SUMMARY
It is often presumed that, in vivo, the initiation of RNA synthesis by DNA-dependent RNA polymerases occurs using NTPs alone. Here, using the model Gram-negative bacterium Pseudomonas aeruginosa, we demonstrate that depletion of the small-RNA-specific exonuclease, Oligoribonuclease, causes the accumulation of 2- to ~4-nt RNAs, “nanoRNAs”, which serve as primers for transcription initiation at a significant fraction of promoters. Widespread use of nanoRNAs to prime transcription initiation is coupled with global alterations in gene expression. Our results, obtained under conditions in which the concentration of nanoRNAs is artificially elevated, establish that small RNAs can be used to initiate transcription in vivo, challenging the idea that all cellular transcription occurs using NTPs alone. Our findings further suggest that nanoRNAs could represent a distinct class of functional small RNAs that can affect gene expression through direct incorporation into a target RNA transcript rather than through a traditional antisense-based mechanism.
INTRODUCTION
Transcription initiation in all cells is often presumed to occur using NTPs alone. However, it is well established that prokaryotic and eukaryotic RNA polymerase (RNAP) can utilize 2- to ~8-nt RNAs to prime transcription initiation in vitro (Chamberlin and Ring, 1973; Di Nocera et al., 1975; Grachev et al., 1984; Hoffman and Niyogi, 1973; Learned and Tjian, 1982; Minkley and Pribnow, 1973; Niyogi and Stevens, 1965; Ruetsch and Dennis, 1987; Samuels et al., 1984; Smagowicz and Scheit, 1978). Nevertheless, whether 2- to ~8-nt RNAs can also prime transcription initiation in vivo has not previously been tested. Here we demonstrate that conditions can be achieved in vivo such that a significant fraction of transcription involves the use of 2- to ~4-nt “nanoRNAs” (Mechold et al., 2007) that serve as primers to initiate transcription. We further demonstrate that widespread use of nanoRNAs to prime transcription initiation is coupled with global alterations in gene expression. Our findings indicate that nanoRNA-mediated priming may represent a previously unrecognized mechanism to control gene expression.
RESULTS
Experimental strategy: determine the effect of depleting Oligoribonuclease
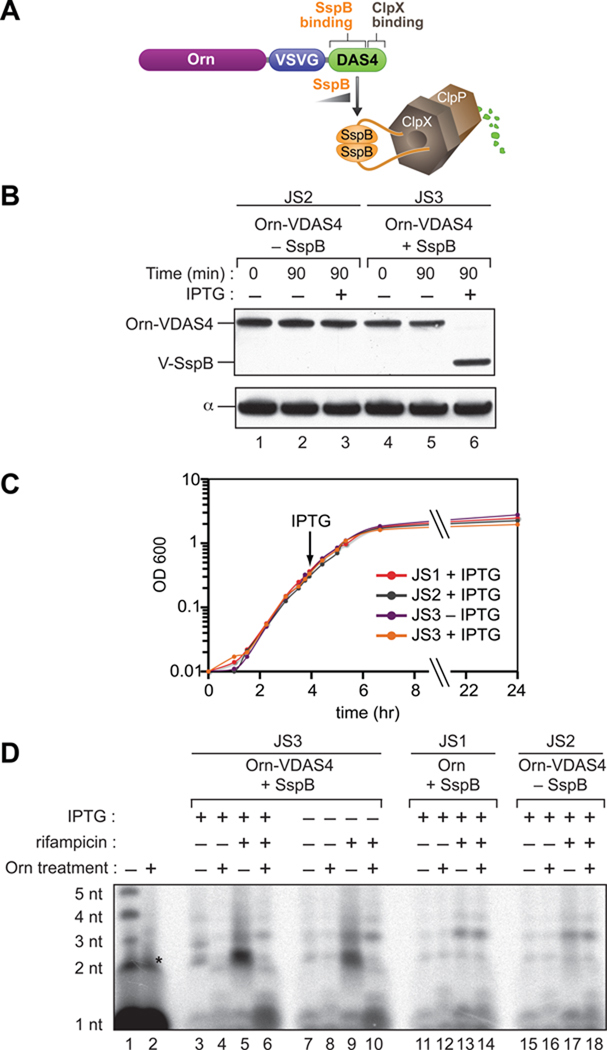
In Escherichia coli, the conversion of 2- to ~5-nt RNAs to mononucleotides is accomplished through the action of a highly conserved 3' to 5' exonuclease, Oligoribonuclease (Orn) (Ghosh and Deutscher, 1999). Thus, inactivation of Orn in E. coli results in the accumulation of 2- to ~5-nt RNAs (Ghosh and Deutscher, 1999); these species have been termed “nanoRNAs” to distinguish them from other classes of small RNAs (Mechold et al., 2007). Thus, to establish whether nanoRNAs can act as primers for transcription initiation in vivo we sought to genetically engineer cells such that we could increase the intracellular concentration of nanoRNAs in a controlled manner through depletion of Orn. Because Orn is essential in E. coli (Ghosh and Deutscher, 1999) we chose to study the effects of depleting Orn in a closely related bacterium, Pseudomonas aeruginosa, which remains viable in the absence of Orn (Jacobs et al., 2003). To deplete Orn in a controlled manner in P. aeruginosa cells we employed a method that exploits the ClpXP protease (Figure 1A) (Castang et al., 2008; McGinness et al., 2006). In particular, proteins can be targeted for ClpXP-dependent degradation by addition of an 11-residue peptide, the so-called DAS4 tag, to their C-termini. DAS4 tagged proteins are targeted to the ClpXP protease through the action of an adaptor protein, SspB. Thus, to construct a P. aeruginosa strain in which Orn can be targeted for depletion specifically in the presence of SspB we placed a DAS4 degradation tag along with a VSV-G epitope tag at the C-terminus of the chromosomal copy of orn in a strain in which sspB was under the control of an IPTG-inducible promoter (creating strain JS3) (Figure 1A). To determine whether induction of SspB in JS3 cells results in the depletion of the DAS4-VSV-G tagged Orn (Orn-VDAS4) we monitored the abundance of Orn-VDAS4 using an antibody against VSV-G. The abundance of Orn-VDAS4 in JS3 cells grown in the absence of IPTG was slightly lower (~2-fold) than the abundance of Orn-VDAS4 in cells of a control strain that lacked sspB (strain JS2) (Figure 1B, compare lanes 1 and 2 with lanes 4 and 5). In contrast, when JS3 cells were grown in the presence of IPTG, the Orn-VDAS4 was fully depleted (Figure 1B, lane 6). Furthermore, depletion of Orn did not result in an alteration in the cellular growth rate (Figure 1C).
Figure 1. Controlled depletion of Orn in P. aeruginosa.
A. Strategy to enable controlled depletion of Orn. Schematic depicts how the presence of SspB targets Orn-VDAS4 for ClpXP-mediated degradation.
B. Controlled depletion of Orn. JS2 cells (lanes 1–3), which express Orn-VDAS4, or JS3 cells (lanes 4–6), which express Orn-VDAS4 and carry VSV-G-tagged SspB (V-SspB) under the control of an IPTG-inducible promoter, were first grown to mid-log (time 0), then grown for a further 90 minutes either in the absence (−) or presence of IPTG (+). Cells were harvested at the indicated time points and analyzed for protein content by Western blot using an antibody against the VSV-G tag (upper panel), or, to control for sample loading, an antibody against the α subunit of RNAP (bottom panel).
C. Depletion of Orn does not affect the cellular growth rate. Growth of JS3 cells, JS2 cells and JS1 cells (which express untagged Orn and carry V-SspB under the control of an IPTG-inducible promoter) was monitored over a 24 hour period. Arrow indicates the time point at which IPTG (when present) was added to cells.
D. Accumulation of nanoRNAs in vivo. JS3 cells, JS2 cells or JS1 cells, were grown to mid-log in the presence of 32Pi, then grown for a further 90 minutes either in the absence (−) or presence of IPTG (+). Cells were either harvested immediately (–) or grown in the presence of rifampicin (+) for an additional 60 minutes prior to harvesting. The acid soluble fraction obtained from cell extracts was either treated with purified Orn (+) or untreated (–) prior to electrophoresis on a 22.5% denaturing acrylamide gel. Sizes of labeled nanoRNAs were estimated based upon comparison to RNA standards that carry a 5' triphosphate group and a 3' hydroxyl group (lane 1). RNA standards carrying a 5' monophosphate group and a 3' hydroxyl group migrate within a similar size range as the 5' triphosphate-carrying standards (Figure S5). The asterisk in lane 2 indicates an Orn-insensitive impurity in the RNA standards that migrates at the same position as the 2 nt transcript. [We note that non-depleted JS3 cells carry ~2-fold less Orn-VDAS4 than JS2 cells (panel B). Furthermore, non-depleted JS3 cells accumulate 2 nt nanoRNAs upon treatment with rifampicin whereas JS2 cells do not (compare lanes 9 and 17). Thus, even a modest reduction in the abundance of Orn appears to enable the accumulation of detectable quantities of 2 nt nanoRNAs in rifampicin treated P. aeruginosa cells.]
Depletion of Orn in P. aeruginosa results in the accumulation of 2- to ~4-nt nanoRNAs
Prior studies indicate that 2- to ~5-nt nanoRNAs accumulate in E. coli cells lacking Orn activity (Ghosh and Deutscher, 1999). We next determined whether nanoRNAs also accumulate in P. aeruginosa cells lacking Orn activity. To do this, we grew cells in the presence of radioactive inorganic phosphate (32Pi) and isolated radiolabeled small RNAs by acid extraction. To detect nanoRNAs we subjected the extract to electrophoresis on a high-percentage urea-polyacrylamide gel and visualized radiolabeled species by autoradiography (Figure 1D). To distinguish nanoRNAs from other radiolabeled species we treated half of each extract with purified Orn prior to electrophoresis. Thus, radiolabeled nanoRNAs should be present in untreated extracts and absent from Orn-treated extracts, while the abundance of other radiolabeled species should be unaffected by Orn treatment.
The untreated extract isolated from Orn-depleted JS3 cells contained prominent radiolabeled species that migrated near the same positions as 2-nt, 3-nt, and 4-nt RNA standards (Figure 1D, lane 3). These species were absent from Orn-treated extracts (lane 4) and were not detected in extracts obtained from non-depleted JS3 cells (lane 7), extracts obtained from a control strain in which Orn lacks the VDAS4 tag (strain JS1) (lane 11), or extracts obtained from a control strain that lacked sspB (strain JS2) (lane 15). These results suggest that depletion of Orn in P. aeruginosa results in the accumulation of 2- to ~4-nt nanoRNAs.
As an additional test of whether depleting Orn prevents the degradation of nanoRNAs in P. aeruginosa we determined the effect of treating Orn-depleted cells with the transcription inhibitor rifampicin. Rifampicin inhibits transcription in vitro by blocking the addition of the 3rd nt to the nascent RNA (Campbell et al., 2001; McClure and Cech, 1978). Thus, if depleting Orn compromises the cells’ ability to degrade nanoRNAs, addition of rifampicin to Orn-depleted cells should result in a pronounced accumulation of 2-nt nanoRNAs compared with cells that were not treated with rifampicin. Consistent with this hypothesis, extracts isolated from JS3 cells grown in the presence of rifampicin contained prominent Orn-sensitive species that migrated near the same position as a 2- nt RNA standard (Figure 1D, lanes 5 and 9). These species were not detected in extracts obtained from a control strain in which Orn lacks the VDAS4 tag (strain JS1) (lane 13), or a control strain that lacked sspB (strain JS2) (lane 17). The pronounced accumulation of 2-nt nanoRNAs in Orn-depleted cells treated with rifampicin provides further evidence that depleting Orn in P. aeruginosa severely compromises the cells’ ability to degrade nanoRNAs.
Priming of transcription initiation with 2- to 4-nt RNAs can cause transcription start site shifting by P. aeruginosa RNAP in vitro
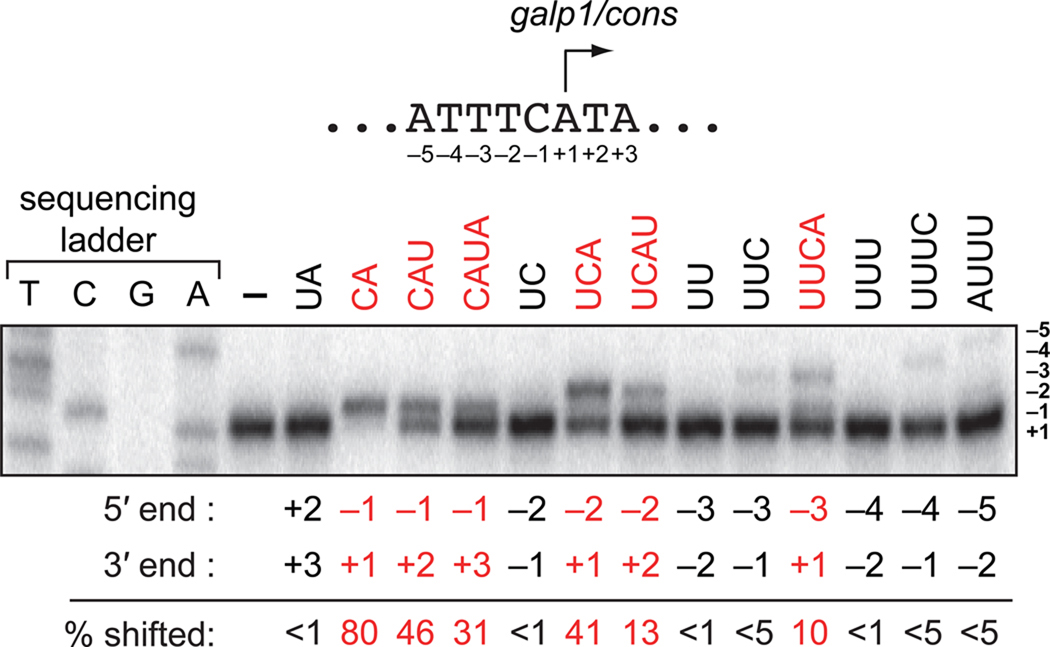
Prior studies with E. coli RNAP indicate that use of 2- to 4-nt RNAs to prime transcription initiation in vitro can, in certain cases, result in transcription start site (TSS) shifting to positions −3, −2, or −1 (Di Nocera et al., 1975; Grachev et al., 1984; Hoffman and Niyogi, 1973; Minkley and Pribnow, 1973; Ruetsch and Dennis, 1987; Smagowicz and Scheit, 1978). In particular, studies with E. coli RNAP suggest that 2- to 4-nt RNAs are effective primers during transcription initiation in vitro provided the 5′ end of the RNA is complementary to sequences between positions −3 and +1 and the 3′ end is complementary to position +1, +2 or +3. To determine if, as expected, these same “rules” apply to P. aeruginosa RNAP we performed in vitro transcription experiments using purified P. aeruginosa RNAP and a linear DNA template carrying a well-characterized E. coli derived promoter (galP1/cons) (Figure 2). Reactions were performed in the presence or absence of a series of 2- to 4-nt RNAs that were complementary to promoter sequences spanning positions −5 to +3 with respect to the TSS observed in the presence of NTPs only (position +1). Reactions were performed by adding a mixture of the small RNA and NTPs (each present at 100 µM) to preformed RNAP-promoter open complexes; in this manner, we could assess the extent to which each of the complementary RNAs could effectively compete with NTPs for use as primers during transcription initiation.
Figure 2. Priming of transcription initiation with 2- to 4-nt RNAs can alter the TSS in vitro.
Top shows galP1/cons promoter sequences extending from position −5 to +3 (the galP1/cons promoter is a derivative of the E. coli galP1 promoter with a consensus extended −10 element). Bottom shows results of primer extension analysis of RNA transcripts produced during in vitro transcription assays performed using a DNA fragment containing the galP1/cons promoter (10 nM). Assays were done using P. aeruginosa RNAP (50 nM) in the presence of 100 µM NTPs in the absence (−) or presence of 100 µM of the indicated 2- to 4-nt RNA (see Supplemental Information for details). The position of the 5′ and 3′ end of each small RNA is indicated below the gel along with the percentage of transcripts shifted by each RNA (averages of duplicate measurements). Highlighted in red are RNAs that effectively compete with NTPs and shift the TSS of >10% of transcripts initiated from galP1/cons.
The results presented in Figure 2 indicate that six of the RNAs we tested (the 2-nt RNA complementary to −1/+1, the 3-nt RNA complementary to −1/+2, the 4-nt RNA complementary to −1/+3, the 3-nt RNA complementary to −2/+1, the 4-nt RNA complementary to −2/+2, and the 4-nt RNA complementary to −3/+1) could shift the TSS of at least 10% of the transcripts initiated from galP1/cons. Furthermore, three of these species (the 2-nt RNA complementary to −1/+1, the 3-nt RNA complementary to −1/+2, and the 3-nt RNA complementary to −2/+1) were the most effective, shifting the TSS of greater than 40% of the transcripts initiated from galP1/cons. Thus, the data presented in Figure 2 indicate that 2- to 4-nt RNAs that are complementary to the DNA template can effectively compete with NTPs for use as primers by P. aeruginosa RNAP during transcription initiation in vitro provided the 5′ end of the RNA is complementary to sequences between positions −3 and +1 and the 3′ end is complementary to position +1, +2 or +3. As anticipated, these findings are consistent with prior in vitro analysis of E. coli RNAP (Di Nocera et al., 1975; Grachev et al., 1984; Hoffman and Niyogi, 1973; Minkley and Pribnow, 1973; Ruetsch and Dennis, 1987; Smagowicz and Scheit, 1978).
Accumulation of 2- to ~4-nt nanoRNAs causes widespread transcription start site shifting in vivo
As illustrated by the analysis presented in Figure 2, priming of transcription initiation with 2- to 4-nt RNAs in vitro can result in TSS shifting to positions −3, −2, or −1. Therefore we hypothesized that if the 2- to 4-nt nanoRNAs that accumulate in Orn-depleted P. aeruginosa can prime transcription initiation, TSS shifting to positions −3, −2, or −1 would be observed at a significant fraction of promoters in Orn-depleted cells compared with non-depleted cells. To test this hypothesis we used high-throughput sequencing to analyze the sequences of the 5′ ends of primary transcripts isolated from Orn-depleted cells and non-depleted cells. As a control to establish that any TSS shifting we observed upon depletion of Orn was a direct consequence of the accumulation of nanoRNAs, we also analyzed the 5′ ends of primary transcripts isolated from Orn-depleted cells carrying a plasmid that directed the synthesis of either wild-type Orn or a heterologous nanoRNA-degrading enzyme, the NrnB protein from Bacillus subtilis (Fang et al., 2009); unlike Orn, NrnB is a member of the DHH family of predicted phosphoesterases (Aravind and Koonin, 1998; Fang et al., 2009).
We note that the identification of the 5′ ends of primary transcripts is typically performed by analyzing only those transcripts carrying a 5′ triphosphate [see, for example, (Cho et al., 2009)]. As 5′ triphosphate-carrying transcripts cannot arise as products of RNA degradation, analysis of 5′ triphosphate-carrying transcripts provides a way of identifying bona-fide promoters and their associated TSSs. However, given that the 5′ phosphorylation status of nanoRNAs is unknown we reasoned that a significant fraction of the primary transcripts that result from nanoRNA-primed initiation might not carry a 5′ triphosphate. For this reason we sequenced the 5′ ends of transcripts carrying a 5′ triphosphate group (to identify bona-fide promoters) and, in parallel, we sequenced the 5′ ends of all transcripts regardless of the phosphorylation status of the 5′ end (to allow us to observe priming events that involved nanoRNAs that did not carry a 5′ triphosphate).
We generated cDNAs derived from RNA 5′ ends (see Figures S1 and S2) and sequenced these cDNAs using a Sequence by Oligonucleotide Ligation and Detection (SOLiD) Analyzer. Using the sequencing data obtained from the analysis of transcripts carrying a 5′ triphosphate group isolated from non-depleted cells we first defined a set of 842 putative promoters along with their associated primary TSS (i.e. position +1) (data not shown). To do this, we aligned sequencing reads against the P. aeruginosa PAO1 genome and identified reads that could be placed at a unique genomic location within ~300 bp upstream of an annotated open reading frame (or reads that aligned directly to the 5′ end of an annotated non-coding RNA) (see Experimental Procedures for details on the identification of TSSs). Next, in order to make meaningful comparisons between the various samples, we parsed our list of 842 candidate promoters down to a set of 148 “high quality” promoters. This high quality set represented candidate promoters for which we obtained sufficient sequencing depth in all of the samples (Tables S1, S2, and S3).
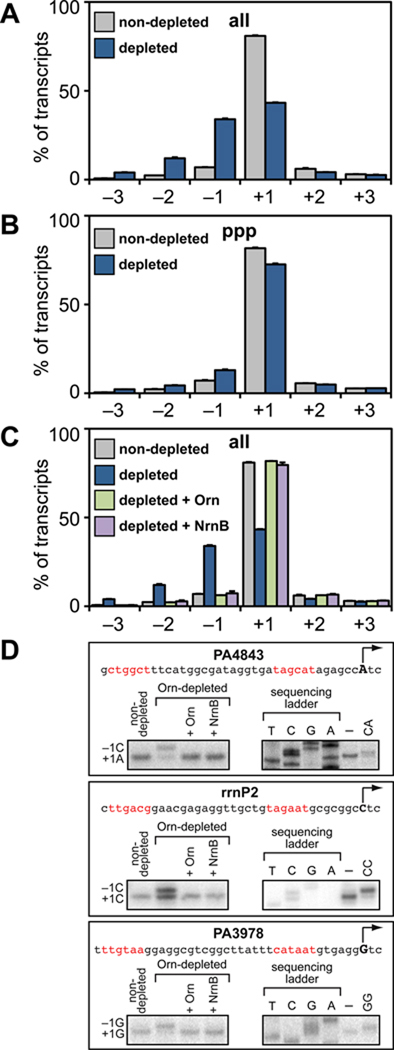
Using the data obtained from the 148 high quality promoters we compared the average distribution of TSSs between positions −3 to +3 relative to the primary TSS in Orn-depleted cells with non-depleted cells. Analysis of the 5′ ends of all transcripts revealed prominent TSS shifting to positions −1, −2, and −3 in Orn-depleted cells compared with non-depleted cells (Figure 3A, Tables S1, S2, and S3). In particular, the average percentage of transcripts initiated at position +1 decreased from 80.9% (in non-depleted cells) to 43.3% (in Orn-depleted cells), whereas the average percentage of transcripts initiated at position −1 increased from 6.9% to 34%, the average percentage of transcripts initiated at position −2 increased from 2.4% to 12%, and the average percentage of transcripts initiated at position −3 increased from 0.7% to 4%. Furthermore, individual analysis of each of the 148 promoters revealed that 83 of these (56%) showed a greater than 30% reduction in the percentage of transcripts initiated at position +1 coupled with a corresponding increase in the percentage of transcripts initiated at positions −1, −2, or −3 upon depletion of Orn (Table S1).
Figure 3. NanoRNAs prime transcription initiation in vivo.
A. – C. Accumulation of nanoRNAs leads to TSS shifting as detected by high-throughput sequencing. Graphs show average percentage of all transcripts (panels A and C) or 5' triphosphate carrying transcripts (panel B) initiated at positions −3 to +3 relative to the primary TSS for 148 promoters. Data were derived from the analysis of transcripts isolated from cells of strain JS1.F harboring plasmid pPSV35 (non-depleted), and cells of strain JS3.F harboring plasmids pPSV35 (depleted), pPAOrn (depleted + Orn), or pNrnB (depleted + NrnB). Plotted are the averages and standard deviations for two independent measurements (Table S3).
D. TSS shifting observed in vivo can be recapitulated in vitro using 2 nt RNAs. Shown is the sequence of the rrnP2 promoter, the promoter associated with PA4843, and the promoter associated with PA3978. The primary TSS (position +1) is indicated by the arrow and putative promoter elements are in red. The box on the bottom left of each panel shows the results of primer extension analysis performed using transcripts isolated from the indicated cells. The box on the bottom right of each panel shows primer extension analysis of RNA transcripts produced during in vitro transcription assays performed in the absence (–) or presence of the indicated 2 nt RNA (see Supplemental Information for details).
In contrast to the analysis of all 5′ ends, TSS shifting was significantly less prominent upon Orn depletion when only transcripts carrying a 5′ triphosphate group were analyzed (Figure 3B, Tables S1, S2, and S3). Specifically, the average percentage of transcripts initiated at position +1 decreased slightly from 81.7% (in non-depleted cells) to 72.6% (in Orn-depleted cells), whereas the average percentage of transcripts initiated at position −1 increased from 7.1% to 13%, the average percentage of transcripts initiated at position −2 increased from 2.3% to 4.4%, and the average percentage of transcripts initiated at position −3 increased from 0.5% to 2.2%. Furthermore, individual analysis of each of the 148 promoters revealed that only 11 (7%) showed a greater than 30% reduction in the percentage of transcripts initiated at position +1 (Table S1). Based on the 5′ end analysis presented in Figures 3A and 3B we conclude that depletion of Orn leads to widespread TSS shifting to upstream positions. In addition, given that TSS shifting was less pronounced in Orn-depleted cells when only 5′ triphosphate ends were analyzed (Figure 3B), we infer that a significant fraction of transcripts initiated from positions −1, −2 and −3 in Orn-depleted cells do not carry a 5′ triphosphate.
We next determined whether the TSS shifting observed upon depletion of Orn was a direct consequence of the accumulation of nanoRNAs and not simply due to an unrelated consequence of Orn depletion. To do this, we calculated the average distribution of TSSs when Orn-VDAS4 was depleted in the presence of a plasmid that directed the synthesis of wild-type Orn. As expected, the average distribution of TSSs observed when Orn-VDAS4 was depleted in the presence of wild-type Orn was essentially identical to that observed in non-depleted cells (Figure 3C, Tables S1, S2, and S3). Next, we determined the average distribution of TSSs when Orn-VDAS4 was depleted in the presence of a plasmid that directed the synthesis of a heterologous nanoRNA-degrading enzyme, the Bacillus subtilis NrnB protein (Fang et al., 2009). The average distribution of TSSs observed when Orn-VDAS4 was depleted in the presence of NrnB was also essentially identical to that observed in non-depleted cells (Figure 3C, Tables S1, S2, and S3). Control experiments established that the presence of either wild-type Orn or NrnB prevented the accumulation of nanoRNAs in cells in which Orn-VDAS4 was depleted (Figure S3). We therefore conclude that the TSS shifting observed upon depletion of Orn (Figures 3A and 3B) is a direct consequence of the accumulation of nanoRNAs and not simply due to an unrelated consequence of Orn depletion.
The widespread transcription start site shifting observed upon depletion of Orn is a result of widespread nanoRNA-mediated priming
Several lines of evidence support the conclusion that the widespread TSS shifting observed in Orn-depleted cells are the result of widespread use of 2- to 4-nt nanoRNAs as primers for transcription initiation. First, as illustrated in Figure 2, for any given promoter, three distinct 2–4 nt RNAs can effectively compete with NTPs for use as primers during transcription initiation and shift the TSS to position −1, two distinct 2–4 nt RNAs can shift the TSS to position −2, and one distinct 2–4 nt RNA can shift the TSS to position −3. (Note that 2- to 4-nt RNAs that, in principle, could cause a TSS shift to downstream positions are not effective primers.) Thus, on average, priming with a mixture of 2- to 4-nt nanoRNAs of random sequence would be predicted to shift TSSs primarily to position −1, followed by positions −2 and −3; this pattern of TSS shifting is precisely what is observed in Orn-depleted cells compared with non-depleted cells (Figure 3, Table S3). Second, the TSS shifting to position −1 detected upon depletion of Orn at the rrnP2 promoter, the promoter associated with the PA4843 gene, and the promoter associated with the PA3978 gene could be recapitulated in vitro through the addition of a 2-nt RNA complementary to positions −1/+1 (Figure 3D). Finally, although it is well established that alterations in the intracellular concentrations of NTPs can lead to changes in the position where transcription initiation begins (Liu et al., 1994; Meng et al., 2004; Qi and Turnbough, 1995; Tu and Turnbough, 1997), the TSS shifts observed upon depletion of Orn cannot be explained on the basis of alterations in NTP concentrations for three reasons: (i) If alterations in NTP concentrations were the cause of TSS shifting, then the shifted transcripts would be predicted to carry a 5′ triphosphate group. However TSS shifting was considerably more pronounced when the 5′ ends of all transcripts were analyzed (the average percentage of transcripts initiated from positions −1, −2 and −3 increased from ~10% to ~50%) (Figure 3A, Table S3) compared with the analysis of only transcripts carrying a 5′ triphosphate group (the average percentage of transcripts initiated from positions −1, −2 and −3 increased from ~10% to ~20%) (Figure 3B, Table S3). (ii) If alterations in NTP concentrations were the cause of TSS shifting one would expect to observe TSS shifting to both upstream and downstream positions (Lewis and Adhya, 2004), whereas initiation with nanoRNAs would be expected to result in only upstream TSS shifting (Figure 2). We found that pronounced TSS shifting only to upstream positions (−1, −2 and −3) was observed upon depletion of Orn (Figure 3, Table S3). (iii) The absolute concentration of each NTP was not significantly changed in Orn-depleted cells compared with non-depleted cells (Figure S4).
Widespread use of nanoRNAs to prime transcription initiation is coupled with global alterations in gene expression
To establish the effect of widespread use of nanoRNAs to prime transcription initiation on gene expression we performed a microarray analysis to compare the global gene expression profile of Orn-depleted cells to the global gene expression profile of non-depleted cells. From this comparison we identified a total of 1,158 genes whose expression was significantly altered (P< 0.05) by a factor of 2 or more as a consequence of Orn depletion (Figure 4, Table S4). This represents a change in the abundance of ~20% of all known transcripts in P. aeruginosa. Specifically, expression of 586 genes increased by a factor of 2 or more (by a maximum of 41-fold) upon depletion of Orn, whereas the expression of 572 genes decreased by a factor of 2 or more (by a maximum of 40-fold).
Figure 4. NanoRNA-mediated priming of transcription initiation in vivo leads to changes in gene expression.
A. and B. Effect of Orn depletion on gene expression in P. aeruginosa as determined by DNA microarray. Data were derived from the analysis of transcripts isolated from cells of strain JS1.F harboring plasmid pPSV35 (non-depleted), and cells of strain JS3.F harboring plasmids pPSV35 (Orn-depleted), pPAOrn (Orn-depleted + Orn), or pNrnB (Orn-depleted + NrnB). The table in panel A shows the number of genes whose expression changes by a factor of 2 or more upon depletion of Orn-VDAS4, or upon depletion of Orn-VDAS4 in the presence of either wild-type Orn (supplied by plasmid pPAOrn), or NrnB (supplied by plasmid pNrnB). Panel B shows a heatmap representation of the 1,158 genes whose expression changes by a factor of 2 or more upon depletion of Orn-VDAS4. Also shown are the corresponding effects on expression of these genes when either wild-type Orn, or NrnB, are supplied ectopically and Orn-VDAS4 is depleted.
To establish to what extent the changes in gene expression observed upon depletion of Orn were a consequence of the accumulation of nanoRNAs, we compared the global gene expression profile of non-depleted cells with Orn-VDAS4-depleted cells carrying a plasmid that directed the synthesis of either wild-type Orn or NrnB. In contrast to the widespread changes in gene expression observed upon depletion of Orn in the absence of a complementing nanoRNA-degrading enzyme, we identified only 39 genes and 46 genes whose expression was altered by a factor of 2 or more when Orn-VDAS4 was depleted in the presence of wild-type Orn or NrnB, respectively (Figure 4, Table S4). Thus, the data presented in Figure 4 show that depletion of Orn leads to widespread changes in gene expression that can be complemented by providing a nanoRNA-degrading enzyme in trans. We conclude that the global alterations in gene expression observed upon depletion of Orn are a direct consequence of the accumulation of nanoRNAs and not simply due to an unrelated consequence of Orn depletion.
DISCUSSION
NanoRNAs prime transcription initiation in vivo
Here we investigated whether extremely small RNAs, “nanoRNAs”, can act as primers for transcription initiation in vivo. To accomplish this goal, we determined the effect of increasing the intracellular levels of 2- to ~4-nt nanoRNAs in a controlled manner through depletion of Orn (Figure 1). By use of high-throughput sequencing we established that increasing the levels of 2- to ~4-nt nanoRNAs results in dramatic TSS shifting at a significant fraction of promoters (Figure 3, Tables S1, S2, and Table S3). These dramatic TSS shifts are the result of widespread use of 2- to 4-nt nanoRNAs as primers for transcription initiation. Thus, our findings provide evidence that small RNAs can be used to initiate transcription in vivo. In addition, we show by microarray analysis that widespread use of nanoRNAs to prime transcription initiation is correlated with global changes in gene expression (Figure 4, Table S4).
Currently we do not have any information regarding how nanoRNAs are produced in cells. In principle, there are at least three distinct pathways by which nanoRNAs could be generated. First, nanoRNAs could be generated as end-products of RNA metabolism by RNA-degrading enzymes unable to metabolize small RNAs. Second, nanoRNAs could be generated by RNAP during abortive transcription initiation, a process whereby the RNAP-promoter initial transcribing complex engages in cycles of synthesis and release of ~2- to 15-nt RNA transcripts prior to productive initiation (Goldman et al., 2009). Third, nanoRNAs could be generated by endonucleolytic cleavage of the nascent RNA during transcription. Such cleavage events might occur during transcription proofreading (Zenkin et al., 2006) or during transcription elongation to enable backtracked elongation complexes to resume RNA synthesis (Borukhov et al., 1992; Fish and Kane, 2002; Surratt et al., 1991).
Orn suppresses nanoRNA-mediated priming
Our findings suggest that cellular perturbations that alter the balance between nanoRNA-primed transcription and NTP-initiated transcription can significantly impact global gene expression. In this regard, nanoRNA degrading enzymes such as Orn serve to suppress nanoRNA-primed transcription. Furthermore, cellular factors that influence the activities of nanoRNA degrading enzymes would be predicted to alter the extent of nanoRNA-priming and, in turn, influence gene expression.
Orn homologues have been identified in β Proteobacteria, γ Proteobacteria, Actinomycetes and eukaryotes (Mechold et al., 2007; Zhang et al., 1998; Zuo and Deutscher, 2001). In Firmicutes such as Bacillus subtilis, nanoRNA degradation is performed by functional analogues of Orn, NrnA and NrnB, which carry no sequence similarity to Orn (Fang et al., 2009; Mechold et al., 2007). Orn is not essential in P. aeruginosa (Jacobs et al., 2003) (Figure 1) and Streptomyces (Ohnishi et al., 2000). Furthermore, loss of both NrnA and NrnB does not alter the viability of Bacillus subtilis (Fang et al., 2009). In contrast, Orn is essential in E. coli, likely due to the toxic effects of the nanoRNAs that accumulate in cells lacking Orn function (Fang et al., 2009; Ghosh and Deutscher, 1999; Mechold et al., 2007). Our findings suggest that altered gene expression as a consequence of widespread nanoRNA-priming of transcription initiation could, in part or wholly, account for the apparent toxic effects of nanoRNAs in E. coli.
NanoRNA-mediated priming as a mechanism to alter gene expression
The finding that widespread use of nanoRNAs to prime transcription initiation is coupled with global changes in gene expression suggest that nanoRNAs might represent a distinct class of regulatory small RNAs that alter gene expression through a mechanism of direct incorporation into the 5' end of a target RNA transcript rather than through an antisense-based mechanism. We propose at least three mechanistically distinct ways that nanoRNA-priming could affect gene expression. First, the phosphorylation status of the 5' end is a key determinant of RNA transcript stability (Deana et al., 2008; Mackie, 1998). Therefore, nanoRNA-mediated priming could alter transcript stability by altering the phosphorylation status of the 5' ends of primary transcripts. Second, priming with nanoRNAs can alter the TSS, and thus alter the sequence of the 5′ end of the resultant transcript. Alterations in the sequence of the 5′ end of a nascent transcript can impact gene expression through effects on the stability of the RNA/DNA hybrid in the initial transcribing complex or through effects on RNA secondary structure that influence transcript stability, transcription elongation, or transcription attenuation (i.e. premature transcription termination) (Goliger et al., 1989; Liu et al., 1994; Meng et al., 2004; Qi and Turnbough, 1995; Telesnitsky and Chamberlin, 1989; Turnbough and Switzer, 2008). Third, nanoRNA-primed transcription initiation can potentially be more efficient than de novo initiation. For example, the half-maximal effective concentration for initiating transcription of a 2-nt RNA complementary to positions +1/+2 is ~10-fold lower than that of a mononucleotide (Ruetsch and Dennis, 1987; Smagowicz and Scheit, 1981). Therefore, nanoRNAs could act as “activators” of transcription initiation from certain promoters, particularly promoters that would otherwise require high concentrations of the initiating NTP for efficient initiation.
The results of high-throughput sequencing (Figure 3, Tables S1, S2, and S3) indicate that TSS shifting to positions −1 and −2 was significantly less pronounced in Orn-depleted cells when only 5' triphosphate ends were analyzed. Thus, our findings suggest that the pool of 2- to 4-nt nanoRNAs that prime transcription initiation in Orn-depleted cells are a mixture of transcripts that carry a 5' monophosphate or a 5' hydroxyl. Therefore, we consider it likely that a significant fraction of the observed alterations in gene expression occur due to changes in the phosphorylation status of the 5′ ends of primary transcripts that result from incorporation of a nanoRNA carrying a 5' monophosphate or a 5' hydroxyl into the 5' ends of primary transcripts during transcription initiation, which, in turn, impacts transcript stability.
In summary, as expected from results obtained in vitro, our results demonstrate that small RNA primers, “nanoRNAs”, can be used to initiate transcription in vivo. Our results were obtained in bacterial cells in which the intracellular concentration of nanoRNAs was increased artificially by depleting Orn. Therefore, future work will focus on determining to what extent nanoRNA-mediated priming of transcription occurs in bacterial cells under physiological growth conditions.
EXPERIMENTAL PROCEDURES
Plasmids and strains
Details of plasmid and strain construction is provided in the Supplemental Information.
Growth media
P. aeruginosa cells were grown in low Ca2+ type three secretion media (TTSM) which contains 5 g tryptone, 2.5 g yeast extract, 9.4g NaCl per 500 ml; following autoclaving MgCl2, CaCl2, and EGTA are added to final concentrations of 10 mM, 0.5 mM, and 20 mM respectively.
Proteins
All enzymes were purchased from New England Biolabs, unless otherwise noted. His-tagged E. coli Orn was purified as described in (Young Park et al., 2008) after overproduction from plasmid pSG133 in BL21(DE3) cells.
Tandem Affinity Purification (TAP) of P. aeruginosa RNA Polymerase
PAO1 β′-TAP cells were grown in LB to the mid-logarithmic phase of growth and RNAP (β′) was purified by TAP essentially as described previously (Rietsch et al., 2005). Following TAP, RNAP was recovered in a buffer consisting of 25 mM Tris pH 8, 0.1 M NaCl, 0.1 mM EDTA, 1 mM 2-mercaptoethanol, 10 µM ZnCl2, and 50% glycerol.
Orn depletion
For the experiments shown in Figures 1B and C, 5 ml cultures were grown with aeration overnight at 37°C then back diluted to a starting OD600 of 0.01 in 200 ml fresh TTSM. Cultures were grown with aeration (at 300 rpm) at 37°C. IPTG (when present) was added to 20 mM when cultures reached an OD600 of ~0.3. 1 ml samples were removed at the indicated time points to measure the OD600 and/or to analyze protein content.
Western blot analysis
For the experiment of Figure 1B proteins were separated on 4–12% Bis-Tris NuPage gels (Invitrogen) and VSV-G-tagged proteins (Orn-VDAS4 and V-SspB) were detected by Western blotting essentially as described (Castang et al., 2008), using an anti-VSV-G antibody (Sigma-Aldrich). As a loading control, the membrane was probed with a monoclonal antibody that recognizes the α subunit of RNAP (Neoclone).
Detection of nanoRNAs in vivo
For the experiment of Figure 1D, overnight cultures of JS1, JS2 and JS3 were diluted to an OD600 of 0.01 into 20 ml of TTSM containing 20 µCi/ml of 32Pi (as orthophosporic acid in water). One JS1 culture, one JS2 culture and two JS3 cultures were grown at 37°C in 125 ml flasks shaking at 300 rpm. When the OD600 reached ~0.3, IPTG (20 mM final concentration) was added to the JS1 culture, the JS2 culture and one of the JS3 cultures. Cultures were grown for an additional 90 minutes at which time 200 µl of each culture was harvested into chilled tubes containing 20 µl of 98% formic acid. Rifampicin (1 mg/ml final concentration) was added to the remainder of each culture. After shaking for an additional 60 minutes, 200 µl of the rifampicin-treated cultures were harvested into chilled tubes containing 20 µl of 98% formic acid. The lysates were incubated at 4°C for 30–60 minutes, then centrifuged at 4°C (5 minutes at 21,000 g) in order to remove acid insoluble material. The supernatant (~200 µl) was precipitated with ethanol and the precipitate was collected by centrifugation at 4°C (30 minutes at 21,000 g), resuspended in 40 µl 1 X Orn-RB [50 mM Tris (pH 8), 150 mM NaCl and 1 mM MnCl2], and divided into two 20 µl aliquots. One of these aliquots was treated with 250 pmol of His-Orn for 30 minutes at 25°C while the other aliquot was not treated. Reactions were extracted with acid phenol:chloroform (Ambion), precipitated with ethanol and the precipitate was resuspended in formamide loading dye [95% deionized formamide, 18 mM EDTA (pH 8.0), 0.025% SDS, bromophenol blue, xylene cyanol, amaranth]. Samples were electrophoresed on a 22.5% 7 M urea slab gel in a buffer gradient comprising 1 X TBE (upper reservoir) and 1 X TBE containing 0.3 M NaOAc (lower reservoir). Radiolabeled species were detected by storage-phosphor imaging.
To generate the 2- to 5-nt radiolabled RNA standards shown in Figure 1D (lanes 1 and 2), transcription reactions were performed as described in the presence of γ32P ATP (PerkinElmer) using a PCR-generated template corresponding to a KpnI-KpnI segment of plasmid pSG10 (Goldman et al., 2009), which carries the bacteriophage T5 N25 promoter derivative N25anti. During transcription from the N25anti promoter, 2–15 nt abortive RNA transcripts are generated (Goldman et al., 2009). The γ32P ATP contained a contaminant that co-migrated with the 2-nt abortive transcripts and was insensitive to addition of Orn (Figure 1D, lane 2). A contaminant in commercial preparations of radiolabled NTPs that co-migrates with 2-nt RNA transcripts has also been noted in a prior study (Hsu, 2009).
RNA isolation
Duplicate overnight cell cultures of JS1.F carrying pPSV35 (an ‘empty vector’), JS3.F carrying pPSV35, JS3.F carrying pPAOrn (that directs the synthesis of wild-type Orn), or JS3.F carrying pNrnB (that directs the synthesis of NrnB) were diluted to an OD600 of 0.01 into 20 ml of TTSM supplemented with gentamicin (30 µg/ml) and grown at 37°C in 125 ml flasks shaking at 300 rpm. When the OD600 reached ~0.3, IPTG was added to 20 mM and cells were grown for an additional 90 minutes. At this time, total RNA was harvested from 7 ml of culture as described in the Supplemental Information. These RNA preparations were used for the global 5′ end analysis (Figure 3A, B, and C), the primer extension analysis (Figure 3D) and the microarray analysis (Figure 4).
Determination of TSS by high-throughput sequencing
Preparation of RNA transcripts
Total RNA was passed through both an RNeasy Mini Kit (Qiagen), to remove transcripts less than ~200 nt, and through a MICROBExpress Kit (Ambion) to remove rRNAs.
Preparation of cDNA libraries derived from all 5′ ends (Figure S1) was accomplished by first treating the RNA with Antarctic Phosphatase and then treating with OptiKinase (USB Corporation). The treated transcripts were next ligated to a 5′ RNA adaptor and cDNAs were generated by reverse transcription using an RT primer that contained 9 degenerate nucleotides at the 3′ end. Next, the samples were treated with RNase H (Ambion) and cDNAs were isolated after electrophoresis on a 6% 7 M urea slab gel (equilibrated and run in 1 X TBE). Amplification of cDNAs was performed using reagents from the SOLiD Total RNA-Seq Kit and primers from the SOLiD RNA Barcoding Kit (Applied Biosystems). After amplification, cDNAs were isolated after electrophoresis on a 6% 7 M urea slab gel (equilibrated and run in 1 X TBE) and sequenced using an Applied Biosystems SOLiD system (Version 4.0). Preparation of cDNA libraries derived from 5′ triphosphate ends (Figure S2) was accomplished by first treating the RNA with Terminator 5′-Phosphate-Dependent Exonuclease (Epicentre) and then treating with 5′-RNA Polyphosphatase (Epicentre) The treated transcripts were used to prepare cDNAs compatible with the SOLiD sequencer as described above (beginning with the ligation of the 5′ RNA adaptor). (See the Supplemental Information for a detailed description of the library construction.)
Identification of 148 high quality promoters and their associated TSS
We first identified 35 bp sequencing reads that mapped uniquely with zero mismatches to a single position in the P. aeruginosa PAO1 genome using Bioscope 1.3 (Applied Biosystems). Next, using the data obtained from the analysis of 5′ triphosphate-carrying transcripts isolated from non-depleted cells (i.e. JS1.F carrying pPSV35), we identified a set of 842 candidate primary TSSs; these TSSs represented genomic positions that met the following two criteria: (i) the genomic position was located within a 300 bp window upstream of an annotated open reading frame or within a 10 bp window upstream of the 5′ end of an annotated non-coding RNA and (ii) the genomic position aligned to the 5′ end of at least 50 individual sequencing reads and represented a peak of enrichment within a 11 bp window spanning 5 bp upstream and 5 bp downstream.
Next, for each of the 842 candidate primary TSSs we counted the number of individual sequencing reads whose 5′ ends aligned within a window spanning from 3 bp upstream to 2 bp downstream (i.e. positions −3 to +3) using the data obtained from each of the 12 cDNA libraries. We identified a set of 148 “high quality” TSSs along with their associated promoters as genomic positions for which the 5′ end of at least 50 individual sequencing reads aligned within the −3 to +3 window in each of the cDNA libraries (Table S2). This “high quality set” represented loci that had sufficient sequencing depth in each of the 12 samples, thus enabling us to draw meaningful comparisons between the TSS distributions of the same set of promoters in all samples. For each of the 148 “high quality” candidate promoters we calculated the percentage of transcripts initiated at each position between −3 to +3 (Table S1); these values were averaged to generate the graphs shown in Figure 3.
[We note that 124 out of the 148 “high quality” candidate promoters (~84%) carried a hexameric sequence element that resembled a σ70-dependent promoter −10 element (consensus sequence TATAAT) located ~6 bp upstream of the primary TSS (Table S1). This suggests that the vast majority of the high quality candidate promoters identified from our sequencing analysis are indeed bona-fide promoters.]
Microarray experiments
cDNA synthesis, cDNA fragmentation and labeling were performed as described (Wolfgang et al., 2003). Labeled cDNA was hybridized to Affymetrix GeneChip P. aeruginosa genome arrays (Affymetrix) and processed as described (Wolfgang et al., 2003). Data were analyzed and filtered for statistically significant changes in gene expression (present call in all samples, p < 0.05, and fold change > 2) using GeneSpring GX (Agilent Technologies) (Table S4).
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Hochschild, R. Ebright, B. Bochner, R. Gourse, C. Turnbough, S. Garrity, K. McFarland and K. Turner for discussion and R. Hellmiss for artwork. Work was supported by an NRSA fellowship (S.R.G.), training grant T32-AI07061-32 (J.S.S.), NIH grant AI069007 (S.L.D.), NIH grant GM096454 (B.E.N. and S.L.D.), NIH grant GM088343 (B.E.N.), and a Pew Scholars Award (B.E.N.). We thank the Waksman Institute Genomic Core Facility for assistance with high-throughput sequencing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aravind L, Koonin EV. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem Sci. 1998;23:17–19. doi: 10.1016/s0968-0004(97)01162-6. [DOI] [PubMed] [Google Scholar]
- Borukhov S, Polyakov A, Nikiforov V, Goldfarb A. GreA protein: a transcription elongation factor from Escherichia coli. Proc Natl Acad Sci U S A. 1992;89:8899–8902. doi: 10.1073/pnas.89.19.8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- Castang S, McManus HR, Turner KH, Dove SL. H-NS family members function coordinately in an opportunistic pathogen. Proc Natl Acad Sci U S A. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M, Ring J. Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J Biol Chem. 1973;248:2235–2244. [PubMed] [Google Scholar]
- Cho BK, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BO. The transcription unit architecture of the Escherichia coli genome. Nat Biotechnol. 2009;27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A, Celesnik H, Belasco J. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- Di Nocera PP, Avitabile A, Blasi F. In vitro transcription of the Escherichia coli histidine operon primed by dinucleotides. Effect of the first histidine biosynthetic enzyme. J Biol Chem. 1975;250:8376–8381. [PubMed] [Google Scholar]
- Fang M, Zeisberg WM, Condon C, Ogryzko V, Danchin A, Mechold U. Degradation of nanoRNA is performed by multiple redundant RNases in Bacillus subtilis. Nucleic Acids Res. 2009;37:5114–5125. doi: 10.1093/nar/gkp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochimica et biophysica acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci USA. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman SR, Ebright RH, Nickels BE. Direct detection of abortive RNA transcripts in vivo. Science. 2009;324:927–928. doi: 10.1126/science.1169237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goliger JA, Yang XJ, Guo HC, Roberts JW. Early transcribed sequences affect termination efficiency of Escherichia coli RNA polymerase. J Mol Biol. 1989;205:331–341. doi: 10.1016/0022-2836(89)90344-6. [DOI] [PubMed] [Google Scholar]
- Grachev M, Zaychikov E, Ivanova E, Komarova N, Kutyavin I, Sidelnikova N, Frolova I. Oligonudeotides complementary to a promoter over the region −8… + 2 as transcription primers for E. coli RNA polymerase. Nucleic Acids Res. 1984;12:8509–8524. doi: 10.1093/nar/12.22.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DJ, Niyogi SK. RNA initiation with dinucleoside monophosphates during transcription of bacteriophage T4 DNA with RNA polymerase of Escherichia coli. Proc Natl Acad Sci U S A. 1973;70:574–578. doi: 10.1073/pnas.70.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LM. Monitoring abortive initiation. Methods. 2009;47:25–36. doi: 10.1016/j.ymeth.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learned RM, Tjian R. In vitro transcription of human ribosomal RNA genes by RNA polymerase I. J Mol Appl Genet. 1982;1:575–584. [PubMed] [Google Scholar]
- Lewis DE, Adhya S. Axiom of determining start points by RNA polymerase in Escherichia coli. Mol. Microbiol. 2004;54:692–701. doi: 10.1111/j.1365-2958.2004.04318.x. [DOI] [PubMed] [Google Scholar]
- Liu C, Heath LS, Turnbough CL., Jr Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes & Development. 1994;8:2904–2912. doi: 10.1101/gad.8.23.2904. [DOI] [PubMed] [Google Scholar]
- Mackie GA. Ribonuclease E is a 5'-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- McClure WR, Cech CL. On the mechanism of rifampicin inhibition of RNA synthesis. J Biol Chem. 1978;253:8949–8956. [PubMed] [Google Scholar]
- McGinness K, Baker T, Sauer R. Engineering Controllable Protein Degradation. Molecular Cell. 2006;22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Mechold U, Fang G, Ngo S, Ogryzko V, Danchin A. YtqI from Bacillus subtilis has both oligoribonuclease and pAp-phosphatase activity. Nucleic Acids Res. 2007;35:4552–4561. doi: 10.1093/nar/gkm462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Turnbough CL, Jr, Switzer RL. Attenuation control of pyrG expression in Bacillus subtilis is mediated by CTP-sensitive reiterative transcription. Proc Natl Acad Sci U S A. 2004;101:10943–10948. doi: 10.1073/pnas.0403755101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkley EG, Pribnow D. Transcription of the early region of bacteriophage T7: selective initiation with dinucleotides. J Mol Biol. 1973;77:255–277. doi: 10.1016/0022-2836(73)90335-5. [DOI] [PubMed] [Google Scholar]
- Niyogi SK, Stevens A. Studies of the Ribonucleic Acid Polymerase from Escherichia Coli. IV. Effect of Oligonucleotides on the Ribonucleic Acid Polymerase Reaction with Synthetic Polyribonucleotides as Templates. J Biol Chem. 1965;240:2593–2598. [PubMed] [Google Scholar]
- Ohnishi Y, Nishiyama Y, Sato R, Kameyama S, Horinouchi S. An oligoribonuclease gene in Streptomyces griseus. J Bacteriol. 2000;182:4647–4653. doi: 10.1128/jb.182.16.4647-4653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Turnbough CL., Jr Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J Mol Biol. 1995;254:552–565. doi: 10.1006/jmbi.1995.0638. [DOI] [PubMed] [Google Scholar]
- Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2005;102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruetsch N, Dennis D. RNA polymerase. Limit cognate primer for initiation and stable ternary complex formation. J Biol Chem. 1987;262:1674–1679. [PubMed] [Google Scholar]
- Samuels M, Fire A, Sharp PA. Dinucleotide priming of transcription mediated by RNA polymerase II. J Biol Chem. 1984;259:2517–2525. [PubMed] [Google Scholar]
- Smagowicz W, Scheit K. A minimal mechanism for abortive initiation of transcription of T7 DNA. Nucleic Acids Res. 1981;9:5845–5854. doi: 10.1093/nar/9.21.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagowicz WJ, Scheit KH. Primed abortive initiation of RNA synthesis by E. coli RNA polymerase on T7 DNA. Steady state kinetic studies. Nucleic Acids Res. 1978;5:1919–1932. doi: 10.1093/nar/5.6.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surratt CK, Milan SC, Chamberlin MJ. Spontaneous cleavage of RNA in ternary complexes of Escherichia coli RNA polymerase and its significance for the mechanism of transcription. Proc Natl Acad Sci U S A. 1991;88:7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky AP, Chamberlin MJ. Sequences linked to prokaryotic promoters can affect the efficiency of downstream termination sites. J Mol Biol. 1989;205:315–330. doi: 10.1016/0022-2836(89)90343-4. [DOI] [PubMed] [Google Scholar]
- Tu AH, Turnbough CL., Jr Regulation of upp expression in Escherichia coli by UTP-sensitive selection of transcriptional start sites coupled with UTP-dependent reiterative transcription. J Bacteriol. 1997;179:6665–6673. doi: 10.1128/jb.179.21.6665-6673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbough C, Switzer R. Regulation of Pyrimidine Biosynthetic Gene Expression in Bacteria: Repression without Repressors. Microbiol Mol Biol Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MC, Lee VT, Gilmore ME, Lory S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell. 2003;4:253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- Young Park A, Elvin CM, Hamdan SM, Wood RJ, Liyou NE, Hamwood TE, Jennings PA, Dixon NE. Hydrolysis of the 5'-p-nitrophenyl ester of TMP by oligoribonucleases (ORN) from Escherichia coli, Mycobacterium smegmatis, and human. Protein Expr Purif. 2008;57:180–187. doi: 10.1016/j.pep.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Zenkin N, Yuzenkova Y, Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313:518–520. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhu L, Deutscher MP. Oligoribonuclease is encoded by a highly conserved gene in the 3'-5' exonuclease superfamily. J Bacteriol. 1998;180:2779–2781. doi: 10.1128/jb.180.10.2779-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.