Abstract
A spectrum of complex oligogenic disorders called the ciliopathies have been connected to dysfunction of cilia. Among the ciliopathies are Nephronophthisis (NPHP), characterized by cystic kidney disease and retinal degeneration, and Meckel–Gruber syndrome (MKS), a gestational lethal condition with skeletal abnormalities, cystic kidneys and CNS malformation. Mutations in multiple genes have been identified in NPHP and MKS patients, and an unexpected finding has been that mutations within the same gene can cause either disorder. Further, there is minimal genotype–phenotype correlation and despite recessive inheritance, numerous patients were identified as having a single heterozygous mutation. This has made it difficult to determine the significance of these mutations on disease pathogenesis and led to the hypothesis that clinical presentation in an individual will be determined by genetic interactions between mutations in multiple cilia-related genes. Here we utilize Caenorhabditis elegans and cilia-associated behavioral and morphologic assays to evaluate the pathogenic potential of eight previously reported human NPHP4 missense mutations. We assess the impact of these mutations on C. elegans NPHP-4 function, localization and evaluate potential interactions with mutations in MKS complex genes, mksr-2 and mksr-1. Six out of eight nphp-4 mutations analyzed alter ciliary function, and three of these modify the severity of the phenotypes caused by disruption of mksr-2 and mksr-1. Collectively, our studies demonstrate the utility of C. elegans as a tool to assess the pathogenicity of mutations in ciliopathy genes and provide insights into the complex genetic interactions contributing to the diversity of phenotypes associated with cilia disorders.
INTRODUCTION
Recent advances in genome sequencing technologies along with the HapMap project have led to a rapid expansion in the identification of genetic changes associated with human diseases (1). However, in many cases it is difficult to assess the significance of these genetic alterations on gene function and their potential contribution to disease initiation or progression. This is particularly relevant in complex polygenic or oligogenic disorders where overall genetic mutational load is thought to influence severity of disease presentation. Among human disorders with polygenic/oligogenic inheritance are diseases collectively referred to as ciliopathies—disorders caused by abnormal function of the cilium, a cellular appendage that is essential for development and postnatal homeostasis in almost all tissues (2). To date, at least 35 different ciliopathy genes have been identified; however, their functions and how mutations in these genes collectively contribute to disease phenotypes remain unknown (2–4).
Nephronophthisis (NPHP) and Meckel–Gruber syndrome (MKS) represent the two ends of a spectrum of clinical diseases caused by disruption of at least three related molecular ciliary modules (5–7), where NPHP is the least and MKS the most severe form of the disease (2). Multiple genes involved in both of these disorders have been identified and in several cases they share common loci; these include NPHP3/MKS7, NPHP6/MKS4, NPHP8/MKS5 and NPHP11/MKS3 (4,8–12). Nearly all NPHP/MKS genes encode proteins that localize to the base of the cilium (basal body/transition zone (TZ) region) or to the cilium axoneme itself (6,9,13). Most of these proteins associate together in one or more macromolecular complexes and therefore are likely to participate collectively in particular cellular functions (5,14).
MKS/NPHP patients exhibit a wide spectrum of phenotypes that can include mid-gestation lethality, left–right body asymmetry defects, skeletal abnormalities, cystic kidney and liver disease, retinal degeneration, CNS malformation and cognitive deficits (2,13,15). In the case of NPHP4 patients, phenotypes can be limited to renal dysfunction or may additionally include retinal degeneration, with no apparent genotype–phenotype correlation between the two presentations (16,17). This phenotypic variability may be influenced by additional background mutations, potentially in other known or unknown ciliopathy genes (2,16,18–20). This is further supported by studies in C. elegans revealing that there are significant genetic interactions between NPHP and MKS genes that result in the appearance of new phenotypes that are not observed in any of the single mutants alone (7,21). Similarly, data generated in Zebrafish using Morpholino antisense oligonucleotides to impair expression of combinations of ciliopathy genes have shown synergistic interactions between MKS/NPHP genes and genes implicated in Bardet–Biedl Syndrome (BBS) that result in a more severe phenotype (22,23).
Numerous missense mutations have been identified in the NPHP4 gene in NPHP patients; however, despite being a recessive disorder in a majority of these cases, the genetic variants were found as a single heterozygous mutation (16,24). The pathogenicity of these mutations was suggested based on the observation that they were not present in healthy control cohorts analyzed (16,24). Thus, it remains unresolved in these patients whether the heterozygous missense mutations represent benign polymorphisms or are pathogenic in nature—perturbing NPHP4 function, localization and protein interactions or potentially having an influence on clinical outcome through genetic interactions with a mutation in another ciliopathy gene. Addressing these issues in mammalian cell lines or using mouse models (25) has been hindered since Nphp4 mutants have only retinal and male sterility defects, cells appear to be normally ciliated and there are currently no easily quantifiable assays to evaluate the effects of mutations on gene function.
In previous studies, we characterized the C. elegans homologs of NPHP1 and NPHP4 (NPHP-1 and NPHP-4, respectively) and found that these proteins are expressed in amphid (head) and phasmid (tail) sensory neurons and localize specifically to the TZ at the base of cilia that form at the tips of the dendrites of the sensory neurons. We found that NPHP-4 was required for the proper localization of NPHP-1 at the TZ (26). Similar to that seen in mammalian cells (25,27,28), single nphp-1 or nphp-4 mutant worms have mild ciliary ultrastructural defects. The mutant worms also have abnormal cilia-mediated signaling activity as demonstrated by blunted response to chemo-attractants and a behavioral defect in which mutants spend significantly less time foraging on a bacterial lawn compared with wild-type controls (7,26). A similar but more severe foraging phenotype was observed previously in C. elegans with mutations in genes required for assembly of the cilium, such as intraflagellar transport (IFT) genes (29–31). Finally, strong synergistic effects were found between mutations in either nphp-1 or nphp-4 and mutations in mks-associated genes (mks-1, mksr-1, mksr-2, mks-3, mks-5 or mks-6) such that double nphp;mks mutants exhibited severe synthetic foraging and ciliogenesis defects (7,14). As a consequence of cilia structural defects, nphp;mks double mutants are not able to absorb lipophilic dye into the sensory neurons (Dyf phenotype) (7). Thus, in contrast to the relatively limited assays currently available in mammalian Nphp mutants, nphp mutations in C. elegans cause easily quantifiable phenotypes that provide an opportunity to evaluate the consequence of the missense mutations on gene function.
To understand how mutational load may affect clinical presentation, it is important to establish approaches to assess the consequence of genetic variants identified in human patients on the activity of the corresponding protein and how genetic variants may interact with mutations in other ciliopathy genes. In this manuscript, we utilize the nematode C. elegans and a series of behavioral and phenotypic assays to evaluate eight previously identified human NPHP4 missense mutations (16,32,33) and characterize them as being functionally perturbing and likely disease causative or benign. Expressing each of the conserved NPHP-4 missense mutations identified in human NPHP patients in nphp-4 null animals revealed that six of the eight are unable to restore normal foraging behavior that is regulated through the cilium and is disrupted in the nphp-4 null mutants. Further, three of the eight mutations could not rescue the cilia morphology defects when expressed in null nphp-4; mksr-2 double mutants and one variant was found to disrupt normal cilia formation in mksr-2 mutants even in the context of a wild-type nphp-4 background. These data demonstrate that some but not all of the variants have severe consequences on NPHP-4 function and are likely to be disease causative, at least with respect to the genetic interactions tested and the phenotypes analyzed. Moreover, these analyses highlight the utility of C. elegans as a model system to screen for deleterious missense alleles and assess their contribution to severity of disease presentation in human patients.
RESULTS
Missense mutations in human NPHP4 occur in conserved domains
To initiate this study, human NPHP4 was aligned with its orthologs from multiple species. Since NPHP4 does not have any known functional domains, the alignment was used to select human NPHP4 mutations that occur in conserved domains in C. elegans. On the basis of this conservation, eight missense mutations were deemed suitable for further analysis. These mutations were spread across the entire length of the NPHP4 protein, including a domain that mediates NPHP4's interaction with NPHP1 in mammalian systems (Supplementary Material, Fig. S1, Fig. 1A). With the exception of human NPHP-4(F991S) (F903S in C. elegans), which was identified as a homozygous change in a human NPHP patient, all the variants selected were identified as single heterozygous substitutions (16). Thus, in the absence of further functional analyses, the significance of these variants and their potential contribution to disease pathogenesis is unclear.
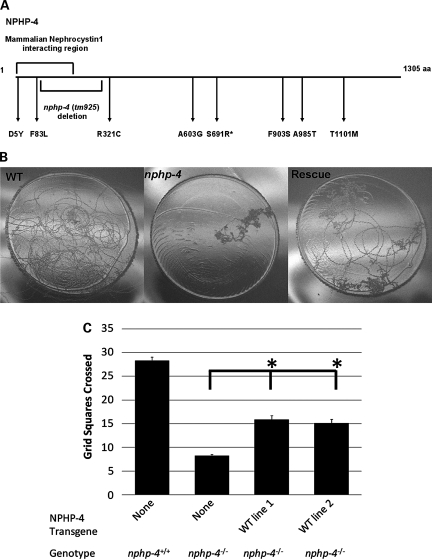
Figure 1.
Location of tm925 deletion and missense mutations in C. elegans NPHP-4 and rescue of the nphp-4(tm925) foraging defect. (A) Graphical representation of NPHP-4 protein showing the location of the tm925 genomic deletion and the amino acid positions corresponding to human NPHP4 missense mutations characterized in this study. Asterisk indicates amino acid which was not conserved in C. elegans NPHP-4 but located in a highly conserved domain. (B) nphp-4(tm925) mutants exhibit a foraging behavior defect which was partially rescued in transgenic lines expressing wild-type NPHP-4. Photographs show single worm tracks on the uniformly sized round bacterial lawns. (C) Quantification of the nphp-4(tm925) foraging defect rescue (n = 50 for each line). The asterisk indicates a statistically significant difference from nphp-4(tm925) P < 0.05. Rescue lines 1 and 2 are not significantly different from each other (P = 0.9). Bars represent SEM.
The potential pathogenic nature of these substitutions was initially assessed using Screening for Non-Acceptable Polymorphisms (SNAP) (http://cubic.bioc.columbia.edu/services/SNAP/) (34) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) which predict the effect of single-nucleotide changes on protein function. On the basis of SNAP, two out of the eight mutations were predicted to be neutral, and six were non-neutral while PolyPhen-2 predicted four out of the eight mutations as being functionally perturbing (Table 1).
Table 1.
SNAP and PolyPhen-2 neutrality prediction
| SNAP |
PolyPhen-2 | ||||
|---|---|---|---|---|---|
| Human NPHP4 mutation | C. elegans NPHP-4 mutation | C. elegans mutation prediction | Predicted accuracy, % | C. elegans mutation prediction | PSIC** |
| D3Y | D5Y | Non-neutral | 78 | Probably damaging | 2.025 |
| F91L | F83L | Non-neutral | 70 | Benign | 1.350 |
| R342C | R321C | Non-neutral | 82 | Possibly damaging | 1.800 |
| A654G | A603G | Neutral | 53 | Benign | 0.675 |
| G754R | S691R* | Non-neutral | 82 | Benign | 1.125 |
| F991S | F903S | Non-neutral | 70 | Possibly damaging | 1.800 |
| A1098T | A985T | Neutral | 85 | Benign | 0.000 |
| T1225M | T1101M | Non-neutral | 70 | Benign | 1.350 |
Prediction of pathogenicity of the mutations in NPHP-4 is based on Screening for Non-Acceptable Polymorphisms (SNAP, http://cubic.bioc.columbia.edu/services/SNAP/) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/). The predicted accuracy reflects SNAP's assignment confidence.
*Amino acid which was not conserved in C. elegans NPHP-4 but located in a highly conserved domain.
**Position-Specific Independent Counts (PSIC) and is an assessment of the likelihood of given amino acid occurring at a particular position to the likelihood of this amino acid occurring at any position (background frequency) (see http://genetics.bwh.harvard.edu/pph2/).
Rescue of the nphp-4(tm925) mutant phenotype
nphp-4(tm925) mutants have a deletion starting in intron 2 that extends through intron 6, and analysis of nphp-4 expression in these mutants indicated that only short transcripts are produced that result in prematurely terminated proteins (largest predicted open reading frame is 107 amino acids) (26). Thus, the tm925 allele is thought to be a null mutation.
Normal function of sensory cilia in C. elegans is necessary for the animal to properly respond to food and other environmental stimuli (29). Indicative of NPHP-4 functioning in cilia signaling, nphp-4(tm925) mutant adult worms have reduced foraging activity in the presence of food (Fig. 1B and C) (7). To confirm that this phenotype is due to the loss of NPHP-4 function, we generated nphp-4(tm925) transgenic worms expressing fluorescent protein-tagged wild-type NPHP-4 (NPHP-4::YFP) driven by the nphp-4 promoter. L4 stage animals were analyzed in two independent foraging experiments. The wild-type NPHP-4 construct significantly rescued the foraging defect from 29% of wild-type in nphp-4(tm925) to 55 and 51% of the mean in the two independent rescue lines, respectively (P < 0.001) (Fig. 1B and C). This partial rescue is likely due to somatic mosaicism in transgenic worms as reported in similar studies previously (35,36). Additionally, in both rescue lines, expression of NPHP-4::YFP restored localization of fluorescent protein-tagged NPHP-1 (NPHP-1::CFP) to the ciliary TZ (Fig. 2A–D and data not shown).
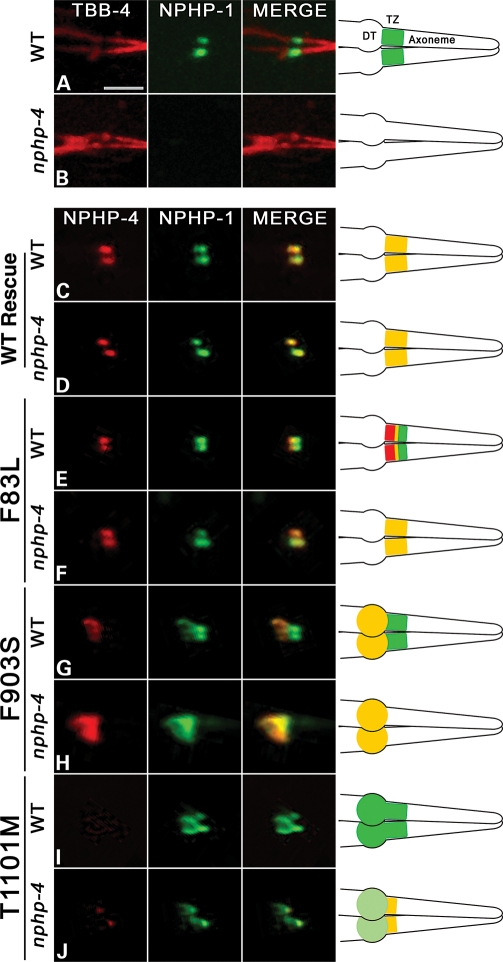
Figure 2.
Consequence of NPHP-4 missense mutations on NPHP-4 localization and the interaction with NPHP-1. (A–J) Confocal images of phasmid (tail) cilia expressing various fluorescent cilia and transition zone (TZ) markers. Depictions of cilia structures and protein localizations are provided on the right. Expression of β tubulin (TBB-4::GFP (pseudocolored red) and NPHP-1::CFP (pseudocolored green) in (A) wild-type and (B) nphp-4(tm925) mutant backgrounds. (A) TBB-4 marks cilia axonemes while NPHP-1 occupies the transition zone (TZ) in wild-type animals. Note the absence of NPHP-1 at the base of the cilium in (B) nphp-4(tm925) mutants. Expression of NPHP-4::YFP (pseudocolored red) and NPHP-1::CFP (pseudocolored green) in (C) wild-type and (D) nphp-4(tm925) mutant backgrounds showing that wild-type NPHP-4 can restore localization of NPHP-1 at the base of the cilium. The effect of the NPHP-4 missense mutations on NPHP-4 and NPHP-1 localization was analyzed in (E,G,I,J) wild-type and in (F,H,K) nphp-4(tm925) mutant backgrounds. In the merged panel in (E) note the shift of NPHP-4(F83L) more proximal toward the dendritic tip (DT) and that this shift does not occur in (F) nphp-4(tm925) mutant background. In (G) wild-type animals, NPHP-4(F903S) is localized to the distal end of the dendrite and fails to enter the TZ region of the cilium where NPHP-1 is localized. In (H) the absence of the endogenous NPHP-4 protein,
Effect of NPHP-4 missense mutations on NPHP-4 and NPHP-1 localization
Using site-directed mutagenesis, the sequence of the wild-type NPHP-4 expression construct was altered to match the corresponding point mutations identified in human patients. Transgenic lines were then generated in the nphp-4(tm925) mutant background by injecting the NPHP-4 construct containing a missense mutation along with a wild-type NPHP-1 expression construct. At least two independent transgenic lines were obtained for each of the eight missense mutations. For comparison of these proteins in the absence and presence of functional endogenous NPHP-4, we also backcrossed the new transgenic animals to remove the nphp-4(tm925) null allele.
We first analyzed the effects that the missense mutations had on both NPHP-4 targeting to the base of the cilium and the ability of the mutant proteins to restore NPHP-1 localization to the TZ. In contrast to the complete absence of NPHP-1 signal at the TZ at the base of the cilium in the nphp-4(tm925) null background, each of the eight variants were able to restore NPHP-1 localization in both amphid (data not shown) and phasmid neurons (Fig. 2 and Supplementary Material, Fig. S2). These data indicate that none of the missense mutations examined abolish NPHP-4 binding with NPHP-1. Five of the eight NPHP-4 mutations (D5Y, R321C, A603G, S691R and A985T) showed normal localization of both the NPHP-4 and NPHP-1 proteins at the TZ (Supplementary Material, Fig. S2). Intriguingly, NPHP-4 with F83L, F903S or T1101M mutations revealed protein localization defects of different varieties (Fig. 2). The most subtle effect on localization was observed for NPHP-4(F83L) (Fig. 2E and F). In the nphp-4(tm925) null background, NPHP-4(F83L) appeared to be in the region of the TZ and co-localized with NPHP-1 (Fig. 2F). However, in the wild-type background, NPHP-4(F83L) localized only to the most proximal end of the TZ where it had a small domain of overlap with NPHP-1, which by comparison localized more distally (Fig. 2E). This more distal localization of NPHP-1 is attributed to the presence of wild-type NPHP-4 in the TZ. The fact that NPHP-1 and NPHP-4(F83L) were found to completely co-localize in the nphp-4(tm925) mutant background indicates that despite the F83L mutation occurring within the NPHP-1 interaction domain, it does not disrupt binding between the two proteins.
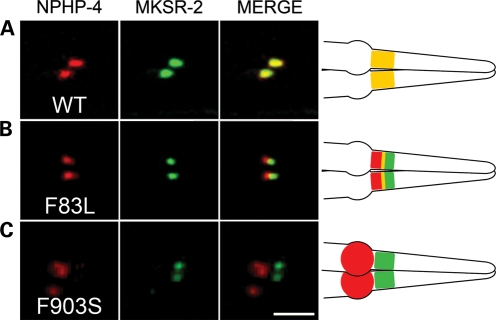
To address this small mislocalization defect further, we generated transgenic worms expressing NPHP-4(F83L) together with another TZ marker, MKSR-2, whose localization does not depend on NPHP-4. There was nearly complete overlap between MKSR-2::CFP and NPHP-4::YFP (Fig. 3A). In contrast, for NPHP-4(F83L) there was a marked reduction in the extent of co-localization between MKSR-2 and NPHP-4(F83L), with the latter being displaced more proximally (Fig. 3B). To quantify the degree of MKSR-2 and NPHP-4(F83L) separation, we calculated the Pearson's co-localization coefficient (PCC) using three-dimensional confocal stacks. The mean PCC for MKSR-2/NPHP-4(F83L) was 0.731 (n = 20 phasmid neurons) compared with 0.906 (n = 20 phasmid neurons) for MKSR-2/NPHP-4. Comparison of PCC revealed that MKSR-2/NPHP-4(F83L) co-localization is statistically different from MKSR-2/NPHP-4 co-localization (P < 0.001).
Figure 3.
NPHP-4(F83L) and NPHP-4(F903S) localization relative to MKSR-2 transition zone marker in wild-type genetic background. (A) Confocal images of phasmid (tail) cilia expressing TZ markers NPHP-4 and MKSR-2 show complete co-localization of these proteins. (B) Expression of NPHP-4(F83L)::YFP (pseudocolored red) and MKSR-2::CFP (pseudocolored green) in wild-type background. A clear separation of these two proteins is evident. (C) Expression of NPHP-4(F903S)::YFP (pseudocolored red) and MKSR-2::CFP (pseudocolored green) in wild-type background. Right panels show a schematic representation of the results. Scale bar is 3 µm. At least 20–25 transgenic worms were examined per line.
In contrast to F83L, the F903S mutation caused a more severe defect in localization of NPHP-4; although NPHP-4(F903S) was properly targeted to the dendritic tip, it completely failed to enter into the TZ at the base of the cilium in both wild-type and nphp-4(tm925) mutant backgrounds (Fig. 2G and H). Interestingly, in the nphp-4(tm925) mutant background, accumulation of NPHP-4(F903S) at the dendritic tip was accompanied by the relocation of NPHP-1 to this region outside of the TZ as well (Fig. 2H). This indicates that NPHP-4(F903S) is still able to interact with NPHP-1 but is unable to incorporate into the TZ. This is also evident in the wild-type background in which NPHP-1 was present in the TZ as normal (due to the presence of wild-type NPHP-4 at the TZ), but a portion of NPHP-1 also co-localized with NPHP-4(F903S) at the dendritic tip (Fig. 2G). To further confirm these data, we analyzed NPHP-4(F903S) localization in wild-type worms expressing fluorescent protein-tagged MKSR-2. Indeed, we observed complete separation of NPHP-4(F903S) from TZ-localized MKSR-2, verifying that the F903S mutation impairs the ability of NPHP-4 to be assembled into the TZ (Fig. 3C).
In the case of the NPHP-4(T1101M) variant, the protein was detected very faintly in the dendritic tip in nphp-4(tm925) mutant animals and occupied only the proximal end of the TZ (Fig. 2J). In wild-type animals, NPHP-4(T1101M) similarly showed very faint localization in the dendritic tip, but fluorescence signal was barely detected at the proximal end of the TZ (Fig. 2I). The reduction of NPHP-4(T1101M) in the TZ in wild-type animals compared with the nphp-4(tm925) mutants may be due to competition from wild-type NPHP-4 protein. In the cell body of the sensory neurons, the level of expression of NPHP-4(T1101M) was similar to that of wild-type NPHP-4 protein (data not shown). These data suggest that the T1101M mutation may inhibit the ability of NPHP-4 to be targeted efficiently to the dendritic tip and to be fully incorporated into the TZ or that this mutation may impair NPHP-4 stability. In addition, the NPHP-4(T1101M) mutation does not prevent NPHP-4 and NPHP-1 interactions since NPHP-1 is both restored to the proximal end of the TZ in nphp-4(tm925) mutant animals expressing NPHP-4(T1101M) and abnormally present along with NPHP-4(T1101M) in the dendritic tip (Fig. 2I and J).
To address the possible degradation issue, we generated constructs for the mouse wild-type NPHP4 and its equivalent of theT1101M (T1154M) variant fused with mCherry in a retrovirus and infected inner medullary collecting duct (IMCD) cells. Both wild-type NPHP4 and mutant NPHP4(T1154) were expressed at similar levels in the cytosol of most transfected cells, arguing against a general increase in NPHP4(T1154) protein instability (Supplementary Material, Fig. S3). In addition, both the control and mutant proteins were present at the base of the cilium and qualitatively appeared at lower levels in the case of the mutant protein. Although the mutant protein was present around the basal body region, more subtle defects in its localization as seen with NPHP-4(T1101M) in C. elegans will require higher resolution analysis.
Functional analyses of the NPHP-4 missense mutations on the nphp-4(tm925) foraging defect
Since most of the mutations analyzed did not overtly alter or had only minor effects on NPHP-4 or NPHP-1 localization, we next wanted to evaluate whether any of the eight mutations altered NPHP-4 function in regulating cilia-initiated sensory-dependent behavior. This was assessed by analyzing whether the NPHP-4 mutations were able to rescue the nphp-4(tm925) mutant foraging defect.
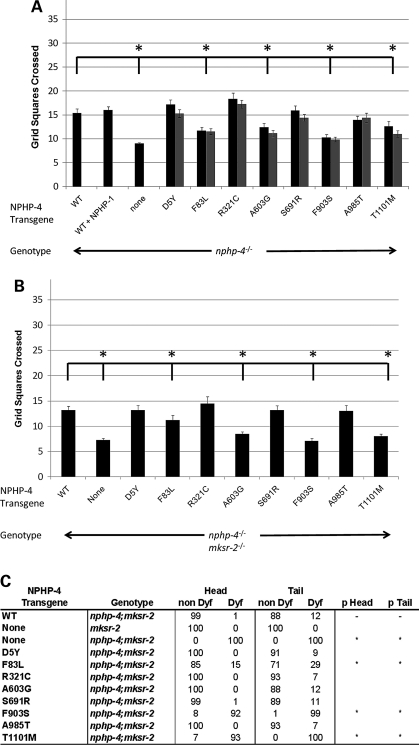
For these studies, we used at least two independent nphp-4(tm925) transgenic lines for each mutation (the same lines that were analyzed for protein localization defects). Compared with rescue-lines expressing wild-type NPHP-4, four out of eight mutations (F83L, A603G, F903S and T1101M) failed to correct the nphp-4(tm925) mutant foraging defect (P < 0.05). Analysis of the other mutations (D5Y, R321C, S691R and A985T) was not statistically different from wild-type (P > 0.05) (Fig. 4A). Importantly, three of the mutations that failed to rescue the foraging defect (F83L, F903S and T1101M) were also identified as the mutations affecting localization of NPHP-4 and/or the restoration of NPHP-1 at the TZ. Intriguingly, the only mutant protein that localized normally and restored NPHP-1 (Supplementary Material, Fig. S2F) yet failed to correct the foraging defect was NPHP-4(A603G) (Fig. 4A). This suggests that A603G impairs foraging behavior through subtle alterations to ciliary signaling activities rather than through gross mislocalization of NPHP complex proteins.
Figure 4.
Functional analysis of the NPHP-4 missense mutations on nphp-4(tm925) foraging phenotype and mksr-2(tm2452);nphp-4(tm925) foraging and dye-filling phenotypes. (A) Each of the NPHP-4 missense mutations were analyzed for the ability to rescue the nphp-4(tm925) foraging defect. Four mutations were determined to be statistically different (asterisks) from the data obtained with nphp-4 transgenic worms expressing wild-type NPHP-4 (P < 0.05). Two independent transgenic lines (black and grey adjacent bars) were analyzed for each mutation (n = 50 for each line) and the data obtained from the same mutation in different lines were not statistically different from each other (P > 0.05). (B) The NPHP-4 missense mutations were evaluated for their ability to rescue the synthetic foraging defective phenotype in double mksr-2(tm2452);nphp-4(tm925) mutants (n = 40 for each line). Four mutations were determined to be statistically different (asterisks) from the data obtained with nphp-4;mksr-2 transgenic worms expressing the wild-type NPHP-4 (P < 0.05). Bars represent SEM. (C) The NPHP-4 missense mutations were evaluated for their ability to rescue the synthetic dye-filling defect (Dyf) phenotype in double mksr-2(tm2452);nphp-4(tm925) mutants (n = 100 for each line). Three mutations were determined to be statistically different (asterisks) from the data obtained with mksr-2(tm2452);nphp-4(tm925) transgenic worms expressing wild-type NPHP-4 (P < 0.001).
Assessing the functional activity of NPHP-4 mutations on the synthetic foraging phenotype of mksr-2(tm2452);nphp-4(tm925) double mutants
Another possibility that we wanted to address is whether these mutations could contribute to disease pathogenesis and severity through interactions with additional mutations in other ciliopathy genes. To begin assessing this possibility, the NPHP-4 mutations were evaluated for their ability to rescue the enhanced foraging defect observed in nphp;mks double mutants. Although nphp-4(tm925) and mksr-2(tm2452) mutants are moderately and mildly defective in foraging, respectively, mksr-2(tm2452);nphp-4(tm925) double mutants have a synergistically more severe phenotype (7). We generated mksr-2(tm2452);nphp-4(tm925) double mutants expressing either wild-type or a missense NPHP-4 variant and assessed the extent of rescue of the foraging defect. Our analysis revealed that the same four mutations (F83L, A603G, F903S and T1101M) that failed to rescue the foraging defect in nphp-4(tm925) mutants also had impaired rescue of the synthetic foraging phenotype in the double mutants (Fig. 4B). This was particularly evident in the A603G, F903S and T1101M mutants, while F83L, which has a relatively more subtle effect on NPHP-4 and NPHP-1 localization, was less impaired (Fig. 4B). The other four mutations (D5Y, R321C, S691R and A985T) were not significantly different from the data obtained with the wild-type NPHP-4 protein (P > 0.05) (Fig. 4B).
Assessing the functional activity of NPHP-4 missense mutations on the synthetic Dyf phenotype in mksr-2(tm2452);nphp-4(tm925) double mutants
Single mutations in nphp-4 or mksr-2 do not noticeably disrupt ciliogenesis and thus mutant worms have cilia that are able to uptake hydrophobic fluorescent dye into the ciliated sensory neurons (7,26). In contrast, double mksr-2(tm2452);np hp-4(tm925) mutants lack properly formed cilia, due to their inability to form stable attachments between TZ microtubules and the surrounding membrane (14), and are completely dye-filling defective (Dyf) in both head and tail neurons. To further assess potential interactions between nphp-4 and mksr-2, mksr-2(tm2452);nphp-4(tm925) double mutants expressing the wild-type or mutant forms of NPHP-4 were analyzed for correction of the Dyf phenotype. In double mutants expressing wild-type NPHP-4, nearly all animals had restored dye filling (Fig. 4C). We found that only the three mutations altering NPHP-4 and NPHP-1 localization (F83L, F903S and T1101M; Fig. 2) failed to rescue the synthetic Dyf phenotype. In the case of F903S and T1101M, greater than 90% of worms were Dyf in both head and tail neurons, statistically no different than the mksr-2(tm2452);nphp-4(tm925) double mutant phenotype. F83L showed a significant and partial rescue compared with non-transgenic double mutants (Fig. 4C). In contrast to the results with the foraging assay, A603G appeared to correct the Dyf phenotype (Fig. 4C).
These data from the functional analysis correlate well with our protein localization data; the F83L, F903S and T1101M mutations are all responsible for NPHP-4 and NPHP-1 mislocalization, and these variants were the least efficient in rescuing the foraging and dye-filling defects. In contrast, D5Y, R321C, S691R and A985T mutations did not affect NPHP-4 or NPHP-1 localization and also largely rescued both foraging and dye-filling defects, comparable to the level of rescue conferred by wild-type NPHP-4. The one outlier in this analysis is NPHP-4(A603G), which localized normally and was able to rescue dye-filling but could not rescue foraging defects.
Analyzing potential dominant negative effects of NPHP-4 missense mutations
Our foraging and localization data suggest that in the homozygous condition, four of the mutations that we analyzed would likely be pathogenic, at least based on the phenotypes and genetic interactions analyzed here. However, with the exception of NPHP-4(F903S), all of these mutations were heterozygous when identified in human patients (16). In that study the possibility was raised that these mutations may have dominant effects interfering with the functions of the wild-type protein, although this is not supported by the genetics of the disease (4,37). To explore whether the presence of any of these missense alleles can alter the activity of wild-type NPHP-4, we performed foraging and dye-filling experiments on the transgenic lines expressing the eight NPHP-4 variants in an nphp-4 wild-type background. We theorized that if a variant is capable of exerting dominant negative effects, we might be able to utilize the artificially higher level of mutant NPHP-4 expression conferred by the transgenic arrays to uncover potential interferences with wild-type protein function. In foraging assays, we observed in most cases no differences between the lines expressing mutant protein versus those expressing wild-type NPHP-4; however, in NPHP-4(D5Y) and NPHP-4(S691R) transgenic lines, we saw a small decrease in foraging activity (P < 0.05) (Supplementary Material, Fig. S4). Similarly, none of the analyzed mutations affected the ability of the transgenic lines to dye-fill as compared with the wild-type transgene (P > 0.05) (Supplementary Material, Fig. S5). These data indicate that the mutant proteins do not have overt strong dominant negative effects.
Assessing potential modifying effects of NPHP-4 missense mutations on the mksr-2(tm2452) and mksr-1(ok2092) phenotypes
An unresolved question is why NPHP4 patients with a single heterozygous mutation nevertheless develop disease. We found that certain mutations in the heterozygous state have negligible dominant negative effects based on our assays, suggesting that alone they are insufficient to cause disease. These data lead to the hypothesis that the NPHP-4 heterozygous mutations could modify or have synergistic effects on a more minor phenotype resulting from mutations in another ciliopathy gene.
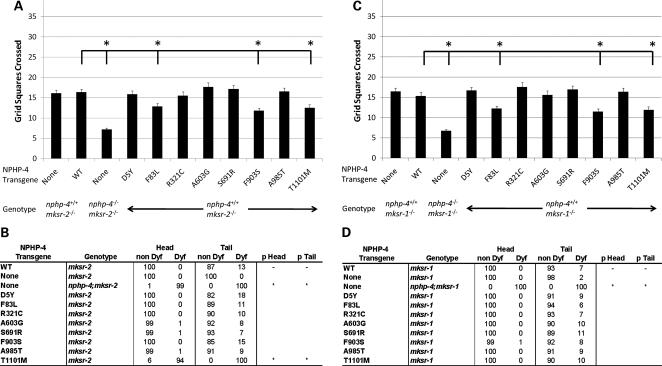
To test this possibility, transgenic lines expressing NPHP-4 proteins with missense mutations were generated in mksr-2(tm2452);nphp-4(wild type) and mksr-1(ok2092);np hp-4(wild type) backgrounds. MKSR-2 and MKSR-1 were chosen for this purpose because they are two anchor proteins required for most of the known MKS proteins to be properly localized (14) and thus mutations in these genes would likely produce more severe synthetic phenotypes with NPHP-4 missense mutations than would mutations in more peripheral MKS genes such as mks-1 or mks-3. We rationalized that this would be similar to the heterozygous condition in human patients if their phenotype indeed resulted from genetic interactions with other ciliopathy mutations. Intriguingly, our analysis of foraging behavior showed that the three mutations affecting NPHP-4 and NPHP-1 localization (F83L, F903S and T1101M) significantly worsened the mksr-2(tm2452) and mksr-1(ok2092) foraging defects (P < 0.001) (Fig. 5A and C). Although our approach does not precisely mimic a true heterozygous condition, these results suggest that the NPHP-4 variants can in fact modify the expressivity of a phenotype caused by a mutation in another ciliopathy-related gene.
Figure 5.
Modifying effect of NPHP-4 missense mutations on mksr-2(tm2452) and mksr-1(ok2092) foraging and dye-filling phenotypes. (A) NPHP-4 missense mutations were evaluated for their ability to modify the foraging phenotype in mksr-2(tm2452) mutants in the context of the endogenous wild-type nphp-4 background (n = 40 for each line). Compared with mksr-2(tm2452) mutants expressing wild-type NPHP-4, three mutations (asterisks) were found to enhance the foraging defect of mksr-2(tm2452) mutants (P < 0.001). Bars represent SEM. (B) NPHP-4 missense mutations were evaluated for their potential to produce synthetic dye-filling defective phenotypes in mksr-2(tm2452);nphp-4(wild-type) (n = 100 for each line). Compared with mksr-2(tm2452) worms expressing wild-type NPHP-4, one mutation (asterisk) was found to produce a significant synthetic dye-filling defect in mksr-2(tm2452) mutant background compared with wild-type NPHP-4 transgene (P < 0.001). (C) NPHP-4 missense mutations were evaluated for their ability to modify the foraging phenotype in mksr-1(ok2092) mutants in the context of the endogenous wild-type nphp-4 background (n = 40 for each line). Compared with mksr-1(ok2092) mutants expressing wild-type NPHP-4, three mutations (asterisks) were found to enhance the foraging defect of mksr-1(ok2092) mutants (P < 0.001). Bars represent SEM. (D) NPHP-4 missense mutations were evaluated for their potential to produce synthetic dye-filling defective phenotypes in mksr-1(ok2092);nphp-4(wild-type) (n = 100 for each line). Compared with mksr-1(ok2092) worms expressing wild-type NPHP-4, none of the mutations was found to produce a significant synthetic dye-filling defect in mksr-1(ok2092) mutant background compared with wild-type NPHP-4 transgene (P > 0.05).
We additionally evaluated in these animals whether the NPHP-4 missense mutations were able to produce a synthetic dye-filling defect in the mksr-2(tm2452) and mksr-1(ok2092) mutant backgrounds in the presence of normal endogenous NPHP-4. In contrast to data obtained with all of the other variants, expression of NPHP-4(T1101M) in an mksr2(tm24 52);nphp-4(wild type) background resulted in a complete Dyf phenotype (Fig. 5B). These data indicate that NPHP-4(T1101M) is able to enhance a ciliogenesis defect in the context of an mksr-2(tm2452) mutation even when there is wild-type NPHP-4 present. Interestingly, none of the analyzed mutations including T1101M produced any synthetic dye-filling defects in mksr-1(ok2092);nphp-4(wild type) background (Fig. 5D). This difference may reflect the hierarchy with which MKSR-2 and MKSR-1 differentially participate in the assembly of the MKS complex (7,14).
DISCUSSION
The number of mutations associated with human ciliopathies is rapidly expanding as a consequence of deep resequencing efforts. However, as demonstrated in human patients with mutations in NPHP4, the significance of these mutations on disease presentation can be greatly limited without additional assessment of the physiological impact. Many of the missense mutations were heterozygous with no evident mutation in the other allele. Thus the disease causality of these mutations and how they may affect NPHP4 protein function remained unknown (16).
We addressed this problem using C. elegans as a model system to characterize the effect of eight mutations found in human patients on NPHP-4 function. In contrast to rodent NPHP models or human NPHP cell lines, C. elegans nphp-1 or nphp-4 mutants exhibit a series of easily quantifiable phenotypes that are amenable to the analysis of NPHP protein function. On the basis of our morphological and functional analysis, we are able to categorize these missense mutations as functionally null, hypomorphic or neutral (Table 2). Overall, six out of eight mutations impaired NPHP-4 function to a variable degree. Our assessments of the mutation severity correspond closer to SNAP's predictions than to PolyPhen-2's predictions (Table 2). A caveat of these assignments is that they are only based on the phenotypic traits and specific mks mutation that we have analyzed. Thus, we may uncover pathogenic effects of the other nphp-4 alleles with different assays or in combination with other mks or nphp alleles not evaluated here.
Table 2.
Classification of NPHP-4 missense mutations
| Human NPHP4 mutation | C. elegans NPHP-4 mutation | C. elegans mutation SNAP prediction | C. elegans mutation PolyPhen-2 prediction | Assignment |
|---|---|---|---|---|
| D3Y | D5Y | Non-neutral | Probably damaging | Weak hypomorphic/neutral |
| F91L | F83L | Non-neutral | Benign | Hypomorphic |
| R342C | R321C | Non-neutral | Possibly damaging | Neutral |
| A654G | A603G | Neutral | Benign | Hypomorphic |
| G754R | S691R* | Non-neutral | Benign | Weak hypomorphic/neutral |
| F991S | F903S | Non-neutral | Possibly damaging | Null |
| A1098T | A985T | Neutral | Benign | Neutral |
| T1225M | T1101M | Non-neutral | Benign | Strong hypomorphic/null |
Classification of NPHP-4 missense mutations based on the collective effects of the mutations on protein localization, dye-filling and foraging behavior activity.
*Amino acid which was not conserved in C. elegans NPHP-4 but located in a highly conserved domain.
Four out of eight mutations (D5Y, R321C, S691R, A985T) were able to rescue nphp-4 mutant foraging defects as well as nphp-4;mksr-2 double mutant synthetic foraging and dye-filling defects. These were each able to fully restore NPHP-1 localization at the TZ. Thus, we predict that in the homozygous state these mutations are functionally neutral. The NPHP-4(F83L) and NPHP-4(A603G) mutations impaired NPHP-4 function as evidenced by reduced ability to rescue the foraging defect in both nphp-4 and nphp-4;mksr-2 mutants. Nevertheless, NPHP-4(A603G) completely restored dye-filling in nphp-4;mksr-2 mutants, unlike NPHP-4(F83L). These data suggest that NPHP-4(A603G) possibly disrupts the function of a signaling pathway controlling worm foraging activity. In contrast, the NPHP-4(F83L) mutation probably alters a structural or scaffolding function of NPHP-4 needed for the protein's role in normal ciliogenesis. On the basis of these phenotypic effects, we conclude that NPHP-4(F83L) and NPHP-4(A603G) represent hypomorphic alleles that can be pathogenic in the absence of wild-type protein or in the presence of an additional ciliopathy mutation (Table 2). Our data also suggest that NPHP-4(F83L) can function as a genetic modifier in the heterozygous state. In support of this possibility, Otto et al. (24) recently identified an additional NPHP patient carrying a single copy of the human F91L allele (C. elegans counterpart F83L), in addition to the original two heterozygous F91L carriers identified by Hoefele et al. (16).
The most severe variants identified here are NPHP-4(F903S) and NPHP-4(T1101M), neither of which was able to rescue foraging or dye-filling defects. In addition, F903S and T1101M caused NPHP-4 and NPHP-1 mislocalization, suggesting that these are functional null mutations. NPHP-4(F903S) corresponds to human F991S, the only allele out of the eight examined here that was found in the homozygous state in a patient that was enrolled in the original positional cloning of NPHP4 (32). On the basis of these lines of evidence, F903S can be classified as a causative NPHP mutation.
In contrast to NPHP-4(F903S), all of the other variants analyzed in this study were found as heterozygous changes in human patients. Thus, we sought to address the question of whether the presence of these mutations in only one NPHP4 copy could be pathogenic. We envisioned two possible genetic mechanisms by which this could occur. One possibility was that these missense mutations have dominant negative effects; however, NPHP pedigree analyses do not support a classical dominant negative mechanism. This possibility was further diminished based on our foraging and dye-filling experiments where over-expression of the NPHP-4 variants in the wild-type genetic background failed to induce overt phenotypes. Only two out of eight variants analyzed (D5Y and S691R) caused minimal reduction in foraging activity. In contrast to our findings with NPHP-4, a dominant negative effect has been reported as a genetic mechanism underlying a BBS mutation based on similar studies using morpholino knockdowns in Zebrafish (38). The possibility still remains that the mutant nphp-4 alleles exert more profound dosage-dependent dominant negative effects in the presence of another nphp or mks mutant alleles.
An alternative possibility is that these alleles have modifying or synergistic effects with another mutation in a ciliopathy gene. Similar mechanisms have been described previously with NPHP, MKS and BBS mutations (22,38–40). The data obtained from our C. elegans analysis here show that expression of NPHP-4(F83L), NPHP-4(F903S) or NPHP-4(T1101M) can enhance mksr-2 and mksr-1 mutant foraging defects. Further, NPHP-4(T1101M) caused a synthetic Dyf phenotype with mksr-2(tm2452), even in the presence of endogenous wild-type NPHP-4 protein. Thus, based on our data, we propose that these mutations are capable of producing a modifying or synergistic effect in worms and that a similar mechanism with another ciliopathy gene could be occurring in human patients.
In addition to functional analysis, we also observed several mutations that disrupt normal targeting or localization of NPHP-4. These include NPHP-4(F83L), NPHP-4(F903S) and NPHP-4(T1101M). In the case of NPHP-4(F83L) and NPHP-4(F903S), the mutant proteins were targeted correctly to the end of the dendrite but were unable to localize correctly within the TZ region. This was particularly evident in the case of NPHP-4(F903S). In contrast, NPHP-4(T1101M) was localized at the TZ but at lower levels than wild-type NPHP-4 in worm neurons. A similar effect was observed with the corresponding mammalian mutation T1154M in IMCD cells. Interestingly, the levels of wild-type and mutant proteins were comparable in the cell cytosol in both systems suggesting defects in efficient targeting of the mutant protein rather than decreased protein stability. These data argue that NPHP-4(F83L), NPHP-4(F903S) and NPHP-4(T1101M) mutations are functionally perturbing as a consequence of their incorrect localization and that they impair NPHP-4 interactions necessary for movement into the TZ domain.
Two of the mutations analyzed in this study (DY5 and F83L) occur within the region known to mediate the interaction between NPHP4 and NPHP1. However, our data show that all of the mutant proteins were able to influence the localization of NPHP-1, indicating that none of the mutations completely ablate the NPHP-1 and NPHP-4 interaction.
Although frequently occurring variants in the NPHP4 gene have been functionally analyzed in a human association study (41), similar approaches to characterize the effect of other known rare variants on NPHP4 function are not feasible because of the small experimental groups' sizes. The analysis conducted here in C. elegans represents a powerful in vivo approach that can be used to assess the pathogenic nature of missense mutations in ciliopathy genes. There are, however, a few caveats of this approach. One of these is that the analysis is relatively restricted to conserved domains and thus will not be applicable to all mutations identified in human patients. In addition, one is not able to truly assess the effect in a heterozygous condition as observed in human patients with mutations in NPHP-4, since gene targeting in C. elegans is limited to transposon-based strategies and thus is not always applicable. However, we found that synergistic or modifying effects could be uncovered by expressing the mutant protein from a transgene in a wild-type background and that through this approach we could explore complex genetic interactions between mutations in two ciliopathy genes. We could additionally characterize the consequence of these mutations on protein localization, interactions and function. Thus, our analysis was able to provide important insights into whether the mutations in NPHP4 identified in human patients are likely to be pathogenic or simply innocuous polymorphisms.
Similar studies as conducted here have been performed in Zebrafish focusing on proteins involved in BBS. As done in our analysis, these studies were able to catalog mutations as being benign or pathogenic and also identified a potential modifying effect for a particular BBS gene mutation on the phenotype resulting from other ciliopathy gene mutations (22). On the basis of data obtained from this analysis in C. elegans and similar studies in Zebrafish, it is clear that these models will serve as important tools for dissecting the complex genetic and physical interactions between genes and proteins involved in human ciliopathies.
MATERIALS AND METHODS
General molecular biology methods
General molecular biology was conducted following standard procedures as described (42). C. elegans genomic DNA, C. elegans cDNA, single worms and cloned worm DNA were utilized for PCR amplifications, direct sequencing and subcloning as described (42). PCR conditions and reagents are available on request. DNA sequencing was performed by the UAB Genomics Core Facility of the Heflin Center for Human Genetics.
DNA and protein sequence analyses
Genome sequence information was obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) or from the Celera Database (http://www.celera.com), Gene sequences were identified using the C. elegans database Wormbase and references therein (http://www.wormbase.org). Sequence alignments were performed using ClustalW (http://www.ebi.ac.uk/clustalw/).
C. elegans strains
Worm Strains were obtained from Caenorhabditis Genetics Center, C. elegans Knock-Out Consortium and the National BioResource Project in Japan. The strains were grown using standard C. elegans growth methods (43) at 20°C unless otherwise stated. The wild-type strain was N2 Bristol. The following mutant strains were used for experiments: FX925 nphp-4(tm925)V, FX2452 mksr-2(tm2452)IV, RB1682 mksr-1(ok2092)X, YH482 nphp-4(tm925)V; mksr-2(tm24 52)IV, YH496 mksr-1(ok2092)X;nphp-4(tm925)V. FX925, FX2452, RB1682 were out-crossed at least three times and genotyped by PCR before analysis. The following transgenic strains were utilized to determine the effects of the conserved patient mutations on protein localization and function: YH330 yhEx201 (tl: nphp-4::YFP) and YH332 yhEx203 (tl :nphp-4::YFP), YH793 yhEx441 (tl: nphp-4(D5Y)::YFP; tl: nphp-1::CFP), (YH1020 yhEx504 (tl: nphp-4(D5Y)::YFP; tl: nphp-1::CFP), YH1008 yhEx492 (tl: nphp-4(F83L)::YFP; tl: nphp-1::CFP), YH1009 yhEx493 (tl: nphp-4(F83L)::YFP; tl: nphp-1::CFP), Yh1010 yhEx494 (tl: nphp-4(R321)C::YFP; tl: nphp-1::CFP), YH1011 yhEx495 (tl: nphp-4(R321C)::YFP; tl: nphp-1::CFP), YH791 yhEx440 (tl: nphp-4(A603G)::YFP; tl: nphp-1::CFP), YH1012 yhEx496 (tl: nphp-4(A603G)::YFP; tl: nphp-1::CFP), YH787 yhEx437 (tl: nphp-4(S691R)::YFP; tl: nphp-1::CFP), Yh788 yhEx438 (tl: nphp-4(S691R)::YFP; tl: nphp-1::CFP), Yh1013 yhEx497 (tl: nphp-4(F903S)::YFP; tl: nphp-1::CFP), YH1014 yhEx498 (tl: nphp-4(F903S)::YFP; tl: nphp-1::CFP), YH1015 yhEx499 (tl: nphp-4(A985T)::YFP; tl: nphp-1::CFP), YH1016 yhEx500 (tl: nphp-4(A985T)::YFP; tl: nphp-1::CFP), YH1017 yhEx501 (tl: nphp-4(T1101M)::YFP; tl: nphp-1::CFP), YH1018 yhEx 502 (tl: nphp-4(T11 01M)::YFP; tl: nphp-1::CFP), YH1019 yhEx503 (tl: nphp-4::YFP; tl: nphp-1::CFP) in FX925. YH1021 yhEx441 (tl: nphp-4(D5Y)::YFP; tl: nphp-1::CFP), YH1026 yhEx493 (tl: nphp-4(F83L)::YFP; tl: nphp-1::CFP), YH1023 yhEx494 (tl: nphp-4(R321)C::YFP; tl: nphp-1::CFP), YH1024 yhEx496 (tl: nphp-4(A603G)::YFP; tl: nphp-1::CFP), YH1025 yhEx438 (tl: nphp-4(S691R)::YFP; tl: nphp-1::CFP), YH1028 yhEx479 (tl: nphp-4(F903S)::YFP; tl: nphp-1::CFP), YH1029 yhEx499 (tl: nphp-4(A985T)::YFP; tl: nphp-1::CFP), YH1032 yhEx502 (tl: nphp-4(T1101M)::YFP; tl: nphp-1::CFP), YH1033 yhEx503 (tl: nphp-4::YFP; tl: nphp-1::CFP), YH924 yhEx497 (tl: nphp-4(F903S)::YFP; tl: mksr-2::CFP), YH925 yhEx480 (tl: nphp-4(F903S)::YFP; tl: mksr-2::CFP), YH1052 yhEx504 (tl: onc-122::GFP),YH1064 yhEx511 (tl: nphp-4(F83L)::YFP, tl: mksr-2::CFP), YH1067 yhEx514 (tl: nphp-4::YFP, tl: mksr-2::CFP) in N2. YH1034 yhEx441 (tl: nphp-4(D5Y)::YFP; tl: nphp-1::CFP), YH1035 yhEx493 (tl: nphp-4(F83L)::YFP; tl: nphp-1::CFP), YH1036 yhEx494 (tl: nphp-4(R321)C::YFP; tl: nphp-1::CFP), YH1037 yhEx496 (tl: nphp-4(A603G)::YFP; tl: nphp-1::CFP), YH1038 yhEx438 (tl: nphp-4(S691R)::YFP; tl: nphp-1::CFP), YH1039 yhEx479 (tl: nphp-4(F903S)::YFP; tl: nphp-1::CFP), YH1040 yhEx499 (tl: nphp-4(A985T)::YFP; tl: nphp-1::CFP), YH1041 yhEx502 (tl: nphp-4(T1101M)::YFP; tl: nphp-1::CFP), YH1042 yhEx503 (tl: nphp-4::YFP; tl: nphp-1::CFP) in FX2452. YH1072 yhEx441 (tl: nphp-4(D5Y)::YFP; tl: nphp-1::CFP), YH1073 yhEx493 (tl: nphp-4(F83L)::YFP; tl: nphp-1::CFP), YH1074 yhEx494 (tl: nphp-4(R321C)::YFP; tl: nphp-1::CFP), YH1075 yhEx496 (tl: nphp-4(A603G)::YFP; tl: nphp-1::CFP), YH1076 yhEx438 (tl: nphp-4(S691R)::YFP; tl: nphp-1::CFP), YH1048 yhEx479 (tl: nphp-4(F903S)::YFP; tl: nphp-1::CFP), YH1078 yhEx499 (tl: nphp-4(A985T)::YFP; tl: nphp-1::CFP), YH1079 yhEx502 (tl: nphp-4(T1101M)::YFP; tl: nphp-1::CFP), YH1080 yhEx503 (tl: nphp-4::YFP; tl: nphp-1::CFP) in RB1682. YH1043 yhEx441 (tl: nphp-4(D5Y)::YFP; tl: nphp-1::CFP), YH1044 yhEx493 (tl: nphp-4(F83L)::YFP; tl: nphp-1::CFP), YH1045 yhEx494 (tl: nphp-4(R321C)::YFP; tl: nphp-1::CFP), YH1046 yhEx496 (tl: nphp-4(A603G)::YFP; tl: nphp-1::CFP), YH1047 yhEx438 (tl: nphp-4(S691R)::YFP; tl: nphp-1::CFP), YH1048 yhEx479 (tl: nphp-4(F903S)::YFP; tl: nphp-1::CFP), YH1049 yhEx499 (tl: nphp-4(A985T)::YFP; tl: nphp-1::CFP), YH1050 yhEx502 (tl: nphp-4(T1101M)::YFP; tl: nphp-1::CFP), YH1051 yhEx503 (tl: nphp-4::YFP; tl: nphp-1::CFP) in mksr-2(tm2452);nphp-4(tm925). The rescue and site-directed mutagenesis lines were generated using UNC-122::GFP (44) as an injection marker.
Generation of C.elegans constructs and strains
The translational NPHP-1::CFP (pCJ148) and NPHP-4::YFP (pCJ146) vectors have been described previously (26). The pCJ146 vector was modified by site-directed mutagenesis to generate each nphp-4 missense mutation identified in human patients. MKSR-2::CFP (pCJ305) was generated by amplifying via PCR the MKSR-2 promoter and coding region from N2 genomic DNA and inserting it into pCJF6 CFP vector (described in 21). The site-directed mutagenesis was performed using the QuikChange™ Site-Directed Mutagenesis Kit according to manufacturer's instructions. The presence of the desired mutations was confirmed by sequencing. All PCR was performed using AccuTaq-LA DNA Polymerase (Sigma, St Louis, MO) according to manufacturer's instructions. The resulting constructs p278 - NPHP-4(D5Y)::YFP, p279 - NP HP-4(F83L)::YFP, p384 - NPHP-4(R321C)::YFP, p280 - NP HP-4(A603G)::YFP, p219 - NPHP-4(S691R)::YFP, p220 - NPHP-4(F903S)::YFP, p281- NPHP-4(A985T)::YFP and p282 - NPHP-4(T1101M)::YFP were co-injected at 5 ng/μl with the pCJ148 vector into the FX925 strain. p305 - MK SR-2::CFP was co-injected with p279 - NPHP-4(F83L)::YFP, p220 - NPHP-4(F903S)::YFP and p282 - NPHP-4 (T1101M)::YFP at 5 ng/μl into wild-type N2 worms.
The WT N2 transgenic strains were generated by crossing nphp-4(tm925) transgenic hermaphrodite worms with WT males. The mksr-2 transgenic strains were generated by crossing WT transgenic hermaphrodite worms with mksr-2(tm2452) males. The mksr-1 transgenic strains were generated by crossing WT transgenic hermaphrodite worms with mksr-1(ok2092) males. The double mutant nphp-4;mksr-2 transgenic strains were generated by crossing mksr-2(tm2452) transgenic hermaphrodites with nphp-4(tm925) males. For each series of out-crosses the resulting F2 offspring obtained from self-fertilization were screened by PCR to identify strains containing required mutations.
Generation of retroviral constructs
For mouse NPHP4 constructs, the open-reading frame of mouse NPHP4 was PCR amplified from a full-length EST (Open Biosystems; Huntsville, AL, USA Clone ID: 8861390). During PCR amplification, attB sites were added to the 5′ and 3′ ends of the NPHP4 ORF to facilitate cloning using Gateway Technology (Invitrogen) and the stop codon was removed to facilitate creation of a C-terminal fusion protein. NPHP4 Gateway PCR product was recombined into pDONR221 (Invitrogen) for Entry clone formation. NPHP4 C-terminal mCherry fusion Destination clones were created by recombination into pQCXIP-mCherryC Dest, a gift from the Hildebrandt Lab (University of Michigan). NPHP4 T1154M was created by site-directed mutagenesis of the wild-type NPHP4 Entry clone using Quickchange (Stratagene) and recombined into a Destination clone as described earlier. All sequences were verified for accuracy and frame usage by sequencing.
Cell culture and transduction
IMCD3 cells were maintained in DMEM/F12 1:1 media (Hyclone; Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco-Invitrogen) and 1% Penicillin–Streptomycin (Hyclone). A plasmid containing GFP-SSTR3 as a primary cilia marker was a gift from Kirk Mykytn (Ohio State University). IMCD3 cells were transduced with GFP-SSTR3 using the Virapower Lentiviral system (invitrogen). Stable clones were isolated by GFP expression. IMCD3 GFP-SSTR3 cells were then subject to retroviral transduction of mouse NPHP4 constructs. Briefly, HEK293FT cells (Invitrogen) were transfected with 1 μg each of pVSVG, pGagPol and expression plasmid using Fugene 6 (Roche; Madison, WI, USA). After 48 h of virus production, viral supernatants were collected, filtered and allowed to infect IMCD3 GFP-SSTR3 cells for 24 h.
Imaging
Worms were anesthetized using 10 mm Levamisole, immobilized on a 10% agar pad coated with polystyrene beads. For imaging of cells, transduced cells were ceded on coverslips coated with 0.1% gelatin and allowed to adhere for 48 h. Coverslips were then washed with PBS and fixed with 4% paraformaldehyde for 20 min at room temperature followed by washes with phosphate buffer saline supplemented with 2% donkey serum (PBSD). Nuclei were counterstained with Hoescht in PBSD for 10 min at room temperature followed by three washes with PBSD. Coverslips were then mounted with Dabco for imaging. Fluorescence imaging was performed using a Nikon 2000U inverted microscope (Melville, KY, USA) outfitted with a PerkinElmer UltraVIEW ERS 6FE-US spinning disk laser apparatus (Shelton, CT, USA). Confocal images were captured and analyzed with Volocity 5.3 software (Improvision Inc., Waltham, MA, USA) and processed using Photoshop 7.0 (Adobe Systems, Inc., San Jose, CA, USA).
Assays
Foraging behavior analysis was performed as described with slight modifications (29). Briefly, synchronized populations of worms were grown until L4 stage. A single L4 worm was placed in the center of a uniformly sized lawn of bacteria on a 6 cm plate and was allowed to move freely for 18 h. After this time, the worm was removed from the plate by aspiration. The tracks were quantified by counting the number of 3 mm squares on a grid the worm tracks entered. In the case of transgenic lines, only GFP marker-expressing worms were picked for the assays.
Dye-filling assays were performed using DiI (Molecular Probes, Carlsbad, CA, USA) as described previously (45) with modifications. Briefly, synchronized populations of worms were grown until young adult stage on 6 cm plates. Worms were washed off the plates with M9 medium (0.3% KH2PO4, 0.6%Na2HPO4, 0.5% NaCl, 1 mm MgSO4) into 15 ml conical tubes and allowed to settle for 10 min. DiI was added (2 mg/ml in DMF) 1 µl per 200 µl of M9. Worms were allowed to uptake dye for 20 min at RT, washed two times with 15 ml of M9 and plated onto new plates. One hundred worms were picked at random from each strain and analyzed for dye-filling defects in amphid and phasmid neurons using a Lieca MZ16FA fluorescence stereomicroscope. Worms were scored as Dyf if there was no dye detected. In the case of transgenic lines, GFP marker expressing worms were picked for the assays.
Statistical analysis
For foraging assays, comparisons were performed using the Student's t-test. If the data were not normally distributed, the Mann–Whitney Ranked Sum was used instead. These tests were performed using SigmaStat3.1 software package (Systat Software IC). Dye-filling experiments were analyzed using a 2 × 2 χ2 test in Microsoft Office Excel. For all tests, P-values <0.05 were considered significant. Bars on the foraging assays graphs represent standard errors of the mean.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institutes of Health grants (R01 DK065655 to B.K.Y. and P30DK074038 to G.-W.). C.L.W. and J.N.P were supported in part by NIH T32 DK007545-22 (to D.B.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Croyle, V. Roper and J. Stallworth for technical assistance; D. Landis and S. Henke for revising the manuscript and providing useful comments. We would like to thank Travis Ptacek for assistance with figure generation and statistical analyses. We thank the C. elegans Knockout Consortium and National BioResource Project in Japan for the nphp-4(tm925), mksr-1(ok2092) and mksr-2(tm2452) deletion mutants.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Manolio T.A., Collins F.S. The HapMap and genome-wide association studies in diagnosis and therapy. Annu. Rev. Med. 2009;60:443–456. doi: 10.1146/annurev.med.60.061907.093117. doi:10.1146/annurev.med.60.061907.093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tobin J.L., Beales P.L. The nonmotile ciliopathies. Genet. Med. 2009;11:386–402. doi: 10.1097/GIM.0b013e3181a02882. doi:10.1097/GIM.0b013e3181a02882. [DOI] [PubMed] [Google Scholar]
- 3.Hurd T.W., Hildebrandt F. Mechanisms of nephronophthisis and related ciliopathies. Nephron. Exp. Nephrol. 2010;118:e9–e14. doi: 10.1159/000320888. doi:10.1159/000320888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simms R.J., Eley L., Sayer J.A. Nephronophthisis. Eur. J. Hum. Genet. 2009;17:406–416. doi: 10.1038/ejhg.2008.238. doi:10.1038/ejhg.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murga-Zamalloa C., Swaroop A., Khanna H. Multiprotein complexes of retinitis pigmentosa GTPase regulator (RPGR), a ciliary protein mutated in X-linked retinitis pigmentosa (XLRP) Adv. Exp. Med. Biol. 2010;664:105–114. doi: 10.1007/978-1-4419-1399-9_13. doi:10.1007/978-1-4419-1399-9_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiba D., Manning D.K., Koga H., Beier D.R., Yokoyama T. Inv acts as a molecular anchor for Nphp3 and Nek8 in the proximal segment of primary cilia. Cytoskeleton (Hoboken) 2010;67:112–119. doi: 10.1002/cm.20428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams C.L., Winkelbauer M.E., Schafer J.C., Michaud E.J., Yoder B.K. Functional redundancy of the B9 proteins and nephrocystins in Caenorhabditis elegans ciliogenesis. Mol. Biol. Cell. 2008;19:2154–2168. doi: 10.1091/mbc.E07-10-1070. doi:10.1091/mbc.E07-10-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baala L., Audollent S., Martinovic J., Ozilou C., Babron M.C., Sivanandamoorthy S., Saunier S., Salomon R., Gonzales M., Rattenberry E., et al. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am. J. Hum. Genet. 2007;81:170–179. doi: 10.1086/519494. doi:10.1086/519494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker K., Beales P.L. Making sense of cilia in disease: the human ciliopathies. Am. J. Med. Genet. C. Semin. Med. Genet. 2009;151C:281–295. doi: 10.1002/ajmg.c.30231. doi:10.1002/ajmg.c.30231. [DOI] [PubMed] [Google Scholar]
- 10.D'Angelo A., Franco B. The dynamic cilium in human diseases. Pathogenetics. 2009;2:3. doi: 10.1186/1755-8417-2-3. doi:10.1186/1755-8417-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto E.A., Tory K., Attanasio M., Zhou W., Chaki M., Paruchuri Y., Wise E.L., Wolf M.T., Utsch B., Becker C., et al. Hypomorphic mutations in meckelin (MKS3/TMEM67) cause nephronophthisis with liver fibrosis (NPHP11) J. Med. Genet. 2009;46:663–670. doi: 10.1136/jmg.2009.066613. doi:10.1136/jmg.2009.066613. [DOI] [PubMed] [Google Scholar]
- 12.Baala L., Romano S., Khaddour R., Saunier S., Smith U.M., Audollent S., Ozilou C., Faivre L., Laurent N., Foliguet B., et al. The Meckel-Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. Am. J. Hum. Genet. 2007;80:186–194. doi: 10.1086/510499. doi:10.1086/510499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardenas-Rodriguez M., Badano J.L. Ciliary biology: understanding the cellular and genetic basis of human ciliopathies. Am. J. Med. Genet. C. Semin. Med. Genet. 2009;151C:263–280. doi: 10.1002/ajmg.c.30227. doi:10.1002/ajmg.c.30227. [DOI] [PubMed] [Google Scholar]
- 14.Williams C.L., Li C., Kida K., Inglis P.N., Mohan S., Semenec L., Bialas N.J., Stupay R.M., Chen N., Blacque O.E., et al. MKS and NPHP modules cooperate to establish basal body/transition zone-membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 2011;21:1023–1041. doi: 10.1083/jcb.201012116. doi:10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopp K., Heyer C.M., Hommerding C.J., Henke S.A., Sundsbak J.L., Patel S., Patel P., Consugar M.B., Czarnecki P., Gliem T.J., et al. B9D1 is revealed as a novel Meckel syndrome (MKS) gene by targeted exon-enriched next-generation sequencing and deletion analysis. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr151. Epub ahead of print April 14, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoefele J., Sudbrak R., Reinhardt R., Lehrack S., Hennig S., Imm A., Muerb U., Utsch B., Attanasio M., O'Toole J.F., et al. Mutational analysis of the NPHP4 gene in 250 patients with nephronophthisis. Hum. Mutat. 2005;25:411. doi: 10.1002/humu.9326. doi:10.1002/humu.9326. [DOI] [PubMed] [Google Scholar]
- 17.Wolf M.T., Hildebrandt F. Nephronophthisis. Pediatr. Nephrol. 2010;26:181–194. doi: 10.1007/s00467-010-1585-z. doi:10.1007/s00467-010-1585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khanna H., Davis E.E., Murga-Zamalloa C.A., Estrada-Cuzcano A., Lopez I., den Hollander A.I., Zonneveld M.N., Othman M.I., Waseem N., Chakarova C.F., et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat. Genet. 2009;41:739–745. doi: 10.1038/ng.366. doi:10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coppieters F., Casteels I., Meire F., De Jaegere S., Hooghe S., van Regemorter N., Van Esch H., Matuleviciene A., Nunes L., Meersschaut V., et al. Genetic screening of LCA in Belgium: predominance of CEP290 and identification of potential modifier alleles in AHI1 of CEP290-related phenotypes. Hum. Mutat. 2010;31:E1709–E1766. doi: 10.1002/humu.21336. doi:10.1002/humu.21336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoefele J., Wolf M.T., O'Toole J.F., Otto E.A., Schultheiss U., Deschenes G., Attanasio M., Utsch B., Antignac C., Hildebrandt F. Evidence of oligogenic inheritance in nephronophthisis. J. Am. Soc. Nephrol. 2007;18:2789–2795. doi: 10.1681/ASN.2007020243. doi:10.1681/ASN.2007020243. [DOI] [PubMed] [Google Scholar]
- 21.Williams C.L., Masyukova S.V., Yoder B.K. Normal ciliogenesis requires synergy between the cystic kidney disease genes MKS-3 and NPHP-4. J. Am. Soc. Nephrol. 2010;21:782–793. doi: 10.1681/ASN.2009060597. doi:10.1681/ASN.2009060597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leitch C.C., Zaghloul N.A., Davis E.E., Stoetzel C., Diaz-Font A., Rix S., Alfadhel M., Lewis R.A., Eyaid W., Banin E., et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat. Genet. 2008;40:443–448. doi: 10.1038/ng.97. doi:10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 23.Badano J.L., Leitch C.C., Ansley S.J., May-Simera H., Lawson S., Lewis R.A., Beales P.L., Dietz H.C., Fisher S., Katsanis N. Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature. 2006;439:326–330. doi: 10.1038/nature04370. doi:10.1038/nature04370. [DOI] [PubMed] [Google Scholar]
- 24.Otto E.A., Ramaswami G., Janssen S., Chaki M., Allen S.J., Zhou W., Airik R., Hurd T.W., Ghosh A.K., Wolf M.T., et al. Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. J. Med. Genet. 2010;48:105–116. doi: 10.1136/jmg.2010.082552. doi:10.1136/jmg.2010.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Won J., de Evsikova C.M., Smith R.S., Hicks W.L., Edwards M.M., Longo-Guess C., Li T., Naggert J.K., Nishina P.M. NPHP4 is necessary for normal photoreceptor ribbon synapse maintenance and outer segment formation, and for sperm development. Hum. Mol. Genet. 2010;20:482–496. doi: 10.1093/hmg/ddq494. doi:10.1093/hmg/ddq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winkelbauer M.E., Schafer J.C., Haycraft C.J., Swoboda P., Yoder B.K. The C. elegans homologs of nephrocystin-1 and nephrocystin-4 are cilia transition zone proteins involved in chemosensory perception. J. Cell Sci. 2005;118:5575–5587. doi: 10.1242/jcs.02665. doi:10.1242/jcs.02665. [DOI] [PubMed] [Google Scholar]
- 27.Fliegauf M., Horvath J., von Schnakenburg C., Olbrich H., Muller D., Thumfart J., Schermer B., Pazour G.J., Neumann H.P., Zentgraf H., et al. Nephrocystin specifically localizes to the transition zone of renal and respiratory cilia and photoreceptor connecting cilia. J. Am. Soc. Nephrol. 2006;17:2424–2433. doi: 10.1681/ASN.2005121351. doi:10.1681/ASN.2005121351. [DOI] [PubMed] [Google Scholar]
- 28.Jauregui A.R., Nguyen K.C., Hall D.H., Barr M.M. The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J .Cell. Biol. 2008;180:973–988. doi: 10.1083/jcb.200707090. doi:10.1083/jcb.200707090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujiwara M., Sengupta P., McIntire S.L. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/s0896-6273(02)01093-0. doi:10.1016/S0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi T., Gengyo-Ando K., Ishihara T., Katsura I., Mitani S. IFT-81 and IFT-74 are required for intraflagellar transport in C. elegans. Genes Cells. 2007;12:593–602. doi: 10.1111/j.1365-2443.2007.01076.x. doi:10.1111/j.1365-2443.2007.01076.x. [DOI] [PubMed] [Google Scholar]
- 31.Murayama T., Toh Y., Ohshima Y., Koga M. The dyf-3 gene encodes a novel protein required for sensory cilium formation in Caenorhabditis elegans. J. Mol. Biol. 2005;346:677–687. doi: 10.1016/j.jmb.2004.12.005. doi:10.1016/j.jmb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Mollet G., Salomon R., Gribouval O., Silbermann F., Bacq D., Landthaler G., Milford D., Nayir A., Rizzoni G., Antignac C., et al. The gene mutated in juvenile nephronophthisis type 4 encodes a novel protein that interacts with nephrocystin. Nat. Genet. 2002;32:300–305. doi: 10.1038/ng996. doi:10.1038/ng996. [DOI] [PubMed] [Google Scholar]
- 33.Otto E., Hoefele J., Ruf R., Mueller A.M., Hiller K.S., Wolf M.T., Schuermann M.J., Becker A., Birkenhager R., Sudbrak R., et al. A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am. J. Hum. Genet. 2002;71:1161–1167. doi: 10.1086/344395. doi:10.1086/344395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bromberg Y., Yachdav G., Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24:2397–2398. doi: 10.1093/bioinformatics/btn435. doi:10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDowall J.S., Rose A. Alignment of the genetic and physical maps in the dpy-5 bli-4 (I) region of C. elegans by the serial cosmid rescue of lethal mutations. Mol. Gen. Genet. 1997;255:78–95. doi: 10.1007/s004380050476. doi:10.1007/s004380050476. [DOI] [PubMed] [Google Scholar]
- 36.Stinchcomb D.T., Shaw J.E., Carr S.H., Hirsh D. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol. Cell. Biol. 1985;5:3484–3496. doi: 10.1128/mcb.5.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otto E.A., Helou J., Allen S.J., O'Toole J.F., Wise E.L., Ashraf S., Attanasio M., Zhou W., Wolf M.T., Hildebrandt F. Mutation analysis in nephronophthisis using a combined approach of homozygosity mapping, CEL I endonuclease cleavage, and direct sequencing. Hum. Mutat. 2008;29:418–426. doi: 10.1002/humu.20669. doi:10.1002/humu.20669. [DOI] [PubMed] [Google Scholar]
- 38.Zaghloul N.A., Liu Y., Gerdes J.M., Gascue C., Oh E.C., Leitch C.C., Bromberg Y., Binkley J., Leibel R.L., Sidow A., et al. Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet-Biedl syndrome. Proc. Natl Acad. Sci. USA. 2010;107:10602–10607. doi: 10.1073/pnas.1000219107. doi:10.1073/pnas.1000219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tory K., Lacoste T., Burglen L., Moriniere V., Boddaert N., Macher M.A., Llanas B., Nivet H., Bensman A., Niaudet P., et al. High NPHP1 and NPHP6 mutation rate in patients with Joubert syndrome and nephronophthisis: potential epistatic effect of NPHP6 and AHI1 mutations in patients with NPHP1 mutations. J. Am. Soc. Nephrol. 2007;18:1566–1575. doi: 10.1681/ASN.2006101164. doi:10.1681/ASN.2006101164. [DOI] [PubMed] [Google Scholar]
- 40.Badano J.L., Kim J.C., Hoskins B.E., Lewis R.A., Ansley S.J., Cutler D.J., Castellan C., Beales P.L., Leroux M.R., Katsanis N. Heterozygous mutations in BBS1, BBS2 and BBS6 have a potential epistatic effect on Bardet-Biedl patients with two mutations at a second BBS locus. Hum. Mol. Genet. 2003;12:1651–1659. doi: 10.1093/hmg/ddg188. doi:10.1093/hmg/ddg188. [DOI] [PubMed] [Google Scholar]
- 41.Konta T., Takasaki S., Ichikawa K., Emi M., Toriyama S., Satoh H., Ikeda A., Suzuki K., Mashima Y., Shibata Y., et al. The novel and independent association between single-point SNP of NPHP4 gene and renal function in non-diabetic Japanese population: the Takahata study. J. Hum. Genet. 2010;55:791–795. doi: 10.1038/jhg.2010.113. doi:10.1038/jhg.2010.113. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J., Fritsch E.F., Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 43.Brener S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loria P.M., Hodgkin J., Hobert O. A conserved postsynaptic transmembrane protein affecting neuromuscular signaling in Caenorhabditis elegans. J. Neurosci. 2004;24:2191–2201. doi: 10.1523/JNEUROSCI.5462-03.2004. doi:10.1523/JNEUROSCI.5462-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perkins L.A., Hedgecock E.M., Thomson J.N., Culotti J.G. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. doi:10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.