Summary
Increased mitochondrial biogenesis by activation of PPAR- or AMPK/PGC-1α-dependent homeostatic pathways has been proposed as a treatment for mitochondrial disease. We tested this hypothesis on three recombinant mouse models characterized by defective cytochrome c-oxidase (COX) activity: a knockout (KO) mouse for Surf1, a knockout/knockin mouse for Sco2, and a muscle-restricted KO mouse for Cox15. First, we demonstrated that double-recombinant animals overexpressing PGC-1α in skeletal muscle on a Surf1 KO background showed robust induction of mitochondrial biogenesis and increase of mitochondrial respiratory chain activities, including COX. No such effect was obtained by treating both Surf1−/− and Cox15−/− mice with the pan-PPAR agonist bezafibrate, which instead showed adverse effects in either model. Contrariwise, treatment with the AMPK agonist AICAR led to partial correction of COX deficiency in all three models, and, importantly, significant motor improvement up to normal in the Sco2KO/KI mouse. These results open new perspectives for therapy of mitochondrial disease.
Highlights
► AICAR partially corrects COX deficiency in three mouse models ► Bezafibrate, a pan-PPAR activator, failed to correct COX deficiency in vivo ► Motor performance improves under AICAR in one mouse model ► Activation of the AMPK/Pgc1α axis can correct OXPHOS defects in vivo
Introduction
Present in virtually all eukaryotes, mitochondria are the major source of the high-energy phosphate molecule adenosine triphosphate (ATP), which is synthesized by the mitochondrial respiratory chain (MRC) through the process of oxidative phosphorylation (OXPHOS) (Wallace, 2005). ATP is the principal energy substrate required for all active processes within the cell, and ATP deficiency leads to cellular dysfunction and ultimately cell death. A host of pathways and enzymatic functions residing within mitochondria can modify and influence OXPHOS, and OXPHOS abnormalities can in turn generate signals that trigger either homeostatic pathways, e.g., mitochondrial biogenesis, or execution programs, e.g., mitophagy, which eliminates single dysfunctional organelles, or apoptosis, which eliminates the whole dysfunctional cell (Goldman et al., 2010).
A complex, finely tuned homeostatic system senses the dynamic balance between energy demand and expenditure and regulates the biogenesis, activity, and coupling of mitochondria, according to type, function, and metabolic status of cells and tissues (Wallace and Fan, 2010). In mammals, a master regulator of mitochondrial biogenesis is PPAR-gamma Coactivator 1 alpha (PGC-1α), a transcriptional coactivator that is physiologically induced or activated by conditions of shortage of, or increased demand for, energy, such as cold, physical activity and fasting (Handschin and Spiegelman, 2006). PGC-1α acts by interacting with, and increasing the activity of, a number of transcription factors that in turn induce genes directly or indirectly related to mitochondrial proliferation, oxidative catabolism, and respiration. For instance, PGC-1α interacts with, and potentiates the function of, nuclear respiratory factors (NRF1 and NRF2) and α, β/δ, and γ isoforms of the peroxisomal proliferator activated receptors (PPARs). Both NRF1 and NRF2 induce the expression of several nucleus-encoded subunits of MRC complexes, and of genes involved in replication and transcription of mtDNA, including Tfam (Kelly and Scarpulla, 2004). PPARs are tissue-specific, or ubiquitously expressed, nuclear receptors of fatty acids, which act as their activating ligands. The principal action of PPARs is to induce transcription of genes encoding enzymes involved in cellular and organelle uptake of fatty acids and fatty acid oxidation (FAO), in both mitochondria and peroxisomes (Lefebvre et al., 2006; Barish et al., 2006). PGC-1α is activated by posttranslational modifications, including phosphorylation and deacetylation. Phosphorylation at PGC-1α T177 and S538 residues is operated by the AMP-dependent kinase (AMPK) (Jäger et al., 2007). Again, this activation responds to a physiological homeostatic mechanism, since ATP and AMP are part of the same nucleotide pool, their levels being inversely related, so that when shortage of the former occurs, the latter increases, and vice versa. As a consequence of increased AMP levels, reflecting low ATP availability and low energy reserve, AMPK is activated, resulting in a cascade of phosphorylation-dependent adaptive modifications of several factors, including PGC-1α, that ultimately determine the shutdown of anabolic, energy-consuming processes, whereas catabolic, energy-producing pathways, such as FAO and MRC, are switched on. AMPK activates PGC-1α not only by direct phosphorylation but also by indirect means, for instance by activating the NAD+-dependent deacetylase sirtuin 1, Sirt1, which in turn increases the amount of deacetylated, active form of PGC-1α (Cantó et al., 2009). Sirt1 activation by AMPK is mainly due to increased NAD+/NADH ratio, a condition that stimulates Sirt1, as a consequence of AMPK-mediated increased expression of FAO (and OXPHOS) genes, and also of the Nampt gene, encoding the enzyme that converts nicotinamide into NAD+ (Cantó et al., 2009). AMPK is a heterotrimer consisting of a catalytic α subunit and two regulatory subunits, β and γ. The latter subunits exist as multiple isoforms, giving 12 different possible combinations of holoenzyme with different tissue distribution and subcellular localization. AMPK is allosterically stimulated by AMP and is itself regulated by phosphorylation via two main upstream AMPK kinases (Cantó and Auwerx, 2010; Steinberg and Kemp, 2009). The major regulatory phosphorylation site of AMPK has been identified as T172 within the catalytic loop between the DFG and APE motifs of the α subunit (Fogarty et al., 2010). Besides physiological conditions, PGC-1α and PPARs can also be stimulated or induced pharmacologically. For instance, bezafibrate is a pan-PPAR agonist that is widely used to treat the metabolic syndrome and hyperlipidemias (Tenenbaum et al., 2005). Resveratrol, a natural polyphenol compound, activates PGC-1α via Sirt1 (Lagouge et al., 2006), and 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR) does so by generating inosine monophosphate, which acts as an AMPK agonist by mimicking AMP (Corton et al., 1995; Merrill et al., 1997).
Mitochondrial disorders are clinical entities associated with defects of OXPHOS, which are ultimately genetically determined (Wallace, 2010). Mitochondrial disorders can be determined by mutations in mtDNA, or in OXPHOS-related nuclear genes (Zeviani and Di Donato, 2004). Many pathogenic mtDNA mutations are heteroplasmic, i.e., they coexist with a variable percentage of wild-type mtDNA. Clinical symptoms typically ensue when the amount of mutant mtDNA offsets a critical threshold, usually >50%–60% of total mtDNA, below which the mutation has virtually no clinical impact. Likewise, most of the mitochondrial syndromes due to mutations in nuclear genes behave as recessive traits and in many cases are determined either by mutations in “ancillary” proteins that play an arguably relevant, but nevertheless not essential, often partially redundant, role in OXPHOS, or by hypomorphic mutant alleles encoding a crippled but still partially functional protein. These observations have suggested the hypothesis that the deleterious effects of either mtDNA- or nuclear-gene mutations could be overcome, at least partially, by increasing the total amount of mitochondria and/or of functionally active “MRC units,” via activation of the mitochondrial biogenesis program in critical tissues (Bastin et al., 2008; Wenz et al., 2008). This idea has first been tested in cells from mitochondrial disease patients, by overexpressing recombinant PGC-1α or by adding bezafibrate in the medium (Bastin et al., 2008; Srivastava et al., 2009). Both treatments showed mutant cells to partially recover MRC activities and increase ATP production. Oral bezafibrate has later been reported to ameliorate the clinical conditions and the biochemical impairment of a cre-lox murine model of COX-defective myopathy, determined by muscle-specific ablation of Cox10, the heme b farnesyl transferase, by expressing the cre recombinase under the myosin light-chain 1F (MLC1F) promoter. Here, we report the results of experimental treatments carried out on three other mouse models of COX deficiency: (1) a Surf1 constitutive knockout (KO) mouse, (2) a muscle-specific Cox15 KO mouse, and (3) a constitutive knockout/knockin Sco2 mouse (Sco2KO/KI) (Yang et al., 2010) carrying a null Sco2 allele and a second Sco2 allele with a missense mutation E129K, corresponding to the E140K recurrent, pathogenic change in SCO2 mutant patients. Absence of Surf1, an assembly factor of COX, causes severe COX deficiency and fatal Leigh syndrome in children, but milder COX deficiency, and hardly any clinical sign, in mice (Dell'Agnello et al., 2007). Mutations in Cox15, the key enzyme for the conversion of heme o into heme a (Khalimonchuk and Rödel, 2005), lead to fatal encephalocardiomyopathy in children (Antonicka et al., 2003; Bugiani et al., 2005) and severe myopathy with marked COX deficiency in muscle-restricted KO mice (this paper). Sco2 is a metallochaperone involved in copper metallation of COX (Leary et al., 2009), mutations in which lead to infantile fatal encephalocardiomyopathy (Papadopoulou et al., 1999).
Results
Overexpression of PGC-1α Increases MRC Activities
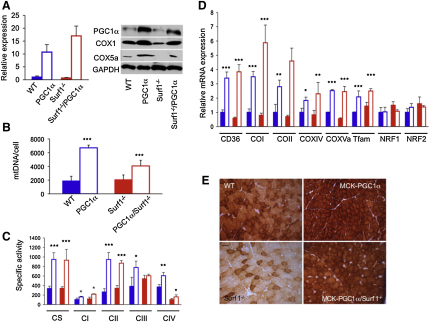
We first investigated a transgenic mouse that overexpresses PGC-1α under the promoter of the skeletal-muscle-specific creatine-kinase (MCK) gene (MCK-PGC-1α). Muscle-restricted overexpression of the PGC-1α gene in the transgenic individuals was 10.7 ± 2.9-fold that of controls, resulting in a 4-fold steady-state increase in the amount of the PGC-1α protein (Figure 1A). The MCK-PGC-1α mice are vital, with hardly any clinically relevant features; however, they did show a 3- to 4-fold increase in both muscle mtDNA content (from 1900 ± 674 to 6706 ± 394 copies/cell) and citrate synthase (CS) activity in muscle homogenate (from 337.9 ± 48.6 to 948.4 ± 139.3 nmol/min/mg of protein), two indexes of mitochondrial mass, compared to WT littermates (Figures 1B and 1C). We also determined the expression levels of the immediate targets of PGC-1α, i.e., Nrf1 and Nrf2, and of several downstream targets of the latter factors, including Tfam, COI, COII, COXIV, and COX5a (Figure 1D). The mRNA levels of both Nrf genes remained unchanged, whereas the downstream genes were all overexpressed 2- to 3-fold in MCK-PGC-1α muscle samples, compared to those of WT littermates. These findings are fully compatible with the role of PGC-1α as a coactivator that increases the transcriptional activity of several factors, including the Nrf's, but is not a transcription factor by itself. Increased expression of COX subunit transcripts was correlated to increased amount of the corresponding proteins, demonstrated by quantitative western blot (WB) immunovisualization (Figure 1A), which resulted in a 1.5- to 2-fold increase in the specific activities of MRC complexes, cI cII, cIII, and cIV (Figure 1C).
Figure 1.
Effects of PGC-1α Overexpression in Surf1−/− Mice
(A) Expression analysis of PGC-1α. (Left panel) The PGC-1α transcript from muscle of each mouse genotype (three animals/genotype) was retrotranscribed into cDNA, normalized to that of the Hprt gene, and expressed as time-fold variations relative to the values obtained from wild-type (WT) animals. Solid blue, WT; blue outline, PGC-1α: MCK-PGC-1α transgenic mouse; solid red, Surf1−/−, constitutive Surf1 knockout mouse; red outline, Surf1−/−/PGC-1α: Surf1−/−/MCK-PGC-1α double mutant mouse. Error bars represent the standard deviation (SD). (Right panel) Western blot immunovisualization of skeletal muscle proteins of the different genotypes, listed as above. Densitometric analysis of each band, normalized against that of GAPDH, revealed the following variations, relative to WT. (1) PGC-1α, 4.3 in MCK-PGC-1α; 0.5 in Surf1−/−; 2.2 in Surf1−/−/PGC-1α. (2) COX1, 1.8 in MCK-PGC-1α; 0.2 in Surf1−/−; 1.0 in Surf1−/−/PGC-1α. (3) COX5a, 5.0 in MCK-PGC-1α; ∼0.0 in Surf1−/−; 1.0 in Surf1−/−/PGC-1α.
(B) MtDNA analysis in different genotypes (three animals/genotype). Color code is as in (A). MtDNA is expressed as number of DNA molecules per cell (Cree et al., 2008). Error bars represent SD.
(C) MRC activities in the different genotypes (three animals/genotype), expressed as nmoles/min/mg of protein. Note that the activity of cII has been multiplied by 10 for visualization clarity. Color code is as in (A). CS, citrate synthase; CI-IV, MRC complexes I–IV. Error bars represent the standard deviation (SD). The asterisks represent the significance levels calculated by unpaired, Student's two-tailed t test: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(D) Expression analysis of FAO- and OXPHOS-related genes in WT, Surf1−/−, MCK-PGC-1α, MCK-PGC-1α/Surf1−/− muscles of mice. The levels of the gene transcripts, retrotranscribed into cDNA, were normalized to that of the Hprt gene transcript and expressed as time-fold variation relative to the WT. Error bars represent SD. The asterisks represent the significance levels calculated by unpaired, Student's two-tailed t test: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(E) COX staining in muscles from different mouse lines including wild-type (WT), Surf1−/−, MCK-PGC1α, and MCK-PGC1α/Surf1−/− individuals. Note that the reaction is increased in the PGC1α overexpressing mouse models compared to the corresponding naive models. Scale bar, 25 μm, magnification 20×.
PGC-1α is also known to activate factors, e.g., PPARs, that increase the cellular and organelle uptake, and the catabolic oxidation, of fatty acids (FAO). This effect was reflected by a 3.5-fold increased expression in MCK-PGC-1α mouse muscle of CD36/FAT, a gene encoding the plasma membrane transporter of fatty acids (Figure 1D).
In order to test whether PGC-1α overexpression may correct specific MRC defects in vivo, we then carried out suitable crosses that transferred the MCK-PGC-1α allele into a COX-defective model due to the constitutive ablation of Surf1. Surf1 is a putative assembly factor specific to COX, its absence being associated with profound COX deficiency in humans (≤10%–15% of the lower normal limit in different cell types) and, albeit milder, in mice as well (typically 30%–40% of the lower normal limit). While the loss of Surf1 causes early onset, invariably fatal Leigh encephalopathy in patients, recombinant null mice showed hardly any clinically deleterious consequence, and standard treadmill test showed no locomotor impairment.
The skeletal muscle of Surf1−/−/MCK-PGC-1α double mutant mice did show the same effects detected in MCK-PGC-1α transgenic muscles: increased mtDNA content (from 2078 ± 693 to 4079 ± 824 copies/cell) and CS activity (from 343 ± 33 to 934 ± 226 nmol/min/mg) (Figures 1B and 1C), increased levels of CD36/FAT transcript, and increased levels of COX-specific mRNA and protein subunits (Figures 1A and 1D). These changes resulted in (1) partial but significant recovery of COX specific activity (1.6-fold, from 104 ± 24 to 162 ± 42 nmol/min/mg; p = 0.02) (Figure 1C), which nevertheless remained well below the normal range (370 ± 49 nmol/min/mg); and (2) increased histochemical reaction to COX (Figure 1E). Taken together, these data suggest that overexpression of PGC-1α induces mitochondrial biogenesis and can partially correct a specific defect of COX in vivo.
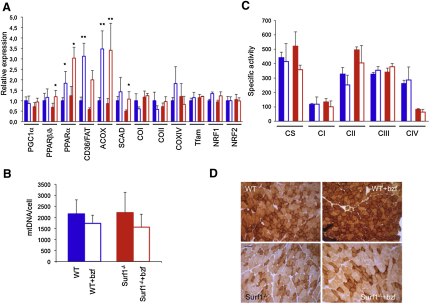
Bezafibrate Induces Expression of FAO- but Not OXPHOS-Related Genes
In a second set of experiments, we then tested the effects of PPAR activation in the Surf1−/− mouse model. To this end, the same genes, proteins, and activities measured in the MCK-PGC-1α and Surf1−/−/MCK-PGC-1α mouse models were also measured in Surf1−/− and WT littermates exposed to 0.5% bezafibrate, mixed in standard food for 1 month (Wenz et al., 2008). We observed significant body weight loss during the treatment and marked hepatomegaly at the end of treatment, in both Surf1−/− and WT animals (see Figure S1 available online), indicating toxicity of this drug in mice. As expected, the expression of PPAR β/δ and PPAR α, the two PPAR isoforms present in skeletal muscle, was significantly increased in treated animals, along with FAO-related genes, including CD36/FAT, ACOX, encoding acylCoA oxidase, and SCAD, encoding short-chain acylCoA dehydrogenase (Figure 2A). Taken together, these data clearly demonstrated that bezafibrate treatment was indeed effective on the main targets specific for this drug in vivo. Contrariwise, no significant change in mtDNA content, CS, and MRC activities was measured in treated versus untreated Surf1−/− or WT muscles (Figures 2B and 2C), and the histochemical reaction to COX was equally defective in both treated and untreated Surf1−/− specimens (Figure 2D). Accordingly, the expression of PGC-1α, both NRFs, and Tfam remained unchanged among the different groups of animals, as did several COX subunit transcripts, from either nuclear (e.g., COXIV) or mtDNA (COI and COII) genes (Figure 2A).
Figure 2.
Bezafibrate Treatment on the Surf1−/− Mouse Model
(A) Expression analysis of FAO- and OXPHOS-related genes in Surf1−/− and WT muscles of bezafibrate (bzf)-treated and untreated mice. Solid blue, WT, untreated; blue outline, bzf-treated WT; solid red, Surf1−/−, untreated; red outline, bzf-treated Surf1−/−. The levels of the gene transcripts, retrotranscribed into cDNA, were normalized to that of the Hprt gene transcript and expressed as time-fold variation relative to the WT. Error bars represent SD. The asterisks represent the significance levels calculated by unpaired, Student's two-tailed t test: ∗p < 0.05; ∗∗p < 0.01.
(B) MtDNA content analysis in the different genotypes. The color codes are as in (A). Error bars represent SD. MtDNA is expressed as number of DNA molecules per cell (Cree et al., 2008).
(C) MRC activities in the different genotypes, expressed as nmoles/min/mg of protein. Note that the activity of cII has been multiplied by 10 for visualization clarity. Color code is as in (A).
(D) COX staining in muscles from Surf1−/− and WT muscles of bzf-treated and untreated mice. Scale bar, 25 μm, magnification 20×.
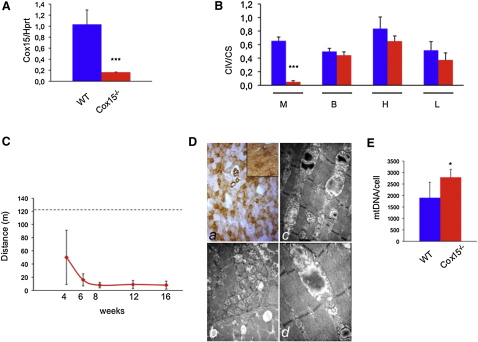
A Muscle-Specific Cox15 KO Mouse Displays COX-Defective Mitochondrial Myopathy
Since our results failed to confirm those on bezafibrate treatment reported in the MLC1F-Cox10−/− mouse model (Wenz et al., 2008), we created a KO mouse for the Cox15 gene, using the cre-lox technology (Figures S2A and S2B). The Cox15 gene product is the enzyme that comes next to Cox10 in the biosynthetic pathway of heme a, the COX-specific heme moiety. Cox10 adds a farnesyl chain to heme b, converting it into heme o, which can then be inserted into the protein backbone of nascent COX. Cox15 eventually converts heme o into functionally active heme a by oxidation of the C8-methyl group (Khalimonchuk and Rödel, 2005). The constitutive Cox15−/− animals, obtained by crossing Cox15loxP/loxP mice with a general cre-deleter transgenic mouse, showed consistent embryonic lethality at ≤7.5 dpc (not shown). We then produced a muscle-specific Cox15−/− model by crossing Cox15loxP/loxP with a transgenic mouse expressing the cre recombinase under the control of the human skeletal muscle-specific α-actin (ACTA1) promoter (ACTA-Cox15−/− mouse).
Recombination of the Cox15 locus by the cre recombinase was detected only in skeletal muscle and, in trace amount, in the heart, whereas no recombination occurs in other tissues (Figure S2C). Accordingly, the amount of Cox15 mRNA in ACTA-Cox15−/− mouse skeletal muscle was ≤16% than that of WT muscle (Figure 3A), and so was the COX/CS ratio (0.05 ± 0.02 in ACTA-Cox15−/− versus 0.66 ± 0.06 in WT), whereas there was no significant difference in other tissues (Figure 3B). Albeit born at the expected mendelian ratio, the ACTA-Cox15−/− mice were smaller than WT littermates at 30 days after birth (data not shown) and had significantly reduced motor performance measured by a standard treadmill test, initially administered to 4-week-old individuals, and then repeated at 6, 8, 12, and 16 weeks (Figure 3C). Histochemical analysis in skeletal muscle confirmed severe reduction of COX and compensatory proliferation of aberrant mitochondria (Figure 3D), which was confirmed by increased mtDNA content (Figure 3E), increased cII and CS activities (Table S1), and increased SDH histochemical reaction (data not shown), in ACTA-Cox15−/− muscles. COX histochemistry was normal in other tissues (data not shown). Except for cII activity, which increased from 24.2 ± 4.2 in WT to 34.9 ± 7.8 in ACTA-Cox15−/− muscle homogenate, other MRC activities, including cI and cIII, in muscle (Table S1), and all the MRC activities in nonmuscle tissues (data not shown), of ACTA-Cox15−/− were comparable to those of WT littermates.
Figure 3.
Characterization of the ACTA-Cox15−/− Mouse Model
(A) Cox15 transcript levels in skeletal muscle. The Cox15-specific mRNA content is ∼15% in ACTA-Cox15−/− relative to the parental Cox15lox/lox mouse line, considered as wild-type (WT). The levels of the Cox15 gene transcripts, retrotranscribed into cDNA, were normalized to that of the Hprt gene transcript and expressed as time-fold variation relative to the WT. Three animals were used for each genotype. Error bars represent SD. The asterisks represent the significance levels calculated by unpaired, Student's two-tailed t test: ∗∗∗p < 0.001.
(B) COX (CIV)-specific activity normalized to that of citrate synthase (CS) in tissues from ACTA-Cox15−/− mice. M, muscle; B, brain; H, heart; L, liver. Error bars represent SD. The asterisks represent the significance levels calculated by unpaired, Student's two-tailed t test, ∗∗∗p < 0.001.
(C) Treadmill analysis of ACTA-Cox15−/− mice at different time points. The dotted line indicates the lower limit of the motor performance of the parental Cox15lox/lox mouse line, considered as WT. Note the rapidly progressive reduction of motor endurance in the ACTA-Cox15−/− mouse line over time. Six animals were used for each genotype. Error bars represent SD.
(D) (Top) COX staining from ACTA-Cox15−/− skeletal muscle. The inset shows the reaction in a Cox15lox/lox (WT) muscle, for comparison. Magnification, 20×. (Bottom) Electron microscopy analysis of ACTA-Cox15−/− skeletal muscle. Note the subsarcolemmal accumulation of mitochondria in (Db) (magnification, 7000×) and the presence of an abnormal mitochondria characterized by disruption of the inner membrane and accumulation of electron dense material in (Dc) and (Dd) (magnification, 16,000× and 20,000×, respectively).
(E) MtDNA content in Cox15lox/lox (WT) and ACTA-Cox15−/− skeletal muscles. Error bars represent SD. The asterisk represents the significance levels calculated by unpaired, Student's two-tailed t test, ∗p < 0.05.
Bezafibrate Is Highly Toxic to ACTA-Cox15−/− Mice
We then tested the response of eight ACTA-Cox15−/− female mice to bezafibrate, according to the same protocol used on the Surf1−/− and MLC1F-Cox10−/− models. However, after 24–48 hr from the treatment start, the ACTA-Cox15−/− animals became critically ill, prompting us to euthanize them. We found worsening of the mitochondrial myopathy (Figures S3A and S3B), and massive apoptosis, demonstrated by numerous TUNEL-positive nuclei (Figures S3C and S3D), corresponding ultrastructurally to chromatin condensation and apoptotic bodies (Figures S3E and S3F). No increased levels of plasma creatine kinase, urea, creatinine, or lactate were detected in plasma of euthanized animals, as observed in massive rhabdomyolysis and acute kidney failure, an adverse effect occasionally reported in patients under bezafibrate treatment (Charach et al., 2005; Wu et al., 2009).
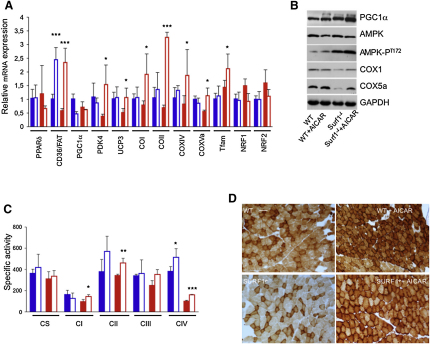
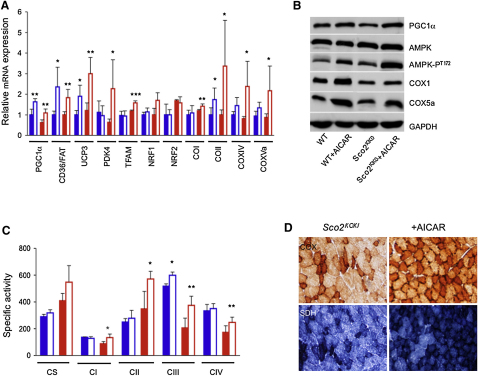
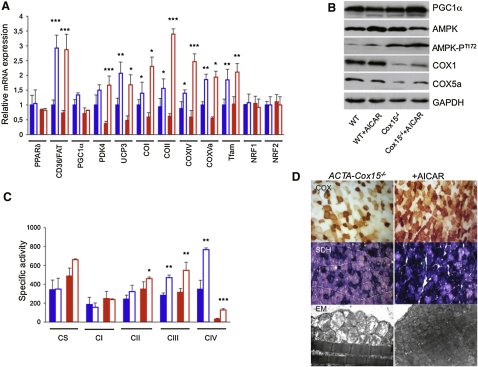
AICAR Induces FAO and OXPHOS Genes in Surf1−/−, Sco2KO/KI, and ACTA-Cox15−/− Muscle
In a third set of experiments, we tested the capacity of AICAR, an AMPK agonist, to activate PGC-1α in our animal models. AICAR was given for 1 month in daily subcutaneous injections of 0.25 mg/day/gm of body weight to 2-month-old Surf1−/−, Sco2KO/KI, and WT littermates, and of 0.5 mg/day/gm to 2-month-old ACTA-Cox15−/− animals and WT littermates, each group being composed of three/four individuals (see legends of Figures 4–6 for details). At the end of treatment, the mtDNA content (data not shown) and CS activity in skeletal muscle (Table S2) were similar in each treated group versus the corresponding untreated group. Analysis of selected transcripts (Figures 4A, 5A, and 6A) showed that, similar to the MCK-PGC-1α overexpressing mice, Nrf1 and Nrf2 gene expression was unchanged in all groups; however, both Tfam and the COX subunit encoding genes were significantly increased in treated versus untreated Surf1−/− (p < 0.05), Sco2KO/KI (p < 0.01), and ACTA-Cox15−/− muscles (p < 0.01). Increased expression of these genes was observed in treated WT mice exposed to the higher (0.5 mg/gm) but not to the lower (0.25 mg/gm) dosage (p < 0.01). The expression of the FAO-related CD36/FAT gene was significantly increased in all treated groups. PDK4 and UCP3, two genes that are activated by AMPK independently from PGC-1α, were also increased under AICAR treatment. Immunovisualization of selected proteins (Figures 4B, 5B, and 6B) showed that increased expression of COX subunit genes was paralleled by increased amount of COX1 and COX5a proteins in treated Surf1−/−, Sco2KO/KI, and ACTA-Cox15−/− mice, resulting in significantly increased COX-specific activity (Figures 4C, 5C, and 6C and Tables S2–S4; p < 0.01). We also immunovisualized T172-phosphorylated (AMPK-PT172) versus total muscle AMPK (Figures 4B, 5B, and 6B). Interestingly, the levels of AMPK-PT172 were already 8-fold higher in untreated Surf1−/− than in untreated WT mice, whereas total AMPK was unchanged. A similar result was obtained in the untreated ACTA-Cox15−/− and, albeit less pronounced, in Sco2KO/KI muscles. AICAR further increased AMPK phosphorylation in all three COX-defective models, and in WT littermates as well. The quantitative biochemical results obtained by spectrophotometric assays (Figures 4C, 5C, and 6C and Tables S2, S3, and S4) were paralleled by a similar, albeit merely qualitative, trend shown by histochemical reactions to COX (Figures 4D, 5D, and 6D), SDH (Figures 5D and 6D), and, in treated ACTA-Cox15−/− mice, by marked decrease of ultrastructural abnormalities in mitochondria (Figure 6D). Taken together, these results indicate a compensatory activation of the AMPK-Pgc1α axis in conditions of defective COX activity.
Figure 4.
AICAR Treatment in Surf1−/− Mice
(A) Expression analysis of FAO- and OXPHOS-related genes in Surf1−/− and WT muscles of AICAR-treated (0.25 mg/day/gm) and untreated mice. Solid blue, WT, untreated; blue outline, AICAR-treated WT; solid red, Surf1−/−, untreated; red outline, AICAR-treated Surf1−/−. The levels of the gene transcripts, retrotranscribed into cDNA, were normalized to that of the Hprt gene transcript and expressed as time-fold variation relative to the WT. Error bars represent SD. The asterisks represent the significance levels calculated by unpaired, Student's two-tailed t test: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(B) Western blot immunovisualization of skeletal muscle proteins of the different genotypes, listed as above. Densitometric analysis of each band, normalized against that of GAPDH, revealed the following variations, relative to untreated WT. (1) PGC-1α: 1.1 in WT+AICAR; 1.5 in Surf1−/−; 1.8 in Surf1−/−+AICAR. (2) Total AMPK: 0.8 in WT+AICAR; 1.0 in Surf1−/−; 0.9 in Surf1−/−+AICAR. (3) AMPK-PT172: 2.3 in WT+AICAR; 8.2 in Surf1−/−; 9.5 in Surf1−/−+AICAR. COX1: 1.0 in WT+AICAR; 0.2 in Surf1−/−; 0.6 in Surf1−/−+AICAR. (4) COX5a: 1.1 in WT+AICAR; 0.04 in Surf1−/−; 0.6 in Surf1−/−+AICAR.
(C) MRC activities in the different genotypes, AICAR-treated or untreated, expressed as nmoles/min/mg of protein. Note that the activity of cII has been multiplied by 10 for visualization clarity. Error bars represent SD.
(D) COX staining in skeletal muscles from AICAR-treated and untreated Surf1−/− and WT littermates. Note the increased staining of the Surf1−/−+AICAR sample. Magnification, 20×.
Figure 5.
AICAR Treatment in Sco2KO/KI Mice
(A) Expression analysis of FAO- and OXPHOS-related genes in Sco2KO/KI and WT muscles of AICAR-treated (0.25 mg/day/gm) and untreated mice. Solid blue, WT, untreated; blue outline, AICAR-treated WT; solid red, Sco2KO/KI, untreated; red outline, AICAR-treated Sco2KO/KI. The levels of the gene transcripts, retrotranscribed into cDNA, were normalized to that of the Hprt gene transcript and expressed as time-fold variation relative to the WT. Error bars represent SD. The asterisks represent the significance levels calculated by unpaired, Student's two-tailed t test: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(B) Western blot immunovisualization of skeletal muscle proteins of the different genotypes, listed as above. Densitometric analysis of each band, normalized against that of GAPDH, revealed the following variations, relative to untreated WT. (1) PGC-1α: 1.7 in WT+AICAR; 1.0 in Sco2KO/KI; 1.7 in Sco2KO/KI+AICAR. (2) Total AMPK: 1.0 in WT+AICAR; 1.2 in Sco2KO/KI; 1.2 in Sco2KO/KI+AICAR. (3) AMPK-PT172: 1.8 in WT+AICAR; 1.6 in Sco2KO/KI; 3.2 in Sco2KO/KI+AICAR. COX1: 1.1 in WT+AICAR; 0.6 in Sco2KO/KI; 0.8 in Sco2KO/KI+AICAR. (4) COX5a: 1.0 in WT+AICAR; 0.5 in Sco2KO/KI; 0.7 in Sco2KO/KI+AICAR.
(C) MRC-specific activities in the different genotypes, AICAR-treated or untreated, expressed as nmoles/min/mg of protein. Note that the activity of cII has been multiplied by 10 for visualization clarity. Error bars represent SD.
(D) COX and SDH staining in skeletal muscles from AICAR-treated and untreated Sco2KO/KI and WT littermates. Note the increased staining of the Sco2KO/KI+AICAR sample. Magnification, 20×.
Figure 6.
AICAR Treatment in ACTA-Cox15−/− Mice
(A) Expression analysis of FAO- and OXPHOS-related genes in ACTA-Cox15−/− and WT muscles of AICAR-treated (0.5 mg/day/gm) and untreated mice. Solid blue, WT, untreated; blue outline, AICAR-treated WT; solid red, ACTA-Cox15−/−, untreated; red outline, AICAR-treated ACTA-Cox15−/−. The levels of the gene transcripts, retrotranscribed into cDNA, were normalized to that of the Hprt gene transcript and expressed as time-fold variation relative to the WT. Error bars represent SD. The asterisks represent the significance levels calculated by unpaired, Student's two-tailed t test: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
(B) Western blot immunovisualization of skeletal muscle proteins of the different genotypes, listed as above. Densitometric analysis of each band, normalized against that of GAPDH, revealed the following variations, relative to untreated WT. (1) PGC-1α: 0.9 in WT+AICAR; 1.0 in ACTA-Cox15−/−; 1.8 in ACTA-Cox15−/−+AICAR. (2) Total AMPK: 1.8 in WT+AICAR; 1.2 in ACTA-Cox15−/−; 1.0 in ACTA-Cox15−/−+AICAR. (3) AMPK-PT172: 4 in WT+AICAR; 11.0 in ACTA-Cox15−/−; 18.0 in ACTA-Cox15−/−+AICAR. COX1: 1.1 in WT+AICAR; 0.06 in ACTA-Cox15−/−; 0.4 in ACTA-Cox15−/−+AICAR. (4) COX5a: 1.0 in WT+AICAR; ∼0.1 in ACTA-Cox15−/−; 0.6 in ACTA-Cox15−/−+AICAR.
(C) MRC-specific activities in the different genotypes, AICAR-treated or untreated, expressed as nmoles/min/mg of protein. Note that the activity of cII has been multiplied by 10 for visualization clarity. Error bars represent SD.
(D) COX (upper panel) and SDH (middle panel) staining in AICAR-treated and untreated ACTA-Cox15−/− and WT muscle. Magnification, 20×. Note the increased staining of the ACTA-Cox15−/−+AICAR sections. (Bottom) EM of muscles from untreated and AICAR-treated muscles. Note the reduction of abnormal mitochondria in the AICAR-treated mouse.
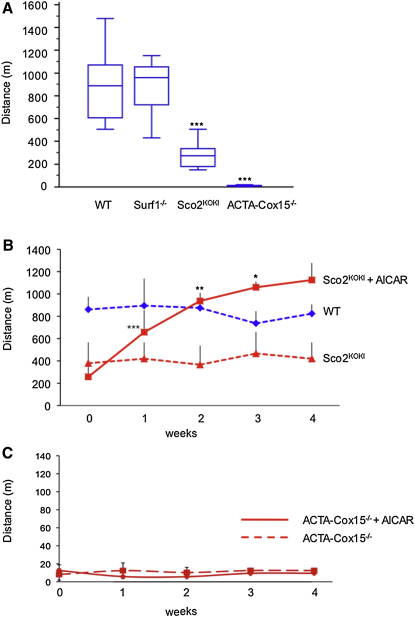
Effect of AICAR Treatment on Motor Performance
In order to evaluate the clinical effect of treatment, we administered a standard, quantitative incremental motor endurance test using a treadmill device. The motor performance was similar to WT littermates in untreated Surf1−/− mice, significantly impaired in Sco2KO/KI mice, and extremely impaired in ACTA-Cox15−/− mice, which were able to walk on the treadmill for just a couple of minutes (Figure 7A). Since no motor phenotype was detected in Surf1−/− mice, we focused our analysis on the other two COX-defective models that did show motor impairment. In treated Sco2KO/KI mice, the biochemical improvement was mirrored by marked amelioration of motor performance, which reached the same score as the untrained WT littermates by the first to second week of treatment (Figure 7B). Contrariwise, and in spite of partial biochemical recovery, we found no change by treadmill test in ACTA-Cox15−/− mice, suggesting that the mitochondrial myopathy was too severe for AICAR treatment to achieve a clinically detectable effect (Figure 7C).
Figure 7.
Treadmill Test
(A) Box-and-whisker plot of treadmill tests administered to WT (n. 16), Surf1−/− (n. 11), Sco2KO/KI (n. 9), and ACTA-Cox15−/− (n. 11) mice at 3 months of age. There is no motor impairment in Surf1−/−; significant motor impairment in Sco2KO/KI and severe motor impairment in ACTA-Cox15−/− mice compared to WT mice. The vertical bars represent the range of values; the boxes represent the values from the lower to upper quartile; the horizontal lines represent the median value for each group of animals. ∗∗∗, unpaired, Student's two-tailed t test: p < 10−6.
(B) Treadmill test in AICAR-treated Sco2KO/KI and in untreated Sco2KO/KI and WT littermates. Each group was composed of four individuals. The scores of motor performance measured week by week showed significant improvement in the AICAR-treated Sco2KO/KI animals during the first 3 weeks of treatment (paired, Student's two-tailed t test ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05), with an additional, albeit not significant, increment at week 4. Notably, the score of the AICAR-treated Sco2KO/KI animals became not significantly different from that of untreated WT littermates at the end of the first week, identical to that of WT at the second week, and significantly higher at the third and fourth weeks of treatment. Note that untreated WT or Sco2KO/KI mice showed no change in motor performance over time, indicating no significant effect of training. Vertical bars show the SDs.
(C) Treadmill test in untreated and AICAR-treated ACTA-Cox15−/− mice. No change was detected in motor performance over time. Vertical bars show the SDs.
Discussion
There is no effective treatment for mitochondrial disorders, and current clinical management is focused on treating complications. Stimulation of mitochondriogenesis and MRC activity is one of the strategies that have been proposed to correct OXPHOS failure leading to these conditions.
Two principal systems, PGC-1α and PPARs, are known to increase mitochondrial aerobic metabolism, acting on different, albeit partially overlapping, sets of genes related to mitochondrial bioenergetic pathways.
PGC-1α functions as a coactivator, i.e., it interacts with a number of transcription factors, potentiating their induction of mitochondrial bioenergetics-related genes, including those of OXPHOS. The mechanisms that regulate the expression and activity of PGC-1α are only partially known, but an important role is certainly played by posttranslational modifications of this protein, such as deacetylation, mediated by Sirt1, and phosphorylation, mediated by AMPK.
The control on energy metabolism by PPARs is centered on the activation of mitochondrial FAO, which in turn feeds both the tricarboxylic acids cycle and the MRC. While PPARs are clearly induced by activation of PGC-1α, the opposite effect, i.e., PPAR-dependent induction of PGC-1α, is questioned. In principle, the activation of Sirt1 by NAD+, produced by FAO, links the PPARs system to mitochondriogenesis and OXPHOS, activated by PGC-1α. Another possible mechanism is the direct PPAR-dependent induction of PGC-1α mediated by a PPAR-responsive element present in the promoter region of the PGC-1α gene. While the latter mechanism has been shown to be active in vitro, several reports have failed to demonstrate a physiologically effective PPAR-dependent control on PGC-1α expression and activity in mammalian organisms (Luquet et al., 2003; Wang et al., 2004; Kleiner et al., 2009). However, these mechanisms have been proposed to explain the recent observation that a pan-PPAR agonist, bezafibrate, was able to correct the COX defect, ameliorate motor impairment, and prolong life span, in MLC1F-Cox10−/−, a mouse model characterized by muscle-selective ablation of Cox10, a heme a biosynthetic enzyme.
To confirm and extend these observations in vivo, we first sought for a proof-of-principle demonstration by showing that (1) marked overexpression of transgenic Pgc1α increases the mtDNA content (Lin et al., 2002) and activities of the MRC complexes (this paper) on a WT background; and (2) is also able to partially, albeit not completely, correct COX deficiency in the skeletal muscle of a mildly COX-defective mammalian model, i.e., a constitutive Surf1−/− recombinant mouse. These data are largely concordant with results previously obtained in the MLC1F-Cox10−/− mouse, and indicate that vigorous stimulation of the Pgc1α axis is effective in increasing OXPHOS in normal and disease conditions in vivo.
Next, we tested the effects of the pharmacological activation of either PPARs- or PGC-1α-regulatory systems in our mouse models. Although these models differ in the severity of biochemical impairment and clinical outcome, all maintain some residual COX activity in skeletal muscle, similar to the corresponding human conditions. This biochemical “leakiness” occurs by different mechanisms in the different mutant organisms. For instance, the function of Surf1 is known to be partially redundant in humans (Tiranti et al., 1999), and even more so in mice (Dell'Agnello et al., 2007). Likewise, while the complete ablation of Sco2 leads to embryonic lethality (Yang et al., 2010), the combination of a null allele with an allele carrying a missense mutation prevents the activity of the Sco2 protein from being completely abolished. In turn, the excision of Cox15 in the ACTA-Cox15−/− mouse affects a high percentage but not all the muscle fibers, therefore allowing a residual Cox15 activity to be maintained; and all of the reported SCO2 and COX15 mutant patients carried missense changes that do not disrupt the corresponding proteins and, presumably, determine a decrease but not the complete loss of their activity. Therefore, all mouse models used in this work carry “permissive” defects that are in principle liable to (partial) correction by stimulating COX biogenesis.
We first stimulated the PPAR axis using bezafibrate, a pan-PPAR agonist, in two COX-defective models: Surf1−/−, associated with mild COX deficiency and no clinical phenotype, and ACTA-Cox15−/−, associated with profound COX deficiency and severe mitochondrial myopathy. In spite of the fact that FAO-related genes were in fact induced in treated animals, thus demonstrating that bezafibrate was pharmacologically active, we found neither stimulation of mitochondriogenesis nor induction of MRC enzyme activities, in both Surf1−/− and WT animals, which developed instead marked hepatomegaly and body weight loss. The toxic effects of bezafibrate were even more dramatic and rapid in the ACTA-Cox15−/− mouse model. Albeit still unknown, the mechanism of the bezafibrate-associated toxicity must be related to the muscle-specific ablation of Cox15, and is possibly linked to the very severe COX deficiency present in the skeletal muscle of these animals, since we showed that bezafibrate did not interfere with the maintenance of the gene in extramuscular tissues of the ACTA-Cox15−/− model (data not shown). Albeit less dramatic than the toxic myopathy observed in the ACTA-Cox15−/− model, adverse effects, such as body weight loss and hepatomegaly, were also observed in other bezafibrate-treated animal models, including mice with milder COX deficiency, such as (1) our constitutive Surf1−/− mouse model (Figure S1); and (2) the Sco2KO/KI mouse (E.A.S., unpublished data); as well as in animals with other OXPHOS defects, such as (3) constitutive Mpv17−/− mice (C.V. and M.Z., unpublished data). On a practical side, we believe that these observations in mice should prompt physicians to be cautious on the use of bezafibrate in patients with severe MRC defects, particularly COX deficiencies.
We then activated the PGC-1α axis by treating our mouse models with the AMPK agonist AICAR. Interestingly, we found that the levels of activated AMPK-P172 were already higher in skeletal muscle of untreated COX-defective models than in WT controls, and further increased upon AICAR treatment. This finding could be occurring because of the homeostatic loop that relates the PGC-1α axis to the activity of MRC (Wallace and Fan, 2010), and can explain why the response of the latter to AICAR was significantly higher in COX-defective muscles than in WT muscles.
The ∼1.5-fold increase of PGC1α transcript and AMPK-dependent activation of the corresponding protein induced by AICAR did not determine an increase of the mtDNA content of skeletal muscle, as observed in the MCK-PGC1α models, where PGC1α is >10-fold expressed compared to WT muscle. This result suggests that the increase of OXPHOS-related proteins and activities, including COX, that was observed in AICAR-treated COX-defective mice, are likely due to increased transcription and translation of the corresponding genes, in agreement with the increased levels of Tfam expression. Importantly, the Sco2KO/KI mice, which are characterized by mild, but significant, motor impairment, did markedly improve their performance under AICAR treatment, in parallel with recovery of COX activity in skeletal muscle. Unfortunately, no motor impairment was measured in untreated Surf1−/− mice (Figure 7A), which prevented us from further investigating the clinical features of this model under AICAR treatment. Contrariwise, the ACTA-COX15−/− mouse model showed very severe impairment of motor performance at 2 months of age, which failed to respond to AICAR, in spite of doubling the daily dosage of the drug (0.5 mg/gm versus 0.25 mg/gm). Further dosage increase, earlier and prolonged treatment, and association with other drugs (e.g., antioxidants) are warranted in this model to possibly improve clinical efficacy. Unlike the results obtained with AICAR, bezafibrate had no effect on OXPHOS, whereas it was clearly effective in inducing FAO-related genes. Acute exposure of MRC-defective cultured fibroblasts to high-dose bezafibrate has given controversial results (Bastin et al., 2008): some cell lines did respond by increasing MRC activities, others, including a Surf1−/− cell line, did not, which suggests that the responsiveness to the drug may depend on a number of still undisclosed variables, possibly intrinsic to the cell type and culturing conditions. Likewise, we have no obvious explanation for the discrepancy between our results and those recently reported in the MLC1F-Cox10−/− mouse model. One possibility is that the MLC1F-Cox10−/− model is characterized by predominant excision of the Cox10 gene in fast-twitch, type 2, glycolytic muscle fibers, whereas the slow-twitch, type 1, oxidative fibers are relatively spared. Therefore, a shift in favor of the latter type induced by bezafibrate-dependent activation of PPARs could explain the improvement in COX activity, exercise performance, and eventually life span of the treated animals. Nevertheless, other effects reported with this model, for instance increased expression of Pgc1α, were not observed in ours. One reason may be related to the fact that the PPAR system acts on Pgc1α via FAO induction and stimulation of NAD+-dependent Sirt1 deacetylase (Gerhart-Hines et al., 2007); however, block of mitochondrial respiration, as occurs in COX deficiency, typically decreases the NAD+/NADH ratio and should in fact reduce the activation of Sirt1 (Wallace, 2005), thus opposing the effect of PPAR activation.
Taken together, the results of our study prove that pharmacological activation of mitochondrial biogenesis is a rational, potentially effective approach to mitochondrial disease. Nevertheless, since both AICAR and activated AMPK determine a host of pleiotropic effects on different regulatory and homeostatic pathways (Cantó et al., 2010), prolonged observation on chronically treated animals is needed to establish treatment safety and eventual translatability to humans.
Experimental Procedures
Reagents and Materials
AICAR was from Toronto Research Chemicals. Anti-PGC-1α antibody was from Abcam; anti-AMPK and anti-AMPK-P antibodies were from Cell Signaling; anti-COI and -COX5a were from Invitrogen; anti-GAPDH was from Millipore. Anti-rabbit and anti-mouse secondary antibodies were from Amersham. Chemicals were from Sigma. MCK-PGC-1α and ACTA-Cre transgenic mice were obtained from Jackson Laboratory, Bar Harbor, USA. Oligonucleotides were from PRIMM, Italy. The sequences of all the primers used in this study are available on request.
Western Blot Analysis
Total homogenates were prepared in RIPA buffer (50 mM Tris HCl [pH 8], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) with the addition of a protease inhibitor cocktail (Roche). Protein concentration was determined by the Lowry method. Aliquots, 80 μg each, were run through a 12% SDS-PAGE and electroblotted onto a nitrocellulose membrane, which was then matched with different antibodies.
In Vivo Experiments
Animal studies were in accordance with the Italian Law D.L. 116/1992 and the EU directive 86/609/CEE. Standard food and water were given ad libitum. Mice were maintained in a temperature- and humidity-controlled animal-care facility, with a 12 hr light/dark cycle and free access to water and food (Standard Diet, Mucedola, Italy). Bezafibrate (Sigma) 0.5% was added to a standard diet (Mucedola, Italy) and administered for 1 month. AICAR or vehicle was administered for 4 weeks with a single daily subcutaneous injection. No difference in daily monitored water and food intake was detected between the treated versus untreated groups of animals during the experiment. Animals were sacrificed by cervical dislocation.
Treadmill Test
A treadmill apparatus (Columbus Instruments, Columbus, OH) was used to measure motor exercise endurance according to the number of falls in the motivational grid during a gradually accelerating program with speed initially at 6.5 m/min and increasing by 0.5 m/min every 3 min. The test was terminated by exhaustion, defined as >10 falls/min into the motivational grid.
Morphological Analysis
Histochemical and ultrastructural analyses were performed as described (Sciacco and Bonilla, 1996; Dubowitz, 1985). TUNEL reaction was performed following the manufacturer's instructions (In Situ Cell Death Detection Kit, Roche, Germany).
Biochemical Analysis of MRC Complexes
Muscle quadriceps samples stored in liquid nitrogen were homogenized in 10 mM phosphate buffer (pH 7.4), and the spectrophotometric activity of cI, cII, cIII and cIV, as well as CS, was measured as described (Bugiani et al., 2004).
Real-Time PCR
For mtDNA content analysis, SYBR Green real-time PCR was performed using primers specific to a mouse mtDNA region in the COI gene and primers specific to RNaseP, a single copy gene taken as a nuclear gene reference, as described (Viscomi et al., 2009). For the analysis of transcripts, total RNA was extracted from liquidnitrogen snap frozen muscle by Trizol, according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). Of total RNA, 2 μg was treated with RNase free-DNase and retrotranscribed using the “cDNA cycle” kit (Invitrogen, Carlsbad, CA, USA). Approximately 2–5 ng of cDNA was used for real-time PCR assay using primers specific for amplification of several genes.
Creation of a Recombinant Cox15 KO Mouse
A mouse genomic region spanning Cox15 exons 1–2 was PCR amplified with suitable oligos to introduce a loxP site, and cloned into a vector containing a Neomycin resistance cassette, flanked by two Flp sites, and a second loxP site. The short and long arms of homology were amplified by PCR using suitable oligos and cloned into the vector containing the floxed region. The final targeting vector was linearized by digestion with XhoI and electroporated in 129Sv cells. A total of 600 clones were PCR screened by PCR (see legend of Figure S2). In one recombinant clone, homologous recombination was ascertained on the long arm by long-range PCR (see Figure S2). Injection into C57Bl/6 blastocysts was followed by selection of chimaeric individuals, which were then backcrossed to a C57Bl/6 female. The recombinant mice were crossed to a general deleter cre transgenic mouse to obtain Cox15+/− heterozygous mice, which were then crossed to a Flpe transgenic mouse in order to eliminate the neomycin-resistance cassette, resulting in Cox15lox/lox individuals. ACTA-Cox15−/− mice were eventually obtained by crossing Cox15lox/lox with ACTA-Cre mice.
Acknowledgments
This work was supported by the Pierfranco and Luisa Mariani Foundation Italy; Fondazione Telethon-Italy grants numbers GGP07019 and GPP10005; Fondazione Tomasello and Associazione Mitocon to M.Z.; the National Institutes of Health (grant HD32062), the Muscular Dystrophy Association, and the Marriott Mitochondrial Disorder Clinical Research Fund to E.A.S.; and the “Associazione Amici del Centro Dino Ferrari” to M.M.
Published: July 5, 2011
Footnotes
Supplemental Information includes four tables and three figures and can be found with this article online at doi:10.1016/j.cmet.2011.04.011.
Supplemental Information
References
- Antonicka H., Mattman A., Carlson C.G., Glerum D.M., Hoffbuhr K.C., Leary S.C., Kennaway N.G., Shoubridge E.A. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2003;72:101–114. doi: 10.1086/345489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish G.D., Narkar V.A., Evans R.M. PPAR delta: a dagger in the heart of the metabolic syndrome. J. Clin. Invest. 2006;116:590–597. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin J., Aubey F., Rotig A., Munnich A., Djouadi F. Activation of peroxisome proliferator activated receptor pathway stimulates the mitochondrial respiratory chain and can correct deficiencies in patients' cells lacking its components. J. Clin. Endocrinol. Metab. 2008;4:1433–1441. doi: 10.1210/jc.2007-1701. [DOI] [PubMed] [Google Scholar]
- Bugiani M., Invernizzi F., Alberio S., Briem E., Lamantea E., Carrara F., Moroni I., Farina L., Spada M., Donati M.A. Clinical and molecular findings in children with complex I deficiency. Biochim. Biophys. Acta. 2004;1659:136–147. doi: 10.1016/j.bbabio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Bugiani M., Tiranti V., Farina L., Uziel G., Zeviani M. Novel mutations in COX15 in a long surviving Leigh syndrome patient with cytochrome c oxidase deficiency. J. Med. Genet. 2005;42:e28. doi: 10.1136/jmg.2004.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C., Auwerx J. AMP-activated protein kinase and its downstream transcriptional pathways. Cell. Mol. Life Sci. 2010;67:3407–3423. doi: 10.1007/s00018-010-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C., Gerhart-Hines Z., Feige J.N., Lagouge M., Noriega L., Milne J.C., Elliott P.J., Puigserver P., Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charach G., Grosskopf I., Rotmensch H.H., Kitzis I., Weintraub M. Bezafibrates cause moderate, reversible impairment in renal function in patients without prior renal disease. Nephron Clin. Pract. 2005;100:c120–c125. doi: 10.1159/000085291. [DOI] [PubMed] [Google Scholar]
- Corton J.M., Gillespie J.G., Hawley S.A., Hardie D.G. 5-aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells? Eur. J. Biochem. 1995;229:558–565. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- Cree L.M., Patel S.K., Pyle A., Lynn S., Turnbull D.M., Chinnery P.F., Walker M. Age-related decline in mitochondrial DNA copy number in isolated human pancreatic islets. Diabetologia. 2008;51:1440–1443. doi: 10.1007/s00125-008-1054-4. [DOI] [PubMed] [Google Scholar]
- Dell'Agnello C., Leo S., Agostino A., Szabadkai G., Tiveron C., Zulian A., Prelle A., Roubertoux P., Rizzuto R., Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum. Mol. Genet. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- Dubowitz V. 2nd ed. Baillière Tindall; London: 1985. Muscle Biopsy, A Practical Approach. [Google Scholar]
- Fogarty S., Hawley S.A., Green K.A., Saner N., Mustard K.J., Hardie D.G. Calmodulin-dependent protein kinase kinase-β activates AMPK without forming a stable complex: synergistic effects of Ca2+ and AMP. Biochem. J. 2010;426:109–118. doi: 10.1042/BJ20091372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z., Rodgers J.T., Bare O., Lerin C., Kim S.H., Mostoslavsky R., Alt F.W., Wu Z., Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S.J., Taylor R., Zhang Y., Jin S. Autophagy and the degradation of mitochondria. Mitochondrion. 2010;10:309–315. doi: 10.1016/j.mito.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C., Spiegelman B.M. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- Jäger S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D.P., Scarpulla R.C. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O., Rödel G. Biogenesis of cytochrome c oxidase. Mitochondrion. 2005;5:363–388. doi: 10.1016/j.mito.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Kleiner S., Nguyen-Tran V., Baré O., Huang X., Spiegelman B., Wu Z. PPAR{delta} agonism activates fatty acid oxidation via PGC-1{alpha} but does not increase mitochondrial gene expression and function. J. Biol. Chem. 2009;284:18624–18633. doi: 10.1074/jbc.M109.008797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Leary S.C., Sasarman F., Nishimura T., Shoubridge E.A. Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1. Hum. Mol. Genet. 2009;18:2230–2240. doi: 10.1093/hmg/ddp158. [DOI] [PubMed] [Google Scholar]
- Lefebvre P., Chinetti G., Fruchart J.C., Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J. Clin. Invest. 2006;116:571–580. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Wu H., Tarr P.T., Zhang C.-Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibers. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Luquet S., Lopez-Soriano J., Holst D., Fredenrich A., Melki J., Rassoulzadegan M., Grimaldi P.A. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. FASEB J. 2003;17:2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- Merrill G.F., Kurth E.J., Hardie D.G., Winder W.W. AICA riboside increases AMP-activated protein kinase, fatty acid oxidation, and glucose uptake in rat muscle. Am. J. Physiol. 1997;273:1107–1112. doi: 10.1152/ajpendo.1997.273.6.E1107. [DOI] [PubMed] [Google Scholar]
- Papadopoulou L.C., Sue C.M., Davidson M.M., Tanji K., Nishino I., Sadlock J.E., Krishna S., Walker W., Selby J., Glerum D.M. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet. 1999;23:333–337. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- Sciacco M., Bonilla E. Cytochemistry and immunocytochemistry of mitochondria in tissue sections. Methods Enzymol. 1996;264:509–521. doi: 10.1016/s0076-6879(96)64045-2. [DOI] [PubMed] [Google Scholar]
- Srivastava S., Diaz F., Iommarini L., Aure K., Lombes A., Moraes C.T. PGC-1alpha/beta induced expression partially compensates for respiratory chain defects in cells from patients with mitochondrial disorders. Hum. Mol. Genet. 2009;18:1805–1812. doi: 10.1093/hmg/ddp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G.R., Kemp B.E. AMPK in health and disease. Physiol. Rev. 2009;89:1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- Tenenbaum A., Motro M., Fisman E.Z. Dual and pan-peroxisome proliferator-activated receptors (PPAR) co-agonism: the bezafibrate lessons. Cardiovasc. Diabetol. 2005;16:4–14. doi: 10.1186/1475-2840-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiranti V., Galimberti C., Nijtmans L., Bovolenta S., Perini M.P., Zeviani M. Characterization of SURF-1 expression and Surf-1p function in normal and disease conditions. Hum. Mol. Genet. 1999;8:2533–2540. doi: 10.1093/hmg/8.13.2533. [DOI] [PubMed] [Google Scholar]
- Viscomi C., Spinazzola A., Maggioni M., Fernandez-Vizarra E., Massa V., Pagano C., Vettor R., Mora M., Zeviani M. Early-onset liver mtDNA depletion and late-onset proteinuric nephropathy in Mpv17 knockout mice. Hum. Mol. Genet. 2009;18:12–26. doi: 10.1093/hmg/ddn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D.C. Mitochondrial energetics and therapeutics. Annu. Rev. Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D.C., Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.X., Zhang C.L., Yu R.T., Cho H.K., Nelson M.C., Bayuga-Ocampo C.R., Ham J., Kang H., Evans R.M. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T., Diaz F., Spiegelman B.M., Moraes C.T. Activation of the PPAR/PGC-1alpha pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 2008;8:249–256. doi: 10.1016/j.cmet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wu J., Song Y., Li H., Chen J. Rhabdomyolysis associated with fibrate therapy: review of 76 published cases and a new case report. Eur. J. Clin. Pharmacol. 2009;65:1169–1174. doi: 10.1007/s00228-009-0723-7. [DOI] [PubMed] [Google Scholar]
- Yang H., Brosel S., Acin-Perez R., Slavkovich V., Nishino I., Khan R., Goldberg I.J., Graziano J., Manfredi G., Schon E.A. Analysis of mouse models of cytochrome c oxidase deficiency owing to mutations in Sco2. Hum. Mol. Genet. 2010;19:170–180. doi: 10.1093/hmg/ddp477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeviani M., Di Donato S. Mitochondrial disorders. Brain. 2004;127:2153–2172. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.