in Tris-buffered saline pH 7.4 containing 0.005% Tween 20 (TBS-T) and probed with 1:1,000 polyclonal anti-IκBα antibody (cat. 9242; Cell Signaling, Danvers, Mass., USA), 1:250 polyclonal anti-IRAK-1 antibody (cat. H-273, Santa Cruz Biotechnology, Santa Cruz, Calif., USA), 1:1,000 monoclonal phospho-p38 (Thr180/Thy182) antibody (cat. AF869; R&D Systems), 1:500 polyclonal phospho-JNK (Thr221/Tyr223) (cat. 07-175; Upstate Biotechnology, New York, N.Y., USA) or 1:250 polyclonal anti-actin antibody (cat. I19; Santa Cruz Biotechnology) for 16 h at 4°C. The membrane was washed with TBS-T and incubated with 1:20,000 donkey anti-rabbit IgG peroxidase conjugate (Jackson Immunoresearch) for IκB and IRAK-1 or 1:20,000 rabbit anti-goat IgG peroxidase conjugate (Jackson Immunoresearch) for actin for 1 h at 25°C. After washing with TBS-T, signals were developed using an ECL Plus Western blot detection system (GE Healthcare).
NF-κB Reporter Assay
HEK293 cells expressing mTLR4/MD2-CD14 (Invivogen) were seeded at 4.0 × 104 cells/well in 96-well plate and transfected with 200 ng of the reporter plasmid pNiFty2SEAP which encodes NF-κB-inducible soluble form of embryonic alkaline phosphatase (AP) with 120 ng of either mCD22-pcDNA3.1(+) or empty vector. After 24 h incubation, 10 μg/ml of LPS were added to wells. After 16 h incubation, AP activity in the supernatant was measured following the manufacturer's instruction.
NF-κB DNA Binding Assay
Nuclear extracts of Cd22–/– and WT B cells were prepared using the Nuclear Extract Kit (Active Motif, Carlsbad, Calif., USA) following the manufacturer's instructions. Protein concentration was determined by BCA assay (Pierce) and 9.4 μg of nuclear extracts per well were subjected to ELISA using oligonucleotides containing the NF-κB-binding site and specific antibodies to determine which NF-κB subunits within the bound complexes were active (TransAM NF-κB Family Kit; Active Motif). Assays were run in triplicates.
Results and Discussion
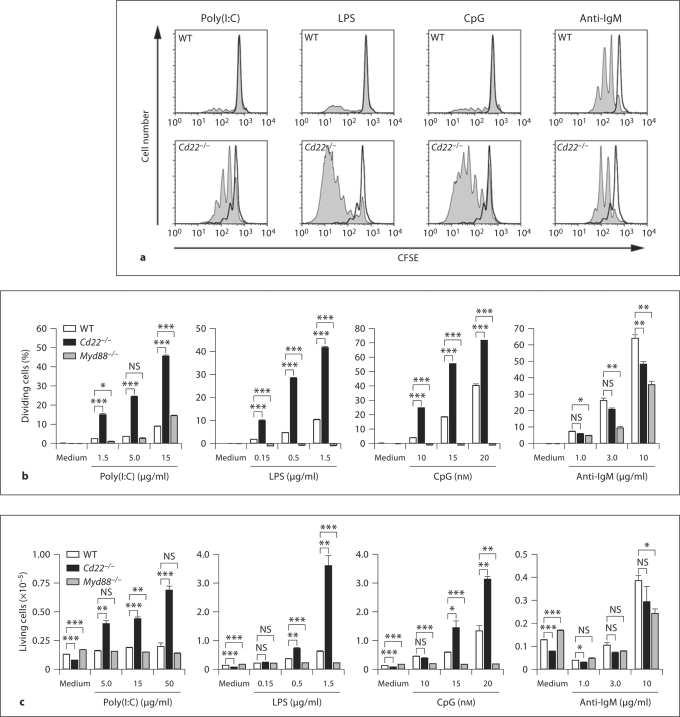
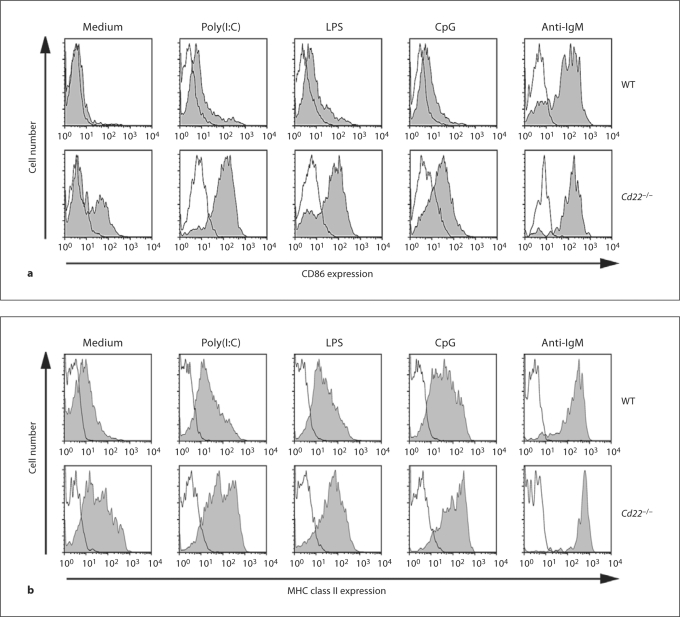
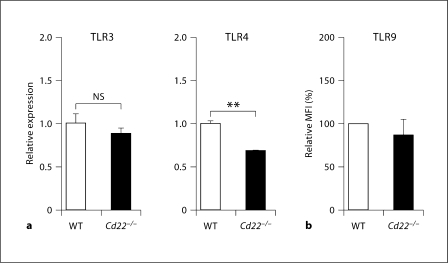
Although CD22 is known as a regulator of the BCR, several earlier reports showed that B cells from Cd22–/– mice exhibit hyperproliferation in response to LPS, a ligand of TLR4 [15,16,17], and more recently to ligands to TLR7 and TLR9 [18], which are localized to endosomes. However, the significance of these findings has been largely overlooked [1,2]. CD22 is known to undergo constitutive endocytosis [19], and is therefore localized to both the cell surface and endosomes, consistent with a potential role in regulation of these TLRs. As shown in figure 1, CFSE-labeled B cells from Cd22–/– mice exhibited hyperproliferation relative to WT B cells in response to ligands for TLRs 3, 4 and 9 (poly(I:C), LPS and CpG, respectively). Paradoxically, Cd22–/– B cells showed slightly impaired proliferation in response to anti-IgM, consistent with several previous reports [15,16,17]. Proliferation by LPS and CpG was completely MyD88 dependent, consistent with known dependence on MyD88 for signaling mediated by TLR4 and TLR9 (fig. 1b) [20], while signaling in response to poly(I:C) was unaffected by the MyD88 deficiency, since TLR3 mediates signaling through the TRIF pathway [20]. The CFSE dilution was concomitant with an increase in the absolute number of living Cd22–/– B cells at 3 days culture (fig. 1c). The augmented proliferation of Cd22–/– B cells was accompanied by increased induction of the co-stimulatory molecule CD86 and MHC class II, required for activation of T cells (fig. 2). The augmented TLRs 3, 4 and 9 signaling in Cd22–/– B cells was not a result of increased expression of TLRs, since expression of TLR3 and 9 in Cd22–/– B cells was similar to that in WT B cells, and Cd22–/– B cells express TLR4 at a lower level than WT B cells (fig. 3; online suppl. fig. 1, www.karger.com/doi/10.1159/000322375). Taken together, these data indicate that CD22 deficiency affects both MyD88-dependent and MyD88-independent TLR signaling in B cells.
Fig. 1.
CD22 deficiency causes hyperproliferation of B cells in response to TLR ligands. a CFSE-labeled B cells were cultured for 3 days in the absence (black) or presence (gray) of 5.0 μg/ml poly(I:C), 1.5 μg/ml LPS, 15 n M CpG and 3.0 μg/ml anti-IgM. Cells were analyzed by flow cytometry. b Percentage of dividing cells from 2 days' culture subtracted with the value in the absence of stimulation are shown. Statistical analyses were performed by Student's t test. * p < 0.05; * * p < 0.01; * * * p < 0.001. c Living cell number in the wells of 3 days' culture is shown. Before the analysis, 1.0 × 104 of beads was added to the culture well. Cells were then harvested and analyzed by flow cytometry. Living cell number was calculated by the formula: living cell number in the well = the number of living cells acquired × number of beads acquired / 1.0 × 104. Data are representative of 3 independent experiments with similar results.
Fig. 2.
Augmented CD86 and class II induction in Cd22–/– B cells in response to TLR ligands. WT and Cd22–/– B cells were stimulated for 2 days with 10 μg/ml poly(I:C), 0.15 μg/ml LPS, 7.5 n M CpG and 0.3 μg/ml anti-IgM. Then cells were harvested and stained with anti-CD86 (a) or anti-MHC class II I-A b (b) (gray) or isotype-matched control Ab (black). Stained cells were washed and analyzed by flow cytometry. Data are representative of 3 independent experiments with similar results.
Fig. 3.
TLR expression in Cd22–/– B cells is not significantly different from that of WT B cells. a Total RNA was extracted from isolated WT and Cd22–/– B cells. Expression of TLR3 and 4 was examined by quantitative real-time PCR. Expression levels were normalized to β-actin. Statistical analyses were performed by Student's t test. b Isolated B cells were fixed, permeabilized and then stained with anti-TLR9 Ab followed by R-phycoerythrin-labeled secondary Ab. Stained cells were washed and analyzed by flow cytometry. Geometric mean fluorescence intensity (MFI) of Cd22–/– B cells relative to WT is shown. Statistical analyses were performed by Student's t test. Data are representative or means of 3 independent experiments with similar results.
Several approaches were taken to investigate the possibility that the increased TLR signaling might be an indirect effect of CD22 on B cell development. We first examined the impact of the expression of CD22 on TLR signaling in the Cd22–/– J2-44 B cell line [21]. MHC class II induction by TLR ligands was reproducibly suppressed by expression of CD22 (fig. 4a; online suppl. fig. 2), consistent with a direct effect of CD22 on TLR signaling. We next sought to demonstrate the impact of CD22 on TLR signaling on WT B cells by antibody-mediated sequestration of CD22, an approach used to demonstrate the importance of CD22 co-localization on the regulation of BCR signaling [22]. In wells containing immobilized anti-CD22 mAb, WT B cells exhibited enhanced proliferation in response to TLR ligands relative to cells exposed to wells with immobilized isotype control Ab (fig. 4b; online suppl. fig. 3), suggesting sequestration of CD22 from TLRs 4 and 9 by the immobilized anti-CD22 mAb augments proliferation by TLR ligands. Furthermore, using a TLR4 reporter cell line, we found that the transcriptional activity of NF-κB, which is a hallmark of TLR signaling [20], is well suppressed by the ectopic expression of CD22 in TLR4 reporter cells (fig. 4c; online suppl. fig. 4), suggesting that CD22 has an intrinsic inhibitory effect on NF-κB activation. Thus, three independent approaches suggest that CD22 directly regulates TLR signaling in B cells. To gain further insight into the mechanism of negative regulation of TLR signaling by CD22, we tested whether hallmarks of TLR signaling pathways including p38, JNK and NF-κB signaling were augmented in Cd22–/– B cells. We found that p38 and JNK phosphorylation was similar in Cd22–/– and WT B cells (online suppl. fig. 5). While the IκBα degradation which results in the release of NF-κB dimers into nucleus, preferably p65/p50, is not altered in Cd22–/– B cells (online suppl. fig. 4), we found that the repertoire of active NF-κB subunits in Cd22–/– B cells in both resting and CpG-stimulated conditions was significantly biased to the nonclassical NF-κB pathways revealed by higher DNA-binding activity of p50, p52 and RelB than that in WT B cells (fig. 5a) [23]. With respect to inhibitory regulators of TLR activation, we found that induction of SOCS1 and 3, known as suppressors of TLR signaling [12,13,14], was impaired in Cd22–/– B cells (fig. 5b). It is interesting to speculate that altered NF-κB activation caused by CD22 deficiency results in poor induction of SOCS1 and 3, leading to the prolonged TLR signaling, which in turn results in the augmented phenotype of Cd22–/– B cells.
Fig. 4.
CD22 negatively regulates TLR signaling in B cells. a Expression of CD22 in J2-44 Cd22–/– B cells suppresses induction of MHC II by TLR ligands. J2-44 cells were retrovirally transduced with mouse CD22 and stimulated with 3.0 μg/ml LPS, 15 n M CpG and 3.0 μg/ml anti-IgM. After 2 days, MHC class II expression was examined by flow cytometry as in figure 2. b Sequestration of CD22 by immobilized anti-CD22 Ab enhances B cell proliferation by TLR ligands. CFSE-labeled WT B cells were stimulated with soluble LPS, CpG and anti-IgM in the presence of immobilized anti-CD22 mAb or isotype-matched control Ab. After 2 days, cells were analyzed by flow cytometry. c Expression of CD22 inhibits NF-κB activation in a TLR4 reporter cell line. mTLR4/ HEK293 cells were transfected with NF-κB-inducible AP and mCD22 or empty vector and stimulated with 10 μg/mL of LPS. The culture supernatant was incubated with the AP substrate at 37°C. Then the enzymatic activity was measured by the absorbance of 405 nm. Background absorbance from culture supernatant only was subtracted. Statistical analyses were performed by Student's t test. Data are representative of 2 or 3 independent experiments with similar results.
Fig. 5.
Signaling analysis of Cd22–/– B cells. a CD22 deficiency results in different spectrum of NF-κB activation. DNA binding activity of the NF-κB subunits, p50, p52, p65 and RelB in the nuclear extracts from WT and Cd22–/– B cells was assayed by ELISA using oligonucleotides containing the NF-κB-binding site and specific antibodies to each subunit. b Impaired induction of SOCS1 and 3 in Cd22–/– B cells upon CpG stimulation. Total RNA was extracted from 1.0 × 107 of B cells from WT or Cd22–/– B cells stimulated with 250 n M CpG for 3 h or unstimulated. Expression of SOCS1 and 3 was examined by quantitative real-time PCR. Expression levels were normalized to β-actin. Statistical analyses were performed by Student's t test. Data are representative of 2 independent experiments with similar results.
Our results suggest that CD22 is a constitutive negative regulator of TLR-activated signaling pathways that are distinct from and not mediated by the BCR [7]. Recently, Jellusova et al. [18] also reported the augmented proliferation of Cd22–/– and Siglecg–/– B cells in response to TLR ligand. Results presented here suggest that the augmented TLR signaling in Cd22–/– B cells is a consequence of the loss of constitutive negative regulation of TLR signaling by CD22 in B cells. This conclusion is relevant to the observation that mice carrying a TLR7 gene duplication (Y-linked autoimmune accelerator) exhibit an anti-DNA antibody autoimmune phenotype when crossed into a Cd22+/– background [24].
Our data document that CD22 is a regulator of receptors that mediate both adaptive and innate immune responses of the B cell. In its regulation of the BCR, CD22 is believed to exert its activity through close physical association [3,4,25]. Since sequestration of CD22 augmented LPS activation of TLR4 signaling, this is also likely the case for CD22 regulation of TLRs. In this regard, since TLRs are located on both the cell surface (TLR4) and endosomal compartments (TLR3 and 9), CD22 regulation of TLRs might be tied to its constitutive endocytic activity, placing it both on the cell surface and in endosomal compartments [19,26].
CD22 is a member of the Siglec family, which comprises 14 human and 9 murine members that are predominantly expressed on leukocytes that mediate the adaptive and innate arms of immune function [1]. A few reports suggest that other Siglecs might regulate TLRs. Recently Siglec-E and H were found to regulate cytokine production in response to TLR ligand in macrophages and plasmacytoid dendritic cells, respectively [27,28]. Similarly, ectopic expression of Siglec-9, 11 and 14 in macrophages has also been observed to influence cytokine production in response to TLR ligands [29,30,31]. Thus as shown here for CD22, it is likely that other Siglecs will be found to regulate TLR signaling in the cells that express them, and that this will prove to be a general property of the Siglec family.
Disclosure Statement
The authors declare no competing financial interests.
Supplementary Material
Supplemental Information
Acknowledgements
We would like to thank Bao Duong, Bruce Beutler, David Nemazee, Henry Wortis, Hua Tian, Katharina Brandl, Kazuo Yamamoto, Lars Nitschke, Takayuki Ota, Toshio Kitamura for mice, reagents and insightful discussions, the flow cytometry core facility in The Scripps Research Institute for cell sorting, and Anna Tran-Crie for her help in manuscript preparation. C.R. was supported by an EMBO long-term fellowship. This work was supported by NIH grant AI050143 to J.C.P.
References
- 1.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 2.Walker JA, Smith KG. CD22: an inhibitory enigma. Immunology. 2008;123:314–325. doi: 10.1111/j.1365-2567.2007.02752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duong BH, Tian H, Ota T, Completo G, Han S, Vela JL, Ota M, Kubitz M, Bovin N, Paulson J, Nemazee D. Decoration of T-independent antigen with ligands for CD22 and Siglec-G can suppress immunity and induce B cell tolerance in vivo. J Exp Med. 2010;207:173–187. doi: 10.1084/jem.20091873. S171–S174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Courtney AH, Puffer EB, Pontrello JK, Yang ZQ, Kiessling LL. Sialylated multivalent antigens engage CD22 in trans and inhibit B cell activation. Proc Natl Acad Sci USA. 2009;106:2500–2505. doi: 10.1073/pnas.0807207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanoue A, Batista FD, Stewart M, Neuberger MS. Interaction of CD22 with α2,6-linked sialoglycoconjugates: innate recognition of self to dampen B cell autoreactivity? Eur J Immunol. 2002;32:348–355. doi: 10.1002/1521-4141(200202)32:2<348::AID-IMMU348>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Meyer-Bahlburg A, Rawlings DJ. B cell autonomous TLR signaling and autoimmunity. Autoimmun Rev. 2008;7:313–316. doi: 10.1016/j.autrev.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patterson HC, Kraus M, Kim YM, Ploegh H, Rajewsky K. The B cell receptor promotes B cell activation and proliferation through a non-ITAM tyrosine in the Igalpha cytoplasmic domain. Immunity. 2006;25:55–65. doi: 10.1016/j.immuni.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Pasare C, Medzhitov R. Control of B-cell responses by Toll-like receptors. Nature. 2005;438:364–368. doi: 10.1038/nature04267. [DOI] [PubMed] [Google Scholar]
- 9.Gavin AL, Hoebe K, Duong B, Ota T, Martin C, Beutler B, Nemazee D. Adjuvant enhanced antibody responses in the absence of toll-like receptor signaling. Science. 2006;314:1936–1938. doi: 10.1126/science.1135299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckl-Dorna J, Batista FD. BCR-mediated uptake of antigen linked to TLR9 ligand stimulates B-cell proliferation and antigen-specific plasma cell formation. Blood. 2009;113:3969–3977. doi: 10.1182/blood-2008-10-185421. [DOI] [PubMed] [Google Scholar]
- 11.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naka T, Fujimoto M, Tsutsui H, Yoshimura A. Negative regulation of cytokine and TLR signalings by SOCS and others. Adv Immunol. 2005;87:61–122. doi: 10.1016/S0065-2776(05)87003-8. [DOI] [PubMed] [Google Scholar]
- 13.Dalpke AH, Opper S, Zimmermann S, Heeg K. Suppressors of cytokine signaling (SOCS)-1 and SOCS-3 are induced by CpG-DNA and modulate cytokine responses in APCs. J Immunol. 2001;166:7082–7089. doi: 10.4049/jimmunol.166.12.7082. [DOI] [PubMed] [Google Scholar]
- 14.Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB, Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131:1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 15.Nitschke L, Carsetti R, Ocker B, Kohler G, Lamers MC. CD22 is a negative regulator of B-cell receptor signalling. Curr Biol. 1997;7:133–143. doi: 10.1016/s0960-9822(06)00057-1. [DOI] [PubMed] [Google Scholar]
- 16.Sato S, Miller AS, Inaoki M, Bock CB, Jansen PJ, Tang ML, Tedder TF. CD22 is both a positive and negative regulator of B lymphocyte antigen receptor signal transduction: altered signaling in CD22-deficient mice. Immunity. 1996;5:551–562. doi: 10.1016/s1074-7613(00)80270-8. [DOI] [PubMed] [Google Scholar]
- 17.Otipoby KL, Andersson KB, Draves KE, Klaus SJ, Farr AG, Kerner JD, Perlmutter RM, Law CL, Clark EA. CD22 regulates thymus-independent responses and the lifespan of B cells. Nature. 1996;384:634–637. doi: 10.1038/384634a0. [DOI] [PubMed] [Google Scholar]
- 18.Jellusova J, Wellmann U, Amann K, Winkler TH, Nitschke L. CD22 × Siglec-G double-deficient mice have massively increased B1 cell numbers and develop systemic autoimmunity. J Immunol. 2010;184:3618–3627. doi: 10.4049/jimmunol.0902711. [DOI] [PubMed] [Google Scholar]
- 19.Shan D, Press OW. Constitutive endocytosis and degradation of CD22 by human B cells. J Immunol. 1995;154:4466–4475. [PubMed] [Google Scholar]
- 20.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Jin L, McLean PA, Neel BG, Wortis HH. Sialic acid binding domains of CD22 are required for negative regulation of B cell receptor signaling. J Exp Med. 2002;195:1199–1205. doi: 10.1084/jem.20011796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doody GM, Justement LB, Delibrias CC, Matthews RJ, Lin J, Thomas ML, Fearon DT. A role in B cell activation for CD22 and the protein tyrosine phosphatase SHP. Science. 1995;269:242–244. doi: 10.1126/science.7618087. [DOI] [PubMed] [Google Scholar]
- 23.Chen LF, Greene WC. Shaping the nuclear action of NF-κB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 24.Mary C, Laporte C, Parzy D, Santiago ML, Stefani F, Lajaunias F, Parkhouse RM, O'Keefe TL, Neuberger MS, Izui S, Reininger L. Dysregulated expression of the Cd22 gene as a result of a short interspersed nucleotide element insertion in Cd22a lupus-prone mice. J Immunol. 2000;165:2987–2996. doi: 10.4049/jimmunol.165.6.2987. [DOI] [PubMed] [Google Scholar]
- 25.Hokazono Y, Adachi T, Wabl M, Tada N, Amagasa T, Tsubata T. Inhibitory coreceptors activated by antigens but not by anti-Ig heavy chain antibodies install requirement of costimulation through CD40 for survival and proliferation of B cells. J Immunol. 2003;171:1835–1843. doi: 10.4049/jimmunol.171.4.1835. [DOI] [PubMed] [Google Scholar]
- 26.Tateno H, Li H, Schur MJ, Bovin N, Crocker PR, Wakarchuk WW, Paulson JC. Distinct endocytic mechanisms of CD22 (Siglec-2) and Siglec-F reflect roles in cell signaling and innate immunity. Mol Cell Biol. 2007;27:5699–5710. doi: 10.1128/MCB.00383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyd CR, Orr SJ, Spence S, Burrows JF, Elliott J, Carroll HP, Brennan K, Ni Gabhann J, Coulter WA, Jones C, Crocker PR, Johnston JA, Jefferies CA. Siglec-E is up-regulated and phosphorylated following lipopolysaccharide stimulation in order to limit TLR-driven cytokine production. J Immunol. 2009;183:7703–7709. doi: 10.4049/jimmunol.0902780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blasius A, Vermi W, Krug A, Facchetti F, Cella M, Colonna M. A cell-surface molecule selectively expressed on murine natural interferon-producing cells that blocks secretion of interferon-α. Blood. 2004;103:4201–4206. doi: 10.1182/blood-2003-09-3108. [DOI] [PubMed] [Google Scholar]
- 29.Ando M, Tu W, Nishijima K, Iijima S. Siglec-9 enhances IL-10 production in macrophages via tyrosine-based motifs. Biochem Biophys Res Commun. 2008;369:878–883. doi: 10.1016/j.bbrc.2008.02.111. [DOI] [PubMed] [Google Scholar]
- 30.Yamanaka M, Kato Y, Angata T, Narimatsu H. Deletion polymorphism of SIGLEC14 and its functional implications. Glycobiology. 2009;19:841–846. doi: 10.1093/glycob/cwp052. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Neumann H. Alleviation of neurotoxicity by microglial human Siglec-11. J Neurosci. 2010;30:3482–3488. doi: 10.1523/JNEUROSCI.3940-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information