Abstract
Susceptibility to testicular germ cell tumors (TGCT) has a significant heritable component, and genome-wide association studies (GWASs) have identified association with variants in several genes, including KITLG, SPRY4, BAK1, TERT, DMRT1 and ATF7IP. In our GWAS, we genotyped 349 TGCT cases and 919 controls and replicated top hits in an independent set of 439 cases and 960 controls in an attempt to find novel TGCT susceptibility loci. We identified a second marker (rs7040024) in the doublesex and mab-3-related transcription factor 1 (DMRT1) gene that is independent of the previously described risk allele (rs755383) at this locus. In combined analysis that mutually conditions on both DMRT1 single nucleotide polymorphism markers, TGCT cases had elevated odds of carriage of the rs7040024 major A allele [per-allele odds ratio (OR) = 1.48, 95% confidence interval (CI) 1.23, 1.78; P = 2.52 × 10−5] compared with controls, while the association with rs755383 persisted (per allele OR = 1.26, 95% CI 1.08, 1.47, P = 0.0036). In similar analyses, the association of rs7040024 among men with seminomatous tumors did not differ from that among men with non-seminomatous tumors. In combination with KITLG, the strongest TGCT susceptibility locus found to date, men with TGCT had greatly elevated odds (OR = 14.1, 95% CI 5.12, 38.6; P = 2.98 × 10−7) of being double homozygotes for the risk (major) alleles at DMRT (rs7040024) and KITLG (rs4474514) when compared with men without TGCT. Our findings continue to corroborate that genes influencing male germ cell development and differentiation have emerged as the major players in inherited TGCT susceptibility.
INTRODUCTION
In the USA, testicular germ cell tumors (TGCT [MIM 273300]) are the most common cancers in young men, with a peak incidence among those aged 25–34 years. The incidence of TGCT among white men in the USA has increased substantially over the past 30 years from 4.1 per 100 000 in 1975 to 7.0 per 100 000 in 2007 (1), and similar increases are seen worldwide (2). The disease incidence varies widely across racial groups, with a 5-fold lower rate among black men in the USA (1). Differences in incidence of TGCT also exist across countries and continents, ranging from Denmark (9.2 per 100 000) to Algeria (0.2 per 100 000), which is consistent with racial differences and lower rates in non-white groups (3).
Familial aggregation of TGCT has been documented since the 1930s, and family history is the strongest known risk factor for these malignancies (4,5). Risk of TGCT repeatedly has been shown to be increased among first-degree relatives of affected men, with risk to brothers (estimates range from 5- to 19-fold) being stronger than that to fathers (estimates range from 2- to 4-fold) (6–14). Both mono- and dizygotic twins of affected men have increased risk of TGCT (15,16). Genetic effects have been estimated to account for 25% of TGCT, the third highest heritability among all cancers (17). These familial data along with the racial disparity in disease occurrence provide evidence of a genetic contribution to TGCT susceptibility.
Supporting the genetic contribution to TGCT susceptibility, initial findings from genome-wide association (GWA) studies implicate variation at the KITLG, SPRY4 and BAK1 as associated with TGCT susceptibility (18,19). Prior to these publications, no common susceptibility alleles had been validated. More recently, a follow-up GWA study from the UK also identified and replicated variation in ATF7IP, DMRT1 and TERT as associated with TGCT (20). Herein, we report the results from our follow-up GWA study in the USA, for which we augmented the number of TGCT cases used in the discovery sample as well as TGCT cases and controls used in the replication sample in order to identify additional TGCT susceptibility loci and to validate the new findings.
RESULTS
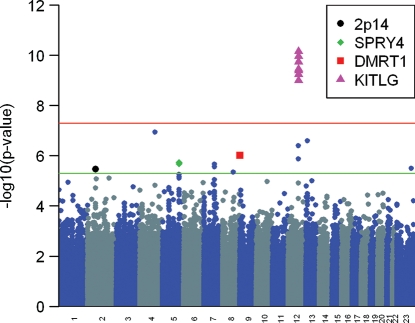
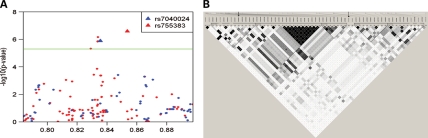
Information on age, risk factors and tumor characteristics for the discovery and replication sample sets are given in Table 1. The calculated genomic control inflation (λ) factor in the discovery set was 0.942, and hence we report unadjusted test statistics (21,22). We again noted our previously reported statistically significant associations with markers at 12q22 in KITLG (P < 5.0 × 10−8), 2p14 and 5q13.3 near SPRY4 (P < 5.0 × 10−6; Fig. 1 and Supplementary Material, Table S1) (18). Six additional markers at five additional autosomal loci also were associated with TGCT (P < 5.0 × 10−6). To screen out hits representing likely false positive associations, we imputed genotypes in the surrounding regions of these six markers using data from 1000 genomes (March 2010 release) (Fig. 2A and Supplementary Material, Fig. S1). After imputation, neither observed nor imputed markers at 8q21.3 and 13q12.3 maintained significance at P < 5 × 10−6 and were excluded from further study, whereas markers at 4q28.2, 7q21.13 and 9p24.3 continued to exceed the P < 5 × 10−6 threshold. The selected markers on chromosomes 4 and 7 that were brought forward into replication included two imputed (rs2279070, rs4834214) that mapped to 4q28.2, as the observed single nucleotide polymorphism (SNP) could not be designed for replication genotyping, and two observed (rs6951213, rs10281060) that mapped within introns 1 and 2 of PFTK1 on 7q21.13. For chromosome 9 markers, the top three imputed markers using 1000 genomes were incompatible with our replication genotyping platform, so we re-imputed markers in this region using data from HapMap (release 22). The top imputed marker (rs755383), which was previously showed to be associated with TGCT by Turnbull et al. (20), was taken into replication along with the top observed marker (rs7040024), both of which exceeded the P < 5 × 10−6 threshold (Supplementary Material, Fig. S2).
Table 1.
Age, family history of TGCT and tumor type in the discovery and replication sample sets
| Discovery | Replication | ||||||
|---|---|---|---|---|---|---|---|
| Status | Case | Control | Case | Control | |||
| Total | n = 349 | n = 919 | n = 439 | n = 960 | |||
| n | % | n | n | % | n | % | |
| Age [median, (interquartile range)] | 31a (25, 39) | 57 (52, 62) | 33 (27, 38) | 34 (30, 39) | |||
| Family history of TGCT | |||||||
| No | 313 | 89.7 | – | 380 | 86.6 | 864 | 90.0 |
| Yes | 32a | 9.2 | – | 11b | 2.5 | 10b | 1.0 |
| Unknown | 4 | 1.2 | – | 48 | 10.9 | 86 | 9.0 |
| Personal history of cryptorchidism | |||||||
| No | 310 | 88.8 | 394 | 89.8 | 942 | 98.1 | |
| Yes | 35 | 10.0 | 42 | 9.6 | 18 | 1.9 | |
| Unknown | 4 | 1.2 | 3 | 0.7 | 0 | 0 | |
| Tumor type | |||||||
| Seminoma | 114 | 32.7 | – | 266 | 60.6 | – | – |
| Non-seminoma | 227 | 65.0 | – | 173 | 39.4 | – | – |
| Unknown | 8 | 2.3 | – | 0 | 0 | – | – |
TGCT, testicular germ cell tumor.
aSixteen cases were recruited based on family history of TGCT. Among the non-selected (n = 333) cases, the proportion reporting any family history of TGCT was 4.8%.
bReflects reported family history of TGCT among first-degree relatives only.
Figure 1.
Manhattan plot of GWA results for 349 TGCT cases and 919 controls. SNP markers that reached significance at P < 5.0 × 10−6 based on Fisher's exact test are plotted above the lower line. Susceptibility loci identified in our original (KITLG, SPRY4, 2p14) and current (DMRT1) analysis are indicated.
Figure 2.
Manhattan plot and linkage disequilibrium structure of the DMRT1 genomic region. (A) SNP markers in the 1.2 Mb region that includes DMRT1 are plotted based on Fisher's exact test. Genotyped SNP markers are indicated in blue and imputed SNP markers are indicated in red. The two markers taken into replication are designated by triangles and labeled. (B) Linkage disequilibrium structure for the 40 Kb region encompassing DMRT1 based on observed and imputed (from HapMap CEU population, release 22) genotypes among the 919 controls in our discovery phase. Pairwise linkage disequilibrium is determined by r2, and methods described by Gabriel et al. are used to define haplotype blocks. Open arrow (left) indicates the position of DMRT1 rs7040024; oval arrow (right) indicates position of DMRT1 rs755383.
We did not replicate associations with rs2279070 (Ptrend = 0.62) and rs4834214 (Ptrend = 0.54) at 4q28.2 or rs6951213 (Ptrend = 0.48) and rs10281060 (Ptrend = 0.68) at PFTK1. However, we observed statistically significant associations with rs7040024 (Ptrend = 2.09 × 10−6) and rs755383 (Ptrend = 2.08 × 10−4) at DMRT1. Using a combined set of discovery and replication samples, TGCT cases had greater odds of carriage of the major A allele in rs7040024 than controls [P = 1.41 × 10−11; odds ratio (OR) = 1.70, 95% confidence interval (CI) 1.46, 1.99] and greater odds of carriage of the major T allele in rs755383 (P = 8.61 × 10−10; OR = 1.50, 95% CI 1.32, 1.7) (Table 2). In addition to the case–control analysis, we performed a case–parent analysis in 179 triads and 135 dyads that showed homozygous carriage of the A allele in rs7042004 was strongly associated with TGCT risk (P = 0.0033; relative risk = 3.42, 95% CI 1.50, 7.76) as was homozygous carriage of the T allele in rs755383 (P = 1.20 × 10−10; relative risk = 4.67, 95% CI 2.92, 7.46). These analyses support the finding of an association between the variation at the DMRT1 locus and TGCT susceptibility.
Table 2.
Associations of TGCT with replicated DMRT1 SNP markers
| Analysis group | Gene markera | Genotype countb |
Phase | OR (95% CI) |
P-valuee | |||
|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Per allele | Heterozygotec | Homozygoted | ||||
| Total | rs7040024 A/C | 493/356/68 | 236/100/11 | Discovery | 1.71 (1.37, 2.14) | 1.74 (0.89, 3.41) | 2.96 (1.54, 5.70) | 3.57 × 10−6 |
| 484/323/57 | 276/110/11 | Replication | 1.69 (1.36, 2.10) | 1.77 (0.89, 3.49) | 2.96 (1.52, 5.73) | 2.09 × 10−6 | ||
| Combined | 1.70 (1.46, 1.99) | 1.75 (1.08, 2.83) | 2.96 (1.86, 4.71) | 1.41 × 10−11 | ||||
| Adjustedf | 1.48 (1.23, 1.78) | 1.39 (0.83, 2.31) | 2.00 (1.25, 3.52) | 2.52 × 10−5 | ||||
| rs755383g T/C | 371/415/133 | 191/133/25 | Discovery | 1.64 (1.35, 1.98) | 1.71 (1.07, 2.73) | 2.74 (1.73, 4.35) | 5.78 × 10−7 | |
| 329/389/144 | 185/172/38 | Replication | 1.40 (1.17, 1.67) | 1.68 (1.12, 2.50) | 2.13 (1.43, 3.18) | 2.08 × 10−4 | ||
| Combined | 1.50 (1.32, 1.71) | 1.68 (1.24, 2.28) | 2.40 (1.78, 3.24) | 8.61 × 10−10 | ||||
| Adjusted | 1.26 (1.08, 1.47) | 1.46 (1.05, 2.03) | 1.71 (1.21, 2.42) | 0.0036 | ||||
| Seminomatous tumors | rs7040024 A/C | 78/31/4 | Discovery | 1.75 (1.22, 2.51) | 1.48 (0.51, 4.33) | 2.69 (0.95, 7.58) | 0.0023 | |
| 156/74/8 | Replication | 1.46 (1.13, 1.88) | 1.66 (0.76, 3.63) | 2.35 (1.10, 5.04) | 0.0036 | |||
| Combined | 1.54 (1.25, 1.89) | 1.59 (0.84, 2.98) | 2.42 (1.31, 4.46) | 4.64 × 10−5 | ||||
| Adjusted | 1.35 (1.06, 1.73) | 1.39 (0.71, 2.72) | 1.87 (0.95, 3.69) | 0.015 | ||||
| rs755383f T/C | 62/40/11 | Discovery | 1.54 (1.14, 2.09) | 1.17 (0.58, 2.34) | 2.02 (1.03, 3.95) | 0.0055 | ||
| 108/104/26 | Replication | 1.31 (1.06, 1.62) | 1.47 (0.92, 2.35) | 1.81 (1.13, 2.90) | 0.012 | |||
| Combined | 1.39 (1.17, 1.65) | 1.37 (0.93, 2.03) | 1.91 (1.30, 2.80) | 2.05 × 10−4 | ||||
| Adjusted | 1.22 (1.00, 1.50) | 1.23 (0.81, 1.88) | 1.49 (0.95, 2.33) | 0.055 | ||||
| Non-seminomatous tumors | rs7040024 A/C | 154/67/6 | Discovery | 1.73 (1.33, 2.26) | 2.13 (0.89, 5.11) | 3.54 (1.51, 8.31) | 4.54 × 10−5 | |
| 120/36/3 | Replication | 2.17 (1.53, 3.09) | 1.94 (0.57, 6.58) | 4.28 (1.30, 14.1) | 1.46 × 10−5 | |||
| Combined | 1.90 (1.54, 2.35) | 2.12 (1.04, 4.30) | 3.94 (1.98, 7.86) | 1.60 × 10−9 | ||||
| Adjusted | 1.64 (1.29, 2.10) | 1.51 (0.72, 3.19) | 2.55 (1.20, 5.39) | 7.42 × 10−5 | ||||
| rs755383f T/C | 126/90/13 | Discovery | 1.71 (1.36, 2.15) | 2.22 (1.20, 4.10) | 3.47 (1.90, 6.36) | 4.63 × 10−6 | ||
| 77/68/12 | Replication | 1.53 (1.18, 1.99) | 2.17 (1.13, 4.16) | 2.84 (1.48, 5.43) | 0.0015 | |||
| Combined | 1.63 (1.37, 1.94) | 2.16 (1.39, 3.37) | 3.16 (2.04, 4.90) | 2.30 × 10−8 | ||||
| Adjusted | 1.29 (1.06, 1.58) | 1.78 (1.11, 2.85) | 2.02 (1.24, 3.30) | 0.012 | ||||
TGCT, testicular germ cell tumor.
adbSNP rsnumber and risk/non-risk alleles.
bNumber of individuals genotyped as homozygous for the risk allele/heterozygous for the risk allele/homozygous for the non-risk allele. MAF for discovery phase markers given in Supplementary Material, Table S1.
cOR for heterozygous carriage of risk allele compared with homozygous carriage of non-risk allele.
dOR for homozygous carriage of risk allele compared with homozygous carriage of non-risk allele.
eTest for trend.
fOR, 95% CI and P-value determined from a logistic regression model of combined data containing main effects of both DMRT1 markers.
gGenotype counts, OR and 95% CI for discovery phase rs755383 estimated from data imputed using HapMap (release 22).
To determine whether the observed associations with rs7040024 and rs755383 were independent, we simultaneously modeled their main effects. Both DMRT1 markers remained statistically significantly associated with TGCT status after mutual adjustment, although more moderately so compared with their respective univariate models. For rs7040024, the P-value was 2.52 × 10−5 (adjusted per allele OR = 1.48, 95% CI 1.23, 1.78), and for rs755383, the P-value was 0.0036 (adjusted per allele OR = 1.26, 95% CI 1.08, 1.47) (Table 2). To further investigate the independence of these two markers, we explored haplotypes in the DMRT1 genomic region using observed and imputed genotypes (based on HapMap CEU population, release 22) among the 919 controls used in our discovery phase. Figure 2B shows that rs7040024 and rs755383 do not reside in the same haplotype block. Haplotype blocks resolved from the HapMap CEU population mirror these findings (Supplementary Material, Fig. S3).
Analyses that additionally adjusted for cryptorchidism or family history of TGCT were limited to the replication sample because information on risk factors was not collected for discovery phase controls. Here, associations with both DMRT1 markers (simultaneously considered) and TGCT were unchanged following further adjustment for either cryptorchidism or family history. Among men without a family history of TGCT or men without cryptorchidism, the ORs were comparable with overall results. This finding suggests that the effects of DMRT1 variants are not based on the pathologic mechanisms leading to these known strong TGCT risk factors.
In the combined discovery and replication analysis and after mutual adjustment, SNP genotypes in DMRT1 were associated with both seminoma and non-seminomatous germ cell tumors (Table 2). While we could not reject homogeneity of the ORs comparing subtypes, odds associated with rs7040024 appeared higher in men with non-seminomatous germ cell tumors than among those with seminomatous tumors.
In the combined set, we explored whether the effects of DMRT1 rs7040024 and rs755383 were modified by genotypes at KITLG rs4474514 and SPRY4 rs6897876, loci we previously reported as independently associated with TGCT status (18). In models that assessed the joint effects of DMRT1 genotypes with KITLG and SPRY4 genotypes, we failed to detect departures from multiplicativity. However, we did observe statistical evidence supporting departure from additivity (i.e. additive synergy) for DMRT1 rs7040024 and KITLG rs4474514 (synergy index, S = 2.2, 95% CI 1.1, 4.2; P = 0.02). That is to say, the OR associated with homozygous carriage of the risk allele at both loci (OR = 14.1, 95% CI 5.12, 38.6; P = 2.98 × 10−7) was greater than the additive effect of the OR for homozygous carriage of the risk allele at either locus alone (OR 2.44, 95% CI 0.62, 9.56, P = 0.20 for DMRT1 rs7040024; OR = 5.59, 95% CI 1.84, 7.0, P = 2.45 × 10−3 for KITLG rs4474514; Fig. 3). Here, the comparison group was individuals who carried a total of zero or one risk allele at both loci (n = 86), which was necessary because only five controls and no TGCT cases were doubly homozygous for the non-risk alleles. We did not find evidence of departure from additivity for DMRT1 rs7040024 and SPRY4 rs6897876 (synergy index, S = 2.2, 95% CI 0.7, 7.2, P = 0.20).
Figure 3.
Joint effects of DMRT1 genotypes with KITLG and SPRY4 genotypes. OR and 95% CI for main effects adjusted for the second SNP marker are given in the row and column headers. Within cells, frequency count (control/case) and OR (95% CI) for joint genotypes are given with referent genotype(s) represented as shaded cells.
Evidence of association with markers at ATF7IP, BAK1 and TERT was inconsistent across our sample sets. Although TGCT status was associated with rs11055991 (ATF7IP), rs210138 (BAK1), rs444697 (BAK1) and rs2736100 (TERT) in our discovery set, no associations were noted in our replication set (Supplementary Material, Table S2). In contrast to reported findings by Turnbull et al. (20), we did not find associations at TERT to be stronger in tumors of the seminoma subtype.
DISCUSSION
We verified the finding that variation at 9p24.3 within DMRT1 is associated with TGCT susceptibility (20) and report the identification of a second risk allele (rs7040024) at this locus that is independent of the one previously reported (rs755383). In combined analysis and after mutual adjustment, TGCT cases had 50% greater odds of carriage per major A allele at rs7040024, while simultaneously having nearly 25% greater odds of carriage per major T allele at rs755383. Of note, rs755383 was the most statistically significant (imputed) marker at this locus and the marker genotyped directly by Turnbull et al. We anticipated that these markers might be independently associated with TGCT because of their low-to-moderate linkage disequilibrium (r2 = 0.29) with each other, and the fact that these two markers do not reside in the same haplotype block. Thus, we have identified a second SNP within DMRT1 that is associated more strongly with TGCT susceptibility than that previously reported. Our finding, together with others recently identified for TGCT susceptibility loci, begins to unravel the complex architecture of inherited genetics of TGCT.
We acknowledge that the use of a comparison group consisting of men who were undergoing routine cardiac catheterization, of whom 76% (n = 700) had angiographically confirmed coronary artery disease (CAD), may have impacted our findings to the degree that there are common genetic factors linking CAD and TGCT. To explore this possibility, we compared genotype frequencies of markers at BAK1, DMRT1 and TERT (Table 2 and Supplementary Material, Table S2) among controls with and without frank CAD and noted no statistically significant differences (0.09 ≤ Ptrend ≤ 0.64 for all comparisons). Further, we compared the observed minor allele frequencies (MAFs) at these markers among CAD controls to those observed in our replication-phase controls, HapMap CEU samples and Coriell CEPH samples. We observed slight fluctuations in allele frequencies among these four groups ranging from a 3 to 8% difference; however, for each marker, the allele frequency among the CAD controls was consistent with that in at least one of the three population-based sample sets. These comparisons lead us to believe that little bias was introduced by using CAD controls. As well, we recognize that 90% of CAD controls were 46 years or older and had already passed the peak age of TGCT development. Based on available age-specific TGCT rates, we estimated that only four TGCT cases would be expected to have arisen in this control group (1). It is unlikely that this potential small misclassification of phenotype would have biased results appreciably.
DMRT1 is a member of a zinc finger-like DNA-binding motif (DM domain) gene family. Genes with the DM domain are highly conserved and play crucial roles in male development and sex determination across the phylogenetic spectrum from flies and nematodes to birds and vertebrates (23). Dmrt1 in mice is expressed only in the gonad and is essential for postnatal germline maintenance and differentiation of germ cells, specifically radial migration, mitotic reactivation and survival (24,25). Dmrt1 is required for normal maturation of Sertoli cells; the cells fail to polarize and stop proliferating when this gene is knocked out (24). Increased dosage of DMRT1 facilitates male development, and decreasing gene dosage leads to feminization of the gonads. In humans, deletion of the region on 9p-containing DMRT1 leads to male-to-female sex reversal and is associated with the development of gonadoblastoma (26,27). Dmrt1 −/− mice also have severely dysgenetic testes, resembling those seen in XY individuals with the loss of 9p, and fail to undergo normal germ cell development, dying by P14 (24,28). Moreover, a loss of Dmrt1 also is associated with a high rate of teratomas in 129Sv mice with 90% of double knockout mice developing tumors (29). In our data, variants at DMRT1 may be slightly more strongly associated with non-seminomatous germ cell tumors, of which teratoma is one type, than with seminomas; but the difference between tumor types was not statistically significant. We do not believe our inability to detect a statistical difference in the association between common DMRT1 variants by histological tumor type should be interpreted as contradicting the evidence from model systems for a role of this gene in human teratoma susceptibility. Rather, the sample size was modest, and consequently there was limited power to detect differences between tumor types.
TGCT is believed to arise from undifferentiated primordial germ cells, which progress to the precursor lesion of TGCT-termed intratubular germ cell neoplasia undifferentiated (ITGCNU) (30–33). All ITGCNU is thought to advance to frank TGCT, i.e. there is no spontaneous regression. Consistent with this hypothesis, predisposition to TGCT is increased among patients with different types of disorders of sexual development and delayed differentiation of primordial germ cells (34). The data from the TGCT GWA studies are strongly supportive of this model of disease development, as male germ cell differentiation is the biological pathway linking DMRT1 to the TGCT susceptibility loci KITLG, SPRY4 and BAK1.
TGCT susceptibility has been linked to infertility or sub-fertility in several studies, and the majority of evidence points to common etiologic risk factors for these conditions. The most convincing studies have followed men with known semen quality for outcomes including TGCT and have examined fertility in brothers of men with TGCT (35–39). Interestingly, Dmrt1 is essential for fertility in mice (24). Thus, the association between common variation at DMRT1 and TGCT provides further support to the notion of shared risk factors for infertility and these malignancies.
Rapley et al. (19) and Turnbull et al. (20) also found evidence in their discovery and replication sets of association of TGCT with rs2900333 and rs4346018 at the ATF7IP locus, with rs210138 in the BAK1 locus and with rs2736100 and rs4635969 in the TERT locus. We genotyped TERT rs31489 (which reached genome-wide significance in the UK study, but was not brought forth into replication) in our replication sample set and looked up results in our discovery set for three other TERT markers in common with the UK study: rs2736100, rs4016810 (which is in linkage disequilibrium with rs4635969, r2 = 0.67) and rs4975616. We genotyped BAK1 rs210138 in our replication sample and looked up results for this and two other BAK1 markers reaching genome-wide significance in the UK study: rs210139 and rs444697 (which is in linkage disequilibrium with rs375555, r2 = 0.72 and rs4713646, r2 = 0.97). At ATF7IP, we genotyped rs2900333 and rs4346018 in our replication sample and looked up results in our discovery set for rs11055991, which was in linkage disequilibrium with both rs2900333 (r2 = 0.72) and rs4346018 (r2 = 0.95). While we were unable to verify previous finding in our replication sample set, in our discovery set we found some evidence of association with ATF7IP rs11055991 (P = 0.0052), BAK1 rs210138 (P = 0.0074), BAK1 rs444697 (P = 0.028) and TERT rs2736100 (P = 0.020) and after adjustment for the false discovery rate (40) observed the following P-values: 0.041, 0.041, 0.077 and 0.073, respectively.
Our inability to verify the associations at TERT and ATF7IP may be due to a difference in power of the UK study and the current study. Given the size of our discovery or replication data sets, and assuming α = 0.05 (uncorrected for multiple comparisons), 1 − β = 0.80, an additive genetic model, and a range of MAF from 40 to 50% that correspond to observed MAF for markers in TERT and ATF7IP, the minimal detectable per allele OR ranged from 1.29 to 1.26 (41), which is greater than those observed at these loci in our study. As the white non-Hispanic population in the USA is more genetically heterogeneous than the white UK population, the associations with specific alleles in our study may be attenuated, exacerbating decreased power.
Only 3% of the TGCT cases did not carry a risk (major) allele at DMRT1 rs7040024; this proportion decreased to 1% for KITLG. Considering these loci jointly, we observed no case who was a double homozygote for the non-risk (minor) alleles at these loci. This finding suggests that carriage of at least one and potentially both major alleles may be necessary for TGCT development. Men with TGCT are 14 times more like to be doubly homozygous for the risk alleles of KITLG and DMRT1 than those who carry a total of only one or zero risk alleles. If associations with genotypes at these loci are further validated, they may identify a group of men at increased risk of developing TGCT; thus in the future, these findings may have implications for screening. At present, screening for TGCT in the general population or in men at increased risk for TGCT such as those with cryptorchidism or a family history of TGCT is not recommended by the US Preventative Services Health Task Force at present (42,43). The relatively small number of individuals in our replication set with these conditions prevented us from assessing the extent to which the risk alleles influenced TGCT development among this subset. Future pooled analyses will increase power for such important analyses.
While in large part, the per-allele ORs identified for TGCT susceptibility alleles are stronger than those identified for other cancers; these tumors are rare and the studies are considerably smaller than GWA studies of many other cancers. Thus, it is likely that further genes remain to be identified, potentially through pooled or meta-analysis (44). Based on these and others'' similar findings, variation in pathways that influence male germ cell development and differentiation, including KITLG, SPRY4, BAK1 and DMRT1, is emerging as the major player in inherited TGCT susceptibility. Still, the biological mechanism and functional impact of the true risk alleles to which our observed risk alleles may be linked will need to be determined in order to better comprehend the genetic basis of TGCT. Other genes that play crucial roles in differentiation of the male gonad or germ cell development, such as SOX9 and STRA8, should be considered potential candidate genes. The identification of this pathway as important in TGCT susceptibility supports the hypothesis that TGCT develops from undifferentiated primordial germ cells and gives further biological basis for the known association between infertility and TGCT.
MATERIALS AND METHODS
Ethics statement
For both the GWA and replication studies, each participant provided written informed consent approved by their local Institutional Review Boards.
Genome-wide association study
In the discovery phase, we included 455 TGCT patients, most of whom were approached while seeking care at the University of Pennsylvania Health System or Fox Chase Cancer Center. TGCT cases not identified through this hospital-based mechanism were identified through the Pennsylvania State Cancer Registry and contacted by mail to solicit for study participation. All TGCT patients were asked to complete a self-administered questionnaire that elicits information on known and presumptive risk factors for TGCT and to provide a biospecimen. Each patient is classified as having seminomatous or non-seminomatous (including yolk sac, choriocarcinoma, embryonal, teratoma and mixed cell, i.e. having both non-seminomatous and seminomatous aspects) TGCT based on histological diagnosis. We obtain this information through medical record review for those participants recruited in person and directly from the Cancer Registry for participants recruited via mail. Only participants with primary disease in the testis are included.
Controls were selected from PennCATH, a multi-institutional hospital-based study of angiographic CAD in almost 4000 subjects undergoing cardiac catheterization. This study investigates the association of biochemical and genetic factors for CAD and its risk factors (45); information on personal history of cancer was not collected. For our comparison group, we selected only males enrolled in PennCATH from the Philadelphia region (n = 932) independent of their disease status.
We used the Affymetrix® Genome-Wide Human SNP Array 6.0 to obtain genotypes for TGCT cases. We used the Birdseed algorithm to determine genotypes for the combined TGCT case and CAD control sample set (46). Among the 455 case samples, 19 subsequently were excluded for not meeting case eligibility (2 Leydig cell tumors, 1 female germ cell tumor erroneously coded as TGCT, 16 non-TGCT samples) and 11 replicate samples with lower genotyping call rates were excluded. Of the 425 unique samples from TGCT cases, 48 (11%) were excluded because of a low (<95%) genotyping call rate. Seven (1.5%) men were excluded because of lower than expected genotypic heterozygosity across called markers (FST ≥ 0.06), and 21 (6.2%) because of non-European ancestry as determined by multidimensional scaling (47); no cases were excluded for cryptic relatedness (proportion of genotypes IBD for all cases was <0.20). The resulting 349 samples represent a 26% increase in the number of TGCT cases analyzed in this report compared with that (n = 277) used in the original analysis. Controls had been genotyped previously using the Affymetrix® Genome-Wide Human SNP Array 6.0 platform and had passed the same genotyping quality measures used for TGCT cases. Among the 932 CAD controls, 13 were excluded because of female or ambiguous sex on genotyping.
After excluding 82 981 (11.3%) markers with a MAF (in the total sample) < 0.05; 1872 (0.3%) that deviated from Hardy–Weinberg equilibrium (P < 1 × 10−7); 38 060 (5.2%) with an individual genotype call rate < 0.95 and 233 (0.03%) invalid markers, 609 482 markers remained in the discovery phase.
Replication study
To independently replicate findings of the discovery phase, we used 439 cases and 960 controls from a population-based case–control study of TGCT in western Washington State. These numbers represent an 18% increase in cases and 12% increase in controls compared with those (n = 371 and n = 860, respectively) used in the original analysis. Briefly, all cases had first primary TGCT diagnosed between 1999 and 2008 and were residents of three urban counties of western Washington aged 18–44 years at diagnosis. Control subjects did not have a personal history of TGCT and were frequency matched on age and ascertained from the general population of the three counties using random digit telephone dialing. Family history of TGCT among first-degree relatives and personal history of cryptorchidism were ascertained by questionnaire. Only cases and controls who self-identified as white, non-Hispanic were included in the replication study. In addition, 314 sets of mothers and fathers of cases (179 case-triads and 135 case-dyads) were used for the case–parent analysis. Methods for recruitment of TGCT cases and parents in this study previously have been published (48).
Genotyping
Replication genotyping was accomplished using the iPLEX Mass Array platform (Sequenom, Inc.) according to the manufacturer's specifications. Genotyping was run in duplicate for 1248 marker pairs (208 sample pairs per each of the six markers in replication). Genotyping calls were made without knowledge of case or duplicate status. In total, 26 (2.1%) calls were discordant, ranging from 0.5 (rs7040024, rs6951213) to 5.3% (rs2279070); the Spearman correlation coefficient was 0.98 (ranging from 0.90 for rs2279070 to >0.99 for rs7040024). We also re-genotyped ∼80% of TGCT cases from the discovery phase for the three observed markers taken into replication. The Spearman correlation coefficient between genotype calls obtained from the Affymetrix® chip and iPLEX platform for these three markers ranged from 0.96 to 0.97.
We also genotyped in our replication sample at least one top hit in BAK1, TERT and ATF7IP, which were previously observed to be associated with TGCT susceptibility (19,20). Where possible, we also looked up associations in our discovery sample for common markers at these loci.
Statistical analysis
For the discovery phase, we used PLINK software to calculate rates of heterozygosity, assess population stratification using multi-dimensional scaling methods and test markers for Hardy–Weinberg equilibrium (49). PLINK also was used to determine allelic associations among the 349 TGCT cases and 919 CAD controls and to assess their statistical significance by Fisher's exact test. For top hits, we determined ORs and 95% CIs for the per allele, heterozygous and homozygous effects of the minor allele using SAS v9.2 (Supplementary Material, Table S1).
Imputation was conducted using a computationally efficient hidden Markov model-based algorithm as implemented in software MACH (50,51). MACH combines our genotyped data with phased chromosomes from the 1000 Genomes CEU samples and then infers the unknown genotypes in the study sample probabilistically by searching for similar stretches of flanking haplotype in the 1000 genomes reference sample. For chromosome 9, we also used data from HapMap as the reference CEU sample for imputing genotypes. We excluded from analysis imputed SNP markers with MAF < 0.05 in both cases and controls or R2 < 0.3, an indicator of suboptimal imputation. To account for uncertainty involved in the imputation, we analyzed case–control associations for imputed SNP markers using software SNPTEST (52).
Analyses were performed using SAS v9.2 (SAS Institute, Cary, NC, USA) for the replication samples and the combined set consisting of both discovery and replication-phase samples. We used unconditional logistic regression to determine per allele associations and associations of homozygous and heterozygous carriage of risk alleles with case status (overall and among specified subgroups). Trend across genotype categories was assessed by the Cochran–Armitage test for trend. We used multinomial logistic models to obtain simultaneously the OR and 95% CI for the association between TGCT subtypes defined on the basis of histology. To determine whether the observed association with a given SNP marker was independent of the effects of other SNP markers, we used logistic regression models that included the main effects of each SNP marker. To investigate the joint effect of two SNP markers on TGCT, an indicator variable was coded to represent the various combinations of genotypes at both markers, and this indicator variable was then entered into a logic regression model as a class variable. The synergy index and its 95% CI were used to assess departure from additivity of genetic effects (53). All analyses using the combined data set incorporated an indicator variable for study phase.
SUPPLEMENTARY MATERIAL
FUNDING
The work was supported by the Abramson Cancer Center at the University of Pennsylvania and National Institute of Health grants (R01CA114478 to P.A.K., K.L.N.; R01CA085914 to C.C., J.R.S., S.M.S.; Fox Chase Cancer Center Support Grant P30 CA006927; and Cancer Biostatistics Training Grant CA093283 to S.V.). Recruitment of controls from the University of Pennsylvania Health System was supported by the Cardiovascular Institute of the University of Pennsylvania, and genotyping of controls was supported by GlaxoSmithKline through an Alternate Drug Discovery Initiative research alliance award (to M.P.R. and D.J.R.) with the University of Pennsylvania School of Medicine. Services provided in the Penn Molecular Diagnosis and Genotyping Facility are supported by the Abramson Cancer Center core grant 5P30CA016520.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank both the men with and without TGCT and parents of TGCT patients, who contributed to the study. We thank D. Pucci, K. Robertson, E. Ellis Ohr, A. Mackley, A. Tran, M. Shellenberger, C. Panks, K. Leach and D. Jacke for patient recruitment and database maintenance. We thank S. Fish for her expert assistance with DNA extraction and genotyping. We would like to thank Drs S. Lesko and G.Bunin, consultants for the ongoing TGCT project at Penn. We acknowledge Drs C. Turnbull and N. Rahman for providing data from their GWA study of TGCT pre-publication. The Pennsylvania Cancer Registry data were supplied by the Bureau of Health Statistics & Research, Pennsylvania Department of Health, Harrisburg, Pennsylvania. The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Altekruse S., Kosary C., Krapcho M., Neyman N., Aminou R., Waldron W., Ruhl J., Howlader N., Tatalovich Z., Cho H., et al. Bethesda, MD: National Cancer Institute; 2010. http://seer.cancer.gov/csr/1975_2007/ based on November 2009 SEER data submission, posted to the SEER web site, 2010. [Google Scholar]
- 2.Huyghe E., Matsuda T., Thonneau P. Increasing incidence of testicular cancer worldwide: a review. J. Urol. 2003;170:5–11. doi: 10.1097/01.ju.0000053866.68623.da. doi:10.1097/01.ju.0000053866.68623.da. [DOI] [PubMed] [Google Scholar]
- 3.Curado M.P., Edwards B., Shin H.R., Ferlay J., Heanue M., Boyle P. Cancer incidence in five continents; 2007. Vol. IX. International Agency for Research on Cancer, Lyon, France. [Google Scholar]
- 4.The International Testicular Cancer Linkage Consortium. Candidate regions for testicular cancer susceptibility genes. APMIS. 1998;106:64–70. discussion 71-62 doi:10.1111/j.1699-0463.1998.tb01320.x. [PubMed] [Google Scholar]
- 5.Raven R.W. Tumours of the testics in two brothers. Lancet. 1934;2:870–871. doi:10.1016/S0140-6736(00)74662-9. [Google Scholar]
- 6.Bromen K., Stang A., Baumgardt-Elms C., Stegmaier C., Ahrens W., Metz K.A., Jockel K.H. Testicular, other genital, and breast cancers in first-degree relatives of testicular cancer patients and controls. Cancer Epidemiol. Biomarkers Prev. 2004;13:1316–1324. [PubMed] [Google Scholar]
- 7.Chia V.M., Li Y., Goldin L.R., Graubard B.I., Greene M.H., Korde L., Rubertone M.V., Erickson R.L., McGlynn K.A. Risk of cancer in first- and second-degree relatives of testicular germ cell tumor cases and controls. Int. J. Cancer. 2009;124:952–957. doi: 10.1002/ijc.23971. doi:10.1002/ijc.23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forman D., Oliver R.T., Brett A.R., Marsh S.G., Moses J.H., Bodmer J.G., Chilvers C.E., Pike M.C. Familial testicular cancer: a report of the UK family register, estimation of risk and an HLA class 1 sib-pair analysis. Br. J. Cancer. 1992;65:255–262. doi: 10.1038/bjc.1992.51. doi:10.1038/bjc.1992.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gundy S., Babosa M., Baki M., Bodrogi I. Increased predisposition to cancer in brothers and offspring of testicular tumor patients. Pathol. Oncol. Res. 2004;10:197–203. doi: 10.1007/BF03033760. doi:10.1007/BF03033760. [DOI] [PubMed] [Google Scholar]
- 10.Heimdal K., Olsson H., Tretli S., Flodgren P., Borresen A.L., Fossa S.D. Familial testicular cancer in Norway and southern Sweden. Br. J. Cancer. 1996;73:964–969. doi: 10.1038/bjc.1996.173. doi:10.1038/bjc.1996.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemminki K., Chen B. Familial risks in testicular cancer as aetiological clues. Int. J. Androl. 2006;29:205–210. doi: 10.1111/j.1365-2605.2005.00599.x. doi:10.1111/j.1365-2605.2005.00599.x. [DOI] [PubMed] [Google Scholar]
- 12.Sonneveld D.J., Sleijfer D.T., Schrafford Koops H., Sijmons R.H., van der Graaf W.T., Sluiter W.J., Hoekstra H.J. Familial testicular cancer in a single-centre population. Eur. J. Cancer. 1999;35:1368–1373. doi: 10.1016/s0959-8049(99)00140-9. doi:10.1016/S0959-8049(99)00140-9. [DOI] [PubMed] [Google Scholar]
- 13.Spermon J.R., Witjes J.A., Nap M., Kiemeney L.A. Cancer incidence in relatives of patients with testicular cancer in the eastern part of The Netherlands. Urology. 2001;57:747–752. doi: 10.1016/s0090-4295(00)01058-x. doi:10.1016/S0090-4295(00)01058-X. [DOI] [PubMed] [Google Scholar]
- 14.Westergaard T., Olsen J.H., Frisch M., Kroman N., Nielsen J.W., Melbye M. Cancer risk in fathers and brothers of testicular cancer patients in Denmark. A population-based study. Int. J. Cancer. 1996;66:627–631. doi: 10.1002/(SICI)1097-0215(19960529)66:5<627::AID-IJC8>3.0.CO;2-V. doi:10.1002/(SICI)1097-0215(19960529)66:5<627::AID-IJC8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 15.Neale R.E., Carriere P., Murphy M.F., Baade P.D. Testicular cancer in twins: a meta-analysis. Br. J. Cancer. 2008;98:171–173. doi: 10.1038/sj.bjc.6604136. doi:10.1038/sj.bjc.6604136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swerdlow A.J., De Stavola B.L., Swanwick M.A., Mangtani P., Maconochie N.E. Risk factors for testicular cancer: a case-control study in twins. Br. J. Cancer. 1999;80:1098–1102. doi: 10.1038/sj.bjc.6690470. doi:10.1038/sj.bjc.6690470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czene K., Lichtenstein P., Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int. J. Cancer. 2002;99:260–266. doi: 10.1002/ijc.10332. doi:10.1002/ijc.10332. [DOI] [PubMed] [Google Scholar]
- 18.Kanetsky P.A., Mitra N., Vardhanabhuti S., Li M., Vaughn D.J., Letrero R., Ciosek S.L., Doody D.R., Smith L.M., Weaver J., et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat. Genet. 2009;41:811–815. doi: 10.1038/ng.393. doi:10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rapley E.A., Turnbull C., Al Olama A.A., Dermitzakis E.T., Linger R., Huddart R.A., Renwick A., Hughes D., Hines S., Seal S., et al. A genome-wide association study of testicular germ cell tumor. Nat. Genet. 2009;41:807–810. doi: 10.1038/ng.394. doi:10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turnbull C., Rapley E.A., Seal S., Pernet D., Renwick A., Hughes D., Ricketts M., Linger R., Nsengimana J., Deloukas P., et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat. Genet. 2010;42:604–607. doi: 10.1038/ng.607. doi:10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bacanu S.A., Devlin B., Roeder K. The power of genomic control. Am. J. Hum. Genet. 2000;66:1933–1944. doi: 10.1086/302929. doi:10.1086/302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dadd T., Weale M.E., Lewis C.M. A critical evaluation of genomic control methods for genetic association studies. Genet. Epidemiol. 2009;33:290–298. doi: 10.1002/gepi.20379. doi:10.1002/gepi.20379. [DOI] [PubMed] [Google Scholar]
- 23.Koopman P. Sex determination: the power of DMRT1. Trends Genet. 2009;25:479–481. doi: 10.1016/j.tig.2009.09.009. doi:10.1016/j.tig.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Kim S., Bardwell V.J., Zarkower D. Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev. Biol. 2007;307:314–327. doi: 10.1016/j.ydbio.2007.04.046. doi:10.1016/j.ydbio.2007.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raymond C.S., Murphy M.W., O'Sullivan M.G., Bardwell V.J., Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. doi:10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvari V., Bertini V., De Grandi A., Peverali G., Zuffardi O., Ferguson-Smith M., Knudtzon J., Camerino G., Borsani G., Guioli S. A new submicroscopic deletion that refines the 9p region for sex reversal. Genomics. 2000;65:203–212. doi: 10.1006/geno.2000.6160. doi:10.1006/geno.2000.6160. [DOI] [PubMed] [Google Scholar]
- 27.Livadas S., Mavrou A., Sofocleous C., van Vliet-Constantinidou C., Dracopoulou M., Dacou-Voutetakis C. Gonadoblastoma in a patient with del(9)(p22) and sex reversal: report of a case and review of the literature. Cancer Genet. Cytogenet. 2003;143:174–177. doi: 10.1016/s0165-4608(02)00849-x. doi:10.1016/S0165-4608(02)00849-X. [DOI] [PubMed] [Google Scholar]
- 28.Fahrioglu U., Murphy M.W., Zarkower D., Bardwell V.J. mRNA expression analysis and the molecular basis of neonatal testis defects in Dmrt1 mutant mice. Sex. Dev. 2007;1:42–58. doi: 10.1159/000096238. doi:10.1159/000096238. [DOI] [PubMed] [Google Scholar]
- 29.Krentz A.D., Murphy M.W., Kim S., Cook M.S., Capel B., Zhu R., Matin A., Sarver A.L., Parker K.L., Griswold M.D., et al. The DM domain protein DMRT1 is a dose-sensitive regulator of fetal germ cell proliferation and pluripotency. Proc. Natl Acad. Sci. USA. 2009;106:22323–22328. doi: 10.1073/pnas.0905431106. doi:10.1073/pnas.0905431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almstrup K., Hoei-Hansen C.E., Wirkner U., Blake J., Schwager C., Ansorge W., Nielsen J.E., Skakkebaek N.E., Rajpert-De Meyts E., Leffers H. Embryonic stem cell-like features of testicular carcinoma in situ revealed by genome-wide gene expression profiling. Cancer Res. 2004;64:4736–4743. doi: 10.1158/0008-5472.CAN-04-0679. doi:10.1158/0008-5472.CAN-04-0679. [DOI] [PubMed] [Google Scholar]
- 31.Hoei-Hansen C.E., Nielsen J.E., Almstrup K., Hansen M.A., Skakkebaek N.E., Rajpert-DeMeyts E., Leffers H. Identification of genes differentially expressed in testes containing carcinoma in situ. Mol. Hum. Reprod. 2004;10:423–431. doi: 10.1093/molehr/gah059. doi:10.1093/molehr/gah059. [DOI] [PubMed] [Google Scholar]
- 32.Rajpert-De Meyts E., Bartkova J., Samson M., Hoei-Hansen C.E., Frydelund-Larsen L., Bartek J., Skakkebaek N.E. The emerging phenotype of the testicular carcinoma in situ germ cell. APMIS. 2003;111:267–278. doi: 10.1034/j.1600-0463.2003.11101301.x. discussion 278–269. [DOI] [PubMed] [Google Scholar]
- 33.van Gurp R.J., Oosterhuis J.W., Kalscheuer V., Mariman E.C., Looijenga L.H. Biallelic expression of the H19 and IGF2 genes in human testicular germ cell tumors. J. Natl Cancer Inst. 1994;86:1070–1075. doi: 10.1093/jnci/86.14.1070. doi:10.1093/jnci/86.14.1070. [DOI] [PubMed] [Google Scholar]
- 34.Hersmus R., de Leeuw B.H., Wolffenbuttel K.P., Drop S.L., Oosterhuis J.W., Cools M., Looijenga L.H. New insights into type II germ cell tumor pathogenesis based on studies of patients with various forms of disorders of sex development (DSD) Mol. Cell Endocrinol. 2008;291:1–10. doi: 10.1016/j.mce.2008.02.028. doi:10.1016/j.mce.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 35.Doria-Rose V.P., Biggs M.L., Weiss N.S. Subfertility and the risk of testicular germ cell tumors (United States) Cancer Causes Control. 2005;16:651–656. doi: 10.1007/s10552-005-0169-x. doi:10.1007/s10552-005-0169-x. [DOI] [PubMed] [Google Scholar]
- 36.Peng X., Zeng X., Peng S., Deng D., Zhang J. The association risk of male subfertility and testicular cancer: a systematic review. PLoS ONE. 2009;4:e5591. doi: 10.1371/journal.pone.0005591. doi:10.1371/journal.pone.0005591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richiardi L., Akre O. Fertility among brothers of patients with testicular cancer. Cancer Epidemiol. Biomarkers Prev. 2005;14:2557–2562. doi: 10.1158/1055-9965.EPI-05-0409. doi:10.1158/1055-9965.EPI-05-0409. [DOI] [PubMed] [Google Scholar]
- 38.Richiardi L., Akre O., Montgomery S.M., Lambe M., Kvist U., Ekbom A. Fecundity and twinning rates as measures of fertility before diagnosis of germ-cell testicular cancer. J. Natl Cancer Inst. 2004;96:145–147. doi: 10.1093/jnci/djh012. doi:10.1093/jnci/djh012. [DOI] [PubMed] [Google Scholar]
- 39.Walsh T.J., Croughan M.S., Schembri M., Chan J.M., Turek P.J. Increased risk of testicular germ cell cancer among infertile men. Arch. Intern. Med. 2009;169:351–356. doi: 10.1001/archinternmed.2008.562. doi:10.1001/archinternmed.2008.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Soc. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 41.Skol A.D., Scott L.J., Abecasis G.R., Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. doi:10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 42.U.S. Preventive Services Task Force. Screening for testicular cancer: U.S. Preventive Services Task Force reaffirmation recommendation statement. Ann. Intern. Med. 2011;154:483–486. doi: 10.7326/0003-4819-154-7-201104050-00006. [DOI] [PubMed] [Google Scholar]
- 43.Lin K., Sharangpani R. Screening for testicular cancer: an evidence review for the U.S. Preventive Services Task Force. Ann. Intern. Med. 2010;153:396–399. doi: 10.7326/0003-4819-153-6-201009210-00007. [DOI] [PubMed] [Google Scholar]
- 44.Park J.H., Wacholder S., Gail M.H., Peters U., Jacobs K.B., Chanock S.J., Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 2010;42:570–575. doi: 10.1038/ng.610. doi:10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehrke M., Millington S.C., Lefterova M., Cumaranatunge R.G., Szapary P., Wilensky R., Rader D.J., Lazar M.A., Reilly M.P. CXCL16 is a marker of inflammation, atherosclerosis, and acute coronary syndromes in humans. J. Am. Coll. Cardiol. 2007;49:442–449. doi: 10.1016/j.jacc.2006.09.034. doi:10.1016/j.jacc.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 46.McCarroll S.A., Kuruvilla F.G., Korn J.M., Cawley S., Nemesh J., Wysoker A., Shapero M.H., de Bakker P.I., Maller J.B., Kirby A., et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. doi:10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 47.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14 000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. doi:10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Starr J.R., Chen C., Doody D.R., Hsu L., Ricks S., Weiss N.S., Schwartz S.M. Risk of testicular germ cell cancer in relation to variation in maternal and offspring cytochrome p450 genes involved in catechol estrogen metabolism. Cancer Epidemiol. Biomarkers Prev. 2005;14:2183–2190. doi: 10.1158/1055-9965.EPI-04-0749. doi:10.1158/1055-9965.EPI-04-0749. [DOI] [PubMed] [Google Scholar]
- 49.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y., Willer C., Sanna S., Abecasis G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. doi:10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. doi:10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. doi:10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 53.Hosmer D.W., Lemeshow S. Confidence interval estimation of interaction. Epidemiology. 1992;3:452–456. doi: 10.1097/00001648-199209000-00012. doi:10.1097/00001648-199209000-00012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.