Abstract
We introduce models for the analysis of functional data observed at multiple time points. The dynamic behavior of functional data is decomposed into a time-dependent population average, baseline (or static) subject-specific variability, longitudinal (or dynamic) subject-specific variability, subject-visit-specific variability and measurement error. The model can be viewed as the functional analog of the classical longitudinal mixed effects model where random effects are replaced by random processes. Methods have wide applicability and are computationally feasible for moderate and large data sets. Computational feasibility is assured by using principal component bases for the functional processes. The methodology is motivated by and applied to a diffusion tensor imaging (DTI) study designed to analyze differences and changes in brain connectivity in healthy volunteers and multiple sclerosis (MS) patients. An R implementation is provided.
87
Keywords and phrases: Diffusion tensor imaging, functional data analysis, Karhunen-Loève expansion, longitudinal data analysis, mixed effects model
1. Introduction
Scientific studies now commonly collect functional or imaging data at multiple visits over time. In this paper we introduce a class of models and inferential methods for the analysis of longitudinal data where each repeated observation is functional.
Our motivating data set comes from a diffusion tensor imaging (DTI) study, which was designed to analyze cross-sectional and longitudinal differences in brain connectivity in healthy volunteers and multiple sclerosis (MS) patients. For each of 112 subjects and each visit, we have fractional anisotropy (FA) measurements along the corpus callosum in the brain. Figure 1 shows an image of the corpus callosum with 7 biological landmarks (denoted 1, 20, 40, 60, 80, 100, 120) used for registration of measurements across subjects. Each visit’s data for a subject is a finely sampled function across the corpus callosum, with the argument of the function being the spatial distance along the tract. For illustration, the FA data is displayed for 2 subjects, one with 5 and one with 6 complete visits. Although change over time may be subtle in comparison with measurement error, accurate quantification of that change is crucial for applications ranging from powering clinical trials to understanding brain development. This data structure is not unique to this study. In fact, the tractography data is an example of many new data sets containing functions or images that are observed repeatedly over time (longitudinal functional data).
Fig 1.
Top: Sagittal image of the corpus callosum in one of the study subjects, a healthy 33-year-old man, showing the segmentation used [following 46] for construction of the tract profile. Values denote the bin number at the boundary point from the splenium (back of the head) to the genu/rostrum (closer to the eyes). Bottom: Two example subjects (both MS patients) from the tractography data with 5 and 6 complete visits, respectively. Shown are the fractional anisotropy along the corpus callosum, measured at the 120 sample points. Different visits for the same subject are indicated by color and overlaid.
The common structure of these studies can be understood using an analogy with classical longitudinal data [8]. Longitudinal data is commonly analyzed using the very flexible class of linear mixed models [22, 44], which explicitly decompose the variation in the data into between- and within- subject variability. Similarly, we decompose the dynamic behavior of functional data into a time-dependent population average, baseline (or static) subject-specific variability, longitudinal (or dynamic) subject-specific variability, subject-visit-specific variability and measurement error. Technically this is achieved by replacing random effects with random functional effects.
We propose an estimation procedure that is based on an eigenanalysis and extends functional principal component analysis (FPCA) to the longitudinal setting. Computation is very efficient, even for very large data sets. The estimation procedure performed well both in an extensive simulation study and in the DTI application, where it uncovered subtle but potentially important subject-specific changes over time in a specific region of the corpus callosum, the isthmus. The character of these changes could conceivably be used as an early gauge of disease progression or response to neuroprotective therapies.
Our approach is different from functional mixed models based on the smoothing of fixed and random curves using splines or wavelets [3, 15, 16, 29]. In contrast to these methods focusing on the estimation of fixed and random curves, our approach is based on functional principal component analysis. In addition to the computational advantages of such an approach [compare also 19], we are able to extract the main differences between subjects in their average profiles and in how their profiles evolve over time. Such a signal extraction, which is not possible using smoothing methods alone, allows the relation of subject-specific scores to other variables such as disease status, age or disease progression. Our approach can be seen as an extension of functional principal component analysis for multilevel functional data [7]. Our methods apply to longitudinal data where each observation is functional, and should thus not be confused with nonparametric methods for the longitudinal profiles of scalar variables [17, 30, 31, 37, 41, 48, 50, 51]. For good introductions to functional data analysis in general, please see [10, 34].
The remainder of the paper is organized as follows. Section 2 introduces the longitudinal functional model and explains how dimension reduction via longitudinal functional principal component analysis (LFPCA) is achieved. Section 3 develops our estimation procedure and provides computational efficiency results. Section 4 shows the performance of our procedure in an extensive simulation study. Section 5 provides the application of LFPCA methods to the tractography data, while Section 6 concludes with a discussion. Theoretical results and proofs are given in the appendix. Supplementary material [13] providing an R function implementing LFPCA, simulation code and additional graphs is available in the archive that the Electronic Journal of Statistics maintains on Project Euclid.
2. The longitudinal functional model
In this section we introduce models for data sets where functional data are
recorded at multiple time points or visits for the same observational unit or
subject. The observed data are
{Yij(d), d
∈  ,
Tij,
Zij,
Vij},
where Yij(·) is a random function in
L2[0, 1] observed at arguments
d in some set
,
Tij,
Zij,
Vij},
where Yij(·) is a random function in
L2[0, 1] observed at arguments
d in some set  ,
Tij is the time of visit j for
subject i, and
Zij and
Vij are vectors of
covariates for subject i = 1, …, I
at visit j = 1, …, Ji,
where the number of visits Ji can vary with the subject,
i. We assume that at least some subjects i
have at least 3 visits, that is Ji ≥ 3. The
multi-level case when Ji ≤ 2 for all
i was fully addressed by [7] and [6].
,
Tij is the time of visit j for
subject i, and
Zij and
Vij are vectors of
covariates for subject i = 1, …, I
at visit j = 1, …, Ji,
where the number of visits Ji can vary with the subject,
i. We assume that at least some subjects i
have at least 3 visits, that is Ji ≥ 3. The
multi-level case when Ji ≤ 2 for all
i was fully addressed by [7] and [6].
2.1. The functional random intercept and random slope model
The data structure in this paper is similar to that of standard longitudinal data, with the exception that instead of observing scalars, Yij, one observes functions, Yij(d), over time. We use this analogy to build up intuition and to introduce the functional equivalent of the standard longitudinal model. For simplicity we first extend the random intercept and slope model [35]. The functional analog is
| (2.1) |
where η(d, Tij) is a fixed main effect surface, Xi,0(d) is the random functional intercept for subject i, Xi,1(d) is the random functional slope for subject i, Tij is the time of visit j for subject i, Uij(d) is the random subject and visit-specific functional deviation, and εij(d) is random homoscedastic white noise. We make the following assumptions:
-
A.1
Xi(d) = {Xi,0(d), Xi,1(d)}, Uij(d) and εij(d) are zero-mean, square-integrable, mutually uncorrelated random processes on [0, 1],
-
A.2
Xi,0(d) and Xi,1(d) have auto-covariance functions K0(d, d′) and K1(d, d′), respectively, and cross-covariance function K01(d, d′),
-
A.3
Uij(d) has covariance function KU (d, d′), and
-
A.4
εij(d) is white noise measurement error with variance σ2.
There are several parallels between the scalar random intercept-random slope model and model (2.1). First, Yij(d) is now a functional observation. Second, Xi,0(d) and Xi,1(d) replace the scalar random effects bi0 and bi1 as functional random intercept and random slope, respectively, capturing subject-to-subject variation. Third, the cross-covariance function K01(d, d′) replaces the covariance between bi0 and bi1. Fourth, the subject- and visit-specific deviation now consists of two parts. Ui,j(d) is a visit-specific functional deviation from the subject-specific functional trend, capturing visit-to-visit functional variation on the same subject. εij(d) is additional white noise measurement error, capturing random uncorrelated variation within each curve. The overall mean trend is allowed to be a smooth surface η(d, Tij), which generalizes the linear mean β0 + β1Tij often assumed in the scalar model.
Model (2.1) encompasses several simpler models that can be obtained as special cases. For example, visits may be of equal number Ji = J per subject or equally spaced, Tij = j for all i and j. The mean function η(d, Tij) may be time constant, η(d), additive or linear in T, η(s, Tij) = η1(d) + η2(Tij), or η(d, Tij) = η0(d) + Tijη1(d). The latter formulation is a direct extension of the linear population trend typically assumed in the scalar model.
Model (2.1) allows the decomposition of the variation in the observed curves into a) differences in subject’s baseline functions; b) differences in subjects’ average changes over time; c) visit-specific variation around these average trends; and d) measurement error. This decomposition is of interest in many applications. For example, in the tractography application we describe in Section 5, it is of interest to study both the population cross-sectional and the dynamic behavior of various measurements along neuronal tracts.
2.2. The general functional mixed model
While model (2.1) is rich enough for our application, it lends itself well to generalization, which could be useful in other applications. A more general form of the longitudinal functional model is
| (2.2) |
where we assume that
-
B.1
Xi(d), Uij(d) and εij(d) are zero-mean, square-integrable, mutually uncorrelated random processes on [0, 1],
-
B.2
Xi(d) is a vector-valued random process with auto-covariance functions for the p components K11(d, d′), …, Kpp(d, d′), and cross-covariance functions K12(d, d′), …, K1,p(d, d′), …, Kp−1,p(d, d′),
-
B.3
Uij(d) is a random process with covariance function KU (d, d′), and
-
B.4
εij(d) is white noise measurement error with variance σ2.
Model (2.1) and its assumptions A.1 to A.4 are obtained as a special case of (2.2) and B.1 to B.4 by setting p = 2, Zij = Tij and Vij = (1, Tij)′. Note that model (2.1) counts the components of X from 0 to 1 rather than from 1 to 2 to stress the analogy to the scalar random intercept-random slope model. The functional mixed-effects ANOVA model [7] results if we set p = 1, Zij = j, Vij = 1, and η(d, Zij) = μ(d) + ηj(d). More generally, Zij and Vij are vectors of known covariates for subject i at time Tij. η(d, Zij) is the fixed main effect surface, which can depend parametrically, semi-parametrically or non-parametrically on the covariates Zij = (Zij,1, …, Zij,m). The simplest parametric form is a linear mean , while the most complex nonparametric form is a (p+1)-dimensional smooth function η(d, Zij). Intermediate semi-parametric models such as η(d, Zij) = η1(d, Zij,1) + ···+ ηm (d, Zij,m) or could also be useful in particular applications.
Model (2.2) is the functional analog of the linear mixed model for longitudinal data [22]. It is similar to models used by [15, 29], but we do not assume Gaussianity and allow for more general fixed effects. The model of [15] also does not admit correlated random functional effects, as are present in (2.1). In addition, we follow quite a different modeling approach, using longitudinal functional principal component analysis instead of smoothing splines or wavelets for nested curves. This has large computational advantages compared to [29], especially when the number of random effects is large, see Section 4.3 and [19]. We are at the same time able to extract the main differences between subjects in how their profiles evolve over time, something not possible in these other approaches.
For notational simplicity in the remainder of the paper we will focus on model (2.1). In Appendix B we point out the small technical differences for fitting the more general model (2.2).
2.3. Dimension reduction via longitudinal FPCA
While models (2.1) and (2.2) are intuitive generalizations of linear mixed effects models, their computational feasibility is not obvious, especially for large numbers of subjects, visits and observations. We here propose an efficient modeling approach, Longitudinal Functional Principal Component Analysis (LFPCA). LFPCA is the longitudinal generalization of functional principal component analysis (FPCA) [34] and multilevel functional principal component analysis (MFPCA) [7]. The main idea of LFPCA is to extract the main directions of variation of the X and U processes using an eigen decomposition of their respective covariance operators on the basis of Mercer’s theorem [27]. The Karhunen-Loève expansion [20, 26] is then used to obtain parsimonious expansions of X and U.
In the notation of model (2.1), we expand the covariance operator of the bivariate process Xi(d) = {Xi,0(d), Xi,1(d)} as
where are the ordered eigenfunctions of KX(d, d′) corresponding to the eigenvalues λ1 ≥ λ2 ≥ ··· ≥ 0. Similarly, let , where are the ordered eigenfunctions of KU (d, d′) corresponding to the eigenvalues ν1 ≥ ν2 ≥ ··· ≥ 0. The eigenfunctions { , k ∈ IN} form an orthonormal basis of L2[0, 1] with respect to the usual L2[0, 1] scalar product. The eigenfunctions { , k ∈ IN} form an orthonormal basis of L2[0, 1] × L2[0, 1] with respect to the additive scalar product
The function pairs or ( ) are not required to be orthogonal in L2[0, 1], nor will ( ) or ( ) be orthogonal in general for k ≠ j. The Karhunen-Loève expansions of the random processes are
where the principal components scores
are uncorrelated random variables with mean zero and variances λk and νk, respectively. Assumption A.1. is ensured by assuming that {ξik, i = 1, …, I, k ∈ IN} and {ζijk, j = 1, …, Ji, i = 1, …, I, k ∈ IN} are mutually uncorrelated. Because working with infinite expansions is impractical, we consider the finite-dimensional approximations of the X and U processes
where NX and NU will be estimated, as described in Section 3.4. Conditional on NX and NU the finite approximation to model (2.1) is
| (2.3) |
Vij = (1, Tij)′, which is a linear mixed model [see also 7]. Here, denotes uncorrelated variables with mean 0 and variance a. We are neither assuming normality of the processes in (2.1) nor of the scores in (2.3). LFPCA extends similarly to the more general model (2.2).
3. Estimation
For reasons of simplicity, we focus the presentation on model (2.1), but estimation is done similarly
for model (2.2). The minor
adjustments for fitting (2.2) are
described in Appendix B. We assume that the
mean, covariance operators and eigenfunctions are smooth. For presentation purposes,
we assume that all functions Yij(d) are
measured at a finite number, D, of grid points  ⊂ [0, 1]. However, the
method can easily handle missing data, both in terms of visits per subject or
observations per visit. Estimation can be done using a few simple steps, which will
be described in more detail in the following.
⊂ [0, 1]. However, the
method can easily handle missing data, both in terms of visits per subject or
observations per visit. Estimation can be done using a few simple steps, which will
be described in more detail in the following.
-
Step 1The fixed effect surface η is estimated using the working independence model
Smoothness selection is by REML, which is more robust to neglecting the correlations in the errors than prediction error methods [21].
-
Step 2
The autocovariance functions for the random processes Xi = (Xi,0, Xi,1) and Uij are estimated from the residuals Yij(d) − η̂ (d, Tij), using a linear regression step.
-
Step 3
The ‘raw’ autocovariance function estimates from step 2 are subjected to bivariate smoothing, yielding also an estimate for σ2.
-
Step 4
Eigen decompositions of the smoothed autocovariance functions provide bases for representing X = (Xi,0, Xi,1) and Uij, which are truncated to achieve parsimony.
-
Step 5
Estimated BLUPs then provide estimates for the subject- and visit-specific scores, which summarize the main differences in the dynamics of functions over time.
3.1. Estimation of the mean
The fixed effect population mean surface η(d, T) can be estimated using a bivariate smoother in d and T under a working independence assumption. For discussions of smoothing for correlated data, see [21, 24]. Possibilities for smoothers include penalized splines [39], smoothing splines [11] and local polynomials [9]. Choice of a smoother and of the smoothing parameter or bandwidth is discussed extensively in the literature and is not the main focus here. It is our experience that most reasonable smoothers used judiciously will provide similar results. For simplicity and efficiency of the implementation for large data sets, we use penalized spline smoothing with REML estimation of the smoothing parameter. This choice has also been found to be relatively robust to misspecification of the error correlation structure in [21].
A bivariate smoother is appropriate when the collection of observations across visits and subjects is relatively dense. This need not be the case in general, and simpler choices might be more sensible. For example, η(d, Tij) = η0(d) + Tijβ might be more appropriate if the Tij form a sparser collection. In the case of equally spaced visits, Tij = Tj, [7] used η(d, Tij) = ηj(d). Choices will depend on the particular application, available data and scientific problem. In most applications, estimating the mean function is quite easy and, even, routine. Once a consistent estimator of the mean function is available, data can be centered as follows Yij(d) − η̂(d, Tij) for all i, j and d. In the following we assume that the Yij(d) are mean zero.
3.2. Estimation of the covariance operators
A crucial point of our proposed methodology is estimating the covariance operators KX(·, ·) and KU (·, ·). To estimate the covariance functions, we focus on the cross-products Yij(d)Yik(d′). Because Yij(d) has zero mean, each product Yij(d)Yik(d′) is an estimator of the covariance between the function observed at time Tij evaluated at location d, and the function observed at time Tik evaluated at location d′. Every subject thus contributes an estimator each for every available pair of observations at time Tij evaluated at location d, and at time Tik evaluated at location d′. Available pairs of observations may vary between subjects in their (d, d′) and (Tij, Tik) combinations. The method described in the following can thus easily handle missing data, both in terms of visits per subject or observations per visit.
Under the assumptions of model (2.1),
| (3.1) |
for all d,
d′, i, j and
k, where δjk is
Kronecker’s delta. Equation
(3.1) suggests a straightforward solution for estimating the
covariance operators: regress linearly the “outcome”
Yij(d)Yik(d′)
on the “covariates” (1, Tik,
Tij,
TikTij,
δjk), where the
“parameters” are
{K0(d,
d′),
K01(d,
d′),
K01(d′,
d), K1(d,
d′), KU
(d, d′); d,
d′ ∈  ;
σ2}.
;
σ2}.
While the intuition behind the method is simple, there are two potential pitfalls that should be carefully avoided. First, σ2 is identifiable only under the assumption that KU (d, d′) is a bivariate smooth function in d and d′. Second, in a straightforward implementation of the linear regression on the basis of (3.1), there are observations and 4D2 + 1 variables. In our moderately sized tract data, this would correspond to 19 million observations and 57, 500 variables. In larger data sets the problem would be even more serious. Thus, careful implementation is required to ensure computational feasibility. We propose the following 3-step estimation procedure that avoids these problems.
-
Step A
{K0(d, d′), K01(d, d′), K01(d′, d), K1(d, d′), KU (d, d′)+ σ2δdd′} is estimated for each pair d ≤ d′ ∈
 using least squares estimation based
on (3.1). Symmetry
constraints yield K0(d,
d′) =
K0(d′,
d),
K1(d,
d′) =
K1(d′,
d) and KU
(d, d′) =
KU (d′,
d) for d >
d′. Denote estimates by
K̃0(d,
d′),
K̃01(d,
d′),
K̃1(d,
d′) and
K̃U (d,
d′).
using least squares estimation based
on (3.1). Symmetry
constraints yield K0(d,
d′) =
K0(d′,
d),
K1(d,
d′) =
K1(d′,
d) and KU
(d, d′) =
KU (d′,
d) for d >
d′. Denote estimates by
K̃0(d,
d′),
K̃01(d,
d′),
K̃1(d,
d′) and
K̃U (d,
d′). -
Step B
Bivariate smoothing in d and d′ over K̃0(d, d′), K̃01(d, d′) and K̃1(d, d′) yields smooth estimates K̂0(d, d′), K̂01(d, d′) and K̂1(d, d′). Bivariate smoothing over K̃U (d, d′), leaving out the diagonal elements as proposed by [41, 50], also yields estimates K̂U (d, d′).
Please see Section 3.1 for a discussion of bivariate smoothing.
-
Step C
σ2 can be estimated as , if positive, and as zero otherwise.
Estimation in Step A can be done using efficient matrix-vector computations as detailed in Theorem 1 in Appendix A. The following is a consequence of that theorem.
Corollary 1
The computational effort for estimation of the covariance functions in Step A for the general model (2.2) is of the order O{max(p6, p2D2g)}, where and p is the dimension of the vector-valued random process Xi(d) in (2.2).
All proofs can be found in Appendix A. For model (2.1), p = 2 is small. Note that p2D2 is the order of the number of unknown parameters in the covariance functions, and g is the number of observation pairs contributing to the estimation. The effort thus is linear in both. Our software implementation is so efficient that the computational effort is dominated by the bivariate smoothing of the mean and covariance functions; see Section 4.3 for a detailed investigation of efficiency.
Our procedure does not guarantee that K̂X(·, ·) and K̂U (·, ·) are positive definite. We correct this problem by trimming the eigenvalue-eigenvector pairs corresponding to negative eigenvalues, a method that has been found to increase the L2 accuracy [17] and has been shown to work well in practice [51].
3.3. Estimation of the eigenfunctions and scores
In the previous section we showed how to obtain the estimated covariance
matrices K̂0 =
{K̂0(d,
d′)}d;d′
∈  ,
K̂01 =
{K̂01(d,
d′)}d;d′
∈
,
K̂01 =
{K̂01(d,
d′)}d;d′
∈  ,
K̂1 =
{K̂1(d,
d′)}d;d′
∈
,
K̂1 =
{K̂1(d,
d′)}d;d′
∈  , and
K̂U
= {K̂U (d,
d′)}d;d′
∈
, and
K̂U
= {K̂U (d,
d′)}d;d′
∈  . Estimates of
the eigenvalues and of the eigenfunctions
, k = 1, 2, …,
D, and
, k = 1, 2, …,
D, at the grid points
. Estimates of
the eigenvalues and of the eigenfunctions
, k = 1, 2, …,
D, and
, k = 1, 2, …,
D, at the grid points  can then be obtained using the spectral
decomposition of KX
and KU,
and
, where
and
, are orthonormal vectors in
IR2D and
IRD, respectively. The estimation of the number
of eigenfunctions retained for further analysis, NX
and NU, is described in Section 3.4.
can then be obtained using the spectral
decomposition of KX
and KU,
and
, where
and
, are orthonormal vectors in
IR2D and
IRD, respectively. The estimation of the number
of eigenfunctions retained for further analysis, NX
and NU, is described in Section 3.4.
In Section 2.3, equation (2.3), we showed that for fixed NX and NU, model (2.1) is a linear mixed model. Thus, we can use best linear unbiased prediction (BLUP) to obtain predictions of the subject- and subject/visit-specific scores, ξik and ζijk, respectively. BLUP calculation does not require a normality assumption and is a generalization of the conditional expectations used by [50].
For given eigenfunctions, mean function η(d, T), and variances λk, k = 1, …, NX, νk, k = 1, …, NU, the BLUP for b = (ξ11, …, ξ1NX, …, ξI1, …, ξINX ζ111, …, ζ11NU, …, ζIJI1, …, ζIJINU) in model (2.3) is given in the usual form by
| (3.2) |
where Z = [ZX|ZU], ZX = EI ⊗ Φ0 + T ⊗ Φ1, ZU = In ⊗ ΦU, EI = (δih)ij=11,…,IJI;h=1,…,I, T = (Tijδih)ij=11,…,IJI;h=1,…,I, , D = blockdiag(DX, DU ) = blockdiag{II ⊗ diag(λ1, …, λNX), In ⊗ diag(ν1, …, νNU)}, Y = {Y11(1), …, Y11(D), …, Y1J1(1), …, Y1J1 (D), …, YIJI(1), …, YIJI(D)}, and η = {η(1, T11), …, η(D, T11), …, η(D, T1J1), …, η(1, TIJI), …, η(D, TIJI)}. Here, ⊗ denotes the Kronecker product of matrices, and (aijh)ij=11,…,IJIh=1,…,I denotes a matrix with entries aijh, rows ij, j = 1, …, Ji, i = 1, …, I, and columns h = 1, …, I.
We can obtain estimated BLUPs (EBLUPs) using the estimated functions and variances η̂(·, ·), σ̂2, , λ̂k, k = 1, …, NX, and , ν̂k, k =1, …, NU, from Sections 3.1 and 3.2. This does not require fitting the model (2.3), which greatly increases computational efficiency. While straightforward implementation of (3.2) requires inverting nD × nD matrices, which would result in computational effort of the order O(n3D3), we make use of common matrix rules and of the model structure to obtain a more efficient representation, as detailed in Theorem 2 in Appendix A. The following result of Theorem 2 confirms the manageable computational effort even for very large data sets.
Corollary 2
Computational effort for calculation of the estimated BLUPs in (3.2) is of the order , where and f= NU + NXI/n.
The proofs are provided in Appendix A. NX and NU are typically small, and much smaller than either D or the number of observed curves n. These results and efficient block matrix manipulation make the models proposed here feasible even for very large data sets. For example, one of the simulation examples in Section 4 uses 1, 000 subjects, who were observed at 8 visits and had 200 observations per visit.
3.4. Decomposition of variance and choice of the number of components
There are several possible ways to choose the numbers of eigenfunctions NX and NU. Two alternatives that have been used before are leave-one-curve-out cross validation [38] and an AIC-type criterion [50]. Alternatively, one can make use of the fact that (2.3) is a linear mixed model, with NX and NU corresponding to the number of random effects. The conditional Akaike information criterion (cAIC), proposed for the selection of random effects in linear mixed models [12, 23, 43] could thus be employed. [40] and [6] point out that choosing the number of eigenfunctions corresponds to step-wise testing for zero variance components. They propose using a Restricted Likelihood Ratio Test (RLRT) for this zero variance. The null distribution can be easily approximated using methods introduced by [14] based on the null distribution derived in [5].
Here we follow a simpler approach based on the proportion of variance explained. This approach has several advantages: a) popularity; b) simplicity and interpretability; c) quantification of the contribution of the different processes to the variability in Yij(d).
To better understand variance partitioning, we give the following result.
Lemma 1
Let Yij(d) ∈ L2[0, 1]
be a process that follows model (2.1) with zero mean, η(d, Tij) ≡ 0.
Let Tij be independently distributed as T for all i and j, where
E(T 2) < ∞, and let Tij be independent of
Xi, Uij and
εij(d), d ∈  . Then, the average variance of
Yij(d) can be written as
. Then, the average variance of
Yij(d) can be written as
The proof can be found in Appendix A. Given the usual interpretation of eigenvalues as variance explained in FPCA, one could be tempted to interpret λk similarly in the longitudinal context. The variance decomposition that we just described indicates that in LFPCA, λk can be interpreted as a variance component only if the time variable is standardized to have zero mean and unit variance. In this case, the two components of the eigenfunction, and , will be on the same scale. We can then directly discuss λk as the “variance explained” by the eigenfunction of KX. Thus, we recommend standardizing the time variable. The variation in Yij(d) then has the following simple additive decomposition.
Corollary 3
In the case when E(Tij) = 0 and Var(Tij) = 1, the expression in Lemma 1 reduces to
Thus, for standardized Tij, the variation in Yij(d) can be decomposed additively into the contributions from the random intercept and random slope process, , from the visit-specific deviation process, , and from the additional random noise, σ2. This leads to a simple decision rule for NX and NU: choose and corresponding to λk and νk in decreasing order, until
where L is a pre-specified proportion of explained variation, such as L = 0.95. and provide quantifications of the relative importance of the X and U processes.
4. Simulations
4.1. Simulation design
To investigate the performance of our estimation procedure, we conduct
an extensive simulation study. The design combines and extends scenarios used by
[7] and [50]. For all settings, we generate
1000 data sets from model (2.3),
where NX = NU
= 4. We set the mean function to
η(d, T )
= 0.5(T/4 − d)2.
The unequally spaced time points Tij are simulated
such that the mean for each subject is zero, and increments
Tij −
Tij−1 are independent draws
from a uniform distribution on [0, 1]. The time variable is then
standardized to have unit variance. The curves
Yij(d) are taken to be observed
for d ∈  = {(k −0.5)/D,
k = 1, …, D},
D = 120, as in the tract data. We set the variances
to be λk =
νk =
21−k, k
= 1, …, 4, and σ = 0.05. This
choice corresponds to 0.07% of the overall average variance explained by
the error variance σ2, higher than in the
tract data (0.02%, please see Table
2).
= {(k −0.5)/D,
k = 1, …, D},
D = 120, as in the tract data. We set the variances
to be λk =
νk =
21−k, k
= 1, …, 4, and σ = 0.05. This
choice corresponds to 0.07% of the overall average variance explained by
the error variance σ2, higher than in the
tract data (0.02%, please see Table
2).
Table 2.
Average variance explained by the different model components in percent.
To obtain the variance explained by and , the corresponding λk is multiplied by and , respectively. The cumulative variance explained for row k is the sum of the row entries up to and including row k. The last row gives the cumulative variance explained for the respective column.
| k |
|
|
|
σ 2 | cumulative | |||
|---|---|---|---|---|---|---|---|---|
| 1 | 37.97 | 0.21 | 22.81 | 0.02 | 61.01 | |||
| 2 | 6.76 | 0.55 | 5.41 | 73.73 | ||||
| 3 | 3.33 | 0.32 | 2.96 | 80.34 | ||||
| 4 | 2.11 | 0.43 | 2.07 | 84.95 | ||||
| 5 | 1.50 | 0.28 | 1.51 | 88.24 | ||||
| 6 | 0.98 | 0.19 | 1.32 | 90.73 | ||||
|
| ||||||||
| 52.65 | 1.98 | 36.08 | 0.02 | 90.73 | ||||
We consider all possible combinations of the following scenarios:
number of subjects (a) I = 50 (b) I = 100 (c) I = 200 and (d) I = 500, including both smaller and larger numbers than in the tract data,
balanced design with Ji = 4 for all i,
unbalanced design with Ji ∈ {1, …, 9}, (a multiple of 8, 8, 9, 6, 5, 5, 4, 3, 2 times, respectively), giving 4 observations per subject on average,
normal scores ξik ~
 (0,
λk) and
ζijk ~
(0,
λk) and
ζijk ~
 (0,
νk) for all
i, j and
k,
(0,
νk) for all
i, j and
k,non-normal scores; ξik drawn from a mixture of two normals, with equal probability from either or from ; ζijk drawn with equal probability from either or ,
eigenfunctions with and orthogonal and of equal norm are not orthogonal to either or ,
eigenfunctions with and non-orthogonal and of unequal norms and . are equal to or for some j for all k,
estimation does not include bivariate smoothing of the covariance functions. In this case, smoothing is only used to obtain an estimate of the diagonal KU (d, d), d ∈
 , and of
σ2,
, and of
σ2,estimation includes bivariate smoothing of the covariance functions.
This gives 64 different combinations overall. The eigenfunctions for setting 4. (a) are
Note that while and are orthogonal, they are not orthogonal to for all k and j ≠ 1. The eigenfunctions for setting 4. (b) are
Note that now and are non-orthogonal, and has a larger norm than . Also, is equal to one of the or , j = 1, …, 4, for each k, making separation of the two processes X and U much more difficult. For bivariate smoothing of the mean and covariance functions, we use tensor product penalized cubic regression splines with 10 knots per dimension, where the smoothing parameters are estimated using REML estimation, as implemented in the R package mgcv [47].
To investigate the sensitivity of our results to our choices for η and σ, we also consider four variations on the balanced design with I = 100 and Ji = 4 for all i (1b and 2a), non-orthogonal and with unequal weight on and (4b), a mixture distribution for the scores ξij and ζijk (3b), and bivariate smoothing of the covariance functions (5b). For these four settings, we vary η(d, T ) = (T/4 − d/D + 1/2)(T/4 + d/D − 1/2), η(d, T) = sin(πT/2)d/D, σ = 0.5 (corresponding to 6.25% of the overall average variance explained by the error variance σ2) or σ = 1 (21.05%), respectively.
For each of the 1000 replications and for each of the 68 settings, our estimation procedure from Section 3 with NX = NU = 4 is used to obtain estimates of the mean function, the covariance functions, the eigenfunctions, the scores, and all variances.
4.2. Simulation results
We now discuss results for one of the 68 settings in detail, and point out differences across settings. The complete simulation results can be found in the supplementary material.
Figure 2 and Table 1 show the main results of simulations based on a balanced design with I = 100 and Ji = 4 for all i (1b and 2a), non-orthogonal and with unequal weight on and (4b), a mixture distribution for the scores ξij and ζijk (3b), and no bivariate smoothing of the covariance functions (5a). A plot of the true and estimated mean functions can be found in the supplementary material, illustrating that the mean is well and unbiasedly estimated.
Fig 2.
True and estimated eigenfunctions and , k = 1, …, 4. The left column gives results for the part corresponding to the random functional intercept Xi,0, the middle column for the part corresponding to the random functional slope Xi,1, and the right column for the component corresponding to the visit-specific functional deviation Uij. Shown are the true function (thick black line), the mean of the estimated functions over 1000 simulations (dashed red line), the pointwise 5th and 95th percentiles of the estimated functions (blue), and the estimated functions from the first 50 simulations (grey). Simulations were based on model (2.3) with NX = NU = 4, a balanced design with I = 100 and Ji = 4 for all i, non-orthogonal and with unequal weight on and , a mixture distribution for the scores ξij and ζijk, and no bivariate smoothing of the covariance functions.
Table 1.
True and estimated subject-specific and visit-specific scores ξik and ζijk. Given are summary statistics of the scaled differences (top) and (bottom), k = 1, …, 4. Simulations were based on model (2.3) with NX = NU = 4, a balanced design with I = 100 and Ji = 4 for all i, non-orthogonal and with unequal weight on and , a mixture distribution for the scores ξij and ζijk, and no bivariate smoothing of the covariance functions.
| Minimum | 1st Quantile | Median | Mean | 3rd Quantile | Maximum |
|---|---|---|---|---|---|
| −2.39 | −0.31 | 0.00 | 0.00 | 0.30 | 2.56 |
| −2.52 | −0.24 | 0.00 | 0.00 | 0.23 | 3.13 |
| −3.42 | −0.31 | 0.00 | 0.00 | 0.31 | 3.39 |
| −4.17 | −0.20 | 0.00 | 0.00 | 0.20 | 4.58 |
|
| |||||
| −2.80 | −0.18 | 0.00 | 0.00 | 0.18 | 3.11 |
| −3.68 | −0.39 | 0.00 | 0.00 | 0.39 | 4.26 |
| −2.77 | −0.30 | 0.01 | 0.01 | 0.31 | 2.54 |
| −3.38 | −0.34 | 0.00 | 0.00 | 0.34 | 4.16 |
Figure 2 shows the true and estimated eigenfunctions and , k = 1, …, 4. Results for and are displayed in the left, middle and right panels, respectively. Shown are the true function (thick black line), the mean of the estimated functions over 1000 simulations (dashed red line), the pointwise 5th and 95th percentiles of the estimated functions (blue), and the estimated functions from the first 100 simulations (grey). Note that the covariance functions, and thus the eigenfunctions, are not smoothed in this setting.
For all functions, the mean of the estimated functions is very close to the true function, and the variability around it is small. It can be noted that the are slightly better estimated. This is due to the fact that estimation of the covariance function KU (d, d′) is based on n = ΣJi visits, while estimation of the covariance function KX(d, d′) is based on only I subjects, with I = n/4 in this setting. In this case, has a larger norm than , making estimation of this component easier. This is noticeable in a smaller variance for . Nevertheless, estimation of the is also remarkably good. Overall, estimation of all functions is very good, even though: a) and are not mutually orthogonal; and b) each is actually identical to either or for some j. Our estimation procedure effectively separates the X and U processes, even in the most difficult of circumstances and with a moderate sample size.
Table 1 provides results for the scores ξik and ζijk, k = 1, …, 4. Shown are summary statistics for the scaled differences between estimated and true scores, and , k = 1, …, 4. The table illustrates that the majority of estimates lies close to the true scores, relative to the standard deviation of the score in question, even if the distribution of the estimates is more heavy tailed than in a normal distribution. This might be expected from the fact that the principal components and in model (2.3) are estimated and not observed.
Further figures in the supplementary material show results for the estimation of the variances, σ2, λk and νk, k = 1, …, 4. The ν̂k are centered at the true values νk, with about 70% within 10% and more than 95% within 20% of the true value. The λ̂k show a slight downward bias and somewhat larger variation, reflecting the smaller effective sample size for estimation of these variance components. σ2 is estimated almost as well as the νk.
Overall, the estimation procedure performed very well in a wide range of scenarios. Across simulations, we found the following similarities and differences. First, results improve for an increasing number of subjects I. As expected, a larger I decreases the variability of the estimated eigenfunctions, mean function, scores and variances. The slight downward bias in the λ̂k disappears with increasing I. Second, a balanced design (2a) improves results compared to an unbalanced design (2b) with the same number of subjects and visits. A balanced design leads to a) decreased variability in the estimated mean η̂(d, T ), as we estimate the mean under a working independence assumption before estimating the complex covariance structure b) decreased variability in the estimated eigenfunctions and decreased variability and small sample bias in the variances λ̂k, k = 1, …, NX. This is similarly due to the fact that we estimate the covariance functions using least squares under a working independence assumption. Estimation of the is not much affected by how balanced the design is, although there is some indication that small estimates λ̂k are compensated for by a slight increase in the ν̂k. Third, results for normal scores (3a) and non-normal scores (3b) where virtually identical. This is expected, as BLUPs do not rely on a normality assumption and are thus robust to departures from normality. Still, it is reassuring to see this confirmed in practice. Forth, non-orthogonality of and (4b) does not affect results compared to orthogonality (4a). Even though in (4b), each is equal to either or for some j, estimation of the is equally good in both cases. The only consistent difference between the two designs is, that as in (4b) has a larger part in the norm of , estimation of improves somewhat, while estimation of deteriorates slightly. Fifth, results excluding (5a) and including (5b) bivariate smoothing of the covariance functions were very similar, with the smooth version more effective at filtering out the measurement errors εij(d) and obtaining smooth eigenfunctions and .
Our sensitivity analyses indicate that results are not very sensitive to the choice of the mean function, with all three considered mean functions estimated well and unbiasedly. Large error variances increase the variability of all estimates. When signal-to-noise ratios become small due to very large error variances and small variances λk or νk, this leads to some underestimation of the magnitude (but not shape) of the eigenfunctions. This is due to the unbiasedness of estimation for the covariance functions, which are quadratic in the eigenfunctions, and the attenuation resulting from large variances in .
4.3. Computational efficiency
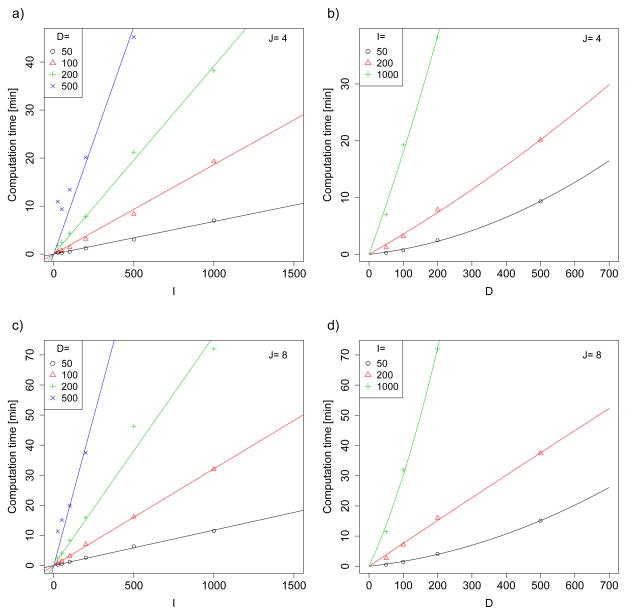
To investigate computation time we considered different combinations of number of subjects I ∈ {25, 50, 100, 200, 500, 1000}, number of observations per subject J ∈ {4, 8}, and number of sample points per curve D ∈ {50, 100, 200, 500}. All other parameters were chosen as for the simulations described in Section 4.1, settings combination 2(a), 3(a), 4(a), 5(a).
Figure 3 provides the computation times. System times were (for practical reasons) measured on three different cluster nodes running 64-bit Red Hat Linux, with 2.3/2.6/3.0 GHz AMD Opteron Processors and 32 GB random access memory. Figures 3 a) and c) for J = 4 and J = 8 display computation time versus the number of subjects I, stratified by D. For example, computation time for I = 100 subjects with J = 4 visits and D = 100 points per curve was just 1.4 minutes, while computation for I = 1, 000, J = 8 and D = 200 took 72 minutes. Figures 3 b) and d) display computation times versus sample points per curve, D, stratified by I. As suggested by Corollaries 1 and 2, computation time is roughly linear in I and between linear and quadratic in D. A linear regression of the log computation time log(C) on log(I), log(J) and log(D) yields . The coefficient for log(D) is indeed between 1 and 2, as expected from Corollaries 1 and 2. The coefficients for log(I) and log(J) are close to 1, but somewhat lower, reflecting that there are some parts of the estimation procedure not depending on I and J. The adjusted R2 for the model is a high 0.98, indicating that good estimates of computation time on comparable machines can be obtained from this regression equation also for parameter combinations not considered here. Note, however, that for very large I and, especially, D, memory might be more of a concern than computational efficiency. In that case, one can replace our efficient matrix computations by less efficient methods that optimize memory usage.
Fig 3.
Computation time for LFPCA for a simulated data set with the given number of subjects I and number of observations per subject J, and with D sample points per curve. Specifics of how computation time was measured are given in Section 4.3.
5. Application
In this section, we use LFPCA to decompose the variability in the tractography data. We first provide the scientific background.
5.1. Background and scientific questions
Multiple sclerosis (MS) is a disorder of the central nervous system (CNS) [e.g. 4]. MS causes typical abnormalities on magnetic resonance imaging (MRI) scans of the brain and spinal cord, and consequently MRI has become the primary diagnostic tool for MS. MRI scanning is increasingly used to monitor disease progression and response to therapy and has become an important surrogate outcome measure in clinical trials.
Diffusion tensor imaging (DTI), in contrast to conventional MRI techniques, is able to resolve individual functional tracts within the CNS white matter, the primary target of MS. DTI is sensitive to diffusion anisotropy, which, in the brain and spinal cord, corresponds to the tendency of water to diffuse along axonal tracts [1]. A focus on one or several tracts with specific functional correlates can then help in understanding the neuroanatomical basis of disability in MS. Quantitative measures derived from DTI data include fractional anisotropy (FA), measuring the degree of anisotropy between 0 and 1 [2]. FA can be decreased in MS due to lesions, loss of myelin and axon damage [42, 45].
Measurement of tract-specific MRI indices has traditionally worked with averages along tracts, ignoring the spatial variation of those indices within tracts [25, 32, 36]. However, that spatial variation can be considerable. The extent to which accounting for this spatial variation can improve detection of abnormality, correlation with disability, or sensitivity to change across time, remains uncertain. The last of these is particularly relevant for monitoring individual patients in the clinic and for the design and powering of clinical trials of new drugs.
We are interested in using the full spatial information to gain a better understanding of differences between subjects both with respect to their mean tract profiles over time (static behavior) and to the changes in those tract profiles over time (dynamic behavior). Our data set includes measurements for 84 MS patients and 28 controls with 1 to 8 complete visits, giving 308 visits overall. At each visit, we have measurements of FA and the diffusivities along several tracts in the brain, which were reconstructed using the tract finding algorithm of [28]. We will focus here on the corpus callosum, a tract connecting the two hemispheres of the brain. The 120 sample points - from the splenium (back of the head) to the genu/rostrum (closer to the eyes) - were chosen equidistantly between certain landmarks on the tract used for registration of curves between subjects [33]. The corpus callosum and its segmentation are illustrated in Figure 1 (top). Figure 1 (bottom) shows example profiles from two MS patients, illustrating the variability of profiles between subjects and within subjects over time. Visual examination of these tract profiles reveals variation within subjects across both space and time but no clear and consistent trend over time.
5.2. Application of LFPCA to the tractography data
As changes in MRI hardware over the five years of the study caused some variation in the measured MRI indices, we use a preprocessing step to remove differences due to variation in scanning technique. For each of the five scanning epochs, we estimate a mean profile for cases alone, using one visit per subject. This avoids confounding of disease status with epoch due to uneven distribution of cases and controls among epochs, and confounding by disease progression. We then subtract the difference of the epoch mean profile to the overall mean profile from all functional observations.
We obtain a decomposition of the variance using LFPCA. The time variable is centered by subject and standardized. For bivariate smoothing of the mean and covariance functions, we use tensor product penalized cubic regression splines with 30 knots per dimension, with smoothing parameters estimated using REML. A graph of the mean function η(d, T) is given in the supplementary material. The mean profile is roughly constant over time, with some variation near areas of high curvature.
For a pre-specified level L = 90% of explained average variance , LFPCA gives NX = NU = 6 principal components and for the X and U processes. The decomposition of the average variance is given in Table 2. 38% of the variation is explained by the first principal component for X, , another 23% by the first principal component for U, . Overall, the first six components , k = 1, …, 6, explain 55% of the average variance, indicating that the X process captures most of the variation in the data. Within X, most of the variation is explained by the random functional intercept Xi,0, but the variance due to the subject-specific random slope is still large compared to the measurement error. Note also that the study period is much shorter than the disease duration for some of the patients, such that Xi,1 might still be of large practical relevance over many years. Within-curve measurement error is negligible due to a smoothing step during profile construction, explaining only 0.02%. Estimated variances were λ̂k = 0.316, 0.060, 0.030, 0.021, 0.015, 0.010, k = 1, …, 6, ν̂k = 0.189, 0.045, 0.025, 0.017, 0.012, 0.011, k = 1, …, 6, and σ̂2 = 0.0002.
Figure 4 shows the first three estimated principal components for the random intercept and slope process X. The left column gives estimates for the , corresponding to the random functional intercept Xi,0. Depicted are estimates for the overall mean η(d) (solid line), and for , k = 1, 2, 3 (+ and −, respectively). The middle column gives the corresponding results for the random functional slope Xi,1. The right column shows boxplots for the estimates of the scores ξik corresponding to ( ), k = 1, 2, 3, by case/control group. Estimated scores for the two example patients with tract profiles shown in Figure 1 are indicated by A and B, respectively.
Fig 4.
The first three estimated principal components for the random intercept and slope process X. The left column gives estimates for the , corresponding to the random functional intercept Xi,0. Depicted are estimates for the overall mean η(d) (solid line), and for , k = 1, 2, 3 (+ and −, respectively). The middle column gives the corresponding results for the random functional slope Xi,1. The right column shows boxplots for the estimates of the scores ξik corresponding to ( ), k = 1, 2, 3, by case/control group. Estimated scores for the two example patients with tract profiles shown in Figure 1 are indicated by A and B, respectively.
Positive loadings ξ̂i1 > 0 on the first component correspond to a lower mean function with a particularly deep FA dip in the isthmus (around 20), but only to small changes of profiles over time. For example, patient A with a much lower dip than B loads positively on this component, while B’s loading is roughly zero. The second component is a mean contrast, with positive scores corresponding to a lower dip around 20 and a higher plateau around 60. The corresponding change over time is similar, if smaller in magnitude, and could explain how the differences in mean profiles evolved over time. The large positive score ξ̂i2 in patient A corresponds to the large contrast between low dip and high plateau in this patient, which is very unpronounced in patient B (roughly zero score). The third component shows a shift of the location of the dip, which might reflect differences in anatomy that affect the tractography. This goes hand in hand with a slight further shifting and deepening (for negative scores) of the dip over time. For example, patient A, in contrast to patient B, exhibits more of a deepening of the dip and a shift to the left, with corresponding negative score ξ̂i3. As mentioned above, these consistent changes over time are not immediately apparent from an examination of the tract profiles in Figure 1 but are clearly revealed by loadings on the principal components derived from the LFPCA analysis. In future work, we plan to examine whether these changes can portend disease course.
Figure 5 shows the corresponding results for the visit-specific functional deviation U. is similar in shape to . Patient A at visit 8, for example, shows a lower profile than would be expected from the average evolution in this patient over time, and consequently has a positive score ζ̂ij1, with the converse being true for patient B at visit 2. Components and seem to pick up variation at the ends of the tract as well as visit-to-visit shifts of the location of the dip, which might be due to measurement error. Note that the U process captures both measurement error and true biological fluctuations, which are impossible to separate without additional subject-matter insight. Filtering out these processes allows us to study the systematic trends modeled by X.
Fig 5.
The first three estimated principal components for the visit-specific deviation process U. The left column gives results for the principal components , depicting estimates for the overall mean η(d) (solid line), and for , k = 1, 2, 3 (+ and −, respectively). The right column shows boxplots for the estimates of the scores ζik corresponding to , k = 1, 2, 3, by case/control group. Estimated scores for example visits of the two patients with tract profiles shown in Figure 1 are indicated by A (visit 8) and B (visit 2), respectively.
Our model allows straightforward inclusion of additional covariates such as case/control status, disease severity, medication or age in the mean function η. In this study, however, we were interested in how the main variations in tract profiles and their changes over time differed by case/control group, i.e. in the covariance part of the model. When fixed effect group specific means are the target of inference, our approach could be used to improve the confidence band estimators on the group-specific mean difference. For example, the group-specific means could be estimated first under independence, then the covariances can be estimated using LFPCA, and then estimates of the group-specific mean differences can be further improved using the estimated covariance structure. The process can even be iterated until convergence.
Focusing on the covariances, we find a statistically significant difference in the distribution of the estimated scores ξ̂i1 between MS patients and controls (p=0.0056 in a Mann-Whitney-Wilcoxon test; also in a linear regression adjusting for age and sex). The patient group in particular seems to have a higher mean and a heavier right tail. This could be an indication of a mixture in this group of patients who are more or less affected by MS along this particular tract. Potential loading-based clustering into patient subgroups will be of interest in future work. Interestingly, FA for this component is not decreased uniformly along the tract, but only posterior to the genu (ca. 1–100), with the decrease being especially pronounced in the area of the isthmus (ca. 20). Our results thus identify the region of the corpus callosum (the isthmus) where MS seems to take its greatest toll and also define the ways in which that portion of the tract changes from one year to the next. In future work, we plan to examine whether these changes can portend disease course. This result could not have been obtained by using the average FA instead of our functional approach.
6. Discussion
We have introduced methods for functional data that is observed at multiple time points for the same subject. Our methods can be viewed as extending longitudinal mixed effects models by replacing the random effects with random processes. Models are designed to decompose the longitudinal functional data into a time-dependent population average, baseline (or static) subject-specific variability, longitudinal (or dynamic) subject-specific variability, subject/visit-specific variability and measurement error. We propose an estimation procedure based on an eigen expansion that is highly computationally efficient and performs well in a wide range of simulations and in our application.
Our work is different from functional data methodology applied to the analysis of longitudinally observed scalar data [41, among others], but builds on methods from both functional and longitudinal data analysis [8, 34]. While the considered model shares similarities with models used by [15, 29], we do not assume Gaussianity and our approach is based on functional principal component analysis. In addition to computational advantages (compare Section 4.3 and [19]), this allows the extraction of the main differences between subjects in the dynamics of their profiles over time, something of interest in many applications including our tractography study. Also, our work is different from methods for the 3-D analysis of subject-specific DTI studies [see for example 18]. It takes a functional data approach to tract data, as has recently been done for non-longitudinal DTI tractography data in [52].
Our approach can serve as a stepping-stone for further developments in the field of longitudinally observed functional data, and lends itself well to extensions. As our estimation procedure performs best when the number of time points per subject is balanced, it might be interesting to investigate further improvements, such as via iterations between mean and covariance estimation. Using an iterative approach, [49] find improvements to the integrated mean squared errors that are most pronounced for sparse functional data, where the number of sample points per curve is small. We will pursue such an approach in the future, in particular if dealing with sparse longitudinal functional data. While the functional random intercept-random slope model was sufficient for our application, it would also be interesting to apply our general model in more complex settings. And as our methods extract the main modes of variation in longitudinal functional data, including differences in mean curves and changes in curves over time, the associated scores could be of interest for further use in regression or classification.
Supplementary Material
Supplementary materials to “Longitudinal functional principal component analysis” by S. Greven, C. Crainiceanu, B. Caffo and D. Reich (doi: 10.1214/10-EJS575SUPP).
Supplementary Material
Acknowledgments
We thank the editor, David Ruppert, for his helpful comments which led to an improved version of the manuscript. The MRI scans used in Section 5 were obtained through a generous grant from the National Multiple Sclerosis Society (TR3760A3) to Dr. Peter Calabresi, whom we gratefully acknowledge.
Appendix A: Theoretical results and proofs
Theorem 1
Estimates of the covariance functions in Step A can be expressed as
and
. Here, β1
is the 5×{D(D−1)/2} matrix with column
{K0(d, d′), K01(d, d′),
K01(d′, d), K1(d, d′),
KU (d, d′)} corresponding to d <
d′ ∈  , and
β2 is the 4 × D matrix with
column {K0(d, d), K01(d, d), K1(d,
d′), KU (d, d) + σ2}
corresponding to d ∈
, and
β2 is the 4 × D matrix with
column {K0(d, d), K01(d, d), K1(d,
d′), KU (d, d) + σ2}
corresponding to d ∈  .
X1 is the
matrix with rows (1, Tik,
Tij, TijTik, δjk),
j, k = 1, …, Ji, i = 1, …, I, and
X2 is the
matrix with rows (1, Tik
+ Tij, TijTik,
δjk), j ≤ k = 1, …,
Ji, i = 1, …, I. c1
is the
matrix with column
(Yij(d)Yik(d′), j, k = 1,
…, Ji, i = 1, …, I) corresponding to d
< d′ ∈
.
X1 is the
matrix with rows (1, Tik,
Tij, TijTik, δjk),
j, k = 1, …, Ji, i = 1, …, I, and
X2 is the
matrix with rows (1, Tik
+ Tij, TijTik,
δjk), j ≤ k = 1, …,
Ji, i = 1, …, I. c1
is the
matrix with column
(Yij(d)Yik(d′), j, k = 1,
…, Ji, i = 1, …, I) corresponding to d
< d′ ∈  , and
c2 the
matrix with column
(Yij(d)Yik(d), j ≤ k = 1,
…, Ji, i = 1, …, I) corresponding to d
∈
, and
c2 the
matrix with column
(Yij(d)Yik(d), j ≤ k = 1,
…, Ji, i = 1, …, I) corresponding to d
∈  .
.
Proof
Consider least squares estimation of
(K0(d,
d′),
K01(d,
d′),
K01(d′,
d),
K1(d,
d′), KU
(d, d′) +
σ2δdd′;
d ≤ d′ ∈
 ) on the basis of
(3.1). First, note
that the design matrix in the corresponding linear regression is block
diagonal, with blocks corresponding to (d,
d′), d <
d′, containing entries (1,
Tik,
Tij,
TijTik,
δjk) in the row
corresponding to
Yij(d)Yik(d′),
and blocks corresponding to (d, d)
containing entries (1, Tik
+Tij,
TijTik,
δjk) in the row
corresponding to
Yij(d)Yik(d).
Second, note that the blocks are identical for all pairs
(d, d′),
d < d′, respectively
all pairs (d, d). Least squares
estimates thus can be expressed as
and
, where
and
are 5 × 5 and 4 × 4
matrices, respectively.
) on the basis of
(3.1). First, note
that the design matrix in the corresponding linear regression is block
diagonal, with blocks corresponding to (d,
d′), d <
d′, containing entries (1,
Tik,
Tij,
TijTik,
δjk) in the row
corresponding to
Yij(d)Yik(d′),
and blocks corresponding to (d, d)
containing entries (1, Tik
+Tij,
TijTik,
δjk) in the row
corresponding to
Yij(d)Yik(d).
Second, note that the blocks are identical for all pairs
(d, d′),
d < d′, respectively
all pairs (d, d). Least squares
estimates thus can be expressed as
and
, where
and
are 5 × 5 and 4 × 4
matrices, respectively.
Proof of Corollary 1
From Theorem 1 and the analogous argument for the general model, only (p2+1)×(p2+1) matrices need to be inverted. Matrix inversion thus is of order O(p6). Matrix multiplication is of order O(p2D2g), giving overall computational effort of order O(max(p6, p2D2g)).
Theorem 2
The estimated BLUPs in (3.2) can be expressed as
with C = EI ⊗ φU′ φ0 + T ⊗ φU′ φ1, R = In ⊗ diag(νk/(νk + σ2)) + GFG′, G = EI ⊗ diag(νk/(νk + σ2)) φU′ φ0 + T ⊗ diag(νk/(νk + σ2)) φU′ φ1, and where B and F are block-diagonal with blocks
i = 1, …, I, denoting , and L = ID − ΦU diag(νk/(νk + σ2)) ΦU′. Here, diag(ak) denotes a diagonal matrix with entries ak, k = 1, …, NU, and we suppress for simplicity hat notation that indicates estimated quantities.
Proof
For simplicity, we suppress hat notation that indicates estimated quantities in the computation of the EBLUPs. Using the Woodbury formula, we obtain
Using the Schur complement S, write
where and .
Using properties of the Kronecker product, we have
Thus, A is a block-diagonal matrix with I blocks Ai of size NX × NX, and B = A−1 can be computed as a block-diagonal matrix with the ith block of size NX × NX of the form
Analogously,
can be computed explicitly, as the columns of ΦU are orthonormal by construction. And finally, using the Woodbury formula again, the Schur complement S can be inverted as
where
and
is again block-diagonal with NX × NX blocks, as is A, such that F = [A−H]−1 can be computed by inverting each block
with L = ID − ΦU diag(νk/(νk + σ2))ΦU′, separately.
Proof of Corollary 2
From Theorem 2, only matrices of size NX × NX need to be inverted to compute the EBLUPs, giving computational effort of order . Usage of the block structure for all matrices reduces computation for the matrix multiplications. For example, multiplication of the (nNU + INX) × nD and nD × 1 matrices Z′ and (Y − η), usually an O(nD(nNU + INX)) operation, here reduces to I multiplications of NX × D with D × 1 matrices and n multiplications of NU × D with D × 1 matrices. Similarly bookkeeping for the other operations leads to the overall effort of order O(nD(NU + NXI/n)).
Proof of Lemma 1 and Corollary 3
Iterated expectations give us
As E{Yij(s)|Tij} = 0,
Now consider the case where E(Tij) = 0 and Var(Tij) = 1. In this case, we have
due to the orthonormality of the eigenfunctions.
Appendix B: Estimation of the general functional mixed model
Estimation for model (2.2) proceeds in the same way as for model (2.1). In this section, we briefly point out the necessary minor adjustments. An estimate of the mean function η(d, Zij) can again be obtained under a working independence assumption and under the specified model. For example, under the specification η(d, Zij) = η1(d, Zij,1) + ···+ ηm(d, Zij,m), bivariate smoothing in an additive model can be used.
In the estimation of the covariance functions, equation (3.1) now becomes
where Vij = (Vij1, …, Vijp), and the three step estimation procedure for the covariance functions extends straightforwardly. The size of the matrix to be inverted during step 1 increases from 5 × 5 to (p2 + 1) × (p2 + 1).
Similarly, estimation of the eigenfunctions using the spectral decomposition, and estimation of the scores using best linear unbiased prediction, proceeds completely analogously, keeping in mind that { , k = 1, 2, …} now form an orthonormal basis for the (L2[0, 1])p.
Choice of NX and NU can again proceed using the proportion of variance explained. Standardization of variables in Vij is recommended. Note, however, an additional complication in the higher-dimensional case. If some of the covariates in Vij are correlated, corresponding additional terms will appear in the expansion of ∫Var{Yij(d)}. The eigenvalues λk might then somewhat over- or underrepresent the relative importance of the corresponding component in explaining the variation in Yij(d). If strong correlations are a concern, additional measures, such as the use of orthogonal polynomials in the case of polynomial Vij, should be taken.
Contributor Information
Sonja Greven, Email: sonja.greven@stat.uni-muenchen.de, Department of Statistics, Ludwig-Maximilians-University Munich, Ludwigstr. 33, 80539 Munich, Germany.
Ciprian Crainiceanu, Email: ccrainic@jhsph.edu, Department of Biostatistics, Johns Hopkins University, 615 N. Wolfe Street, Baltimore, MD 21205, USA.
Brian Caffo, Email: bcaffo@jhsph.edu, Department of Biostatistics, Johns Hopkins University, 615 N. Wolfe Street, Baltimore, MD 21205, USA.
Daniel Reich, Email: reichds@ninds.nih.gov, Translational Neuroradiology Unit, Neuroimmunology Branch, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20814, USA. Departments of Radiology and Neurology, Johns Hopkins Hospital, 600 N. Wolfe Street, Baltimore, MD 21287, USA.
References
- 1.Basser P, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophysical Journal. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. Journal of Magnetic Resonance, Series B. 1996;111:209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- 3.Brumback BA, Rice JA. Smoothing spline models for the analysis of nested and crossed samples of curves. Journal of the American Statistical Association. 1998:961–976. [Google Scholar]
- 4.Calabresi PA. Multiple sclerosis and demyelinating conditions of the central nervous system. In: Goldman L, Ausiello DA, editors. Cecil Medicine. 23. Saunders: Elsevier; 2008. [Google Scholar]
- 5.Crainiceanu C, Ruppert D. Likelihood ratio tests in linear mixed models with one variance component. Journal of the Royal Statistical Society, Series B. 2004;66:165–185. [Google Scholar]
- 6.Crainiceanu CM, Staicu AM, Di CZ. Generalized Multilevel Functional Regression. Journal of the American Statistical Association. 2009;104:1550–1561. doi: 10.1198/jasa.2009.tm08564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di CZ, Crainiceanu CM, Caffo BS, Punjabi NM. Multilevel functional principal component analysis. Annals of Applied Statistics. 2008;3:458–488. doi: 10.1214/08-AOAS206SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diggle P, Heagerty P, Liang KY, Zeger S. Analysis of longitudinal data. Oxford University Press; USA: 2002. [Google Scholar]
- 9.Fan J, Gijbels I. Local polynomial modelling and its applications. CRC Press; 1996. [Google Scholar]
- 10.Ferraty F, Vieu P. Nonparametric functional data analysis: theory and practice. Springer Verlag; 2006. [Google Scholar]
- 11.Green PJ, Silverman BW. Nonparametric Regression and Generalized Linear Models: a Roughness Penalty Approach. Chapman & Hall Ltd; 1994. [Google Scholar]
- 12.Greven S, Kneib T. On the Behaviour of Marginal and Conditional Akaike Information Criteria in Linear Mixed Models. Biometrika. 2010 to appear. [Google Scholar]
- 13.Greven S, Crainiceanu C, Caffo B, Reich D. Supplement to “Longitudinal functional principal component analysis. 2010 doi: 10.1214/10-EJS575SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greven S, Crainiceanu CM, Küchenhoff H, Peters A. Restricted Likelihood Ratio Testing for Zero Variance Components in Linear Mixed Models. Journal of Computational and Graphical Statistics. 2008;17:870–891. [Google Scholar]
- 15.Guo W. Functional mixed effects models. Biometrics. 2002;58:121–128. doi: 10.1111/j.0006-341x.2002.00121.x. [DOI] [PubMed] [Google Scholar]
- 16.Guo W. Functional data analysis in longitudinal settings using smoothing splines. Statistical methods in medical research. 2004;13:49. doi: 10.1191/0962280204sm352ra. [DOI] [PubMed] [Google Scholar]
- 17.Hall P, Müller HG, Yao F. Modelling sparse generalized longitudinal observations with latent Gaussian processes. Journal of the Royal Statistical Society: Series B. 2008;70:703–723. [Google Scholar]
- 18.Heim S, Fahrmeir L, Eilers P, Marx B. 3D space-varying coefficient models with application to diffusion tensor imaging. Computational Statistics & Data Analysis. 2007;51:6212–6228. [Google Scholar]
- 19.Herrick RC, Morris JS. Wavelet-Based Functional Mixed Model Analysis: Computation Considerations. In. Proceedings, Joint Statistical Meetings, ASA Section on Statistical Computing 2006 [Google Scholar]
- 20.Karhunen K. Über Lineare Methoden in der Wahrscheinlichkeit-srechnung. Annales Academiae Scientiarum Fennicae. 1947;37:1–79. [Google Scholar]
- 21.Krivobokova T, Kauermann G. A note on penalized spline smoothing with correlated errors. Journal of the American Statistical Association. 2007;102:1328–1337. [Google Scholar]
- 22.Laird N, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- 23.Liang H, Wu H, Zou G. A note on conditional AIC for linear mixed-effects models. Biometrika. 2008;95:773–778. doi: 10.1093/biomet/asn023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin X, Carroll RJ. Nonparametric Function Estimation for Clustered Data When the Predictor Is Measured Without/With Error. Journal of the American Statistical Association. 2000;95:520–534. [Google Scholar]
- 25.Lin F, Yu C, Jiang T, Li K, Li X, Qin W, Sun H, Chan P. Quantitative analysis along the pyramidal tract by length-normalized parameterization based on diffusion tensor tractography: application to patients with relapsing neuromyelitis optica. NeuroImage. 2006;33:154–160. doi: 10.1016/j.neuroimage.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 26.Loeve M. Fonctions aléatoires du second ordre. Comptes Rendus Académie des Sciences. 1945;220:380. [Google Scholar]
- 27.Mercer J. Functions of positive and negative type, and their connection with the theory of integral equations. Philosophical Transactions of the Royal Society of London Series A. 1909:415–446. [Google Scholar]
- 28.Mori S, Crain BJ, Chacko V, Van Zijl PCM. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Annals of Neurology. 1999;45:265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 29.Morris JS, Carroll RJ. Wavelet-based functional mixed models. Journal of the Royal Statistical Society, Series B. 2006;68:179–199. doi: 10.1111/j.1467-9868.2006.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller HG. Functional modelling and classification of longitudinal data. Scandinavian Journal of Statistics. 2005;32:223–240. [Google Scholar]
- 31.Müller HG, Zhang Y. Time-varying functional regression for predicting remaining lifetime distributions from longitudinal trajectories. Biometrics. 2005;61:1064–1075. doi: 10.1111/j.1541-0420.2005.00378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh JS, Song IC, Lee JS, Kang H, Park KS, Kang E, Lee DS. Tractography-guided statistics (TGIS) in diffusion tensor imaging for the detection of gender difference of fiber integrity in the midsagittal and parasagittal corpora callosa. Neuroimage. 2007;36:606–616. doi: 10.1016/j.neuroimage.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 33.Ozturk A, Smith S, Gordon-Lipkin E, Harrison D, Shiee N, Pham D, Caffo B, Calabresi P, Reich D. MRI of the corpus callosum in multiple sclerosis: association with disability. Multiple Sclerosis. 2009 doi: 10.1177/1352458509353649. to appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramsay JO, Silverman B. Functional data analysis. 2. Springer; 2005. [Google Scholar]
- 35.Rao CR. The theory of least squares when the parameters are stochastic and its application to the analysis of growth curves. Biometrika. 1965;52:447–458. [PubMed] [Google Scholar]
- 36.Reich DS, Smith SA, Zackowski KM, Gordon-Lipkin EM, Jones CK, Farrell JAD, Mori S, van Zijl PCM, Calabresi PA. Multiparametric magnetic resonance imaging analysis of the corticospinal tract in multiple sclerosis. Neuroimage. 2007;38:271–279. doi: 10.1016/j.neuroimage.2007.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice JA. Functional and longitudinal data analysis: Perspectives on smoothing. Statistica Sinica. 2004;14:631–647. [Google Scholar]
- 38.Rice JA, Silverman B. Estimating the mean and covariance structure nonparametrically when the data are curves. Journal of the Royal Statistical Society Series B. 1991;53:233–243. [Google Scholar]
- 39.Ruppert D, Wand MP, Carroll RJ. Semiparametric Regression. Cambridge University Press; 2003. [Google Scholar]
- 40.Staicu AM, Crainiceanu CM, Carroll RJ. Fast Methods for Spatially Correlated Multilevel Functional Data. Biostatistics. 2010 doi: 10.1093/biostatistics/kxp058. to appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staniswalis JG, Lee JJ. Nonparametric Regression Analysis of Longitudinal Data. Journal of the American Statistical Association. 1998;93:1403–1404. [Google Scholar]
- 42.Tievsky AL, Ptak T, Farkas J. Investigation of apparent diffusion coefficient and diffusion tensor anisotropy in acute and chronic multiple sclerosis lesions. American Journal of Neuroradiology. 1999;20:1491–1499. [PMC free article] [PubMed] [Google Scholar]
- 43.Vaida F, Blanchard S. Conditional Akaike information for mixed-effects models. Biometrika. 2005;92:351–370. [Google Scholar]
- 44.Verbeke G, Molenberghs G. Linear mixed models for longitudinal data. Springer; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werring D, Clark C, Barker G, Thompson A, Miller D. Diffusion tensor imaging of lesions and normal-appearing white matter in multiple sclerosis. Neurology. 1999;52:1626–1632. doi: 10.1212/wnl.52.8.1626. [DOI] [PubMed] [Google Scholar]
- 46.Witelson SF. Hand and sex differences in the isthmus and genu of the human corpus callosum: a postmortem morphological study. Brain. 1989;112:799–835. doi: 10.1093/brain/112.3.799. [DOI] [PubMed] [Google Scholar]
- 47.Wood SN. Generalized Additive Models: An Introduction with R. Chapman and Hall/CRC; 2006. [Google Scholar]
- 48.Wu H, Zhang JT. Nonparametric regression methods for longitudinal data analysis: mixed-effects modeling approaches. Wiley-Blackwell; 2006. [Google Scholar]
- 49.Yao F, Lee TCM. Penalized spline models for functional principal component analysis. Journal of the Royal Statistical Society, Series B. 2006;68:3–25. [Google Scholar]
- 50.Yao F, Müller HG, Wang JL. Functional data analysis for sparse longitudinal data. Journal of the American Statistical Association. 2005;100:577–590. [Google Scholar]
- 51.Yao F, Clifford AJ, Dueker SR, Follett J, Lin Y, Buch-holz BA, Vogel JS. Shrinkage estimation for functional principal component scores with application to the population kinetics of plasma folate. Biometrics. 2003;59:676–685. doi: 10.1111/1541-0420.00078. [DOI] [PubMed] [Google Scholar]
- 52.Zhu H, Styner M, Tang N, Liu Z, Lin W, Gilmore J. FRATS: Functional Regression Analysis of DTI Tract Statistics. IEEE Transactions on Medical Imaging. 2010;29:1039–1049. doi: 10.1109/TMI.2010.2040625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.