Abstract
Neural tube defects (NTDs) are common birth defects, occurring in approximately 1/1,000 births; both genetic and environmental factors are implicated. To date, no major genetic risk factors have been identified. Throughout development, cell adhesion molecules are strongly implicated in cell–cell interactions, and may play a role in the formation and closure of the neural tube. To evaluate the role of neural cell adhesion molecule 1 (NCAM1) in risk of human NTDs, we screened for novel single-nucleotide polymorphisms (SNPs) within the gene. Eleven SNPs across NCAM1 were genotyped using TaqMan. We utilized a family-based approach to evaluate evidence for association and/or linkage disequilibrium. We evaluated American Caucasian simplex lumbosacral myelomeningocele families (n=132 families) using the family based association test (FBAT) and the pedigree disequilibrium test (PDT). Association analysis revealed a significant association between risk for NTDs and intronic SNP rs2298526 using both the FBAT test (P=0.0018) and the PDT (P=0.0025). Using the HBAT version of the FBAT to look for haplotype association, all pairwise comparisons with SNP rs2298526 were also significant. A replication study set, consisting of 72 additional families showed no significant association; however, the overall trend for overtransmission of the less common allele of SNP rs2298526 remained significant in the combined sample set. In addition, we analyzed the expression pattern of the NCAM1 protein in human embryos, and while NCAM1 is not expressed within the neural tube at the time of closure, it is expressed in the surrounding and later in differentiated neurons of the CNS. These results suggest variations in NCAM1 may influence risk for human NTDs.
Introduction
Neural tube defects (NTDs) result from failure of neural tube closure and are one of the most common human malformations, occurring at an average rate of 1 per 1,000 human pregnancies (Campbell et al. 1986). Both genetic and environmental components have been implicated; however, no causative genes have been identified. Formation of the neural tube is driven by several morphogenetic cell behaviors, including changes in cell–cell and cell–matrix interactions. During neural tube closure, the neural folds are brought together at the dorsal midline to form the neural tube and adhere to each other. In humans and other mammals, closure of the neural tube is thought to be initiated by sites of fusion at more than one place along the anterior–posterior axis (Van Allen et al. 1993; O’Rahilly and Muller 2002; Sulik et al. 1998). The first fusion occurs in humans when there are 4–6 somite pairs present, at Carnegie stage (C) 10, and closure continues though C12 with the closure of the caudal neuropore (O’Rahilly and Muller 2002).
The cell adhesion molecules (CAMs) are involved in defining the interaction of cell collectives and their borders during morphogenesis. Neural cell adhesion molecule 1 (NCAM1), an integral membrane protein belonging to the immunoglobulin superfamily (Edelman 1983), is involved in cell adhesion-dependent morphogenetic events, including the migration of various cells to the proper sites in neural tissues. NCAM is an important player in cell–cell and cell–matrix adhesion and is involved in many activities, including cell migration, neurite growth, axonal guidance, and synaptic plasticity (Thiery et al. 1982; Edelman 1983; Rutishauser and Jessell 1988).
A diverse group of NCAM molecules can be achieved from a single locus due to both transcriptional and posttranslational modifications. Alterative splicing, which is regulated in a cell and developmental stage-specific manner, produces three major isoforms. Two isoforms, NCAM-140 and NCAM-180, are membrane-spanning with a variable cytoplasmic domain and NCAM-120 is linked to the membrane by a glycosyl phosphatidylinositol lipid anchor (Cunningham et al. 1987). There are two alternate exons that can be included in the final NCAM1 transcript. The VASE or variable alternative spliced exon contains an additional 30-bp insertion that results in an additional ten amino acids in the fourth immunoglobulin-like loop. Inclusion of the SEC exon results in a premature termination of translation and a secreted isoform (Small and Akeson 1990). In mouse cell lines, it has been shown that the binding of transcription factors encoded by Hox- and Pax-gene controls regulation of the NCAM promoter (Jones et al. 1992).
In the mouse, Ncam transcripts are first detected around day 8.5 in the somites and in the forming neural tube. Expression is not uniform along the rostrocaudal axis, with stronger expression in the caudal region of the neural tube and neural plate. Ncam expression continues until day 12.5, but is restricted to postmitotic neurons at later stages (Bally-Cuif et al. 1993). In Ncam-knockout mice with a targeted replacement with a lacZ reporter gene under control of endogenous Ncam regulatory sequences, β-galactosidase staining was seen throughout the spinal cord and dorsal root ganglia from E9.5 to E13.5 (Holst et al. 1998). This expression coincides with the neural tube formation in mouse embryos, around day 8 of gestation, and complete elevation and closure around day 10 (Harris and Juriloff 1999).
NCAM expression is also found in the neural tube in several other species. At the 15-somite stage of chicken embryos, NCAM is found in the neural plate and the adjacent ectoderm near Hensen’s node, where the neural tube is not closed, but is exclusively expressed in the neural tube during and after closure at more rostral trunk levels (Thiery et al. 1982). By immunocytochemical analysis, both cell adhesion molecules N-cadherin and NCAM are detected on the cranial neural folds prior to neural tube closure and on migrating neural crest cells thereafter (Bronner-Fraser et al. 1992). Chicken NCAM is visible in the otic placode (Thiery et al. 1982) and later in differentiating auditory nuclei of the hindbrain, appearing progressively in differentiating neuron groups of the CNS (Hrynkow et al. 1998). The NCAM is also expressed in Xenopus embryos in a radial pattern within the neural tube during and for several hours after neural tube closure (Balak et al. 1987), and both zNCAM and zPCAM in zebrafish are expressed throughout the length of the closing neural tube from 11 to 30 hpf, during somitogenesis (Mizuno et al. 2001).
In the mouse, deletion of exons 3 and 4 of Ncam1 prevents any isoform from being produced. The null mutants have few defects and are otherwise healthy and fertile (Cremer et al. 1994). However, by using homologous recombination to introduce a premature stop codon, a secreted form of NCAM can be produced in the absence of any membrane-associated protein. No heterozygous progeny were obtained from chimera crosses, suggesting dominant lethality. Chimeric embryos (E8.5) with a high ES cell contribution had poorly formed somites with kinking of the neural tube, and by E9.5, the anterior neuropore remained open in the mutant embryos (Rabinowitz et al. 1996). NCAM is a known downstream target of the Pax-3 transcription factor (Moase and Trasler 1991; Neale and Trasler 1994). Pax-3 (splotch) mutant mice display multiple defects, including neural tube closure in the form of both spina bifida and exencephaly. In these mutants, altered NCAM isoform ratio and a decrease in the sialylation of the protein may alter the adhesive properties of NCAM (Epstein et al. 1991; Glogarova and Buckiova 2004). Quail embryos with spontaneous neural tube defects were shown to have disturbed matrix and cell adhesion molecule expression, including NCAM, by immunocytochemistry. The embryos expressed both N-cadherin and NCAM, not normally found at the stage examined (Newgreen et al. 1997). It was proposed that disruption of cell adhesion or extracellular matrix molecules that result in greater adhesion may impede appropriate morphogenetic movements, resulting in NTDs (Newgreen et al. 1997).
Given the importance of cell morphogenic events in neural tube closure, we hypothesized that a genetic variant that compromises the ability of a cell adhesion molecule in neural tissue may be associated with an increased risk for human neural tube defects. For this reason, we investigated 11 SNPs across NCAM1 for possible association with neural tube defects. We evaluated the promoter region, coding sequences, and alternative exons of NCAM1 for polymorphisms that may play a role in NTDs. In addition, we examined the distribution of NCAM1 protein in human embryo sections of the neural tube and lateral tissues by immunohistochemistry.
Materials and methods
Sample population
To identify novel polymorphisms within NCAM1, we screened 230 individuals with lumbosacral myelomeningocele by HPLC and sequencing methods, as described below. The simplex families (n=204) used for frequency and association analysis consisted of a sampled affected individual with lumbosacral myelomeningocele and nuclear families, including unaffected siblings where available. In this study all individuals were American Caucasian. The complete sample set included an initial group of 132 families (Series 1), as well as a replication set of 72 additional families (Series 2) of the same phenotype and ethnicity. The Series 1 families were those first collected and included 107 complete triads and 25 families with one parent; this sample included 121 discordant sibling pairs. Series 2 families are the next set of families collected and included 49 complete triads and 23 families with one parent; this sample included 56 discordant sibling pairs. All data and samples were collected following informed consent of subjects; this study was approved by the Duke University Medical Center Institutional Review Board.
SNP selection and genotyping
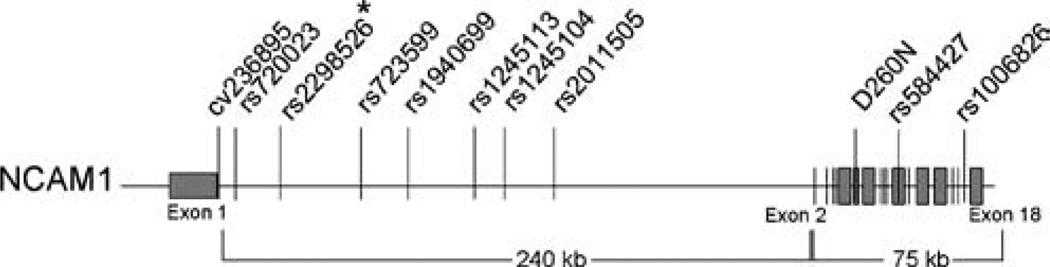
Initially, a set of five SNPs (cv236895, rs2298526, rs2011505, rs584427, and rs1006826) were chosen to characterize association between NCAM1 and human neural tube defects. The SNPs were spaced across the gene, with four being intronic and one coding. Following analysis in the Series 1 families (n=132), an additional five SNPs (rs720023, rs723599, rs1940699, rs1245113, and rs1245104) were typed surrounding the marker that showed association. Where available, SNPs in conserved non-coding sequences were included in the selection. All known SNPs that were genotyped had a heterozygosity of 0.28 or greater. We also initiated dHPLC screening to identify novel SNPs, leading to the identification of one novel coding SNP (see below). SNP locations are shown in Fig.1.
Fig. 1.
Diagram of SNPs analyzed in the NCAM1-association study. The SNP with the asterisk shows the significant association
The TaqMan allelic discrimination assays were used for the genotyping of these 11 SNPs across NCAM1 (Assay-on-demand and Assay-by-Design, Applied Biosystems, Foster City, Calif., USA). The PCR amplification was performed using the GeneAmp PCR system 9700 thermocyclers (Applied Biosystems) according to the assay specifications. Fluorescence detection was performed with the ABI Prism 7900 and analyzed with SDS software (Applied Biosystems). Quality control measures consisted of 24 duplicated individuals per 384-well plate and were blinded from laboratory technicians. In addition, two samples from CEPH individuals were located across all plates for internal control. To pass quality control, data plates had to pass 100% matching for all 26 duplicated samples and 95% overall plate efficiency. The SNPs were tested in the family set for Mendelian inconsistencies.
Variation detection
To search for novel polymorphisms within NCAM1, the genomic sequence was divided into individual segments that included one exon each and minimal surrounding intronic sequence. In addition, the alternative exons VASE and SEC were evaluated in this study. For analysis for the promoter region, the size of the region evaluated was dependent upon regions of mouse homology and presence of predicted promoter element. Primers were designed flanking each region of interest by the use of the program Primer3 (Primer 3 website). For each fragment, a total of 30 ng of pooled DNA was amplified using standard PCR protocols (Table 1). DNA was extracted from blood samples of NTD patients and their families using the PureGene system and the Autopure LS (Gentra Systems, Minneapolis, Minn., USA) according to the manufacturer’s protocol. The DNA samples were prepared and stored by the Duke Center for Human Genetics DNA bank Core. Following visualization and quantitation on a 2% agarose gel, PCR products were heteroduplexed by heating samples to 95°C for 3 min and slowly cooling to 30°C over 40 min. For polymorphism detection by denaturing high performance liquid chromatography (DHPLC), samples were injected into the Transgenomic WAVE Fragment Analysis System (version 4.1) for separation at various melting temperatures, as determined by WAVEMAKER software, version 4.1.42 (Transgenomic, San Jose, Calif., USA). When an apparent variation was noted by DHPLC, the individual samples exhibiting the variable pattern were directly sequenced. The PCR product was purified using QIAquick PCR Purification kit (Qiagen, Valencia, Calif., USA) according to the manufacturer’s directions. The sample was then sequenced using the BigDye Terminator version 3.1 Cycle Sequencing Ready Reaction kit (Applied Biosystems) and purified using Performa DTR Gel Filtration Cartridges (Edge Biosystems, Gaithersburg, Md., USA). In order to determine and confirm putative polymorphisms, sequence analysis was performed using ABI 3100 Data Collection Software Version 1.01 and ABI Sequencing Analysis 3.7.
Table 1.
PCR fragments, primers, and conditions for all NCAM1 fragments analyzed
| Fragment name | Forward primer (5′-3′) | Reverse primer (5′-3′) | HPLC temp (°C) |
|---|---|---|---|
| Exon 1 | ggctgggactgtcactcatt | gcaaaccagattgagaattaaaa | 60 |
| Exon 2 | gggtttcattcttgaacattgg | cctgagggctcctgctctac | 60 |
| Exon 3 | ggggacttattagtcttttcgactt | gcagaagaagaaggaggctct | 61 |
| Exon 4 | gaagcagctgttttccctca | tgaaaaagctagggaacttgg | 60.5 |
| Exon 5 | tgcagatgctctctgactga | ccaaggttgtagcaatgcag | 60 |
| Exon 6 | tgtctcttctccaggccatt | gactttgtgatgccccattc | 59.5 |
| Exon 7 | tggtcgaaatcatgctactttg | attgtggcagagcagtgacc | 60.5 |
| VASE | ctaagggggaaaaaaagctggaca | tcatccactcccaacacagc | 56 |
| Exon 8 | tgcatgccatcatttaaacc | attccaaggccctgaaactc | 56, 60.5 |
| Exon 9 | ccttgggctctgacatgc | catcctgaccctgccttg | 63.5 |
| Exon 10 | aatcatggcagtcatcctga | ttggagcccacctagagtca | 61 |
| Exon 11 | tgaccatcccataggacactt | attgggctggcagggttag | 60 |
| Exon 12 | atggtcttgggccaaactg | caggtggggacatctgagta | 61 |
| SEC | gagggtgatgccgagaaggaa | cacacggagggaacaccaaga | 57 |
| Exon 13 | gaaatagaattgctggaccaaa | aaggtgggctgggaaaag | 57 |
| Exon 14 | cctgtcactccatcccattc | cagggttctggtgaagtctga | 60 |
| Exon 15 | tcccgtaagttttgcctattg | caagcaagttgtcagggttg | 60.5 |
| Exon 16 | gtctggaggtctcgcatctc | caaacctcagcaaggtggac | 63 |
| Exon 17 | gccttgggttgagtcatagc | gggtctctacggagcaggt | 63 |
| Exon 18 | agaccgtggtctcagtggtt | tggaaatgctctggtgaagc | 60.5 |
| Promoter 1 | gagggtttcagtgttctaggc | aagaaaactccgatgtttggaa | 54 |
| Promoter mid | ttttcttcgggttatttctgga | ccagccttccttaatcagca | 56.5 |
| Promoter 2 | ctgattaaggaaggctgggta | tttttgcagaattgtttcctg | 63 |
Statistical analysis
All SNPs were tested for departure from Hardy-Weinberg equilibrium using a single affected and separately using an unaffected individual randomly selected from each family. These tests were conducted using the Genetic Data Analysis (GDA) software with a permutation test to estimate the P value (Lewis Lab Software Website). Pairwise calculations of linkage disequilibrium (LD) were computed with the Graphical Overview of Linkage Disequilibrium (GOLD) software for both the squared correlation coefficient (r2) and Lewontin’s standardized disequilibrium coefficient (D′) (Abecasis and Cookson 2000). Single-locus association analysis was performed using the pedigree disequilibrium test (PDT) for allelic association and the genotype-based version, the geno-PDT (Martin et al. 2000, 2003). We used the PDTave statistic, giving equal weight to all families, for comparison of allele frequencies between affected individuals and their unaffected parents or siblings (Martin et al. 2000). In addition, the family-based association test (FBAT) was performed to test for association in both single loci and in haplotypes (Horvath et al. 2004). Haplotype analysis was performed with the HBAT function, using windows of three adjacent SNPs across NCAM1. Haplotypes with frequencies of <1% were excluded from this analysis.
Bioinformatics
Genomic and protein sequences were obtained from The Human Genome Browser (assembly July 2003) University of California, Santa Cruz (UCSC web site) with additional information from Ensembl at http://www.ensembl.org/. Genomic and mRNA sequences were obtained from NCBI (accession numbers NM_000615 and NM_000615.1). All SNP references are based on NM_00615, with the ATG initiation codon being +1, and are named according to the recommendations of the Nomenclature Working Group (Antonarakis 1998). Protein references are based on NCAM140 (NP_00606). Promoter regions were chosen based on the prediction of promoter elements using Proscan and the Transfac databases (Wingender et al. 2001). Sequence alignment of human and mouse NCAM (Gen-Bank accession numbers NM_000615 and NM_010875, respectively) was performed using PipMaker (Schwartz et al. 2000) to search for conserved non-coding sequences.
Immunohistochemistry
For NCAM expression analysis, unaffected human embryos were obtained from legally terminated pregnancies in agreement with French law 00–800 and with recommendations by the Necker Hospital ethics committee. Sections from embryos ranging from Carnegie stage 9 to 19 and spinal cords from 22 to 27sa fetuses as positive controls were chosen. When available, sections from levels of both open and closed neural tube from the same embryo underwent classical immunohistochemistry using two different primary antibodies: 1:100 dilution of a CD56/NCAM mAb (1B6, Novocastra), recognizing the 120/140-kDa forms or a rabbit pAb to NCAM (Chemicon, AB5032), recognizing all isoforms. Tissues had been fixed with 4% paraformaldehyde, embedded in paraffin blocks, and 5-µm sections were processed in 0.1% Tween-20-containing PBS. Anti-rabbit-555 (Invitrogen/Molecular Probes) was used for visualization at a dilution of 1:400 or anti-mIgG-biotin and ABC-HRP from the Vector MOM kit, PK2200 according to manufacturer’s instructions. Specimens were examined on a Nikon EclipseTE300 fluorescent microscope equipped with a Roper Scientific CCD camera for image capture using Metaview (UIC) software.
Results
SNP detection
No variations were detected by DHPLC in exons 1–4, 8, and 10–18, in addition to two promoter regions and the VASE and SEC exons. However, six novel SNPs were detected and several known SNPs were verified in our population (Table 2). Only two of the SNPs occurred in exonic regions, one resulting in an amino acid change. Of the novel SNPs, one was analyzed in the entire family series via TaqMan allelic discrimination assays based on the frequency and putative correlation with disease status. All other novel SNPs were found to be in five or fewer individuals. Because the variant would have been too rare for identification of linkage disequilibrium in our family sample series (≤ 1%), we pursued no further analysis.
Table 2.
Novel polymorphisms in NCAM1
| Fragment | SNP | Surrounding sequence |
|---|---|---|
| Exon 5 | g.244569G>A | gaatggtgagG/Aagagtccgtt |
| Exon 6 | g.245781C>T | tggctC/TataccttttatcatgG/Aactag |
| g.245797G>A | See above | |
| Exon 7 | c.958G>A; D260N | agaggaagacG/Aatgagaagta |
| c.1032G>A; E284E | acgaggctgaG/Atacatctgca | |
| Exon 9 | g.270021C>A | ccttccccccC/Aacccccggca |
Association studies
No SNP tested in the family-based series showed evidence for deviation from Hardy-Weinberg equilibrium in either the affected or unaffected individuals (data not shown). Pairwise LD across all 11 markers is shown in Table 3. The LD analysis based on r2 (>0.5) revealed strong LD between all intron 1 SNPs cv236895, rs720023, rs2298526, rs723599, and rs1940699, as well as between SNPs rs1245113 and rs2011505, in both the affected and unaffected individuals. Values of D′ were also large for several SNP pairs, suggesting little recombination in the region over evolutionary time. Although novel polymorphism D260N has large values of D′ with several SNPs, the minor allele frequency is very small.
Table 3.
Measures of linkage disequilibrium between NCAM1 SNPs. The r2 values are given above the diagonal and D′ values are given below the diagonal
| cv236895 | rs720023 | rs2298526 | rs723599 | rs1940699 | rs1245113 | rs1245104 | rs2011505 | D260N | rs584427 | rs1006826 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| cv236895 | – | 0.575 | 0.556 | 0.524 | 0.545 | 0.243 | 0.090 | 0.196 | 0.028 | 0.019 | 0.007 |
| rs720023 | 0.991 | – | 0.957 | 0.959 | 0.960 | 0.390 | 0.052 | 0.313 | 0.014 | 0.005 | 0.011 |
| rs2298526 | 0.98 | 0.987 | – | 0.994 | 1 | 0.396 | 0.048 | 0.306 | 0.017 | 0.009 | 0.009 |
| rs723599 | 0.96 | 0.988 | 1 | – | 1 | 0.399 | 0.046 | 0.302 | 0.007 | 0.004 | 0.014 |
| rs1940699 | 0.971 | 0.988 | 1 | 1 | – | 0.406 | 0.045 | 0.310 | 0.014 | 0.005 | 0.385 |
| rs1245113 | 0.695 | 0.669 | 0.667 | 0.664 | 0.673 | – | 0.385 | 0.847 | 0.013 | 0.026 | 0.009 |
| rs1245104 | 0.338 | 0.335 | 0.329 | 0.320 | 0.312 | 0.974 | – | 0.340 | 0.014 | 0.026 | 0.003 |
| rs2011505 | 0.678 | 0.643 | 0.623 | 0.620 | 0.631 | 0.986 | 1 | – | 0.012 | 0.032 | 0.012 |
| D260N | 1 | 1 | 1 | 0.658 | 1 | 1 | 0.616 | 1 | – | 0.013 | 0.002 |
| rs584427 | 0.241 | 0.099 | 0.125 | 0.684 | 0.090 | 0.202 | 0.319 | 0.211 | 1 | – | 0 |
| rs1006826 | 0.236 | 0.217 | 0.213 | 0.239 | 0.974 | 0.187 | 0.154 | 0.195 | 0.775 | 0.001 | – |
The results of the single-locus analysis are summarized in Table 4.The family-based association analysis using FBAT demonstrated strong evidence for association of intronic SNP rs2298526 (P=0.0018; C allele positively associated), and this association was also seen using the PDT test (P=0.0025) with the NTD phenotype in the first data series. Global tests for genotype association were approaching significance (P=0.06) for SNP rs2298526. Upon analysis of the initial five SNPs in the original family set (n=132), we attempted to replicate the findings using an additional 72 families. Single-locus association tests for the five SNPs in this family set did not reveal evidence for association in any marker. To further test for possible influence of NCAM1 on NTDs, we typed five additional SNPs within the large first intronic region where significant SNP rs2298526 is located. In addition, one novel coding SNP found by DHPLC, resulting in the amino acid change D260N, was also followed up in the family set based on its frequency in affected and controls. This secondary screen and all additional SNPs were tested in both sample sets. Analysis of the two data sets as a combined group (n=204) of families, suggested no evidence for association with any of the new SNPs tested. Association with marker rs2298526 in the entire sample set was marginally significant using FBAT (P=0.06). None of the other SNPs tested showed significant evidence for association in any test (Table 4).
Table 4.
Results from single-locus family-based tests of association for NCAM1 SNPs. No P value (n/a) is reported for D260N in the FBAT test due to the low minor allele frequency
| SNP | P value | ||||||
|---|---|---|---|---|---|---|---|
| Series 1 | Series 2 | Combined 204 families | |||||
| FBAT | PDT | FBAT | PDT | FBAT | PDT | GenoPDT | |
| cv236895a | 0.38 | 0.47 | 0.66 | 0.73 | 0.68 | 0.69 | 0.86 |
| rs720023 | 0.041 | 0.035 | 0.31 | 0.17 | 0.28 | 0.37 | 0.28 |
| rs2298526a | 0.0018 | 0.0025 | 0.11 | 0.075 | 0.06 | 0.09 | 0.14 |
| rs723599 | 0.0081 | 0.0059 | 0.12 | 0.077 | 0.22 | 0.37 | 0.20 |
| rs1940699 | 0.012 | 0.012 | 0.16 | 0.11 | 0.23 | 0.27 | 0.10 |
| rs1245113 | 0.64 | 0.57 | 0.78 | 0.81 | 0.59 | 0.77 | 0.60 |
| rs1245104 | 0.46 | 0.49 | 0.83 | 0.55 | 0.49 | 0.36 | 0.51 |
| rs2011505a | 0.33 | 0.22 | 1.00 | 0.75 | 0.43 | 0.41 | 0.87 |
| D260N | n/a | 0.40 | n/a | 0.65 | n/a | 0.41 | 0.41 |
| rs584427a | 0.39 | 0.45 | 0.49 | 0.65 | 0.67 | 0.45 | 0.70 |
| rs1006826a | 0.18 | 0.24 | 0.24 | 0.25 | 0.06 | 0.11 | 0.66 |
The original five SNPs
Results from the haplotype analysis, using a sliding window analysis of three markers, are shown in Table 5 for the haplotype displaying the highest Z-statistic showing association. In the original 132 family sample set, all windows that contain the SNP rs2298526 are significant with the haplotype A-C-G for SNPs rs720023, rs2298526, and rs723599 being the most highly associated (P=0.00028). The transmission of a haplotype window containing up to seven markers remains significant (P=0.05) with inclusion of the SNP rs2298526. In the combined sample set (n=204 families), transmission of this same haplotype remains significant (P=0.02).
Table 5.
Haplotype results for sliding three-marker window across NCAM1
| Window | SNP | Series 1 | Combined set | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cv236895 | rs720023 | rs2298526 | Rs723599 | rs1940699 | rs1245113 | rs1245104 | rs2011505 | D260N | rs584427 | rs1006826 | Haplotype frequency |
P value | Haplotype frequency |
P value | |
| 1 | C | A | C | 0.26 | 0.015 | 0.116 | 0.24 | ||||||||
| 2 | A | C | G | 0.381 | 0.00028 | 0.391 | 0.02 | ||||||||
| 3 | C | G | A | 0.392 | 0.0011 | 0.398 | 0.05 | ||||||||
| 4 | G | A | C | 0.333 | 0.059 | 0.329 | 0.28 | ||||||||
| 5 | A | C | G | 0.191 | 0.102 | 0.177 | 0.012 | ||||||||
| 6 | C | G | G | 0.531 | 0.045 | 0.185 | 0.096 | ||||||||
| 7 | G | G | G | 0.223 | 0.096 | 0.221 | 0.10 | ||||||||
| 8 | G | G | G | 0.333 | 0.28 | 0.332 | 0.31 | ||||||||
| 9 | G | G | T | 0.467 | 0.48 | 0.456 | 0.58 | ||||||||
Immunohistochemistry
The NCAM1 protein expression is present in the paraxial mesoderm at C9 and later in the epithelial somites at stages C10 (data not shown). However, no expression is seen in the open or closed neural tube at these stages. At C11, the first neural tube expression appears, with NCAM1 on a few cells in the ventral rhombencephalon and a faint, diffuse expression dorsally at the contact of the roofplate with the ectoderm (Fig.2b) that is absent at trunk levels. Similar expression is observed at stage C12, characterized by strong expression in discrete ventral areas of the hindbrain (Fig.2d) and individual ventrolateral cells of the spinal cord at all levels (Fig.2e, f). The position of these cells and fiber tracts, distant from the ventricle, is consistent with motoneuron identity. Epithelial somite expression persists (Fig.2f). At C12, human neural tube closure is complete aside from the caudal neuropore (K. Sulik, personal communication, and our observations). At C13, NCAM1 is expressed more robustly in the ventral midbrain, hindbrain, and spinal cord (Fig.2h, j, k) and becomes apparent in the ventral roots as well. As somites mature, expression becomes restricted to a dorsomedial sector in phase with but not surrounding the ventral roots (Fig.2i, j). NCAM1 is also visible in the epithelial mesonephros from C13 (Fig.2i, k). By C16, spinal cord NCAM1 expression has extended to mediolateral axon tracts and continues in the motor roots, but is absent from immature ventricular cells, commissural axons crossing the floorplate or dorsal roots/ganglia (Fig.2l). At C19, this pattern persists; strong annular expression is also seen in a cross-section of spinal nerves (Fig.2m).
Fig. 2.
a Parasagittal section of human embryo stained with DAPI at C11 (24 days), with enlargement in b indicated. b NCAM1 expression is visible on isolated cells of the ventral rhombencephalon (arrowheads). It is also specifically present in its wide, thin roofplate, a region-specific anatomical feature. c Parasagittal-to-transverse section of human embryo at C12 (26 days), DAPI stain, position of d–f indicated. NCAM expression is present in future motor areas of the rhombencephalon (arrowheads, d) and in sparse cells of head mesenchyme, possibly neural crest cells (circled). A few ventrolateral cells of the spinal cord at all levels are NCAM1+ and epithelial somite expression persists (e, f, transverse section). g Parasagittal section of human embryo at C13 (29 days), DAPI stain and photo reconstruction. h Robust NCAM1 expression in the ventral midbrain and hindbrain (arrowheads). i NCAM1 is present in both a trunk-level epithelial somite and in the mesonephros (also see k). j Spinal cord ventral roots are highly immunoreactive and ventral spinal cord expression persists (arrowheads). As somites mature, expression is restricted to a dorsomedial sector in phase with but not in contact with the ventral roots. k NCAM1 is visible in the mesonephros and in ventrolateral spinal cord (arrowheads). l By C16 (37 days), mediolateral axon tracts and ventral roots of the spinal cord are immunoreactive for NCAM1, but not immature ventricular cells, commissural axons crossing the floorplate or dorsal roots or associated ganglia. Dorsal to left. m At C19 (47 days), this pattern persists; strong annular expression is also visible in the cross-section of segmental nerves (asterisks). Dorsal to bottom (drg dorsal root ganglion, h heart, mn mesonephros, op optic vesicle, ot otic vesicle, pa pharyngeal arch(es), pros prosencephalon, s somite, sc spinal cord, v vertebra, vr ventral root). Bar: in a, c, h, i, 0.5 mm; in b, d–f, l, 0.2 mm; in g, 0.71 mm; in j, 0.29 mm; in k, 0.22 mm; in m, 0.27 mm
Discussion
Although rs2298526 appears to be associated with increased risk for neural tube defects, the functional significance of this association remains unclear. Our sample set of American Caucasian simplex lumbosacral myelomeningocele families (n=132 families), revealed a significant association between risk for NTDs and intronic SNP rs2298526 using both the FBAT test (P=0.0018) and the PDT (P=0.0025). Using the HBAT for haplotype association, all pairwise comparisons with SNP rs2298526 were also significant. In a replication study set of 72 additional families, no significant association was detected in this sample set, however; the overall trend for overtransmission of the less common allele of SNP rs2298526 remains significant in the combined sample set.
Our failure to replicate the significant association in the additional Series 2 families added to our screen may be the result of spurious association in the original sample set, or could simply be due to the genetic complexity NTDs in humans. It may also be that the Series 2 families differ in the risk associated with NCAM1 from the initial set of families, or that the smaller sample size (approximately 40% smaller) in Series 2 decreases the power enough so that no association is found. Differences in the family characteristics between the two series may also account for the differences. For instance, in the Series 1 families, 19% of the families had one missing parent, compared with 32% of the families in the Series 2 set. Series 1 families also had a higher proportion of discordant sibling pairs than the Series 2 families (0.92 versus 0.78 per family). Both these differences are statistically significant (P=0.003 and 0.01, respectively). Future follow-up is necessary to assess any role that NCAM1 may have in other ethnic groups or NTD phenotypes.
The significant association could be explained by either some functional significance of the rs2298526 C allele itself, or by LD between this SNP and one that confers a functional role. The SNP is a potential binding site for several transcription factors, some of which are created or eliminated by the C allele, and could explain a possible function. The fact that rs2298526 is within intron 1, a large intron more than 240 kb in size, may suggest a regulatory role. Recent reports have suggested that segments of DNA that are highly conserved across vertebrates from fugu to mouse to humans may represent regions of fundamental importance to vertebrate development (Bejerano et al. 2004; Woolfe et al. 2004). Although none of these are near NCAM1, it is clear that non-coding DNA elements may have a significant functional role.
Alternatively, the association we have found could be due to LD between this marker and some untyped functional variant with an unknown role; several SNPs within this first intron are within a strong LD block. Furthermore, variations that are detected at the nucleotide level may contribute to the development of NTDs only in the presence of other genetic or environmental factors. Such polymorphisms may alter NCAM1 expression or adhesive properties that alone or when combined with other factors may contribute to failure of neural crest elevation and/or neural tube closure. During creation of the neural folds, there is pushing of the presumptive epidermis toward the midline and anchoring of the neural plate to the underlying mesoderm (Alvarez and Schoenwolf 1992). In this process, cells of the epidermis must slide over the mesoderm during neural fold elevation and closure. Newgreen et al. (1997) suggests that the ectopic NCAM expression in the neural tube observed in avian embryos with spontaneous neural tube defects caused a tendency toward greater adhesion, which may impede this process. However, the ectopic expression may be a secondary effect rather than causative. Ectopic expression of NCAM alone in Xenopus embryos does not cause abnormal neural tube closure (Kintner 1988). It is maintained that the differential adhesion between the ectoderm and neural plate plays an important role in neural tube closure, since injection of N-cadherin into a Xenopus embryo results in the failure of the neural tube tissues to separate from the presumptive epidermis (Detrick et al. 1990). Unfortunately, ectopic (in)activation of NCAM was not examined in these experiments.
It has been suggested that NCAM1 may not play an essential role in mammalian neurulation (Copp et al. 2003), based on the Xenopus model and the fact that mice with null mutations in Ncam have normal closure (Cremer et al. 1994). However, interestingly, among the brain defects in null mice is a large reduction in the size of the olfactory bulbs, into which NCAM-expressing neuronal stem cell progeny migrate throughout adult life (Pennartz et al. 2004). Producing only the extracellular domain of the NCAM molecule in mammals results in lethality and serious morphological defects (Rabinowitz et al. 1996), possibly through a dominant negative effect on heterophilic adhesion. Prag et al. (2002) suggest that this puzzling observation may be explained by the fact that both the cytoplasmic and extracellular parts of NCAM are capable of modifying the motility and migration of cells. Indeed, the intracellular domain of NCAM mobilizes integrin signaling through association with the fibroblast growth factor receptor 4 (Cavallaro et al. 2001).
In Xenopus, chicken, and mouse, NCAM appears to be expressed in the immature neural tube during closure, although closer examination reveals that expression in frogs and mice is essentially restricted to migrating neural crest cells or to fiber-projecting, differentiated neurons at later stages. The chicken data, most detailed, vary from our findings in human embryos with respect to expression in the closing avian neural tube, cephalic placodes and derivatives, and the cardiac mesoderm (Thiery et al. 1982). However, our results are concordant with respect to the somites, the presence of positive enteric ganglia and absence of dorsal root ganglion expression. The discordances are probably species-specific, but differences in antibody sensitivity are not to be excluded. We have found that the timing of NCAM1 expression in the dorsal neural tube does not correlate with closure in humans. Nonetheless, it is likely that variations that cause inappropriate or increased NCAM1 expression could incur a risk for NTDs by altering the heterophilic adhesive properties of neural tube cells relative to their NCAM1-expressing somitic environment, rather than homophilic adhesion to each other at the line of closure. In addition, the transitory NCAM1 immunoreactivity we observed in the dorsal roof of the human hindbrain just after neural tube closure may correlate with the propensity of this region to develop occipital encephalocele.
In conclusion, the trend for over-transmission of the rs2298526 C allele is significant in NTD cases. Haplotype analysis in both the original and combined sample sets suggests a role for SNPs located within the first intron of NCAM1. In addition, the expression pattern of the NCAM1 protein in human shows no expression within the neural tube at the time of closure, but it appears to be expressed in the surrounding mesoderm and later in differentiated neurons of the CNS. Our results show a possible involvement of polymorphisms in NCAM1 with the risk for human neural tube defects.
Acknowledgements
The authors wish to thank the families who participated in this study; without their interest, this work could not be performed. We also thank Silke Schmidt for helpful comment on this manuscript and Pat Hurban and Paradigm Genetics for bioinformatics support. We gratefully acknowledge support from grants HD39948, ES11375, NS39818, ES011961, and NS26630.
Appendix
NTD Collaborative Group
Joanna Aben, Children’s Rehabilitation Service, Birmingham, Alabama; Arthur Aylsworth, Cynthia Powell, University of North Carolina, Chapel Hill, North Carolina; Joanne Mackey, Gordon Worley, Duke University Medical Center; Timothy Brei, Connie Buran, Indiana University School of Medicine, Indianapolis, Indiana; Joann Bodurtha, Kathleen Sawin, Virginia Commonwealth University, Richmond, Virginia; Mark S. Dias, Children’s Hospital of Buffalo, Buffalo, N.Y.; Philip Mack, Elli Meeropol, Shriner’s Hospital, Springfield, Massachusetts; Nicole Lasarsky, Carolinas Medical Center, Charlotte, NC; David McLone, Joy Ito, Children’s Memorial Hospital, Chicago, Illinois; W. Jerry Oakes, University of Alabama, Birmingham, Alabama; Marion Walker, University of Utah, Salt Lake City, Utah; Bermans Iskandar, University of Wisconsin Hospitals, Madison, Wisconsin.
Contributor Information
Kristen L. Deak, Center for Human Genetics, Duke University Medical Center, Box 3445, Durham, NC 27710, USA
Abee L. Boyles, Center for Human Genetics, Duke University Medical Center, Box 3445, Durham, NC 27710, USA
Heather C. Etchevers, Département de Génétique Médicale and INSERM U393, Hôpital Necker, Paris, France
Elizabeth C. Melvin, Center for Human Genetics, Duke University Medical Center, Box 3445, Durham, NC 27710, USA
Deborah G. Siegel, Center for Human Genetics, Duke University Medical Center, Box 3445, Durham, NC 27710, USA
Felicia L. Graham, Center for Human Genetics, Duke University Medical Center, Box 3445, Durham, NC 27710, USA
Susan H. Slifer, Center for Human Genetics, Duke University Medical Center, Box 3445, Durham, NC 27710, USA
David S. Enterline, Department of Radiology, Duke University Medical Center, Box 3808, Durham, NC 27710, USA
Timothy M. George, Department of Surgery, Duke University Medical Center, Box 3272, Durham, NC 27710, USA
Michel Vekemans, Département de Génétique Médicale and INSERM U393, Hôpital Necker, Paris, France.
David McClay, Department of Biology, Duke University Medical Center, Box 3445, Durham, NC 27710, USA.
Alexander G. Bassuk, Northwestern University’s Feinberg School of Medicine, Chicago, IL, USA
John A. Kessler, Northwestern University’s Feinberg School of Medicine, Chicago, IL, USA
Elwood Linney, Department of Molecular Genetics and Microbiology, Duke University Medical Center, Box 3020, Durham, NC 27710, USA.
John R. Gilbert, Center for Human Genetics, Duke University Medical Center, Box 3445, Durham, NC 27710, USA
Marcy C. Speer, Center for Human Genetics, Duke University Medical Center, Box 3445, Durham, NC 27710, USA, marcy@chg.duhs.duke.edu, Tel.: +1-919-684-2063, Fax: +1-919-684-0917
References
- Abecasis GR, Cookson WO. GOLD—graphical overview of linkage disequilibrium. BioInformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- Alvarez IS, Schoenwolf GC. Expansion of surface epithelium provides the major extrinsic force for bending of the neural plate. J Exp Zool. 1992;261:340–348. doi: 10.1002/jez.1402610313. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE. Recommendations for a nomenclature system for human gene mutations. Nomenclature Working Group. Hum Mutat. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Balak K, Jacobson M, Sunshine J, Rutishauser U. Neural cell adhesion molecule expression in Xenopus embryos. Dev Biol. 1987;119:540–550. doi: 10.1016/0012-1606(87)90057-1. [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L, Goridis C, Santoni MJ. The mouse NCAM gene displays a biphasic expression pattern during neural tube development. Development. 1993;117:543–552. doi: 10.1242/dev.117.2.543. [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, Wolf JJ, Murray BA. Effects of antibodies against N-cadherin and N-CAM on the cranial neural crest and neural tube. Dev Biol. 1992;153:291–301. doi: 10.1016/0012-1606(92)90114-v. [DOI] [PubMed] [Google Scholar]
- Campbell LR, Dayton DH, Sohal GS. Neural tube defects: a review of human and animal studies on the etiology of neural tube defects. Teratology. 1986;34:171–187. doi: 10.1002/tera.1420340206. [DOI] [PubMed] [Google Scholar]
- Cavallaro U, Niedermeyer J, Fuxa M, Christofori G. NCAM modulates tumour-cell adhesion to matrix by inducing FGF-receptor signalling. Nat Cell Biol. 2001;3:650–657. doi: 10.1038/35083041. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Greene ND, Murdoch JN. The genetic basis of mammalian neurulation. Nat Rev Genet. 2003;4:784–793. doi: 10.1038/nrg1181. [DOI] [PubMed] [Google Scholar]
- Cremer H, Lange R, Christoph A, Plomann M, Vopper G, Roes J, Brown R, Baldwin S, Kraemer P, Scheff S. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature. 1994;367:455–459. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- Cunningham BA, Hemperly JJ, Murray BA, Prediger EA, Brackenbury R, Edelman GM. Neural cell adhesion molecule: structure, immunoglobulin-like domains, cell surface modulation, alternative RNA splicing. Science. 1987;236:799–806. doi: 10.1126/science.3576199. [DOI] [PubMed] [Google Scholar]
- Detrick RJ, Dickey D, Kintner CR. The effects of N-cadherin misexpression on morphogenesis in Xenopus embryos. Neuron. 1990;4:493–506. doi: 10.1016/0896-6273(90)90108-r. [DOI] [PubMed] [Google Scholar]
- Edelman GM. Cell adhesion molecules. Science. 1983;219:450–457. doi: 10.1126/science.6823544. [DOI] [PubMed] [Google Scholar]
- Epstein DJ, Vekemans M, Gros P. Splotch (Sp2H), a mutation affecting development of the mouse neural tube, shows a deletion within the paired homeodomain of Pax-3. Cell. 1991;67:767–774. doi: 10.1016/0092-8674(91)90071-6. [DOI] [PubMed] [Google Scholar]
- Glogarova K, Buckiova D. Changes in sialylation in homozygous Sp2H mouse mutant embryos. Birth Defects Res Part A Clin Mol Teratol. 2004;70:142–152. doi: 10.1002/bdra.20006. [DOI] [PubMed] [Google Scholar]
- Harris MJ, Juriloff DM. Mini-review: toward understanding mechanisms of genetic neural tube defects in mice. Teratology. 1999;60:292–305. doi: 10.1002/(SICI)1096-9926(199911)60:5<292::AID-TERA10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Holst BD, Vanderklish PW, Krushel LA, Zhou W, Langdon RB, McWhirter JR, Edelman GM, Crossin KL. Allosteric modulation of AMPA-type glutamate receptors increases activity of the promoter for the neural cell adhesion molecule, N-CAM. Proc Natl Acad Sci USA. 1998;95:2597–2602. doi: 10.1073/pnas.95.5.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- Hrynkow SH, Morest DK, Bilak M, Rutishauser U. Multiple roles of neural cell adhesion molecule, neural cell adhesion molecule-polysialic acid, and L1 adhesion molecules during sensory innervation of the otic epithelium in vitro. Neuroscience. 1998;87:423–437. doi: 10.1016/s0306-4522(98)00156-0. [DOI] [PubMed] [Google Scholar]
- Jones FS, Prediger EA, Bittner DA, De Robertis EM, Edelman GM. Cell adhesion molecules as targets for Hox genes: neural cell adhesion molecule promoter activity is modulated by cotransfection with Hox-2.5 and –2.4. Proc Natl Acad Sci USA. 1992;89:2086–2090. doi: 10.1073/pnas.89.6.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintner C. Effects of altered expression of the neural cell adhesion molecule, N-CAM, on early neural development in Xenopus embryos. Neuron. 1988;1:545–555. doi: 10.1016/0896-6273(88)90104-3. [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Gilbert JR, Pericak-Vance MA, Hauser ER. Genotype-based association test for general pedigrees: the genotype-PDT. Genet Epidemiol. 2003;25:203–213. doi: 10.1002/gepi.10258. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Kawasaki M, Nakahira M, Kagamiyama H, Kikuchi Y, Okamoto H, Mori K, Yoshihara Y. Molecular diversity in zebrafish NCAM family: three members with different VASE usage and distinct localization. Mol Cell Neurosci. 2001;18:119–130. doi: 10.1006/mcne.2001.1007. [DOI] [PubMed] [Google Scholar]
- Moase CE, Trasler DG. N-CAM alterations in splotch neural tube defect mouse embryos. Development. 1991;113:1049–1058. doi: 10.1242/dev.113.3.1049. [DOI] [PubMed] [Google Scholar]
- Neale SA, Trasler DG. Early sialylation onN-CAM in splotch neural tube defect mouse embryos. Teratology. 1994;50:118–124. doi: 10.1002/tera.1420500206. [DOI] [PubMed] [Google Scholar]
- Newgreen DF, Kerr RS, Minichiello J, Warren N. Changes in cell adhesion and extracellular matrix molecules in spontaneous spinal neural tube defects in avian embryos. Teratology. 1997;55:195–207. doi: 10.1002/(SICI)1096-9926(199703)55:3<195::AID-TERA4>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- O’Rahilly R, Muller F. The two sites of fusion of the neural folds and the two neuropores in the human embryo. Teratology. 2002;65:162–170. doi: 10.1002/tera.10007. [DOI] [PubMed] [Google Scholar]
- Pennartz S, Belvindrah R, Tomiuk S, Zimmer C, Hofmann K, Conradt M, Bosio A, Cremer H. Purification of neuronal precursors from the adult mouse brain: comprehensive gene expression analysis provides new insights into the control of cell migration, differentiation, and homeostasis. Mol Cell Neurosci. 2004;25:692–706. doi: 10.1016/j.mcn.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Prag S, Lepekhin EA, Kolkova K, Hartmann-Petersen R, Kawa A, Walmod PS, Belman V, Gallagher HC, Berezin V, Bock E, Pedersen N. NCAM regulates cell motility. J Cell Sci. 2002;115:283–292. doi: 10.1242/jcs.115.2.283. [DOI] [PubMed] [Google Scholar]
- Rabinowitz JE, Rutishauser U, Magnuson T. Targeted mutation of Ncam to produce a secreted molecule results in a dominant embryonic lethality. Proc Natl Acad Sci USA. 1996;93:6421–6424. doi: 10.1073/pnas.93.13.6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser U, Jessell TM. Cell adhesion molecules in vertebrate neural development. Physiol Rev. 1988;68:819–857. doi: 10.1152/physrev.1988.68.3.819. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SJ, Akeson R. Expression of the unique NCAM VASE exon is independently regulated in distinct tissues during development. J Cell Biol. 1990;111:2089–2096. doi: 10.1083/jcb.111.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulik KK, Zucker RM, Dehart DB, et al. Normal patterns of neural tube closure differ in the human and mouse. Proc Greenwood Genet Ctr. 1998;18:129–130. [Google Scholar]
- Thiery JP, Duband JL, Rutishauser U, Edelman GM. Cell adhesion molecules in early chicken embryogenesis. Proc Natl Acad Sci USA. 1982;79:6737–6741. doi: 10.1073/pnas.79.21.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen MI, Kalousek DK, Chernoff GF, Juriloff D, Harris M, McGillivray BC, Yong SL, Langlois S, Macleod PM, Chitayat D, Freidman JM, Wilson D, McFadden D, Pantzar J, Ritchie S, Hall JG. Evidence for multi-site closure of the neural tube in humans. Am J Med Genet. 1993;47:723–743. doi: 10.1002/ajmg.1320470528. [DOI] [PubMed] [Google Scholar]
- Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, Krull M, Matys V, Michael H, Ohnhauser R, Pruss M, Schacherer F, Thiele S, Urbach S. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, Abnizova I, Gilks W, Edwards YJ, Cooke JE, Elgar G. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2004;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]