Abstract
We present a novel and efficient non-integrating gene expression system in human embryonic stem cells (hESc) utilizing human artificial chromosomes (HAC), which behave as autonomous endogenous host chromosomes and segregate correctly during cell division. HAC are important vectors for investigating the organization and structure of the kinetochore, and gene complementation. HAC have so far been obtained in immortalized or tumour-derived cell lines, but never in stem cells, thus limiting their potential therapeutic application. In this work, we modified the herpes simplex virus type 1 amplicon system for efficient transfer of HAC DNA into two hESc. The deriving stable clones generated green fluorescent protein gene-expressing HAC at high frequency, which were stably maintained without selection for 3 months. Importantly, no integration of the HAC DNA was observed in the hESc lines, compared with the fibrosarcoma-derived control cells, where the exogenous DNA frequently integrated in the host genome. The hESc retained pluripotency, differentiation and teratoma formation capabilities. This is the first report of successfully generating gene expressing de novo HAC in hESc, and is a significant step towards the genetic manipulation of stem cells and potential therapeutic applications.
INTRODUCTION
Human embryonic stem cells (hESc) are an important tool in clinical and basic research. Due to their high replicative lifespan and ability to differentiate into the three different germ layers, they are used for human developmental biology, tissue regeneration, transplant therapies and drug discovery studies (1). Safe and efficient in vitro genetic manipulation of hESc is an essential step in realizing their full potential for clinical applications (2).
Most gene expression studies in hESc utilize lentiviral, adenoviral and adeno-associated viral (AAV) vectors for gene delivery (3–5). However, lentiviral vectors integrate randomly at multiple sites within the host genome leading to insertional mutagenesis (6), and although adenoviral vectors remain episomal, silencing post-transduction may occur. Another disadvantage is that the capacity of AAV and lentiviral vectors is limited to ∼5 and 10 kb of DNA, respectively (7).
An alternative vector type is represented by human artificial chromosomes (HAC), which are autonomous functional chromosomal elements that behave as normal chromosomes. Gene-expressing HAC are generated in cells following delivery of vectors with centromeric alpha-satellite (alphoid, α) DNA, appropriate markers and genes. The maintenance of the HAC does not require integration into the host genome, and they are suitable for the delivery of large gene loci.
Previously, we introduced input HAC DNA (404 kb) containing the hypoxanthine–guanine phosphoribosyl transferase (HPRT) gene into human HPRT-deficient HT1080 fibrosarcoma cells, and the HAC generated successfully complemented the deficiency (8). The input HAC DNA was delivered by standard transfection, but we found this method inefficient for routine large DNA transfer. In more recent studies, we utilized the highly efficient herpes simplex virus-1 (HSV-1) amplicon technology for delivery of the input HAC DNA using infectious amplicons (9). The advantage of this system is that HSV-1 amplicons have a high capacity for large DNA delivery (up to 150 kb) to efficiently introduce HAC input DNA into immortalized cell lines. Following transduction, we observed that the efficiency of input HAC DNA delivery into HT1080 and other cell lines was significantly greater than transfection (by a factor of 104) (9).
In this study, input HAC DNA vectors (ranging from 55 to 115 kb) containing the marker gene for green fluorescent protein (GFP) were delivered into HUES-2 and HUES-10 hESc lines (10) following transduction with HSV-1 amplicons. Mitotically stable, gene-expressing, functional HAC were generated. The HAC were present in up to 70% of the hESc, and gene expression was maintained in the absence of selection over a period of 60 days and following cell differentiation. No DNA integrated into the hESc genome, in contrast to HT1080 cells, where the HAC DNA frequently integrated into the host chromosomes. More importantly, the HSV-1 HAC hESc retained their pluripotency and differentiation capabilities.
This is the first successful study to establish non-integrating, stable, gene-expressing HAC using HSV-1 amplicons in hESc, and is a significant step forward for HAC in gene therapy studies.
RESULTS
Vector construction
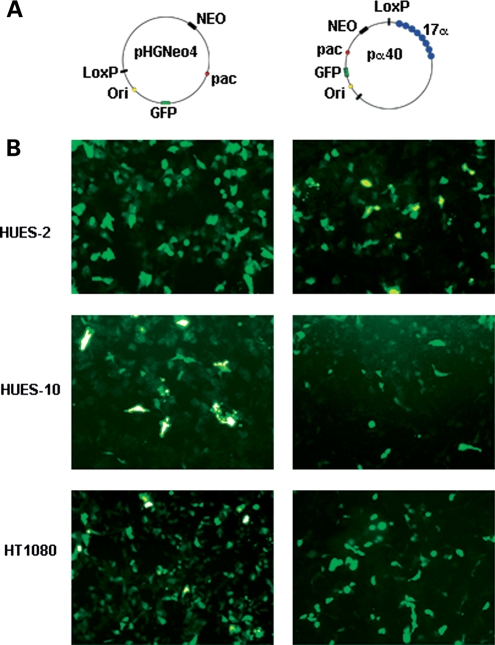
To generate HSV-1-based input HAC DNA vectors that were highly proficient at HAC formation, the BAC hBAC495J24 (containing 220 kb of chromosome 17 core α DNA) used in a previous study to construct an efficient input HAC DNA vector (pJM2256) (11) was modified. As hBAC495J24 was ∼70 kb larger than the 150 kb packaging limit of HSV-1, we aimed to reduce it while retaining its HAC-forming properties. Three derivatives of the BAC hBAC495J24 were generated, two of which arose spontaneously (containing 40 and 100 kb of 17α DNA) during culture, and the third derivative (containing 60 kb of 17α DNA) by utilizing the RED/ET recombination system (12). All three derivatives were modified by LoxP–Cre recombination with pHGNeo4 to include the essential HSV-1 elements, and reporter genes (GFP), thereby generating pα40 (55 kb, including 40 kb of 17α DNA), pα60 (75 kb, including 60 kb of 17α DNA) and pα100 (115 kb, including 100 kb of 17α DNA) (Fig. 1A, and Supplementary Material, Fig. S1A).
Figure 1.
(A) Schematic representation of the vectors used in this study, not drawn to scale. (B) GFP expression in HUES-2, HUES-10 and HT1080 cells 24 h after transduction with pHGNeo4 (left) and pα40 (right) amplicons.
Introduction of input HAC DNA vectors into hES cells
Among the three HSV-1 HAC input DNA vectors we generated, pα40 (40 kb of 17α DNA) was the most useful, as its size allowed up to 100 kb of transgenic DNA to be accommodated within the vector for packaging into HSV-1 amplicons (limit capacity of 150 kb). For this reason, we investigated its HAC-forming efficiency, by delivering pα40 to HUES-2 (at passage 40) or HUES-10 (at passage 29) by HSV-1 amplicon transduction at multiplicity of infection (MOI) 2. In a parallel control experiment, pHGNeo4 amplicons were also delivered to both cell lines. The other 17α input HAC DNA vectors, pα60 and pα100, were delivered only to the HUES-2 line, in the same conditions described above.
The efficiency (%) of transduction, summarized in Table 1, was determined after 24 h by fluorescence-activated cell sorting (FACS) or counting GFP expressing cells. The results are also shown in Figure 1B and Supplementary Material, Figure S1B. In HUES-2, the average transduction efficiency was ∼40% for both pα40 and the control vector pHGNeo4. The two larger vectors, pα100 and pα60, were delivered to HUES-2 with an efficiency of 16 and 20%, respectively (Table 1, Supplementary Material, Fig. S1B). In HUES-10, the delivery efficiency was 27% for both pHGNeo4 and pα40 (Table 1, Fig. 1B).
Table 1.
Average efficiency of HSV-1 amplicon transduction at MOI 2, and HAC formation in HUES-2, HUES-10 and HT1080 cells
| Cell line | Input HAC DNA vector | Efficiency of transduction (%) | No. of clones analysed | HAC positive clones | % of HAC/cells |
|---|---|---|---|---|---|
| HUES-2 | pα40 | 40 | 10 | 5 | 10–70 |
| pα60 | 20 | 9 | 5 | 10–25 | |
| pα100 | 16 | 7 | 3 | 10–20 | |
| pHGNeo4 | 40 | 5 | NA | NA | |
| HUES-10 | pα40 | 27 | 5 | 5 | 35–50 |
| pHGNeo4 | 27 | 1 | NA | NA | |
| HT1080 | pα40 | 19 | 10 | 6 | 5–30 |
| pα60 | 34 | 3 | 1 | 20 | |
| pα100 | 25 | 4 | 1 | 15 | |
| pHGNeo4 | 28 | 3 | NA | NA |
NA, not applicable.
In parallel control experiments, the input HAC DNA amplicon vectors were delivered by HSV-1-mediated transduction to HT1080 cells, which efficiently form HAC (8,9,11). The delivery efficiency of the input HAC DNA vectors was similar to that observed in hESc (Table 1, Fig. 1B, Supplementary Material, Fig. S1A).
Viability of hESc following transduction with HSV-1 amplicons
In addition to the experiments outlined above, to determine whether the HSV-1 amplicon transduction-affected hESc viability, 2.5 × 105 HUES-2 cells were transduced with pHGNeo4 amplicons at MOI 1, 2 and 5. The average efficiency of transduction was determined after 24 h by FACS or counting GFP expressing cells, and found to be ∼27% for MOI 1, 42% for MOI 2 and 48% for MOI 5. Since transduction efficiencies were similar at MOI 2 and 5, the HUES-2 cell lines probably reached transduction saturation in this range.
The cells were monitored for 6 days post-transduction, and the average growth rate was calculated by measuring the rate of population increase divided by the initial number of cells, and compared with that of an untreated control. The growth rate and morphology of HUES-2 were not affected post-HSV-1 amplicon transduction, with a 5% reduction in viability, detected only for MOI 1.
Generation and analysis of stable clones in hES cells
Overall, 10 HUES-2 and 5 HUES-10 clones were isolated following G418 selection, derived from the pα40 transduction (Table 1). In addition, we also isolated 9 and 7 clones from pα60 and pα100, respectively, following transduction into HUES-2 cells. The stable clone formation efficiency of HSV-1 transduction was relatively high for both hESc lines, at 10−4, as calculated by the ratio between the number of stable clones and GFP-positive cells 24 h post-transduction. Chromosome metaphase spreads were prepared from stable clones, and analysed by two colour fluorescence in situ hybridization (FISH) with vector and 17α DNA probes (Table 1 and Fig. 2A, Supplementary Material, Fig. S1B). Following HSV-1 transduction with pα40, HAC were detected in 5 of the 10 stable clones obtained in HUES-2 cells, and in all 5 clones isolated in HUES-10. The HAC were present in up to 70% of the cells from each clone. In the HUES-2 cells transduced with either pα60 or pα100, HAC were detected in approximately half of the clones, with a frequency of up to 25% of the cells of each clone (Supplementary Material, Fig. S1B). The lower HAC frequency per cell observed with pα60 or pα100 indicated that the pα40 vector was the most efficient at HAC formation in HUES-2 following HSV-1 transduction. Most importantly, in none of the 31 clones (HUES-2 or HUES-10) analysed, the HAC DNA had integrated into the host genome. However, in all of the analysed hESc clones derived from the HSV-1 delivery of the control vector pHGNeo4, the exogenous DNA had integrated into the host genome.
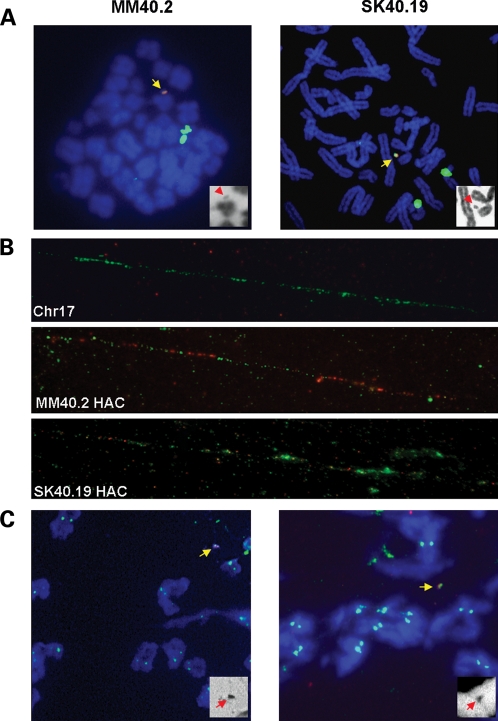
Figure 2.
HAC analysis. (A) FISH with a 17α DNA probe (green), and a vector probe (red). The chromosomes are counterstained in DAPI, blue. The HAC are identified by yellow arrows. The insets show DAPI staining only (HAC, red arrows), in black and white. (B) Fibre FISH on HAC clones, with 17α DNA (green) and vector DNA (red) probes. (C) ImmunoFISH with anti-CENP C antibody (green), and 17α HAC probe (red).
In HT1080, the HSV-1 amplicon transduction successfully generated several hundred clones from each of the 17α HSV-1 HAC input DNA vectors, and several clones were selected for analysis from pα40, pα60 and pα100 (Table 1). The stable clone formation efficiency of HSV-1 transduction was 5 × 10−3. Positive clones were analysed by two colour FISH with vector and 17α DNA probes (Table 1). The HSV-17α input DNA vectors generated HAC in most of the clones following transduction, but were present at a lower frequency in cells (up to 30%), and concomitant integrations in the HT1080 genome were found in all of the clones.
Karyotypic analysis of hESc HAC clones
Two hESc HAC clones were chosen for further studies. Clone MM40.2 derived from the HSV-1-mediated delivery of pα40 into HUES-2, and contained a HAC in 70% of the cells (Fig. 2A). Clone SK40.19 was generated by the transduction of pα40 in HUES-10 cells and contained HAC in 50% of the analysed metaphases (Fig. 2A). The structure of the HAC present in both clones was analysed further by FISH on extended chromatin fibres. The HAC contained repeat units of input DNA, arranged in an alternate fashion as seen in previous studies (8), but the vector backbone DNA was less abundant in clone SK40.19 (Fig. 2B).
The percentage of GFP-positive cells in the two HAC clones was established by FACS analysis, and found to be 33% for SK40.19 and 83% for MM40.2. These values are in concordance with the estimated HAC frequency.
The chromosomal content of the two HAC clones was characterized by chromosome painting for two of the most frequently described chromosomal numerical aberrations in hESc, chromosome 12 and 17 trisomy. In clone MM40.2 (analysed at passage 70), 66% of the cells presented both an extra chromosome 12 and 17. The remaining 34% contained either a chromosome 12 or 17 trisomy.
To investigate whether the HSV-1 transduction and HAC formation processes could have caused the chromosomal instability observed in clone MM40.2, the karyotype of the parental HUES-2 cell line was analysed by chromosome painting at low passage (passage 28) and at the passage when the transduction was undertaken (passage 40). While no extra chromosome 12 or 17 were identified in the parental HUES-2 at passage 28, a double trisomy for chromosome 12 and 17 was detected in 70% of the cells at passage 40, while 30% of cells had either an extra chromosome 12 or 17. These data suggested that the numerical aberration occurred in the parental HUES-2 late passage 40 before the HAC formation and was not the result of the HSV-1 transduction.
In contrast, a similar analysis on clone SK40.19 (at passage 60) showed no extra chromosome 12 or 17. The clone chromosome number was 46 + HAC, and furthermore, the analysis of G-banding pattern indicated that no major rearrangements were present, indicating that SK40.19 was karyotypically normal. The parental HUES-10 line was analysed by whole-genome cytogenetic microarrays. The karyotype of the cell line was found to be completely normal (confidence 0.9, 200 markers).
HAC stability and centromere protein C staining
The mitotic stability of the HAC in MM40.2 and SK40.19 was monitored by FISH for 90 days (corresponding to ∼30 passages) in the absence of selection. In both clones, the HAC frequency did not change significantly over this period, with a daily loss rate of 0.03% for MM40.2 and 0.24% for SK40.19 (calculated by the formula Nn= N0 × (1 − R)n, where N0 is the number of metaphase chromosome spreads showing HAC in the cells cultured under selection, Nn the number of HAC-containing metaphase chromosome spreads after n days of culture in the absence of selection and R the daily rate of loss).
To confirm that an active centromere was present on the HAC, cells from the MM40.2 and SK40.19 clones were stained with anti-centromere protein C (CENP C) antibody, coupled to FISH with HAC-specific probes. A positive CENP C signal was identified on the HAC in both clones at a similar intensity of those observed on endogenous chromosomes (Fig. 2C), thus confirming that an active centromere was present, similar to the HSV-1 HAC in HT1080 cells (9).
Pluripotency studies
To determine whether the MM40.2 and SK40.19 cells expressed pluripotency epitopes compared with the parental lines, cells from each clone (at approximately passages 70 and 60, respectively) were stained with antibodies against the embryonic stem cell markers Oct4, Sox2, Nanog and TRA-1-60 (Fig. 3B). The HAC clones and the parental lines were stained similarly by the antibodies, while HT1080 and mouse embryonic fibroblast (MEF) control cells were completely negative. The result indicated that the HAC-containing hESc clones expressed embryonic stem cell markers.
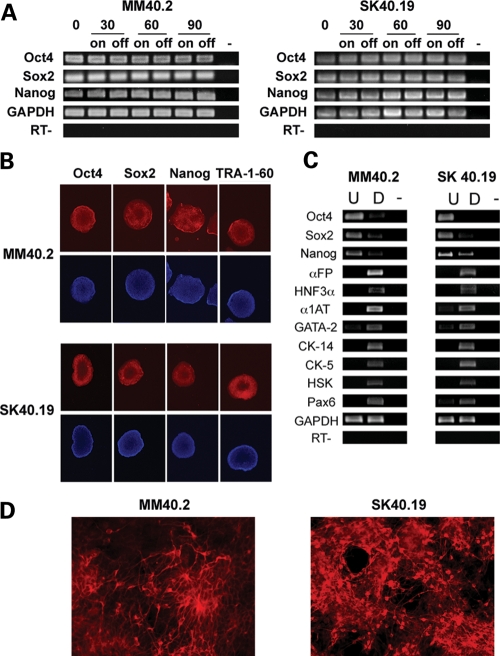
Figure 3.
Pluripotency analysis. (A) RT–PCR with primers specific for pluripotency genes at time points 0, 30, 60 and 90 days in culture (0, 30, 60, 90) in the presence (on) or absence (off) of selection. (RT-) control PCR performed in the absence of the reverse transcriptase, using GAPDH primers, (-) no template control. (B) Immunostaining with pluripotency marker antibodies (red). The cells are counterstained with DAPI, blue. (C) RT–PCR analysis of germinal layer transcripts on cDNA from in vitro differentiated MM40.2 and SK40.19 cells, with primers specific for pluripotency (Oct4, Sox2, Nanog), endoderm (αFP, HFN3α, α1AT), mesoderm (GATA-2), epidermis (CK-14, CK-5, HSK) and neuronal markers (Pax6). U: undifferentiated cells, D: differentiated cells. (RT-): control PCR performed in the absence of the reverse transcriptase, using GAPDH primers. (-) no template control. (D) Neuronal differentiated cells stained with anti-βIII tubulin antibody (red).
The hESc markers expression was sustained over time in both HAC clones, as confirmed by reverse transcription polymerase chain reaction (RT–PCR) analysis on RNA extracted from the MM40.2 and SK40.19 cells, over a period of 90 days, cultivated in the presence and absence of selection (Fig. 3A).
To confirm that the HAC-containing clones were pluripotent, we induced differentiation of the three embryonic germ layers through embryoid body (EB) formation of MM40.2 and SK40.19. Total RNA was extracted from the pool of differentiated cells and analysed for the presence of endoderm (α Feto-protein, HNF3α, α1 anti-trypsin), mesoderm (GATA-2) and ectoderm (CK-5, CK-14, high sulphur keratin, Pax6) specific transcripts. RT–PCR experiments revealed that mRNAs for all the markers were present (Fig. 3C), indicating that both HAC clones retained pluripotency.
HAC clones directed differentiation
Furthermore, neuronal differentiation was induced in clones MM40.2 and SK40.19 by treatment with medium containing noggin and fibronectin (13). After 25 days of directed differentiation, the cells were fixed in formaldehyde and stained with anti-βIII-tubulin antibody, a neuronal cell marker. We detected neuronal cells, highly positive for βIII-tubulin staining, in both HAC clones, ranging approximately between 18 and 40% of the treated cells (Fig. 3D). On average, the SK40.19 was up to two times more efficient at forming neuronal cells, compared with MM40.2. The control cells (untreated MM40.2 and SK40.19; HT1080; MEF) never displayed positive cells. To determine whether the HAC was still present in the cells following neuronal differentiation, we conducted FISH experiments on the interphasic cells stained with the anti-βIII-tubulin antibody. In clone SK40.19, the HAC frequency remained the same (50%), while a positive HAC signal was detected in 44% of the MM40.2 cells (Supplementary Material, Fig. S2).
Teratoma formation assay
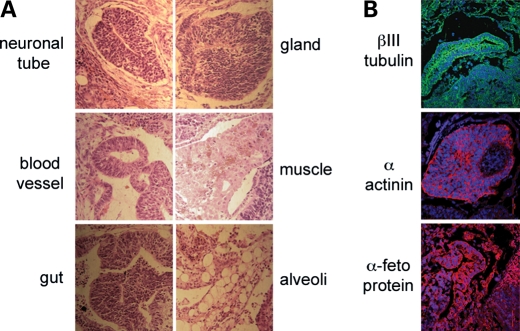
Since the MM40.2 cells contained the highest HAC frequency, teratomas were generated using these cells in immunodeficient mice, as this constituted the most rigorous test of pluripotency for human ES cells. Sub-cutaneous injection into immunodeficient mice generated tumours between the 5th and 7th week post-treatment. The subsequent histological analysis of haematoxylin/eosin-stained tumour sections revealed the presence of ectodermal (neural tube), mesodermal (muscle and blood vessels) and endodermal (gut epithelium, alveoli and glandular epithelium) structures (Fig. 4A). Moreover, immunostaining using suitable antibodies confirmed the presence of all three germinal layers in these sections (Fig. 4B) thereby confirming the tumour growth as a teratoma.
Figure 4.
Teratoma assay. (A) Haematoxylin/eosin-stained MM40.2-teratoma derived sections. (B) immunostaining with antibodies for ectoderm (βIII-tubulin, green), mesoderm (α-actinin, red) and endoderm(α-feto protein, red) markers.
As the sections prepared for the immunological staining were not suitable for FISH experiments, we confirmed the presence of HAC in the teratoma cells by real-time qPCR on total genomic DNA extracted from the teratoma mass. HAC-specific primers (Neo and GFP) were used, and the HAC DNA content in the teratoma cells was normalized against that of the undifferentiated MM40.2 cells (Supplementary Material, Fig. S3). We detected no difference in the abundance of HAC sequences between the undifferentiated MM40.2 and teratoma cells, suggesting that the HAC had not been lost during the in vivo cell replications. A similar analysis was conducted on the neuronal-differentiated MM40.2 cells, previously analysed by FISH. The qPCR results closely reflected the FISH quantification of HAC distribution (Supplementary Material, Fig. S3).
HAC gene expression analysis
The expression level of the reporter gene for GFP from the MM40.2 and SK40.19 HAC was investigated by real-time qPCR experiments on cDNA, in cells grown either on or off selection. The HAC gene expression was compared with that of a ubiquitously expressed gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). In SK40.19, the GFP expression was found to be decreased by about 50% after ∼40 days on or off selection (Table 2). In contrast, in MM40.2, the GFP gene expression level remained constant for a prolonged period of time (60 days) on or off selection (Table 2). To further analyse the reporter gene expression clonal variability, the GFP relative amount in three pHGNeo4 stable clones was measured by real-time qPCR. Compared with the MM40.2, one clone had approximately five times lower levels of GFP, and two had 5 and 10 times more GFP, respectively (Supplementary Material, Table S1). Taken together, these data suggested that stochastic events may have affected the level of expression of the reporter gene in different clones. To confirm that the MM40.2 clone had very stable levels of reporter gene expression, GFP presence was further monitored by FACS over the course of 90 days. The percentage of GFP-expressing cells remained relatively constant and ranged from 81 to 89%. The fold increase in the geometric mean of fluorescence intensity compared with the untreated control ranged from ∼8 to 12, indicating that the average GFP expression per cell also remained relatively constant.
Table 2.
Analysis by qPCR of the GFP reporter gene expression in clones MM40.2 and SK40.19, in the presence or absence of selection
| Day 0 (SD) | Day 30 (SD) | Day 60 (SD) | ||
|---|---|---|---|---|
| MM40.2 | On selection | 1.00 (0.03) | 1.23 (0.03) | 1.34 (0.05) |
| Off selection | 1.00 (0.03) | 1.43 (0.15) | 1.49 (0.07) | |
| SK40.19 | On selection | 1.00 (0.02) | 0.40a (0.01) | NA |
| Off selection | 1.00 (0.02) | 0.52a (0.02) | NA |
The qPCR-fold difference values are expressed in reference to the day 0 time point, using GAPDH as internal control. SD, standard deviation. NA, not analysed.
a37 days.
Furthermore, the GFP reporter gene expression levels of the MM40.2 clone were characterized by qPCR following differentiation, and compared with the levels present in the undifferentiated MM40.2 parental. The GFP gene was still expressed in both EB-derived, and neuronal-differentiated cells, at ∼60–70% of the level present in the undifferentiated parental (Table 3). The level of GFP expression was consistent with the HAC frequency observed in the neuronal-differentiated MM40.2 cells. Based on direct microscopic observation of the EB-derived and neuronal cells, the derivative differentiated cells exhibited a heterogeneous GFP expression: while some cells were still highly GFP positive, in others the GFP fluorescence appeared reduced or absent (Supplementary Material, Fig. S4). This suggested that stochastic events in the early stages of hESc differentiation had an effect in the level of expression of the reporter gene during the later stages.
Table 3.
Analysis by qPCR of the GFP reporter gene expression, in differentiated MM40.2 cells
| Undifferentiated hESc (SD) | EB-derived cells (SD) | Neuronal cells (SD) | |
|---|---|---|---|
| GFP expression | 1.00 (0.03) | 0.70 (0.04) | 0.60 (0.02) |
The values are expressed in reference to the undifferentiated parental, using GAPDH as internal control. SD, standard deviation.
DISCUSSION
hESc have been hailed as an ideal model to study cell differentiation into organ-specific lineages, gene expression, and to eventually establish patient-specific/gene therapy applications (1). Several techniques utilizing viral and non-viral vectors have been used to deliver transgenes to hESc. Most viral vectors are highly efficient at introducing the DNA into the cells, but they are usually limited by the size of the gene they can accommodate and they may randomly integrate into the host genome, possibly resulting in deleterious mutagenic effects (6).
In this study, we describe an alternative efficient approach, based on direct HAC formation in two hESc lines utilizing HSV-1 amplicons for delivery. HAC can accommodate large genomic regions containing potentially therapeutic genes along with their regulatory sequences, and have successfully been used as gene transfer vectors to complement genetic deficiencies in human-cultured cells (8). HAC are also composed of heterochromatic and euchromatic regions in a similar pattern to endogenous chromosomes. While the presence of heterochromatin is necessary for the correct segregation of HAC (14,15), the euchromatin potentially ensures the prolonged transgene expression.
We delivered input HAC DNA vectors to the HUES-2, HUES-10 and HT1080 cells, using HSV-1 amplicon-mediated transduction. This technique is up to 104 times more efficient at delivering large DNA than chemical transfection, in several different cell types (9).
The FISH analysis revealed that HAC were formed in both HUES-2 and HUES-10. This is the first report of de novo HAC formation in a karyotypically normal primary human cell line, and is highly significant for developing HAC as gene expression vectors for gene therapy applications. Most importantly, the input HAC DNA vectors never integrated in the hESc genome, compared with HT1080, where integrations were found in most of the clones. It is possible that the presence in hESc of systems actively guarding genome integrity (16) may prevent or reduce the frequency of large vector integration events, thus giving an advantage to cells where the exogenous DNA forms episomal HAC, in the presence of selective pressure. In this scenario, the smaller vector pHGNeo4 integrated in hES cells, because no other outcome would have allowed the cells to survive selection, as pHGNeo4 is incapable of forming HAC. On the other hand, HT1080 cells are tumour derived, and lack an efficient control of genome integrity, thus explaining why we observed both HAC formation and integration events. Future studies with hESc lines mutant for genome guarding proteins, such as p53, may help clarify this issue.
The hESc HAC were fully stable, and formed an active centromere. This showed that although the HAC were not detected in 100% of the cells, it was not the result of instability. It is possible that the cloning procedure for hESc did not produce pure clones [hESc have a low survival rate as single cells, even when rho-associated kinase (ROCK) inhibitor Y-27632 is used], and hence the HAC frequency in the cells was <100%, as different sublines may have been present within the clones.
The analysis of HAC gene expression showed that the RNA levels of the GFP reporter gene on or off selection were different in the clones characterized, containing either HAC or integrated pHGNeo4 DNA. This confirmed the existence of clonal variability between different lines, possibly due to epigenetic effects. In one of the clones, MM40.2, GFP expression was highly stable, and did not change over prolonged time in culture, both on and off selection. Furthermore, the reporter gene expression was maintained following MM40.2 in vitro differentiation, although we observed heterogeneity in the GFP levels among the differentiated cells. The process of de novo HAC formation generally results in the multimerization of the input DNA, as shown by the alternate pattern of vector and alpha-satellite signals observed in the FISH on chromatin fibres, and the possibility that more than one copy of the transgene is present on the HAC. This ensures that copies of the transgene will be localized away from the centromeric heterochromatic area, and thus escape potential silencing. The spreading of heterochromatin, which is a stochastic event, may explain the variability observed in the GFP reporter gene expression following differentiation in clone MM40.2, and in the prolonged culture in clone SK40.19. Expression studies where the gene on the HAC is under the control of its appropriate promoter and regulatory sequences will verify if physiological expression levels can be achieved. It may also be interesting to study the possibility of loading the transgenes on the HAC via an in vivo LoxP-based system as shown by other reports (17,18), and screening clones where only one copy has been acquired by the HAC.
Importantly, neither the HSV-1 transduction nor the HAC formation led to a loss of pluripotency in the HUES-2 or HUES-10 cells, as suggested by the staining with hESc-specific markers, expression of three germinal layer markers in EB-derived cells, by the differentiation into neuronal types and by MM40.2 teratoma formation. The HAC were present in the neuronal-differentiated cells in both MM40.2 and SK40.19. The HAC frequency in MM40.2 neuronal cells was slightly lower than in the undifferentiated cells, yet the frequency in SK40.19 remained unchanged following differentiation.
The HSV-1 amplicon particles package DNAs up to 150 kb (19). This is far larger than the size which can be accommodated in lentiviral, AAV or adenovirus-based vectors. In this study, the pα40 containing the shortest tract of 17 alpha-satellite DNA (40 kb) was the most efficient vector at HAC formation in hESc, and since its total size is ∼55 kb, it can accommodate a large genomic region containing gene loci of up to 100 kb. HSV-1 HAC replicate at the same rate as the endogenous chromosome, making the gene dosage level more easily controllable.
In summary, we obtained high-capacity, gene-expressing HAC formation in two hES cell lines, using a high-efficiency delivery method based on the HSV-1 amplicon technology. The HAC were stable and sustained long-term gene expression. The lack of integrated DNA in hESc is an important and exciting finding, prompting further work to understand the mechanism in HAC generation. This is the first report of direct de novo HAC formation in human stem cell lines, and shows HAC vectors as a viable alternative to other gene delivery systems in hESc. Our findings are not exclusive to the field of hESc, but may also be relevant for induced pluripotent stem cells.
MATERIALS AND METHODS
Input HAC DNA vector assembly
The pHGNeo4 vector was released from pHSV21αNeo (9) by BstBI digestion, and re-circularized by treatment with T4 DNA ligase. The pHGNeo4 vector carries the HSV-1 amplicon origin of replication (Ori) and packaging signal (pac), the reporter GFP gene, under the control of the I/E promoter from HSV-1, and the G418 resistance gene (Neo), controlled by the SV40 promoter.
Vectors BAC17α40 and BAC17α100, containing, respectively, 40 kb and 100 kb of a satellite DNA from human chromosome 17 (17α), were derived by spontaneous deletions of hBAC495J24 (11,20). PAC17α60 was obtained by RED/ET recombination (12), which transferred 60 kb of 17α DNA from hBAC495J24 to pCYPAC2 vector. Briefly, the pCYPAC2 vector was used as a template to generate a 9.5 kb PCR fragment, using primers containing 50 bp homologous tails to the alpha 17 satellite DNA consensus (Supplementary Material, Table S2). The linear 9.5 kb pCYPAC2 PCR product was then transformed into Escherichia coli cells containing hBAC495J24, and expressing the RED/ET system proteins.
The BAC17α40, PAC17α60 and BAC17α100 constructs were then retrofitted with pHGNeo4 by LoxP-Cre recombination (9), to generate three new HSV-1-based HAC vectors: pα40, pα60 and pα100, respectively.
Cell culture
The hESc lines HUES-2 and HUES-10 (10) were obtained from Douglas Melton (Harvard University, Cambridge, MA, USA) and grown under license from the UK Stem Cell Steering Committee as described previously (10,21), on mitomycin C-inactivated MEFs or SNL76/7 cells. Feeder-independent HUES-2 and HUES-10 cells were grown on Matrigel (BD Biosciences) coated wells using the mTeSR medium (STEMCELL Technologies). TrypLE Express (Invitrogen) was used to enzymatically passage the hESc. Cells were maintained and passaged at high densities on Matrigel. To increase single-cell survival, ROCK inhibitor Y-27632 (Merck Biosciences) was added during each passaging step, at a final concentration of 10 μm. The HT1080 (ATCC-CCL-121) cells were grown using standard techniques in Dulbecco's modified eagle medium (DMEM) medium (Invitrogen), supplemented with 10% foetal bovine serum (FBS) and 1% penicillin/streptomycin.
HSV-1 amplicon preparation
HSV-1 amplicons were prepared as described (9,22). Briefly, the packaging cell line Vero 2-2 was transfected with each input HAC vector DNA, fHSVΔpacΔ270 0+ and pEBHICP27 by Lipofectamine (Invitrogen) and Plus Reagent (Invitrogen). The cells were harvested, sonicated and then amplicons concentrated as described. The pellet was resuspended in 500 µl of phosphate buffered saline (PBS). The titre of the HSV-1 amplicon preparation was determined by transducing the glioma cell line G16-9 (9).
HSV-1 amplicon transduction
Transduction of HUES-2 (passage 40), HUES-10 (passage 29) and HT1080 cells with HSV-1 HAC amplicons was carried out as described (9,22). The HUES-2 and HUES-10 on Matrigel-coated plates, and the HT1080 cells were seeded at 2.5 × 105 cells per 24 well plates the day before transduction. On the day of transduction, HSV-1 HAC amplicons were inoculated at a MOI of 1, 2 or 5 in 250 μl of media (mTeSR or DMEM).
To improve the transduction efficiency, upon addition of the HSV-1 amplicons, the cells were centrifuged under low gravitational forces. The plates were covered with sterile adhesive films, to avoid aerosol escape during centrifugation, and centrifuged at 750g for 45 min (23). Transient expression was monitored at 24 h post-transduction either under the microscope or by flow cytometry. Three days after HSV-1 HAC transduction, the HUES-2 cells were transferred onto G418 resistant-inactivated MEF or SNL-76/7 cells. Two days later, 50 μg/ml of G418 (Invitrogen) was added as selection. After 7 days, individual hESc clones were observed and cells were removed from selection. The clones were allowed to grow for an extra 7 days, then each clone was isolated and expanded on inactivated MEF. Upon reaching confluency in a 24-well dish, the cells were transferred to feeder-free growing conditions on Matrigel and mTeSR. The HT1080 cells were selected with 350 μg/ml of G418.
EB formation and germ layer differentiation
For germ layer differentiation of each hESc line, ∼3 × 107 cells were used to form 192 uniform-sized EBs of size ∼1.5 × 105 cells/EB. The hESc were seeded in non-adherent 96-well V-Bottom plates (Nunc) in mTeSR and 10 μm ROCK inhibitor Y-27632. After 3 days, the EBs were released into suspension on non-adherent plates for 8–10 days in DMEM, 20% FBS. The EBs were then plated on Matrigel-coated plates and left to adhere and expand for a further 15–20 days.
Neuronal differentiation
Neuronal differentiation was carried out as described previously (13) with minor modifications. Uniform-sized EBs composed of ∼4000 cells were formed using AggrewellTM400 plates (STEMCELL Technologies) as described by the manufacturer. After 2 days, the EBs were released and left in suspension in non-adherent plates for 5 days in DMEM F/12 (Invitrogen), 1% N2 supplement (Invitrogen), containing human plasma fibronectin (5 μg/ml) (Sigma) and recombinant human noggin (200 μg/ml) (RDI/Fitzgerald Industries). The EBs were then transferred onto Matrigel-coated plates under the same medium for further 8 days. Recombinant human bFGF (20 ng/ml) (BD Biosciences) was then added to the medium, and the cells were incubated for further 8–10 days for expansion of neuronal rosettes. Neuronal rosettes were lifted using TrypLE Express (Invitrogen) and plated on Matrigel-coated slides with the addition of ROCK Y-27632 inhibitor.
Teratoma formation assay
For the teratoma formation assay, 1 × 106 Matrigel-grown MM40.2 cells were injected subcutaneously into immunodeficient mice (common gamma-chain–/–, RAG2–/–, C5–/–). The mice were sacrificed between 5 and 7 weeks after injection and the teratoma was dissected and processed for haematoxylin/eosin staining and immunostaining with the following primary antibodies: anti-tubulin beta III isoform (Tuj1) (ectodermal derivatives), anti-alpha-actinin (mesodermal derivatives) and anti-alpha fetoprotein (endodermal derivatives) (all from Millipore).
Total genomic DNA was prepared from the teratoma mass by phenol/chloroform extraction. The HAC abundance was estimated by real-time qPCR analysis of the DNA, using the GFP and GAPDH primers listed in Supplementary Material, Table S2, with the kit SYBR Green Supermix IQ (Quanta Biosciences), on an ICycler (Bio-Rad) machine. The DNA relative amount was measured using the 2−ΔΔCt method.
Fluorescence-activated cell sorting (FACS)
Cells were fixed in 4% formaldehyde in PBS. Samples were then run through a FACS Calibur and a minimum of 10 000 events was captured for each sample. Acquired data were analysed using FlowJo 7.6 software. The percentage of GFP-expressing cells was calculated by setting the gate at 0.5% for the negative control.
FISH and immuno-FISH
Chromosome preparation and FISH analyses were carried out as described previously (9,24,25). For each experiment, up to 40 metaphases were scored, and the number of HAC-containing cells was recorded. For HUES-2 karyotype analysis, metaphase spreads were subjected to FISH with whole chromosomes paint probes for chromosomes 12 and 17 (Aquarius Whole Chromosome Paint Probes, Cytocell), according to the manufacturer instructions. The HUES-10 karyotype was analysed on Affymetrix Cytogenetics Whole-Genome 2.7M Arrays, following the manufacturer instructions.
The binding of CENP C to chromosome metaphase spreads was carried out by immuno-FISH as previously described (9), using an anti-CENP C primary antibody.
Cytological preparations were analysed with an Olympus BX-51 epifluorescence microscope coupled to a JAI CVM4 + CCD camera, with CytoVysion software system (Genetix).
Immunofluorescence staining of fixed cells
Actively growing cells were fixed in 2% formaldehyde in PBS. After permeabilization in PBS, 0.1% Triton X-100 the following antibodies were used: mouse-anti-TRA-1-60 (Abcam); rabbit-anti-Oct4 (Abcam); rabbit-anti-Nanog (Abcam); rabbit-anti-Sox2 (Abcam); mouse-anti-βIII tubulin (R&D Systems), followed by TRITC-conjugated anti-rabbit or anti-mouse antibodies (Molecular Probes, Invitrogen).
The cells were analysed with a wide-field-inverted Nikon TE2000U fluorescence microscope. Images were acquired using the IPLab software, and pseudo-coloured using Adobe Photoshop.
RNA preparation and analysis
Total RNA was extracted from ∼6 × 106 cells using the RNeasy kit (Qiagen), following the manufacturer's instructions. The RNA was treated with DNase I (Qiagen) and reverse transcribed into cDNA, using the RETROScript system (Ambion), with random decamer primers. Gene expression levels were quantified by real-time qPCR analysis of the cDNA, using the GFP and GAPDH primers listed in Supplementary Material, Table S2, as described for the genomic DNA analysis. The RNA relative amount was measured using the 2−ΔΔCt method.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Wellcome Trust (075491/Z/04 to Z.L.M. and E.V.V., 082260/Z/07/Z to S.C., 090233/Z/09/Z to A.J.T. and M.P.B.); M.A.M. is a recipient of the PGSD NSERC and EPA Cephalosporin studentship; S.K. is a recipient of the Clarendon Fund/ Keble Sloane Robinson Scholarship; S.C. is a recipient of a Wellcome Career Re-entry Fellowship; D.Y.L.C. is funded by the Clarendon Fund and Oxford China Scholarship; S.M. is supported by a European Union grant (222878).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Richard Wade-Martins, Sara Ahmadi and Elizabeth Hartfield for invaluable technical advice, use of equipment and discussions. We also thank Cathy Browne for technical assistance with the hESc work. We are grateful to Ben Davies for providing the G418 resistant MEF, Allan Bradley for the SNL-76/7 feeder line and William Earnshaw for the kind gift of anti-CENP C antibody. Confocal images of teratoma sections were taken at the Confocal facility, UCL ICH.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Strulovici Y., Leopold P.L., O'Connor T.P., Pergolizzi R.G., Crystal R.G. Human embryonic stem cells and gene therapy. Mol. Ther. 2007;15:850–866. doi: 10.1038/mt.sj.6300125. [DOI] [PubMed] [Google Scholar]
- 3.Mátrai J., Chuah M.K., VandenDriessche T. Recent advances in lentiviral vector development and applications. Mol. Ther. 2010;18:477–490. doi: 10.1038/mt.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giudice A., Trounson A. Genetic modification of human embryonic stem cells for derivation of target cells. Cell Stem Cell. 2008;2:422–433. doi: 10.1016/j.stem.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Gray S.J., Samulski R.J. Optimizing gene delivery vectors for the treatment of heart disease. Expert Opin. Biol. Ther. 2008;8:911–922. doi: 10.1517/14712598.8.7.911. [DOI] [PubMed] [Google Scholar]
- 6.Baum C. Insertional mutagenesis in gene therapy and stem cell biology. Curr. Opin. Hematol. 2007;14:337–342. doi: 10.1097/MOH.0b013e3281900f01. [DOI] [PubMed] [Google Scholar]
- 7.Wu Z., Yang H., Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mejía J.E., Willmott A., Levy E., Earnshaw W.C., Larin Z. Functional complementation of a genetic deficiency with human artificial chromosomes. Am. J. Hum. Genet. 2001;69:315–326. doi: 10.1086/321977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moralli D., Simpson K.M., Wade-Martins R., Monaco Z.L. A novel human artificial chromosome gene expression system using herpes simplex virus type 1 vectors. EMBO Rep. 2006;7:911–918. doi: 10.1038/sj.embor.7400768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowan C.A., Klimanskaya I., McMahon J., Atienza J., Witmyer J., Zucker J.P., Wang S., Morton C.C., McMahon A.P., Powers D., et al. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 2004;350:1353–1366. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 11.Mejía J.E., Alazami A., Willmott A., Marschall P., Levy E., Earnshaw W.C., Larin Z. Efficiency of de novo centromere formation in human artificial chromosomes. Genomics. 2002;79:297–304. doi: 10.1006/geno.2002.6704. [DOI] [PubMed] [Google Scholar]
- 12.Rivero-Müller A., Lajić S., Huhtaniemi I. Assisted large fragment insertion by Red/ET-recombination (ALFIRE)—an alternative and enhanced method for large fragment recombineering. Nucleic Acids Res. 2007;35:e78. doi: 10.1093/nar/gkm250. doi:10.1093/nar/gkm250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iacovitti L., Donaldson A.E., Marshall C.E., Suon S., Yang M. A protocol for the differentiation of human embryonic stem cells into dopaminergic neurons using only chemically defined human additives: studies in vitro and in vivo. Brain Res. 2007;1127:19–25. doi: 10.1016/j.brainres.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano M., Cardinale S., Noskov V.N., Gassmann R., Vagnarelli P., Kandels-Lewis S., Larionov V., Earnshaw W.C., Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spence J.M., Mills W., Mann K., Huxley C., Farr C.J. Increased missegregation and chromosome loss with decreasing chromosome size in vertebrate cells. Chromosoma. 2006;115:60–74. doi: 10.1007/s00412-005-0032-6. [DOI] [PubMed] [Google Scholar]
- 16.Deng W., Xu Y. Genome integrity: linking pluripotency and tumorigenicity. Trends Genet. 2009;10:425–427. doi: 10.1016/j.tig.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazuki Y., Hoshiya H., Takiguchi M., Abe S., Iida Y., Osaki M., Katoh M., Hiratsuka M., Shirayoshi Y., Hiramatsu K., et al. Refined human artificial chromosome vectors for gene therapy and animal transgenesis. Gene Ther. 2010 doi: 10.1038/gt.2010.147. First published on 18 November 2010, doi:10.1038/gt.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iida Y., Kim J.H., Kazuki Y., Hoshiya H., Takiguchi M., Hayashi M., Erliandri I., Lee H.S., Samoshkin A., Masumoto H., et al. Human artificial chromosome with a conditional centromere for gene delivery and gene expression. DNA Res. 2010;17:293–301. doi: 10.1093/dnares/dsq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saeki Y., Breakefield X.O., Chiocca E.A. Improved HSV-1 amplicon packaging system using ICP27-deleted, oversized HSV-1 BAC DNA. Methods Mol. Med. 2003;76:51–60. doi: 10.1385/1-59259-304-6:51. [DOI] [PubMed] [Google Scholar]
- 20.Kim U.J., Birren B.W., Slepak T., Mancino V., Boysen C., Kang H.L., Simon M.I., Shizuya H. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson K.R., Cowley S., Martinez F.O., Shaw M., Minger S.L., James W. Homogeneous monocytes and macrophages from human embryonic stem cells following coculture-free differentiation in m-csf and il-3. Exp. Hematol. 2008;36:1167–1175. doi: 10.1016/j.exphem.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wade-Martins R., Smith E.R., Tyminski E., Chiocca E.A., Saeki Y. An infectious transfer and expression system for genomic DNA loci in human and mouse cells. Nat. Biotechnol. 2001;19:1067–1070. doi: 10.1038/nbt1101-1067. [DOI] [PubMed] [Google Scholar]
- 23.El-Sherbini Y.M., Stevenson M.M., Seymour L.W., Wade-Martins R. Quantitative characterization of cell transduction by HSV-1 amplicons using flow cytometry and real-time PCR. J. Virol. Methods. 2009;159:160–166. doi: 10.1016/j.jviromet.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 24.Moralli D., Monaco Z.L. Simultaneous detection of FISH signals and bromo-deoxyuridine incorporation in fixed tissue cultured cells. PLoS ONE. 2009;4:e4483.. doi: 10.1371/journal.pone.0004483. doi:10.1371/journal.pone.0004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moralli D., Yusuf M., Mandegar M.A., Khoja S., Monaco Z.L., Volpi E.V. An improved technique for chromosomal analysis of human ES and iPS cells. Stem Cell Rev. 2010 doi: 10.1007/s12015-010-9224-4. First published on 29 December 2010. doi:10.1007/s12015-010-9224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.