Abstract
Mutations in the heart and muscle isoform of adenine nucleotide translocator 1 (ANT1) are associated with autosomal-dominant progressive external opthalmoplegia (adPEO) clinically characterized by exercise intolerance, ptosis and muscle weakness. The pathogenic mechanisms underlying the mitochondrial myopathy caused by ANT1 mutations remain largely unknown. In yeast, expression of ANT1 carrying mutations corresponding to the human adPEO ones causes a wide range of mitochondrial abnormalities. However, functional studies of ANT1 mutations in mammalian cells are lacking, because they have been hindered by the fact that ANT1 expression leads to apoptotic cell death in commonly utilized replicating cell lines. Here, we successfully express functional ANT1 in differentiated mouse myotubes, which naturally contain high levels of ANT1, without causing cell death. We demonstrate, for the first time in these disease-relevant mammalian cells, that mutant human ANT1 causes dominant mitochondrial defects characterized by decreased ADP–ATP exchange function and abnormal translocator reversal potential. These abnormalities are not due to ANT1 loss of function, because knocking down Ant1 in myotubes causes functional changes different from ANT1 mutants. Under certain physiological conditions, mitochondria consume ATP to maintain membrane potential by reversing the ADP–ATP transport. The modified properties of mutant ANT1 can be responsible for disease pathogenesis in adPEO, because exchange reversal occurring at higher than normal membrane potential can cause excessive energy depletion and nucleotide imbalance in ANT1 mutant muscle cells.
INTRODUCTION
Familial progressive external ophthalmoplegia (PEO) is an adult onset disorder, clinically characterized by external ocular muscle paralysis, ptosis and exercise intolerance. Ataxia, depression, cardiomyopathy and other clinical symptoms may be present in some patients (1). Typically, muscle biopsies show abnormal accumulation of mitochondria and mild reduction of oxidative phosphorylation enzymes. Muscle mitochondrial DNA (mtDNA) often contains multiple deletions, although the pathogenic significance of such mtDNA abnormalities is still unclear.
Genetically, familial PEO is heterogeneous with autosomal-dominant (ad) and recessive forms, linked to various genes, such as the mtDNA polymerase (POLG1) (2) and the mitochondrial helicase TWINKLE (3). A subset of autosomal-dominant progressive external opthalmoplegia (adPEO) is caused by mutations in the gene encoding for the adenine nucleotide translocator 1 (ANT1) (4).
Two missense mutations in ANT1 were initially identified in a cohort of adPEO Italian patients, one resulting in an Ala114Pro amino acid change in the third transmembrane domain and the other in a Val289Met change in the sixth transmembrane domain (4). Since then, other mutations in ANT1 have been associated with adPEO (5–7).
Adenine nucleotide translocator (ANT) is one of the most abundant mitochondrial proteins. It is encoded by nuclear DNA, synthesized in the cytosol, imported into mitochondria by internal targeting sequences and inserted in the inner membrane (IM) (8–10). ANT assembles in multimeric translocating units (11), whose primary function is to transport cytosolic ADP into mitochondria and ATP generated by oxidative phosphorylation from the matrix to the cytosol. In addition, ANT is thought to have an intrinsic uncoupling property (12), to be a regulatory component of the mitochondrial permeability transition pore (13,14) and to be involved in mitochondria-mediated apoptosis (15).
There are four ANT isoforms in humans, with differential tissue expression (16,17). ANT1 is highly expressed in post-mitotic cells and is the most abundant isoform in heart and muscle. ANT2 is expressed mainly in tissues capable of proliferation, such as kidney and liver. ANT3 is expressed ubiquitously at lower levels. ANT4 is expressed mainly in the testes, liver and brain (17).
The molecular mechanisms underlying mitochondrial dysfunction, mtDNA deletions and the pathogenesis of adPEO remain largely unsolved issues. In particular, the effects of adPEO mutations on the ADP–ATP exchange function of ANT1 in mammalian cells are unknown. This problem has been difficult to address because expression of exogenous ANT1 in most mammalian cultured cell lines results in apoptotic death (18), while myoblasts and fibrobalsts from patients express virtually no ANT1 (4).
In this study, we generated a cell culture system by expressing human mutant ANT1 in disease-relevant differentiated skeletal myocytes. We succeeded in expressing exogenous human ANT1 in these cells, without causing apoptosis. We showed that exogenous human ANT1 localizes to the mitochondrial IM of myotubes. We used this system to study the dominant negative effects of mutant ANT1 on ADP–ATP translocation, demonstrating, for the first time in a mammalian system, that adPEO mutant ANT1 causes reduced ADP–ATP exchange rates as a function of mitochondrial membrane potential and a lower threshold for nucleotide exchange reversal. We also showed that these defects are not caused by simple loss-of-function mechanism, because Ant1 knockdown results in different types of biochemical abnormalities than the ANT1 mutations, suggesting a novel mechanism of adPEO pathogenesis based on abnormal regulation of mutant ANT1 exchange activity.
RESULTS
Cloning and expression of human ANT1 in C2C12 mouse myotubes
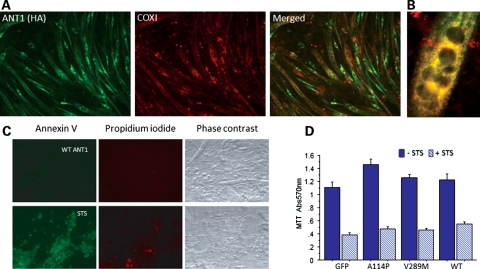
Myotubes, which are multinucleated post-mitotic muscle cells, naturally express high levels of ANT1 (19); thus, we reasoned that transduction of ANT1 in these cells would not cause the apoptotic cell death observed in proliferating cultured cells (18). Therefore, we used an immortalized line of mouse myoblasts, C2C12 cells, which can be efficiently differentiated into myotubes using low-serum culture conditions (20). To express human ANT1 in C2C12 myotubes, we generated adenoviral vectors encoding human ANT1 with an hemagglutinin (HA) epitope tag at the C-terminus to distinguish it from the endogenous mouse Ant1. Human ANT1 was detected in infected myotubes by immunocytochemistry with an HA antibody, showing high transduction efficiency (Fig. 1A). Co-staining with the mtDNA-encoded cytochrome oxidase subunit I protein showed a good degree of co-localization with recombinant ANT1 (Fig. 1B).
Figure 1.
Human ANT1 expression in C2C12 myotubes. (A) WT ANT1 was expressed in C2C12 myotubes (DM4) for 24 h prior to fixation and immunocytochemistry. Cells were immunostained for the HA epitope tag of recombinant ANT1 (in green) and for the COX1 subunit of the mitochondrial respiratory chain (in red). The merged image shows co-localization of these two mitochondrial IM proteins (in yellow). (B) A high-magnification merged image (colors are as in A) of a multinucleated myotube expressing WT ANT1. (C) Expression of WT ANT1 in DM4 myotubes did not cause apoptosis, as determined by negative staining with the apoptotic markers, Annexin V (in green) and propidium iodide (in red). DM4 myotubes treated with 1 μM staurosporine for 24 h caused apoptosis as shown by annexin V and propidium iodide positive staining. (D) Cell viability assessed by MTT assay in DM4 myotubes expressing exogenous WT, or mutant ANT1 (A114P or V289M), or GFP control, for 48 h, and treated with (+STS, 24 h) or without (−STS) 1 μM staurosporine (n = 4 independent experiments).
While we confirmed that wild-type (WT) human ANT1 expression in replicating HEK 293T cells caused cell death (data not shown), ANT1 expression in myotubes did not, as demonstrated by annexin V staining (Fig. 1C) and cell viability assays 48 h post-infection (Fig. 1D). Myotubes transduced with an adenovirus-expressing green fluorescent protein (GFP) were used as controls for these experiments.
Mutant ANT1 cDNA derived from adPEO patients with the Ala114Pro and Val289Met mutations (named A114P and V289M, respectively) were also cloned in adenoviral vectors and expressed in C2C12 myotubes. The expression efficiency and mitochondrial localization of both mutant ANT1 constructs were similar to WT (data not shown). Neither WT nor mutant ANT1 caused cell death or modified the apoptotic cell death induced by staurosporine (Fig. 1D).
Intracellular localization and distribution of recombinant ANT1 in C2C12 myotubes
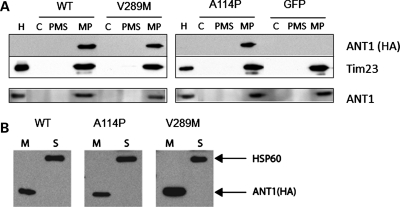
In order to confirm the correct localization of recombinant ANT1, mitochondria were isolated from transduced myotubes and subfractionated into mitoplasts and post-mitoplast supernatants. Immunoblots with the HA antibody confirmed that WT and mutant ANT1 only localized to the mitoplast fractions, which contains the IM proteins but is devoid of outer-membrane proteins (Fig. 2A, top panels). All the exogenous proteins appeared to be imported into mitochondria, since the HA antibody did not detect any recombinant ANT1 in the cytosolic fraction. To compare human ANT1 expression levels with that of the endogenous mouse protein, the blots were stripped and re-probed with an antibody against ANT1. The total levels of ANT1 in the mitoplasts of cells expressing WT or mutant recombinant ANT1 were similar to those of GFP control cells, suggesting that exogenous recombinant ANT1 competes with the endogenous protein for integration in the mitochondrial IM (Fig. 2A, bottom panel). To confirm that recombinant ANT1 was inserted in the IM, mitochondria were subjected to alkaline extraction that removes proteins marginally associated with membranes. By immunoblot with HA antibodies, we found that the recombinant ANT1 was exclusively present in the membrane fraction, while the soluble matrix protein Hsp60 was only detected in the supernatant (Fig. 2B).
Figure 2.
Localization of human ANT1 in the inner mitochondrial membrane. (A) C2C12 myotubes (DM4) expressing ANT1 (WT, A114P and V289M) or GFP were harvested 48 h after transduction to obtain enriched mitochondrial and cytosolic fractions (C). Mitochondria were further subfractionated into mitoplasts (MP), containing IMs and matrix material, and post-mitoplast supernatant (PMS), containing IMS material and outer membranes. Equal amount of proteins from each fraction were resolved by SDS-PAGE, and immunoblotted for exogenous ANT1 with an antibody against HA (top panel). Tim23 was used as a loading control of inner mitochondrial membrane proteins. The membranes were stripped and re-probed with an ANT1 antibody to determine total levels of ANT1. Purified mitochondria from mouse heart (H) were loaded as a positive control. (B) Mitochondrial fractions were treated with alkaline solution to separate membrane-bound (M) and soluble proteins (S). ANT1-HA was exclusively found in the membrane fraction, whereas the soluble matrix protein Hsp60 was completely released in the supernatant.
These results indicate that in myotubes, exogenous ANT1 localizes correctly to the IM and does not increase significantly the total pool of ANT1, likely because it takes the place of the endogenous protein. This suggests that there are different possibilities for ANT multimeric composition in the IM, including only mutant, only endogenous or a combination of mutant and endogenous proteins.
Mitochondrial properties of C2C12 myotubes expressing recombinant ANT1
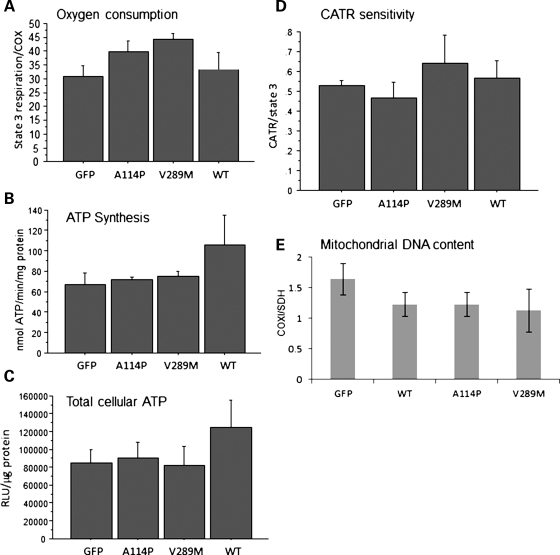
We sought to determine if the introduction of ANT1 mutants in C2C12 mitochondria induced changes in ADP-driven state 3 (phosphorylating) respiration. Mitochondrial oxygen consumption in digitonin-permeabilized myotubes with glutamate and malate as substrates in the presence of ADP did not show statistically significant changes among GFP controls and ANT1-expressing cells (Fig. 3A). In addition, mitochondrial ATP synthesis with the same substrates was not significantly different among ANT1-expressing cells and controls (Fig. 3B). Furthermore, we did not detect changes in the steady-state levels of the total cellular ATP measured in myotubes grown in medium containing galactose as the main carbon source to force cells to utilize oxidative phosphorylation (21) (Fig. 3C). Cell viability measured by MTT assays in myotubes grown in galactose medium for 24 h was unchanged among ANT1-expressing cells and controls (data not shown).
Figure 3.
Mitochondrial properties of human ANT1 expressing myotubes. (A) ADP-driven state 3 respiration in permeabilized C2C12 myotubes expressing GFP, WT, A114P and V289M ANT1 (DM6). State 3 respiration rates were normalized by COX activity to adjust for differences in the mitochondrial content (n = 4). (B) Mitochondrial ATP synthesis in DM6 ANT1 expressing cells with glutamate and malate as substrates (n = 3). (C) Total cellular ATP content (n = 4). (D) The CATR : state 3 respiration ratio is shown, as a measure of the ANT1 sensitivity to CATR (0.5 μM), in permeabilized DM6 myotubes expressing GFP, WT and mutant ANT1. n = 5 for WT, A114P and GFP, n = 3 for V289M. (E) mtDNA content in DM7 myotubes expressing ANT1 for 4 days. Ratio of the mtDNA gene (COXI) to the nuclear DNA-encoded gene (SDH) relative to naive C2C12 cells was determined by real-time PCR (n = 3).
ANT function is inhibited by carboxyatractiloside (CATR), which binds and blocks ANT in its c-state (cytosolic state), with the binding site for nucleotides facing the outer surface of the membrane, and results in inhibition of ADP–ATP translocation (22). Changes in the sensitivity to CATR could indicate modifications of the structural properties of ANT, reflecting on its ability to bind to the inhibitor. First, the concentration of CATR that inhibits state 3 respiration by 50% was assessed in naïve C2C12 myotubes (0.5 µM, data not shown). Then, ADP-driven respiration was measured in ANT1 mutants and GFP controls, before and after the addition of 0.5 µM CATR. The CATR: state 3 respiration ratio was taken as a measure of the ANT1 sensitivity to CATR, which did not differ significantly among ANT1-expressing cells and controls (Fig. 3E). Since all cell lines had similar content of ANT1 (Fig. 2A), this result suggests that the binding of CATR to ANT1 was unmodified by the mutations.
Because ANT1 mutations in humans have been associated with mtDNA instability, characterized by multiple deletions and depletion (23), we studied mtDNA integrity by long-range PCR and copy number by quantitative real-time PCR. Neither WT nor mutant recombinant ANT1 caused detectable mtDNA deletions (data not shown) or depletion (Fig. 3F), in myotubes transduced with ANT1 for 4 days.
These results indicate that under the experimental conditions that we tested, oxidative phosphorylation function was largely preserved in ANT1-mutant myotubes. Furthermore, they suggest that the mutations did not grossly disturb the structure of ANT1. Finally, at least in the time frame of our observation, the mutations did not induce detectable mtDNA abnormalities.
ADP–ATP exchange rate in mutant ANT1-permeabilized myotubes
To better understand the pathogenic consequences of adPEO-linked ANT1 mutations, we studied their effects on the adenine nucleotide exchange function of ANT. We used a recently developed approach to measure ADP–ATP exchange rates, as a function of mitochondrial membrane potential (ΔΨm), in mitochondria in situ in permeabilized myotubes (24).
First, we determined that baseline ΔΨm, measured from Safranine O fluorescence, was not significantly different in myotubes expressing WT and mutant forms of ANT1 (data not shown), confirming that the respiratory chain function was intact in mutant cells.
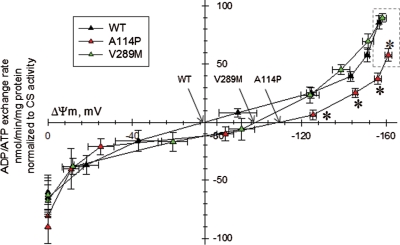
Next, ADP–ATP exchange rate/ΔΨm profiles were generated for WT and mutant ANT1-expressing myotubes. Mitochondria of A114P myotubes, but not V289M, had slower ADP–ATP exchange rates in the −161 to −120 mV ΔΨm range, as compared with WT cells (Fig. 4, asterisks). In addition, A114P myotubes had a 20% smaller ADP-induced depolarization (Fig. 4, ΔΨm values prior to addition of the mitochondrial uncoupler SF6847, in the dashed box), suggesting a reduced efficiency of ADP translocation.
Figure 4.
ADP–ATP exchange rate/ΔΨm profile of C2C12 myotubes expressing WT and mutant ANT1. Plot of ADP–ATP exchange rate mediated by ANT versus ΔΨm in in situ mitochondria of WT, A114P and V289M C2C12 permeabilized cells depolarized to various voltages by increasing amounts of SF 6847, and normalized to specific citrate synthase activity. Each point in the graph represents the average ADP–ATP exchange rate calculated by linear regression of the magnesium green fluorescence, calibrated and converted to ATP appearing in the medium, as a function of calibrated safranine O values. Erev_ANT points are indicated by arrows. Dashed box highlights the ΔΨm values prior to addition of the uncoupler SF6847, which is smaller for A114P myotubes. Each point is the average of five to seven independent experiments. *: Statistically significant by one-way ANOVA followed by Tukey's post-hoc test versus WT, P = 2.0e−3 (ADP), 4.0e−3 (10 sf), 0.043 (20 sf) and 0.045 (30 sf).
The ΔΨm values at which there is no net transfer of ADP–ATP across the IM (reversal potential of the ANT, Erev_ANT) for WT and mutant ANT1 myotubes were extrapolated from the crossing point of the lines connecting the values of ADP–ATP exchange rate (y-axis) with the x-axis (ΔΨm). Both A114P and V289M mutants exhibited right-shifted Erev_ANT towards more polarized values (Fig. 4, arrows; Erev_ANT WT, −75 mV; A114P, −110 mV; V289M, −98 mV). These results indicate that mutant ANT1-expressing cells require more ΔΨm to produce the same amount of ATP. Using the Erev_estimator (http://www.tinyurl.com/Erev-estimator), which allows predicting the matrix adenine nucleotide ratios for a wide range of parameters, we found that at the respective reversal potentials the theoretical matrix ATP/ADP ratio was decreased in ANT1 mutants, as compared with WT (3.6- and 2.2-fold decrease for A114P and V289M, respectively). This suggests that ANT1 mutants are excessively prone to consume ATP in the mitochondrial matrix by reversal of the ATPase function.
ADP–ATP exchange rate in Ant1-silenced myotubes
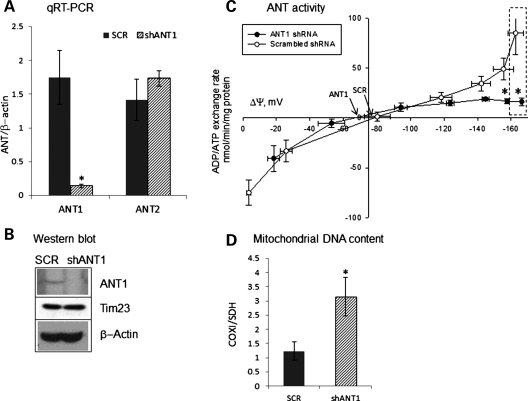
To determine if the effect of ANT1 mutations on the nucleotide translocation function was attributable to loss of functional ANT units, endogenous Ant1 gene was silenced by stable expression of lentiviral shRNA targeted against mouse Ant1, in C2C12 myotubes. In these cells, Ant1 mRNA was decreased by 94% as compared to scrambled shRNA-treated cells, while Ant2 mRNA remained constant (Fig. 5A). Ant1 protein in silenced myoblasts was undetectable by western blot (Fig. 5B), confirming the efficacy of the silencing.
Figure 5.
Biochemical profiles of Ant1-silenced C2C12 myotubes. (A) Ant1 and Ant2 mRNA levels in Ant1-silenced (shAnt1) and scrambled control (SCR) C2C12 myotubes normalized to β-actin mRNA. *: Statistically significant by unpaired, two-tailed Student's t test, P = 0.007. (B) Western blot for Ant1 expression in shAnt1 and SCR C2C12 myotubes. Translocase of the IM (Tim23) and β-actin levels in the lysate were used as loading controls for mitochondrial and cytosolic proteins, respectively. (C) ADP–ATP exchange rate versus ΔΨm in mitochondria of Ant1-silenced C2C12 cells. Dashed box highlights the ΔΨm values prior to addition of the uncoupler SF6847, which is smaller for ANT1-silenced myotubes. *: Statistically significant by unpaired, two-tailed Student's t test, P = 0.02. Each point is the average of four independent experiments. (D) mtDNA content in Ant1-silenced and scrambled control C2C12 myotubes shows 3-fold higher mtDNA in Ant1-silenced cells (n = 5). *: Statistically significant by unpaired, two-tailed Student's t test, P= 0.034.
We generated ADP–ATP exchange rate/ΔΨm profiles in Ant1-silenced myotubes. The ADP–ATP exchange rate in Ant1-silenced cells was reduced compared with scrambled shRNA-treated cells (Fig. 5C). In addition, the silenced cells had a decreased ADP-induced mitochondrial depolarization (22% decrease, dashed box in Fig. 5C), suggesting reduced efficiency of ADP translocation. This was expected, since less Ant1 was available for exchanging adenine nucleotides. However, unlike ANT1-mutant myotubes, the Erev_ANT point for Ant1-silenced cells was minimally affected, as compared with scrambled control cells (Fig. 5C, arrows).
We investigated the mtDNA copy number in Ant1-silenced cells. Ant1 knockdown, but not scrambled shRNA, resulted in increased mtDNA content relative to nuclear DNA (by ∼3-fold, Fig. 5D) compared with untransfected C2C12 cells. These cells did not appear to have increased mitochondrial content, as shown by similar levels of the IM protein Tim23 relative to β-actin (Fig. 5B) and by unchanged levels of the nuclear-encoded mitochondrial matrix enzyme citrate synthase (data not shown).
These results show major differences in the biochemical and genetic effects between Ant1 knockdown and mutant ANT1 expression, indicating that adPEO-linked mutations do not simply cause a loss of functional ANT1 units, but induce functional modifications in the protein.
DISCUSSION
Mutations in ANT1 linked to adPEO were first reported over a decade ago (4). However, the pathogenic biochemical mechanisms underlying the mitochondrial abnormalities in skeletal muscle have remained largely unknown.
So far, the functional consequences of mutant ANT1 associated with adPEO had been investigated in yeast. The A128P mutation in the yeast orthologue AAC2, corresponding to the human A114P pathogenic mutation, caused oxidative phosphorylation deficiency, when expressed in AAC2-null yeast (4). The A128P mutation was shown to form unregulated channels, resulting in swelling and disintegration of mitochondria, when the mutant AAC2 allele was expressed in excess of the WT one (25). In an AAC2-null background, both the A128P and S303M (equivalent to V289M in human ANT1) AAC2 yeast mutants induced ADP–ATP transport impairment, defective oxidative growth and loss of cytochromes (26). However, in hetero-allelic strains, expressing both mutant and WT AAC2, these defects were less severe, with no mitochondrial membrane disintegration (26). The yeast system has also been utilized to mimic the recessive form of myopthy linked to the A123D mutation (A137D in yeast AAC2), which causes a complete loss of translocator activity and a defective oxidative growth that was rescued with antioxidants (17). Furthermore, in these mutants, there was electron transport chain damage and mitochondrial uncoupling (27), suggesting additional pathogenic mechanisms for these recessive mutations, possibly unrelated to ADP–ATP transport.
There are caveats in trying to extrapolate conclusions on human disease from the yeast studies. First, they utilized AAC2 carrying mutations in the amino acids corresponding to the human protein, but yeast AAC2 and human ANT1 are quite dissimilar, having only 54% homology (26). Furthermore, some of the reported abnormalities were only found in yeast, either completely lacking endogenous AAC2 or where the mutant was expressed in excess to the WT (25–27). Therefore, to gain further insight into the effects of mutant ANT1 in mammalian cells our first goal was to generate a viable cell culture system.
The main hurdle in studying ANT1 mutants in mammalian cells is that exogenous ANT1 expression, even WT, results in apoptosis in many replicating cell types (18). Furthermore, in cultured fibroblasts and myoblasts from adPEO patients ANT1 expression is very low, because they express ANT2 predominantly (4). Here, we show that viral expression of ANT1 in differentiated myotubes, which normally express high levels of ANT1, is a viable model to study the pathogenic effects of ANT1 mutations in adPEO. Our findings indicate that mutant ANT1 incorporation in the mitochondrial membrane in the presence of endogenous, normal ANT1, results in a dysfunctional translocator with significant effects on its ADP–ATP exchange rate.
In our myotube model, the decreased exchange rate in mutant A114P ANT1 cells is consistent with a pure defect of ANT function. As baseline ΔΨm values were similar in the ANT1 mutant and WT cells (∼−180 mV), we excluded the possibility of mitochondrial defects other than adenine translocator dysfunction.
Both A114P and V289M ANT1 mutants had a significantly hyperpolarized ΔΨm at Erev_ANT point for ANT1, indicating that the mutant translocator inverted the direction of ADP–ATP exchange (i.e. ADP in–ATP out) at a higher ΔΨm than WT. In other words, in ANT1 mutants, ATP enters the mitochondrial matrix and is hydrolyzed to ADP, despite the fact that ΔΨm is still in the physiological range for ATP synthesis for WT mitochondria. The bioenergetic consequences of this phenomenon are not apparent in cells at rest, respiring with unlimited substrates and oxygen, as shown by normal state 3 respiration and ATP synthesis. However, in the contracting heart or skeletal muscles, ΔΨm fluctuates because of ample changes in intracellular calcium concentration and discontinuous substrate and oxygen supplies (28). In addition to the transient calcium-induced or permeability transition-induced depolarization events, also known as mitochondrial flickers (29), muscle mitochondria undergo frequent transitory depolarization events associated with flashes of superoxide production (30). Under these transient depolarizing conditions, mutant ANT1 mitochondria may be excessively prone to reverse ADP–ATP exchange and consume ATP, possibly leading to spurs of local ATP depletion and nucleotide imbalance.
Pathogenic ANT1 mutations linked with adPEO, including A114P and V289M, lie in the area within helix2–loop–helix3 of the protein (27). Furthermore, both mutants investigated in this study showed hyperpolarized ΔΨm at Erev_ANT. Despite these similarities, ADP–ATP translocation activity was only affected in the A114P mutant. This implies that, although there may be common functional targets affected by different mutations, not all mutations in this region of the protein cause identical defects in ANT. These differences may have implications for the clinical expression of the disease, which will need to be further elucidated.
Ant1 knockout mice show features of mitochondrial myopathy with ragged-red fibers and hypertrophic cardiomyopathy, as well as increased production of reactive oxygen species in muscle, heart and brain (31). In these mice, muscle and heart also contain low levels of mtDNA multiple deletions (32). While this may suggest that Ant1 knockout shares some clinical similarities with the myopathy associated with ANT1 mutations, it is not clear as to whether this holds true at the biochemical level. In our experimental setting, we show that the knockdown of Ant1 in myotubes induces bioenergetic defects different from those caused by expressing mutant ANT1, with decreased translocator activity but normal Erev_ANT. This suggests that the pathogenic mechanisms of adPEO are not simply associated with a loss of ANT1 function, but rather with a modified regulation.
Mutant ANT1 is thought to cause an imbalance of adenine nucleotides in mitochondria that affects dATP synthesis and mtDNA replication/stability, resulting in mtDNA multiple deletions and depletion (33). However, in a study of ANT-defective yeast, it was shown that mtDNA instability may arise without adenine nucleotide imbalance and mitochondrial damage occurs secondarily to mtDNA biogenesis defects (27). In our model system (with expression of mutant ANT1 for up to 4 days), we did not observe mtDNA deletions or depletion. We cannot exclude that mtDNA deletions would eventually occur in this system, but their accumulation may require much longer time. For example, in cultured cells, a large imbalance of mitochondrial dNTPs occurred soon after addition of thymidine to the culture medium; however, deletions in mtDNA were not observed until 8 months later (34). Similarly, imbalance in the pool of dTTP in cultured fibroblasts for 14–64 days did not produce detectable mtDNA alternations (35,36). Another reason why we may have not detected mtDNA deletions could be due to cell culture conditions, where the medium may contain components that mask the effect of mutant ANT1 on nucleotide balance or mtDNA stability. For example, the mounting evidence that RNA/DNA hybrids play a role in mtDNA replication (37) may suggest that, in ANT1-mutant mitochondria, an imbalance in the ratio of dNTP and rNTP could affect mtDNA replication. Nevertheless, our findings suggest a sequence of pathogenic events, where the biochemical impairment in ANT1 function would be the primary cause of mitochondrial dysfunction, in muscle carrying ANT1 mutations. This bioenergetic impairment may be initially transient, and not symptomatically apparent. However, with age, the biochemical impairment in ANT1 would lead to nucleotide imbalance resulting in mtDNA multiple deletions and depletion. The latter may worsen the mitochondrial myopathy and contribute to the pathogenesis of adPEO.
The finding of increased mtDNA content in Ant1 knockdown myotubes, in the absence of increased mitochondrial biogenesis, is intriguing and needs further investigation. However, we could speculate that it may be related to the proposed function of ANT1 in the structure of mtDNA nucleoids (38). Perhaps, mitochondria lacking ANT1, which may be devoid of nucleoid anchoring sites to the IM, could lose the ability to keep mtDNA replication in check. This result further demonstrates the differences between the consequences of having ANT1 mutations and those of a loss of ANT1.
In summary, we have developed a myotube cell culture system for ANT1-associated adPEO with adenoviral transduction of mutant human ANT1. We have identified, for the first time in mammalian cells, the effects of mutant ANT1 on ADP–ATP translocation. We showed that the A114P mutation causes reduced ADP–ATP exchange rates and that both A114P and V289M mutations cause significant effects on the regulation of the translocator by mitochondrial membrane potential. Chronic ADP–ATP exchange defects in ANT1–adPEO may lead to impaired energy metabolism and nucleotide imbalance in muscle and heart mitochondria, resulting in mtDNA abnormalities and mitochondrial myopathy.
MATERIALS AND METHODS
Cell culture
Mouse C2C12 myoblasts (39) were cultured in DMEM containing 10% FBS and antibiotic–antimycotic solution at 37°C in 5% CO2. To induce differentiation of myoblasts into myotubes, culture medium was replaced with differentiation medium (DM) containing DMEM and 2% horse serum when the cells reached above 90% confluence. Fusion of myoblasts into elongated and multinucleated myotubes was evident in ∼4 days in the differentiation medium (DM4). All tissue culture reagents were purchased from Invitrogen (Carlsbad, CA, USA). For growth in galactose, DMEM without glucose was supplemented with 4.5 g/l galactose and 2% dialized horse serum.
ANT1 adenoviral constructs and expression
WT and mutant human ANT1 cDNAs were obtained by RT–PCR of muscle RNA from patients with adPEO and control subjects. Two mutants, A114P in exon 2 and V289M in exon 4, were studied (4). ANT1 cDNA was cloned into the expression vector pCDNA3 (Invitrogen) and an HA-epitope tag was added to the C-terminus of ANT1 for the ease of immunodetection and to discriminate exogenous from endogenous ANT1 expression. Recombinant adenoviral constructs were produced by standard techniques (40). Briefly, WT, A114P or V289M ANT1-HA were subcloned into a shuttle vector (pShuttle, Stratagene, La Jolla, CA) containing a CMV promoter and recombination with pADEasy-1 was achieved following the manufacturer's instructions (Stratagene). All viral constructs were grown to high titer in HEK293 cells (Microbix Biosystems Inc., Ontario, Canada) and purified by cesium chloride density gradient ultracentrifugation. Virus titer was determined by a cytopathic effect assay (41). ANT1 adenoviral constructs were also amplified by ViraQuest Inc. (North Liberty, IA, USA). Adenoviral constructs were added directly to culture media after myotubes were formed (DM4) and used between 200 and 500 MOI. Unless otherwise noted, cells were analyzed for various assays after 48 h of expression by the adenoviral vectors at DM6.
Ant1-silenced C2C12 stable cells
Mouse Ant1 in C2C12 cells were silenced with MISSION shRNA lentiviral transduction particles (Sigma, St Louis, MO). Stable cells were created based on the manufacturer's protocol in C2C12 myoblasts and selected with 1 mg/ml puromycin. Scrambled shRNA lentivirus was used to create the control cell line. Stable cells with shRNA for Ant1 and scrambled differentiated into myotubes in a manner same as that of native C2C12 cells. RNA from DM4 myotubes of Ant1 silenced and controls was extracted with the RNAqueous kit (Ambion, Austin, TX), according to the manufacturer's instructions. Ant1 and Ant2 mRNA content from DM4 myotubes were determined by real-time quantitative PCR with primers for Ant1 and Ant2 (QuantiTect Primer Assays, Slc25a4 and Slc25a5, Qiagen, Valencia, CA) and normalized to β-actin mRNA.
Immunocytochemistry
C2C12 cells were grown on glass coverslips placed in 24-well plates. Forty-eight hours after adenoviral expression, cells were fixed in 4% paraformaldehyde, followed by permeabilization with 0.1% Triton-X, and blocking in phosphate-buffered saline (PBS) containing 1% bovine serum albumin and 10% normal goat serum. Cells were incubated with primary antibodies against HA (1:600, AbCam, Cambridge, MA) and COXI (1:500, Invitrogen) diluted in blocking buffer for 2 h with gentle shaking. Fluorescently labeled secondary antibodies (1:400) were diluted in blocking buffer and applied to cells for 1 h. After three washes in PBS, the coverslips were mounted onto glass slides and dried. All steps were performed at room temperature.
Cell fractionation and mitochondrial subfractionation
For mitochondrial localization of recombinant ANT1, C2C12 cells were grown in 100 mm plates and adenovirus was added at DM3. 48 h later, cells were harvested, resuspended in sucrose isolation buffer, and fractionated by differential centrifugation into cytosolic and enriched mitochondrial fractions, according to established protocols (42). Mitochondria-rich fractions were washed with isolation buffer containing 150 mM KCl to remove proteins peripherally associated with mitochondrial membranes by electrostatic interactions. Enriched mitochondria (50 μg protein) were subfractionated to mitoplasts (IM and matrix) and post-mitoplast fractions (containing outer membrane, intermembrane space contents) by swelling, according to previously published protocols (43). Proteins in the post-mitoplast fraction were precipitated with 12% TCA.
For alkaline extraction of mitochondrial proteins, enriched mitochondria (50 μg protein) were treated with 0.1 M Na2CO3 for 30 min on ice and centrifuged at 100 000 g for 25 min at 4°C to obtain the membrane (pellet) and soluble (supernatant) fractions. Soluble proteins in the supernatant were precipitated with 12% TCA prior to western blot analyses.
Western blot analyses
SDS-polyacrylamide gel electrophoresis (PAGE) was performed in 12% gels and proteins were transferred to PVDF membranes (Bio-Rad, Hercules, CA) using standard techniques. For ANT1 localization experiments, fractions containing 5 μg of proteins were loaded per well. Primary antibodies used were: polyclonal HA (1:6000, Abcam), Hsp60 (1:10 000, Stressgen, Plymouth Meeting, PA), Tim23 (1:2000, BD Bioscience, Sparks, MD), COXI (1:5000, Invitrogen), ANT1 anti-serum (1:1000) and monoclonal ANT1 (1:1000, MitoSciences, Eugene, OR).
Cell viability and apoptosis assays
Cell viability was assessed by the MTT assay using thiazolyl blue tetrazolium bromide (MTT, Sigma) and Cell Death ELISA Plus (Roche Life Sciences, Indianapolis, IN), following the manufacturer's protocols. Apoptotic cell death was determined by Annexin-V-Fluos staining kit (Roche) following the manufacturer's protocol.
MtDNA studies
For mtDNA studies, DNA was extracted from myotubes (DM6 for Ant1-silenced and control cells, DM7 for cells expressing exogenous ANT1 or GFP for 4 days) with a standard phenol/chloroform method. The presence of mtDNA deletions was investigated using the Expand Long Template PCR system (Roche), according to the manufacturer's protocol. Primers sets for mouse mtDNA used for the PCR were previously described (44).
MtDNA content was quantified by real-time PCR using the LightCycler FastStart DNA Master SYBR Green I (Roche). The amount of mtDNA-encoded COXI gene (F-gcccacttcgccatcatattcg, R-ggatatgagattggcttgaaacc) was compared among the different cell lines, with nuclear-encoded SDH (F-tactacagccccaagtct, R-tggacccatcttctatgc) as the reference gene (44). Relative amount of DNA were determined based on the Pfaffl model (45).
Oxygen consumption
WT ANT1, mutant ANT1 or control GFP expressing C2C12 myotubes (DM6) were harvested and permeabilized with digitonin in isolation buffer containing: 20 mM imidazole, 20 mM taurine, 49 mM K-MES, 3 mM KH2PO4, 9.5 mM MgCl2, 2.7 mM CaCl2, 15 mM phosphocreatine, 5 mM ATP and 10 mM EGTA, pH 7.1. Oxygen consumption was measured in a Clark-type electrode oxygraph (Hansatech Inc., UK) in 0.5 ml of respiration buffer containing: 110 mM mannitol, 60 mM KCl, 10 mM KH2PO4, 5 mM MgCl2, 0.5 mM EDTA, 60 mM Tris–HCl, 10 mg/ml fatty acid-free BSA, pH7.4. Glutamate (10 mM) and malate (5 mM) were used as substrates and state 3 respiration was stimulated by 2 mM ADP. In order to inhibit the ADP-induced state 3 respiration in C2C12 cells to 50% of maximum, we used 0.5 μM carboxyatractyloside.
Enzymatic assays
COX activity was measured in cells used for respiration experiments with a spectrophotometric method described previously (46).
Citrate synthase activity was measured as described previously (46), with minor modifications. Briefly, 20 μl aliquots (∼30 μg of protein) were added to a 0.18 ml medium containing 20 mM Hepes pH 7.8, 0.5 mM oxaloacetate and 0.1 mM dithionitrobenzoic acid. The reaction was started after 3 min preincubation time by adding 0.36 mM acetyl-CoA. Changes in the absorbance at 412 nm due to 5-thio-2-nitrobenzoic acid formation were monitored in a Spectramax M5 plate reader (Molecular Devices, Sunnyvale, CA) at 25°C. Activity was calculated as nmol/min/mg protein assuming an extinction coefficient for 5-thio-2-nitrobenzoic acid, of εM = 7075 M−1 cm−1. The light path for 0.2 ml volume in the well of a 96 plate is 0.5 cm. Protein content was measured by the bicinchoninic acid assay using bovine serum albumin protein as standards and calibrating by a 3 parameter power function, f = y0 + a × xb, where y0 is background absorbance in the absence of protein, a and b are constants, and x is the amount of protein in the unknown samples.
For total ATP content measurement DM4 myotubes were transduced with ANT1 or GFP adenovirus. 24 h after transduction, the culture medium was replaced by DM containing galactose instead of glucose and cultured for further 48 h. Cells were washed in PBS and harvested with 4% TCA on ice. ATP content was measured with the ENLITEN ATP Assay System (Promega, Madison, WI) following the manufacturer's protocol.
For mitochondrial ATP synthesis assays DM4 C2C12 myotubes were infected with the ANT1 or GFP virus and harvested at DM6. ATP synthesis was measured by a luciferase/luciferin-based assay as described previously (47).
ADP–ATP exchange rate/membrane potential measurements
DM4 C2C12 myotubes were transduced with WT, A114P or V289M ANT1 adenovirus. At DM6, ADP–ATP exchange rate by magnesium green (Invitrogen) and membrane potential with safranine O (Sigma) were determined in permeabilized myotubes as described previously (24) on a Spectramax M5 plate reader at 37°C. ADP–ATP exchange rates were calculated by linear regression of the magnesium green fluorescence calibrated and converted to ATP appearing in the medium, as a function of calibrated safranine O values (48). ADP–ATP exchange rates were normalized to protein levels and/or to citrate synthase activity and the average rates were plotted as a function of calibrated safranine O values. In Ant1-silenced and scrambled cells, the assays were performed as described above, in DM5 myotubes.
Erev_ANT is defined as the ΔΨm value at which there is no net transfer of ADP–ATP across the IM. By thermodynamic deduction, Erev_ANT derives from the following equation:
where ‘out' signifies outside the matrix, ‘in' inside the matrix, R is the universal gas constant 8.31 J mol−1 K−1, F is the Faraday constant 9.64 × 104 C mol−1 and T is temperature (in Kelvin) (49).
FUNDING
This work was supported by NIH/NINDS F31NS054554 to H.K., NIH Grant R01 GM088999 to G.M. and Országos Tudományos Kutatási Alapprogram (OTKA)-Nemzeti Kutatási és Technológiai Hivatal (NKTH) Grant NF68294, OTKA Grant NNF78905 and NNF2 85658, and an Egeszsegügyi Tudományos Tanács (ETT) grant 55160 to C.C.
ACKNOWLEDGEMENTS
We would like to thank Dr Anatoly Starkov for discussions and advice.
Conflict of Interest statement. The authors have no conflict of interest with the work presented in this manuscript.
REFERENCES
- 1.Suomalainen A., Kaukonen J. Diseases caused by nuclear genes affecting mtDNA stability. Am. J. Med. Genet. 2001;106:53–61. doi: 10.1002/ajmg.1379. doi:10.1002/ajmg.1379. [DOI] [PubMed] [Google Scholar]
- 2.Van Goethem G., Dermaut B., Lofgren A., Martin J.J., Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat. Genet. 2001;28:211–212. doi: 10.1038/90034. doi:10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 3.Spelbrink J.N., Li F.Y., Tiranti V., Nikali K., Yuan Q.P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. doi:10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 4.Kaukonen J., Juselius J.K., Tiranti V., Kyttala A., Zeviani M., Comi G.P., Keranen S., Peltonen L., Suomalainen A. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–785. doi: 10.1126/science.289.5480.782. doi:10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- 5.Deschauer M., Hudson G., Muller T., Taylor R.W., Chinnery P.F., Zierz S. A novel ANT1 gene mutation with probable germline mosaicism in autosomal dominant progressive external ophthalmoplegia. Neuromuscul. Disord. 2005;15:311–315. doi: 10.1016/j.nmd.2004.12.004. doi:10.1016/j.nmd.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Komaki H., Fukazawa T., Houzen H., Yoshida K., Nonaka I., Goto Y. A novel D104G mutation in the adenine nucleotide translocator 1 gene in autosomal dominant progressive external ophthalmoplegia patients with mitochondrial DNA with multiple deletions. Ann. Neurol. 2002;51:645–648. doi: 10.1002/ana.10172. doi:10.1002/ana.10172. [DOI] [PubMed] [Google Scholar]
- 7.Napoli L., Bordoni A., Zeviani M., Hadjigeorgiou G.M., Sciacco M., Tiranti V., Terentiou A., Moggio M., Papadimitriou A., Scarlato G., et al. A novel missense adenine nucleotide translocator-1 gene mutation in a Greek adPEO family. Neurology. 2001;57:2295–2298. doi: 10.1212/wnl.57.12.2295. [DOI] [PubMed] [Google Scholar]
- 8.Endres M., Neupert W., Brunner M. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J. 1999;18:3214–3221. doi: 10.1093/emboj/18.12.3214. doi:10.1093/emboj/18.12.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfanner N., Hoeben P., Tropschug M., Neupert W. The carboxyl-terminal two-thirds of the ADP/ATP carrier polypeptide contains sufficient information to direct translocation into mitochondria. J. Biol. Chem. 1987;262:14851–14854. [PubMed] [Google Scholar]
- 10.Ryan M.T., Muller H., Pfanner N. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 1999;274:20619–20627. doi: 10.1074/jbc.274.29.20619. doi:10.1074/jbc.274.29.20619. [DOI] [PubMed] [Google Scholar]
- 11.Faustin B., Rossignol R., Rocher C., Benard G., Malgat M., Letellier T. Mobilization of adenine nucleotide translocators as molecular bases of the biochemical threshold effect observed in mitochondrial diseases. J. Biol. Chem. 2004;279:20411–20421. doi: 10.1074/jbc.M314259200. doi:10.1074/jbc.M314259200. [DOI] [PubMed] [Google Scholar]
- 12.Brand M.D., Pakay J.L., Ocloo A., Kokoszka J., Wallace D.C., Brookes P.S., Cornwall E.J. The basal proton conductance of mitochondria depends on adenine nucleotide translocase content. Biochem. J. 2005;392:353–362. doi: 10.1042/BJ20050890. doi:10.1042/BJ20050890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoratti M., Szabo I. The mitochondrial permeability transition. Biochim. Biophys. Acta. 1995;1241:139–176. doi: 10.1016/0304-4157(95)00003-a. [DOI] [PubMed] [Google Scholar]
- 14.Kokoszka J.E., Waymire K.G., Levy S.E., Sligh J.E., Cai J., Jones D.P., MacGregor G.R., Wallace D.C. The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature. 2004;427:461–465. doi: 10.1038/nature02229. doi:10.1038/nature02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marzo I., Brenner C., Zamzami N., Jurgensmeier J.M., Susin S.A., Vieira H.L., Prevost M.C., Xie Z., Matsuyama S., Reed J.C., et al. Bax and adenine nucleotide translocator cooperate in the mitochondrial control of apoptosis. Science. 1998;281:2027–2031. doi: 10.1126/science.281.5385.2027. doi:10.1126/science.281.5385.2027. [DOI] [PubMed] [Google Scholar]
- 16.Doerner A., Pauschinger M., Badorff A., Noutsias M., Giessen S., Schulze K., Bilger J., Rauch U., Schultheiss H.P. Tissue-specific transcription pattern of the adenine nucleotide translocase isoforms in humans. FEBS Lett. 1997;414:258–262. doi: 10.1016/s0014-5793(97)01000-4. doi:10.1016/S0014-5793(97)01000-4. [DOI] [PubMed] [Google Scholar]
- 17.Dolce V., Scarcia P., Iacopetta D., Palmieri F. A fourth ADP/ATP carrier isoform in man: identification, bacterial expression, functional characterization and tissue distribution. FEBS Lett. 2005;579:633–637. doi: 10.1016/j.febslet.2004.12.034. doi:10.1016/j.febslet.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 18.Bauer M.K., Schubert A., Rocks O., Grimm S. Adenine nucleotide translocase-1, a component of the permeability transition pore, can dominantly induce apoptosis. J. Cell Biol. 1999;147:1493–1502. doi: 10.1083/jcb.147.7.1493. doi:10.1083/jcb.147.7.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stepien G., Torroni A., Chung A.B., Hodge J.A., Wallace D.C. Differential expression of adenine nucleotide translocator isoforms in mammalian tissues and during muscle cell differentiation. J. Biol. Chem. 1992;267:14592–14597. [PubMed] [Google Scholar]
- 20.Pownall M.E., Gustafsson M.K., Emerson C.P., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. doi:10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- 21.D'Aurelio M., Gajewski C.D., Lin M.T., Mauck W.M., Shao L.Z., Lenaz G., Moraes C.T., Manfredi G. Heterologous mitochondrial DNA recombination in human cells. Hum. Mol. Genet. 2004;13:3171–3179. doi: 10.1093/hmg/ddh326. doi:10.1093/hmg/ddh326. [DOI] [PubMed] [Google Scholar]
- 22.Buchanan B.B., Eiermann W., Riccio P., Aquila H., Klingenberg M. Antibody evidence for different conformational states of ADP, ATP translocator protein isolated from mitochondria. Proc. Natl. Acad. Sci. USA. 1976;73:2280–2284. doi: 10.1073/pnas.73.7.2280. doi:10.1073/pnas.73.7.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Limongelli A., Tiranti V. Inherited Mendelian defects of nuclear-mitochondrial communication affecting the stability of mitochondrial DNA. Mitochondrion. 2002;2:39–46. doi: 10.1016/s1567-7249(02)00043-0. doi:10.1016/S1567-7249(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 24.Kawamata H., Starkov A.A., Manfredi G., Chinopoulos C. A kinetic assay of mitochondrial ADP–ATP exchange rate in permeabilized cells. Anal. Biochem. 407:52–57. doi: 10.1016/j.ab.2010.07.031. doi:10.1016/j.ab.2010.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X.J. Induction of an unregulated channel by mutations in adenine nucleotide translocase suggests an explanation for human ophthalmoplegia. Hum. Mol. Genet. 2002;11:1835–1843. doi: 10.1093/hmg/11.16.1835. doi:10.1093/hmg/11.16.1835. [DOI] [PubMed] [Google Scholar]
- 26.Fontanesi F., Palmieri L., Scarcia P., Lodi T., Donnini C., Limongelli A., Tiranti V., Zeviani M., Ferrero I., Viola A.M. Mutations in AAC2, equivalent to human adPEO-associated ANT1 mutations, lead to defective oxidative phosphorylation in Saccharomyces cerevisiae and affect mitochondrial DNA stability. Hum. Mol. Genet. 2004;13:923–934. doi: 10.1093/hmg/ddh108. doi:10.1093/hmg/ddh108. [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Salinas K., Zuo X., Kucejova B., Chen X.J. Dominant membrane uncoupling by mutant adenine nucleotide translocase in mitochondrial diseases. Hum. Mol. Genet. 2008;17:4036–4044. doi: 10.1093/hmg/ddn306. doi:10.1093/hmg/ddn306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wittenberg B.A., Wittenberg J.B. Transport of oxygen in muscle. Annu. Rev. Physiol. 1989;51:857–878. doi: 10.1146/annurev.ph.51.030189.004233. doi:10.1146/annurev.ph.51.030189.004233. [DOI] [PubMed] [Google Scholar]
- 29.Duchen M.R., Leyssens A., Crompton M. Transient mitochondrial depolarizations reflect focal sarcoplasmic reticular calcium release in single rat cardiomyocytes. J. Cell Biol. 1998;142:975–988. doi: 10.1083/jcb.142.4.975. doi:10.1083/jcb.142.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pouvreau S. Superoxide flashes in mouse skeletal muscle are produced by discrete arrays of active mitochondria operating coherently. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013035. pii, e13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham B.H., Waymire K.G., Cottrell B., Trounce I.A., MacGregor G.R., Wallace D.C. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat. Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. doi:10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 32.Esposito L.A., Melov S., Panov A., Cottrell B.A., Wallace D.C. Mitochondrial disease in mouse results in increased oxidative stress. Proc. Natl. Acad. Sci. USA. 1999;96:4820–4825. doi: 10.1073/pnas.96.9.4820. doi:10.1073/pnas.96.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spinazzola A., Marti R., Nishino I., Andreu A.L., Naini A., Tadesse S., Pela I., Zammarchi E., Donati M.A., Oliver J.A., et al. Altered thymidine metabolism due to defects of thymidine phosphorylase. J. Biol. Chem. 2002;277:4128–4133. doi: 10.1074/jbc.M111028200. doi:10.1074/jbc.M111028200. [DOI] [PubMed] [Google Scholar]
- 34.Song S., Wheeler L.J., Mathews C.K. Deoxyribonucleotide pool imbalance stimulates deletions in HeLa cell mitochondrial DNA. J. Biol. Chem. 2003;278:43893–43896. doi: 10.1074/jbc.C300401200. doi:10.1074/jbc.C300401200. [DOI] [PubMed] [Google Scholar]
- 35.Ferraro P., Pontarin G., Crocco L., Fabris S., Reichard P., Bianchi V. Mitochondrial deoxynucleotide pools in quiescent fibroblasts: a possible model for mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) J. Biol. Chem. 2005;280:24472–24480. doi: 10.1074/jbc.M502869200. doi:10.1074/jbc.M502869200. [DOI] [PubMed] [Google Scholar]
- 36.Pontarin G., Ferraro P., Valentino M.L., Hirano M., Reichard P., Bianchi V. Mitochondrial DNA depletion and thymidine phosphate pool dynamics in a cellular model of mitochondrial neurogastrointestinal encephalomyopathy. J. Biol. Chem. 2006;281:22720–22728. doi: 10.1074/jbc.M604498200. doi:10.1074/jbc.M604498200. [DOI] [PubMed] [Google Scholar]
- 37.Pohjoismaki J.L., Holmes J.B., Wood S.R., Yang M.Y., Yasukawa T., Reyes A., Bailey L.J., Cluett T.J., Goffart S., Willcox S., et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 2010;397:1144–1155. doi: 10.1016/j.jmb.2010.02.029. doi:10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogenhagen D.F., Wang Y., Shen E.L., Kobayashi R. Protein components of mitochondrial DNA nucleoids in higher eukaryotes. Mol. Cell Proteomics. 2003;2:1205–1216. doi: 10.1074/mcp.M300035-MCP200. doi:10.1074/mcp.M300035-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Blau H.M., Chiu C.P., Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 1983;32:1171–1180. doi: 10.1016/0092-8674(83)90300-8. doi:10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- 40.He T.C., Zhou S., da Costa L.T., Yu J., Kinzler K.W., Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. doi:10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Carroll S.J., Hall A.R., Myers C.J., Braithwaite A.W., Dix B.R. Quantifying adenoviral titers by spectrophotometry. Biotechniques. 2000;28:408–410. doi: 10.2144/00283bm03. 412. [DOI] [PubMed] [Google Scholar]
- 42.Pallotti F., Lenaz G. Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol. 2001;65:1–35. doi: 10.1016/s0091-679x(01)65002-7. doi:10.1016/S0091-679X(01)65002-7. [DOI] [PubMed] [Google Scholar]
- 43.Vijayvergiya C., Beal M.F., Buck J., Manfredi G. Mutant superoxide dismutase 1 forms aggregates in the brain mitochondrial matrix of amyotrophic lateral sclerosis mice. J. Neurosci. 2005;25:2463–2470. doi: 10.1523/JNEUROSCI.4385-04.2005. doi:10.1523/JNEUROSCI.4385-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams S.L., Huang J., Edwards Y.J., Ulloa R.H., Dillon L.M., Prolla T.A., Vance J.M., Moraes C.T., Zuchner S. The mtDNA mutation spectrum of the progeroid Polg mutator mouse includes abundant control region multimers. Cell Metab. 2010;12:675–682. doi: 10.1016/j.cmet.2010.11.012. doi:10.1016/j.cmet.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. doi:10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trounce I.A., Kim Y.L., Jun A.S., Wallace D.C. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. doi:10.1016/S0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 47.Manfredi G., Yang L., Gajewski C.D., Mattiazzi M. Measurements of ATP in mammalian cells. Methods. 2002;26:317–326. doi: 10.1016/S1046-2023(02)00037-3. doi:10.1016/S1046-2023(02)00037-3. [DOI] [PubMed] [Google Scholar]
- 48.Chinopoulos C., Vajda S., Csanady L., Mandi M., Mathe K., Adam-Vizi V. A novel kinetic assay of mitochondrial ATP-ADP exchange rate mediated by the ANT. Biophys. J. 2009;96:2490–2504. doi: 10.1016/j.bpj.2008.12.3915. doi:10.1016/j.bpj.2008.12.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chinopoulos C., Gerencser A.A., Mandi M., Mathe K., Torocsik B., Doczi J., Turiak L., Kiss G., Konrad C., Vajda S., et al. Forward operation of adenine nucleotide translocase during F0F1-ATPase reversal: critical role of matrix substrate-level phosphorylation. FASEB J. 2010;24:2405–2416. doi: 10.1096/fj.09-149898. doi:10.1096/fj.09-149898. [DOI] [PMC free article] [PubMed] [Google Scholar]