Abstract
SHANK3 is a synaptic scaffolding protein enriched in the postsynaptic density (PSD) of excitatory synapses. Small microdeletions and point mutations in SHANK3 have been identified in a small subgroup of individuals with autism spectrum disorder (ASD) and intellectual disability. SHANK3 also plays a key role in the chromosome 22q13.3 microdeletion syndrome (Phelan–McDermid syndrome), which includes ASD and cognitive dysfunction as major clinical features. To evaluate the role of Shank3 in vivo, we disrupted major isoforms of the gene in mice by deleting exons 4–9. Isoform-specific Shank3e4–9 homozygous mutant mice display abnormal social behaviors, communication patterns, repetitive behaviors and learning and memory. Shank3e4–9 male mice display more severe impairments than females in motor coordination. Shank3e4–9 mice have reduced levels of Homer1b/c, GKAP and GluA1 at the PSD, and show attenuated activity-dependent redistribution of GluA1-containing AMPA receptors. Subtle morphological alterations in dendritic spines are also observed. Although synaptic transmission is normal in CA1 hippocampus, long-term potentiation is deficient in Shank3e4–9 mice. We conclude that loss of major Shank3 species produces biochemical, cellular and morphological changes, leading to behavioral abnormalities in mice that bear similarities to human ASD patients with SHANK3 mutations.
INTRODUCTION
Shank3 [SH3 and multiple ankyrin (ANK) repeat domain 3] is a member of the highly conserved Shank/ProSAP family of synaptic scaffolding proteins, first identified as a component of the PSD-95/GKAP/Homer1 complex enriched in the postsynaptic densities (PSDs) of excitatory synapses (1–3). Shank3 is a large protein containing multiple conserved motifs, including an ANK repeat, a proline-rich cluster and SH3, PDZ and SAM (sterile α-motif) domains (4–6). In vitro studies have shown that the ANK domain can interact with cytoskeletal proteins in the PSD (7), the proline-rich domain of Shank3 can interact with Homer1b/c protein (2) and the PDZ domain can interact with the GluA1 subunit of the AMPA receptor (AMPAR) directly, and with the NMDA receptor (NMDAR) through GKAP and the PSD-95 complex (1,2,8). In hippocampal cultures, expression of Shank3 in aspiny neurons can recruit AMPARs for the formation of new synapses, and siRNA-induced knockdown of Shank3 reduces spine densities and increases the lengths of spines (9). It remains unclear whether these interactions occur in vivo and whether they can influence synaptic function.
SHANK3 maps to the critical region of chromosome 22q13.3 deletion syndrome (also known as Phelan–McDermid syndrome or PMS) (10–13). PMS patients present with prominent autistic features, including severe language delay and moderate-to-severe intellectual disability (14,15). Heterozygous microdeletions and point mutations, including de novo missense and frame-shift mutations of SHANK3, are reported in a small subgroup of individuals with autism spectrum disorder (ASD) with comorbidity for intellectual disability (16–21). These and other reports suggest that haploinsufficiency of SHANK3 may play a key role in PMS and ASD (11,12,22,23). Interestingly, mutations in SHANK3 have been reported also in schizophrenia, suggesting that SHANK3 deficiency may contribute to both neurobehavioral disorders (20,21).
ASD is characterized by impairments in social behavior, communication and language development, as well as by repetitive and stereotyped behaviors (24–26); comorbidities in intellectual disabilities and movement disorders are common (27–29). A diagnosis of ASD in humans is purely based on behavioral assessment. Although the condition is diagnosed more often in males than in females, the basis for this sex difference is unknown (30,31). While a genetic component is strongly implicated in the etiology of ASD, the underlying molecular and neuropathophysiological basis of the condition is unknown in most ASD cases. Nevertheless, de novo missense mutations in exons 5–9 encoding the ANK domain of SHANK3 and microdeletions of the entire SHANK3 gene have been reported in ASD (16–19). To investigate the possible role of Shank3 in ASD, we generated mutant mice lacking exons 4–9 which encode the ANK domain, disrupting expression of the major isoforms of Shank3 (Shank3e4–9). These Shank3e4–9 homozygous mutants were subjected to biochemical, cellular, electrophysiological and behavioral analyses to elucidate the functional significance of these major Shank3 proteins.
RESULTS
Generation of Shank3 mutant mice
We generated Shank3 mutant mice using a conventional gene-targeting approach (32) where exons 4–9 of the Shank3 gene were deleted (Fig. 1A, B and C and Supplementary Material, Fig. S1A1 and A2). The naturally occurring Disc1 (Disrupted in Schizophrenia 1) mutation in the 129SvEv line of ES cells was segregated from Shank3e4–9 at the beginning of backcrossing to avoid any possible confounding effects in our experiments (Supplementary Material, Fig. S1A3) (33–35). All Shank3 mice used in the experiments had been backcrossed for more than seven generations onto a C57BL/6J background.
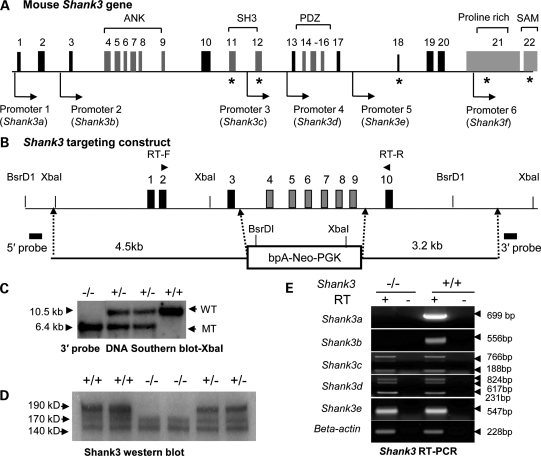
Figure 1.
Generation and characterization of Shank3e4–9 mice. (A) A diagram of the structure of the murine Shank3 gene showing promoters and alternative splicing sites (the asterisk indicates alternatively spliced exons). Five intragenic promoters were identified (see also Supplementary Material, Fig. S1B): ANK, ankyrin repeats; SH3, Src homology 3 domain; PDZ, postsynaptic density protein, Drosophila disk large tumor suppressor (DlgA) and Zonula occludens-1 protein (Zo-1) domain proline-rich domain; SAM, sterile α-motif domain. (B) Genomic map and structure of the Shank3 gene covering exons 1–10 and the targeting construct for the deletion of Shank3 exons 4–9. RT-F = forward primers and RT-R = reverse primer for RT-PCR analysis in Supplementary Material, Fig. S1A4. (C) Genotyping of Shank3e4–9 mice by Southern blot with XbaI and the 3′ flanking probe. The 10.5 kb fragment is the wild-type band; the 6.4 kb fragment is the mutant band. (D) Western blot analysis with Shank3e4–9 (–/–) brain samples; the 190, 170 and 140 kDa bands are seen in the Shank3+/+ (+/+) brain and only the 140 and 170 kDa bands are in Shank3e4–9 samples. (E) RT-PCR analysis for specific Shank3 transcripts in Shank3e4–9 mice. Shank3a and Shank3b transcripts were absent in Shank3e4–9 mice. Other transcripts (Shank3c–e) from promoters 3–5 were present in Shank3e4–9 mice.
Shank3 mRNA species and Shank3 isoform-specific deletion
As predicted, a 940 bp RT-PCR product was identified in Shank3 wild-type (Shank3+/+) mice, using primers anchoring exons 2 and 10 (Fig. 1B). In contrast, two smaller products (330 and 260 bp) were detected in Shank3e4–9 brains (Supplementary Material, Fig. S1A4). Sequencing confirmed that the 330 bp product represented a fragment joining exons 2, 3 and 10, whereas the 260 bp product represented cryptic splicing from exons 2 to 10 (Supplementary Material, Fig. S1A5). Both transcripts resulted in truncated proteins, with premature stop codons in exon 11. Interestingly, the 260 bp transcript appears to be brain-specific, since it is not found in Shank3e4–9 kidney (Supplementary Material, Fig. S1A4).
Western blot was performed with an antibody raised against amino acids 1431–1590 of the human SHANK3 protein (Q9BYB0) (Supplementary Material, Table S4). According to GenBank (AB231013), the full-length mouse Shank3 protein (i.e. isoform 3a) should be ∼190 kDa. A band of this size was found in Shank3+/+ but it was completely absent in Shank3e4–9 brain (Fig. 1D). Although the Shank3b protein (AJ245904, ∼77 kDa) was predicted to be disrupted in Shank3e4–9 mice, we were unable to confirm this, since exon 21 is spliced out of this isoform and our antibody only binds this portion of Shank3. However, we observed bands at ∼140 and ∼170 kDa in both genotypes with our antibody. These bands presumably represent different isoforms of Shank3, although we were not able to determine the corresponding mRNA transcripts.
The Shank3 gene has 22 exons and spans ∼58 kb of genomic DNA. Alternative splicing of the SHANK3 gene has been suggested (11,16,36) and we have identified five intragenic promoters as well as extensive alternative splicing in mouse Shank3 (Fig. 1A). These promoters and splice forms were confirmed for both humans and mice by 5′ RACE, RT-PCR, promoter–reporter assays and in silico analyses (Fig. 1E and Supplementary Material, Fig. S1). Sequences of these new transcripts are deposited in GenBank (HQ405757-HQ405766 and HO7636 62-HO763663) (structures are found in Supplementary Material, Fig. S1B1–B8). We have confirmed at least 11 different Shank3 mRNA species in the mouse brain. The total number of mRNA species produced by combinations of alternatively spliced exons and multiple promoters is expected to be sizable. Due to the limitations of our antibody, we were unable to determine whether proteins are produced from all the mRNA transcripts we have identified. Nonetheless, by sequencing RT-PCR products, we have confirmed that transcripts (Shank3a–b) initiated by promoters 1 and 2 are absent, whereas transcripts (Shank3c–e) from promoters 3–5 are present in the Shank3e4–9 brain (Fig. 1E). Transcript-specific expression from promoter 6 (Shank3f) could not be verified, because this promoter is embedded in exon 21, which is shared by other isoforms. Nevertheless, we conclude that Shank3e4–9 mice possess isoform-specific deletions in which the major Shank3a and Shank3b species, as well as other novel short transcripts (Shank3a1–2 and Shank3b1–2), are disrupted (Supplementary Material, Fig. S1B).
Social behavior is abnormal in Shank3e4–9 mice
Shank3e4–9 mice had normal Mendelian ratios at weaning, had no apparent developmental defects and were viable and fertile with a normal life span. The body weights of Shank3e4–9 mice at 8–12 months of age were slightly higher than those of Shank3+/+ animals (Supplementary Material, Fig. S5B), consistent with the mild overgrowth seen in some human PMS patients (14). However, the body weights of the mice at 3–4 months of age were not significantly different at the time of behavioral testing (see Supplementary Material, Behavioral studies). There were no differences for brain weights between adult Shank3e4–9 and Shank3+/+ mice at 2–4 months of age (Supplementary Material, Fig. S5C).
We conducted extensive behavioral analyses on adult Shank3e4–9 mice (3–6 months old and 4 cohorts of a total of 50 Shank3e4–9 and 50 wild-type littermates from 21 litters of heterozygous breeding); detailed statistical analyses for all behavioral results are provided in Supplementary Material. Since social behavior is dependent upon olfaction, this sense was evaluated in the Shank3 mice. No genotype differences were observed for olfactory preferences for urine from estrus females versus saline (Shank3+/+: 0.50 ± 0.02; Shank3e4–9: 0.41 ± 0.2) or for urine from male breeders versus saline (Shank3+/+: 0.51 ± 0.04; Shank3e4–9: 0.43 ± 0.4) or for total interaction times with the paired stimuli (Shank3+/+: 236 ± 4.2 and 235 ± 7.2 s; Shank3e4–9: 242 ± 4.3 and 228 ± 6.7 s, respectively). Hence, both genotypes are comparably able to discriminate urine from saline. In a sociability test, both social affiliation and social preference were examined. No stimulus preferences were observed for either genotype during the identical non-social 1–non-social 1 (NS1–NS1) pairing (Fig. 2A and B). In the NS1–social 1 (NS1–Soc1) pairing, Shank3+/+ controls showed higher affiliation for the novel social stimulus than Shank3e4–9 mice. Shank3e4–9 females interacted with the social stimulus more than Shank3e4–9 males. In the familiar Soc1–novel social 2 (Soc1–Soc2) pairing, both genotypes—regardless of sex—showed similar social preferences for the novel animal. The reduced social affiliation in the NS1–Soc1 pairing in Shank3e4–9 mice was not due to decreased interaction times, since both genotypes spent similar amounts of time investigating both stimuli, regardless of pairings (Supplementary Material, Table S1). Additionally, the numbers of contacts between NS1 and NS1 stimuli did not differ between genotypes (Supplementary Material, Fig. S2A and B). Although contacts declined for Shank3+/+ mice when social stimuli were present, habituation was not observed for Shank3e4–9 mice. Notably, Shank3e4–9 females made more NS1–Soc1 contacts than the Shank3+/+ controls. Together, these results indicate that when given a choice of interacting with a social or non-social stimulus, Shank3e4–9 mice fail to show social affiliation. However, when forced to choose between two social stimuli in the social preference test, Shank3 mutants demonstrate social reference by preferring the novel animal.
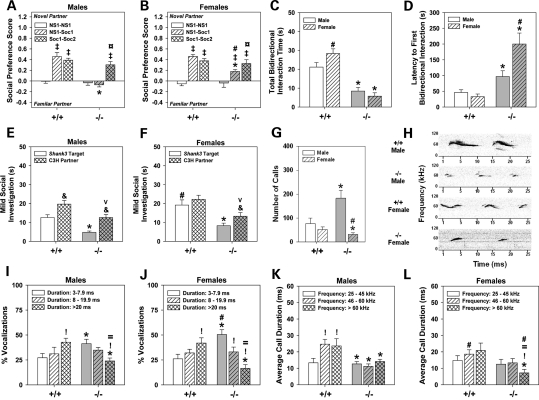
Figure 2.
Sociability and interaction as well as ultrasonic communications are abnormal in Shank3e4–9 mice. (A and B) No genotype or sex differences are evident for preferences between the identical NS objects. In the NS1–Soc1 pairing, Shank3+/+ (+/+) males and females prefer the novel Soc stimulus; Shank3e4–9 (–/–) males and females show no or reduced preferences for this stimulus. When presented with familiar and novel Soc stimuli, all mice prefer the novel social partner. (C) In the dyadic test, Shank3e4–9 mice participate in bidirectional social exchanges with C3H partners for shorter periods of time than Shank3+/+ controls and their partners. (D) Shank3e4–9 mice take longer to initiate their first social interaction than Shank3+/+ mice. (E and F) Both male (E) and female (F) Shank3e4–9 mice and their respective C3H partners show reduced times in social interactions than the respective Shank3+/+ targets and their C3H partners; n = 10 mice/genotype/sex. *P< 0.05, Shank3+/+ versus Shank3e4–9 mice; #P< 0.05, females versus males within genotype; ‡P< 0.05, compared with the NS1–NS1 test within genotype; ¤P< 0.05, compared with the NS1–Soc1 test within genotype; &P< 0.05, target versus partner within genotype; vP< 0.05, Shank3+/+ social partner versus Shank3e4–9 partner (see Supplementary Material, Fig. S2 and Table S1). (G) Shank3e4–9 males emit more and Shankee4–9 females produce fewer calls relative to Shank3+/+ controls. (H) Representative spectrographs of ultrasonic calls for male and female Shank3+/+ and Shank3e4–9 mice. (I and J) The calls emitted by Shank3+/+ mice are primarily of long duration, whereas those by Shank3e4–9 mice are mostly of short duration. (K and L) Shank3+/+ males produce more mid-range and high-frequency calls of longer duration than Shank3e4–9 males. Durations did not vary across frequencies for Shank3e4–9 mice; however, durations of high-frequency calls by Shank3e4–9 females were shorter than those by other groups; n = 10 mice/genotype/sex. *P< 0.05, Shank3+/+ versus Shank3e4–9 mice; #P< 0.05, females versus males within genotype; !P< 0.05, compared with short duration or low-frequency calls; =P< 0.05, compared with moderate duration or mid-range frequency calls.
Social behaviors were examined also in a dyadic test. Pre-test responses were not differentiated by genotype (Supplementary Material, Table S1). Shank3+/+ controls spent more time in bidirectional social interactions than Shank3e4–9 mice (Fig. 2C). Latency to the first bidirectional social interaction was shorter in Shank3+/+ than in Shank3e4–9 mice, and was most protracted in mutant females (Fig. 2D). None of the Shank3e4–9 mice initiated the first social interaction; all initiations were by C3H partners. By comparison, ∼50% of the social exchanges between Shank3+/+ targets and C3H partners were initiated by targets, regardless of sex.
A dyadic test was used to analyze a broad range of social responses. Shank3+/+ males spent less time investigating their partners than Shank3+/+ females (Fig. 2E and F). Nonetheless, investigation times by Shank3e4–9 mice were lower than Shank3+/+ mice and their C3H partners. Possibly due to this reduction, C3H partners of Shank3e4–9 mice engaged in fewer social investigations than pairings with Shank3+/+ controls. Although investigations were reduced, Shank3e4–9 mice spent more time self-grooming and sifting through bedding materials than Shank3+/+ mice (Supplementary Material, Fig. S2C and D); Shank3+/+ controls and their C3H partners rarely engaged in these behaviors. Collectively, these data show that Shank3e4–9 mice engage in fewer social interactions and are more likely to participate in non-social self-focused behaviors than Shank3+/+ mice. Additionally, social investigations are decreased in C3H partners to Shank3e4–9 mice, indicating that the partners are less likely to sustain interactions with non-socially responsive mice.
Ultrasonic vocalizations are aberrant in Shank3e4–9 mice
Ultrasonic communication was examined by exposing Shank3 mice to a non-familiar C3H mouse where no physical interaction could occur. Prior to mouse presentation, no ultrasonic calls were recorded. At the presentation of the C3H mice, the numbers of vocalizations by Shank3+/+ males and females were similar (Fig. 2G). However, Shank3e4–9 males made significantly more calls, and Shank3e4–9 females made significantly fewer calls, than Shank3+/+ mice. The vocal spectrograms revealed that Shank3+/+ mice produced more complex patterns of calls than Shank3e4–9 mice (Fig. 2H). Vocalizations for Shank3+/+ males were longer in duration, with more frequency bands and greater modulation of frequencies within each call, than Shank3e4–9 males. Shank3+/+ females made less complex calls than Shank3+/+ males, but they were longer in duration with more modulation of frequency bands than Shank3e4–9 females. Shank3e4–9 females produced calls as single ‘chirps’ that were devoid of the multiple frequency bands and complexities of Shank3+/+ controls. When vocalizations were analyzed as percent durations, Shank3+/+ mice made more long than short duration calls (Fig. 2I and J); the converse was observed for Shank3e4–9 mice—especially females. When the duration of calls was analyzed as a function of frequencies, Shank3+/+ males and females emitted mid-range and high-frequency vocalizations for longer periods of time than low-frequency calls (Fig. 2K and L). In contrast, the durations of calls by Shank3e4–9 males did not change across frequencies, whereas Shank3e4–9 females emitted high-frequency calls for shorter durations than all other vocalizations and all other mice. Thus, Shank3e4–9 mice emit aberrant ultrasonic vocalizations that are altered in both duration and frequency.
Motor behaviors are aberrant in Shank3e4–9 mice
In a neurophysiological screen, Shank3e4–9 mice displayed mild motor abnormalities as exemplified in tests of vertical placement and delayed climbing down a vertical pole for males and females and in contact righting for males (Supplementary Material, Table S2). In the foot-misplacement test, foot-faults were increased in Shank3e4–9 mice (Fig. 3A). Although the distance traveled in the apparatus was similar for Shank3e4–9 (160.6 ± 19.3 cm/5 min) and Shank3+/+ mice (171.1 ± 21.6 cm/5 min), the ratio of foot-misplacement errors to distance traveled was higher for mutants (0.23 ± 0.2) than for controls (0.13 ± 0.01). Shank3e4–9 mice moved more slowly than Shank3+/+ animals, and the Shank3e4–9 males were slowest of all (Fig. 3B). These data indicate that Shank3e4–9 mice have difficulties in fine motor coordination, and mutant males are more affected than mutant females.
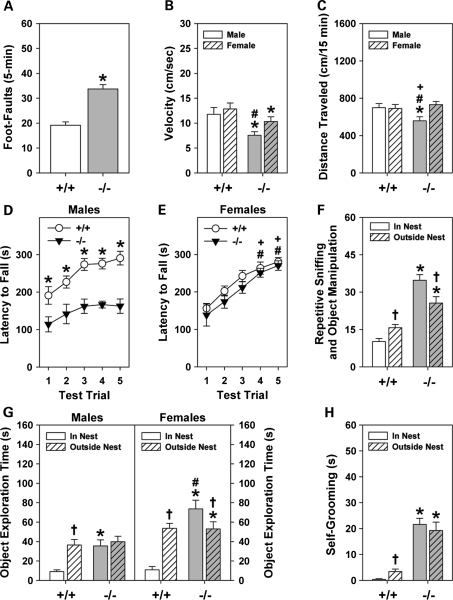
Figure 3.
Shank3e4–9 mice display abnormalities in motor performance. (A) Shank3e4–9(–/–) mice make more foot-misplacements than Shank3+/+ (+/+) controls. (B) Shank3e4–9 mice move more slowly than Shank3+/+ mice; Shank3e4–9 males are slower than mutant females. (C) Spontaneous locomotor activity in the open field is lower in Shank3e4–9 males than in other groups. (D and E) Motor learning on the accelerating rotorod is deficient in Shank3e4–9 males compared with Shank3+/+ males over all five trials and it is perturbed on trials 4 and 5 compared with Shank3+/+ and Shank3e4–9 females. (F) Repetitive frequencies of object explorations from inside and outside the nest were higher for Shank3e4–9 than Shank3+/+ mice. (G) Shank3+/+ males and females typically left the nest to explore the novel object. Shank3e4–9 males spent similar amounts of time exploring the objects from inside and outside the nest than Shank3+/+ males; Shank3e4–9 females spent the most time exploring objects from the nest. (H) Shank3e4–9 spent more time self-grooming than Shank3+/+ controls; n = 10 mice/genotype/sex. *P< 0.05, Shank3+/+ versus Shank3e4–9 mice; #P< 0.05, females versus males within genotype; +P< 0.05, Shank3+/+ females versus Shank3e4–9 males; †P< 0.05, in nest compared with out of the nest.
Mice were tested in an open field to determine whether their motor activities were aberrant. Locomotion was decreased in Shank3e4–9 males compared with all other groups (Fig. 3C). Additional indices, including time spent in movement, rearing activity and machine-scored stereotypical activities did not differ between genotypes (Supplementary Material, Table S2). Nevertheless, ethological scoring found that both male and female Shank3e4–9 mice participated in more self-grooming than Shank3+/+ controls. Together, these findings indicate that Shank3e4–9 males engage their environment less, and that grooming is more frequent in mutants than in Shank3+/+ controls.
Motor learning was evaluated with the accelerating rotorod (Fig. 3D and E). The latency to fall was increased across trials for Shank3+/+ males, indicating motor learning. In contrast, this latency was unchanged for Shank3e4–9 males and was abbreviated across test trials. No genotype differences were observed for females. Hence, Shank3e4–9 males appear impaired on motor learning, whereas females are unaffected.
Shank3e4–9 mice display repetitive behaviors
Repetitive responses were examined in a novel environment with a hole-board test and in a familiar environment with a novel object. With the hole-board, Shank3e4–9 mice engaged in more head pokes (129.4 ± 8.1) than Shank3+/+ controls (101.5 ± 6.6); no sex differences were seen. This enhancement in head pokes was not due to reduced anxiety, because genotype responses were similar in the light–dark emergence and zero maze tests (Supplementary Material, Table S2). Moreover, increased head pokes by mutants cannot be ascribed to novelty-seeking behaviors, because activity in the center of the open field, and stretch-attend postures or head-dipping behaviors in the zero maze were similar between genotypes.
In a home cage, novel object exploration was scored according to investigating the object from inside or outside the nest. Shank3+/+ controls contacted the object more often from outside than inside the nest (Fig. 3F). Conversely, Shank3e4–9 mice contacted the object more often from the nest than outside it, and contacts from both locations were higher than the Shank3+/+ mice. Maps were constructed to analyze the patterns of object interactions. Regardless of sex, Shank3e4–9 mice made repeated contacts, but only on certain locations on the object (Fig. 3G and Supplementary Material, Fig. S3). By comparison, object contacts by Shank3+/+ controls were not repetitive. When Shank3e4–9 mice were not investigating the object, they were engaged in self-grooming—a behavior that rarely occurred in Shank3+/+ controls (Fig. 3H). Thus, upon encountering a novel object in their familiar environment, Shank3e4–9 mice engage in stereotyped investigations and participate in self-stimulatory behavior.
Learning and memory are deficient in Shank3e4–9 mice
Cognitive performance was examined in Shank3 mice by prepulse inhibition, Morris water maze, novel object recognition and social transmission of food preference (STFP) tests. For prepulse inhibition, no genotype differences were observed for null activity, startle responses or inhibition to the three prepulses (Supplementary Material, Table S2). In the water maze, swim distance declined across acquisition testing for both genotypes; this reduction occurred less rapidly over the first 3 days for Shank3e4–9 mice (Fig. 4A, left). When the hidden platform was moved from the NE to the SW quadrant, swim distances initially increased for both genotypes on day 7 (Fig. 4A, right). Subsequently, it decreased for Shank3+/+ controls, but remained higher for Shank3e4–9 mice on days 8–10. The results for swim time were similar (Supplementary Material, Fig. S4A). Representative tracings of swim paths during acquisition supported the swim distance results (Supplementary Material, Fig. S4F, top left). Over reversal testing, tracings revealed that Shank3e4–9 mice swam in more circular search patterns (e.g. day 12) than the Shank3+/+ mice (Supplementary Material, Fig. S4F, top right).
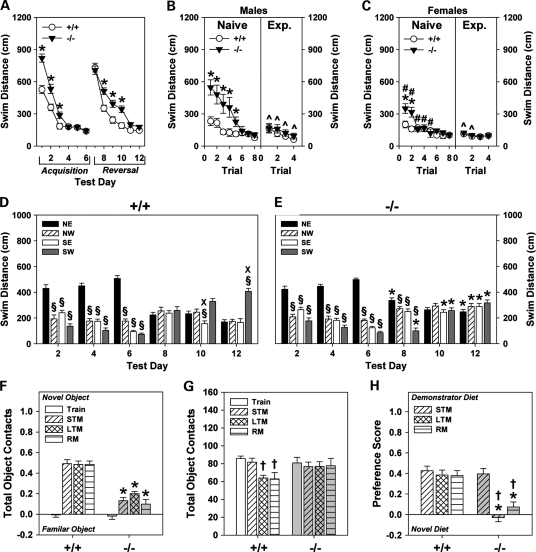
Figure 4.
Shank3e4–9 mice are deficient in learning and memory. (A) To reach the hidden platform in water maze, Shank3e4–9 mice swam over longer distances on the first 3 days of acquisition testing than Shank3+/+ mice. When the hidden platform was moved to a new location, swim distance was increased on days 9–11 for Shank3e4–9 mice. (B and C) Visible platform testing with naïve (left) and water-maze experienced (Exp.) (right) Shank3+/+ and Shank3e4–9 mice. Naïve Shank3e4–9 males (B, left) and females (C, left) swam over longer distances to reach the visible platform than respective Shank3+/+ controls. Swim distances for experienced Shank3e4–9 males (B, right) and females (C, right) were similar to those of the respective Shank3+/+ controls. (D and E) Swim distances for Shank3+/+ (D) and Shank3e4–9 (E) mice during acquisition (days 2, 4 and 6) and reversal probe tests (days 8, 10 and 12). During acquisition, all mice showed a marked preference for the NE quadrant. At reversal, Shank3+/+ mice, but not Shank3e4–9 mice, developed a significant preference for the SW quadrant. (F) Shank3 mice were evaluated in the novel object recognition test consisting of training (Train), and tests for STM, LTM and RM. During training, neither genotype showed any preference for either of the identical objects. Shank3+/+ mice preferred the novel object on the STM, LTM and RM tests; preferences were reduced for Shank3e4–9 mice. (G) Total numbers of object contacts were similar between genotypes. Object contacts declined for Shank3+/+ mice over LTM and RM testing; contacts remained high for Shank3e4–9 mice. (H) In the STFP test, no genotype differences were noted for STM. Shank3e4–9 mice showed a dramatically reduced preference for the demonstrator diet during the LTM and RM tests. For all tests, n = 10 mice/genotype/sex. *P< 0.05, Shank3+/+ versus Shank3e4–9 mice; #P< 0.05, females versus males within genotype; ^P< 0.05, naïve Shank3e4–9 mice versus experienced Shank3e4–9 mice; §P< 0.05, versus NE quadrant; XP< 0.05, versus SW quadrant; †P< 0.05, compared with the STM test.
Probe tests for acquisition testing (days 2, 4, 6) failed to detect significant genotype differences in swim distance (Fig. 4D and E); all mice swam further in the NE than in the other three quadrants. When the hidden platform was moved to the SW quadrant, neither genotype displayed any quadrant preference on day 8. However, by days 10 and 12, Shank3+/+ mice swam further in the SW than the other quadrants. In contrast, Shank3e4–9 mice failed to show any quadrant preference. An examination of swim times revealed similar results (Supplementary Material, Fig. S4D and E). Although tracings of the swim paths during acquisition probe tests were similar between genotypes (Supplementary Material, Fig. S4F, bottom), the wide and circling patterns of Shank3e4–9 mice became more prominent by the last day of reversal. Together, these results suggest that spatial memory and plasticity may be deficient in Shank3e4–9 mice. The impaired performance in reversal training may also indicate inflexibility of behaviors in Shank3e4–9 mice.
Over the first five trials of the visible platform test, naïve Shank3e4–9 males swam further to reach the platform than naïve Shank3+/+ males; naïve mutant females swam further on the first two trials than naïve female controls (Fig. 4B and C, left). Notably, naïve Shank3e4–9 females swam over shorter distances on the first six trials than naïve Shank3e4–9 males. To determine whether the protracted swim times of naïve Shank3e4–9 mice could be attributed to delayed learning or to performance issues, naïve and experienced mice were compared. Although swim distances for naïve and experienced Shank3+/+ controls did not differ (Fig. 4B and C, right), experienced Shank3e4–9 males and females swam over shorter distances than respective naïve mutants. Analyses of swim time revealed similar results (Supplementary Material, Fig. S4B and C). Hence, deficiencies of naïve Shank3e4–9 mice appear to be due to test familiarization or delayed learning, rather than specific sensory, motor or motivational abnormalities.
Episodic short-term (STM), long-term (LTM) and remote memory (RM) were analyzed using a novel object recognition test. Preferences for the novel object by Shank3+/+ mice were high and unchanged across the STM, LTM and RM tests (Fig. 4F). In contrast, novel object preference was decreased for Shank3e4–9 mice. Since poor performance could be attributed to object neophobia, the duration of object contacts and the numbers of object contacts were examined. The duration of object contacts was similar for both genotypes (Supplementary Material, Table S3). Shank3+/+ and Shank3e4–9 mice also contacted the objects similar numbers of times during training and the STM test (Fig. 4G). However, Shank3+/+ mice made fewer object contacts than mutants during the LTM and RM tests, and these contact numbers were less than those during training and the STM test. In contrast, object contacts were not reduced across tests for Shank3e4–9 animals.
In the STFP test, Shank3+/+ controls maintained a strong preference for the demonstrator diet during the STM, LTM and RM tests, with little change across test-days (Fig. 4H). However, this preference was lost by Shank3e4–9 mice during the LTM and RM tests. Despite impairments in LTM and RM in mutants, no genotype differences were discerned for time spent interacting with the demonstrator mouse, the total amount of diets eaten or times investigating the diets on each test-day (Supplementary Material, Table S3). Collectively, the object recognition and STFP results indicate that at least LTM and RM are impaired in Shank3e4–9 mice.
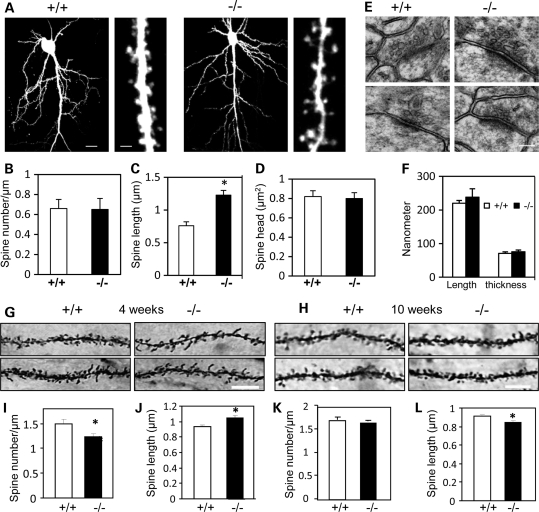
Dendritic spine morphology is altered in Shank3e4–9 mice
Brain anatomy of Shank3e4–9 mice at 2–4 months of age appeared normal by gross, histological and ultrastructural analyses with light and electron microscopy (Fig. 5E and F and Supplementary Material, Fig. S5A). To examine dendritic spine morphology, hippocampal slice cultures were prepared from pups 6–7 days old and neurons were transfected biolistically with a GFP plasmid (Supplementary Material, Fig. S5D). Although no genotype differences were detected in dendritic branching (Fig. 5A) or in spine densities and spine-head sizes (Fig. 5B and D), dendritic spines were significantly longer in Shank3e4–9 (1.23 ± 0.07 μm, 678 spines from 7 mice) than Shank3+/+ mice (0.76 ± 0.06 μm, 1183 spines from 8 mice; P< 0.002) (Fig. 5C). Interestingly, overall, Shank3e4–9 mice had more thin spines than Shank3+/+ controls; however, this difference did not achieve statistical significance (Shank3+/+ mice: stubby 151, mushroom 283 and thin spines 691; Shank3e4–9 mice: stubby 77, mushroom 172 and thin spine 432, P> 0.05). The ratio of stubby- and mushroom-shaped spines to thin spines was borderline lower in Shank3e4–9 mice [Shank3+/+ mice: (151 + 283)/691 = 0.63 ± 0.06; Shank3e4–9 mice: (77 + 172)/432 = 0.58 ± 0.07), P= 0.052]. We used Golgi impregnation to examine dendritic spine morphology in CA1 hippocampus of 4- and 10-week-old Shank3e4–9 mice (Fig. 5G and H). Spine density in the CA1 hippocampus was reduced and spine length was significantly increased in 4-week-old Shank3e4–9 mice compared with Shank3+/+ mice (spine density: +/+, 1.49 ± 0.09/μm; –/–, 1.24 ± 0.07/μm, P= 0.03; spine length: +/+, 0.94 ± 0.02 μm; –/–, 1.05 ± 0.03 μm, P< 0.003) (Fig. 5I and J). Cumulative frequencies for spine length are shown in Supplementary Material, Fig. S5E and F. In contrast, spine density was not significantly reduced but spine length was increased in 10-week-old Shank3e4–9 compared with Shank3+/+ mice (spine density: +/+, 1.70 ± 0.04/μm; –/–, 1.63 ± 0.03/μm, P> 0.05; spine length: +/+, 0.91 ± 0.02 μm; –/–, 0.85 ± 0.01 μm, P= 0.01) (Fig. 5K and L). No gender difference was noted. The reduced density and increased spine length in 4-week-old Shank3e4–9 mice are consistent with the observations of siRNA-mediated knockdown of Shank3 in cultured neurons (9). The discrepancy for the spine density between cultured neurons of newborns and Golgi impregnation of 4-week-old mice and between 4- and 10-week-old mice may be due to differences in the sensitivity of the methods—although it could also reflect developmental differences.
Figure 5.
Dendritic spine morphology was altered in Shank3e4–9 mice. (A) Low (left) and high (right) resolution images of EGFP-expressing pyramidal neurons from Shank3+/+ (+/+) and Shank3e4–9 (–/–) mice. Quantification of spine density (B), spine length (C) and spine-head area (D). A significant difference was revealed in spine length but not for spine density or spine-head area (spine length: 1.23 ± 0.07 μm, 678 spines from 7 Shank3e4–9 mice; and 0.76 ± 0.06 μm, 1183 spines from 8 Shank3+/+ mice; *P< 0.002, Shank3+/+ versus Shank3e4–9 mice). (E and F) Representative electron micrographs of hippocampal CA1 striatum radiatum synapses from Shank3+/+and Shank3e4–9 mice. The PSD is visible as an electron-dense layer adjacent to the post-SPM (scale bar = 0.1 μm). A total of 71 synapses from 3 Shank3+/+ mice and 69 synapses from 3 Shank3e4–9 mice were measured. There were no significant genotype changes in PSD length or thickness. (G and H) Representative images from Golgi staining of 4-week (G) and 10-week-old (H) Shank3+/+ and Shank3e4–9 mice. Scale bar = 5 μm. (I and J) Spine density (I) and spine length (J) in 4-week-old Shank3e4–9 mice (n = 15 cells from 3 +/+ mice and n = 14 cells from 3 –/– mice). A total of 731 spines from Shank3+/+ and 649 spines from Shank3e4–9 mice were measured (*P= 0.03 for spine density and P< 0.003 for spine length). (K and L) Spine density (K) and spine length (L) in 10-week-old Shank3e4–9 mice (n = 40 cells from 3 +/+ mice; n = 31 cells from 3 –/– mice). A total of 992 spines from Shank3+/+ and 707 spines from Shank3e4–9 mice were measured (*P= 0.01 for spine length).
Altered PSD proteins in Shank3e4–9 mice
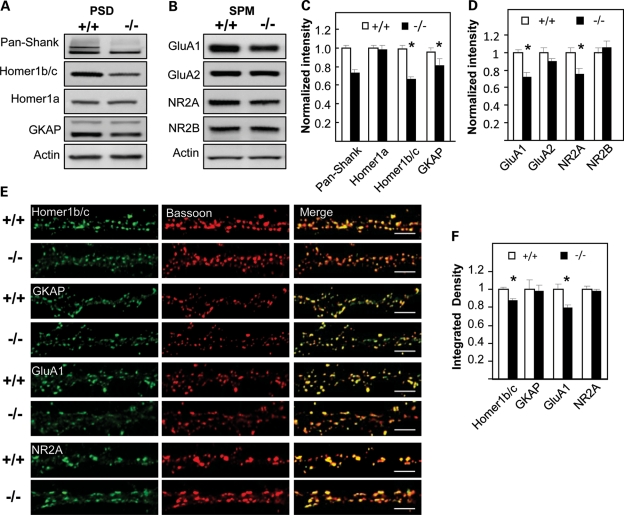
To examine whether the composition of PSD proteins was altered in Shank3e4–9 animals, we performed quantitative immunoblot analyses for candidate Shank3-interacting proteins (Supplementary Material, Table S4). No genotype differences were detected in the levels of cytosolic proteins extracted from brain (Supplementary Material, Fig. S6A and B). In contrast, when levels of synaptic proteins from the PSD-I fraction were examined, GKAP and Homer1b/c were reduced in Shank3e4–9 compared with Shank3+/+ control PSD fractions (85.2 ± 6.1% of Shank3+/+ for GKAP, P= 0.03, n = 11 mice/genotype; 66.1 ± 4.2% of Shank3+/+ for Homer1b/c, P< 0.001, n = 16 mice/genotype); no genotype distinctions were observed for other synaptic proteins at the PSD (Fig. 6A and C and Supplementary Material, Fig. S6C and D). No significant sex differences were noted. The AMPAR subunit GluA1 and the NMDAR subunit NR2A were significantly reduced in membrane (SPM) fractions from Shank3e4–9 mice (72.0 ± 6.2% of Shank3+/+ for GluA1, P< 0.002, n = 8 mice/genotype; 75.0 ± 8.3% of Shank3+/+ for NR2A, P= 0.02, n = 8 mice/genotype). Levels of other receptor subunits, including the AMPAR GluA2 and the NMDAR NR2B subunits, were not significantly altered in SPM fractions from Shank3e4–9 mice (Fig. 6B and D). No sex differences were observed. Reduced levels of GluA1 and Homer1b/c were observed in Shank3e4–9 heterozygotes compared with Shank3+/+ controls, but the differences were more subtle than between the homozygous mutant Shank3e4–9 and Shank3+/+ controls. In addition, we also detected an interaction between Shank3 and Homer1b/c by co-immunoprecipitation (Supplementary Material, Fig. S6E and F).
Figure 6.
Altered protein composition in PSD and SPM fractions of Shank3e4–9 mice. Immunoblot analyses of PSD (A) and SPM (B) fractions from individual Shank3+/+ and Shank3e4–9 mice for the indicated proteins. Pan-Shank (Shank1–3) antisera revealed an absence of the Shank3 band in Shank3e4–9 mice. Homer1b/c and GKAP in the PSD and GluA1 and NR2A in the SPM are reduced in Shank3e4–9 mice. (C and D) Quantification of proteins in PSD fractions based on results shown in (A) and (B), respectively, normalized using actin for protein quantification. There is a significant reduction in the levels of the indicated proteins in Shank3e4–9 compared with Shank3+/+ samples. Homer1b/c (n = 16 each/genotype, *P< 0.001), GKAP (n = 11 each/genotype,*P= 0.03), GluA1 (n = 8 each/genotype,*P< 0.002) and NR2A (n = 8 each/genotype, *P= 0.02). No sex difference was observed. There is no significant differences for other proteins including Homer1a (n = 11 each/genotype), GluA2 (n = 8 each/genotype) and NR2B (n = 8 each/genotype). (E) Hippocampal neurons in dissociated culture (16–18DIV) from Shank3+/+ or Shank3e4–9 mice (n = 8 each/genotype) were immunostained with bassoon antibody (red) and with GKAP, Homer1b/c, GluA1(C-terminus) or NR2A antibodies (green). Merged images shown on the right. Scale bar = 5 µm. (F) Normalized integrated density for the indicated proteins (Shank3+/+= 1.00). Significant differences were found for Homer1b/c and GluA1 proteins between Shank3+/+ and Shank3e4–9 samples (*P< 0.01 for Homer1b/c and *P< 0.001 for GluA1). No significant differences were found for GKAP and NR2A.
The distribution of synaptic PSD-I proteins was further analyzed by immunostaining of hippocampal neuronal cultures from Shank34–9 mice. Consistent with the quantitative immunoblots, immunostaining of Homer1b/c and GluA1 were reduced in Shank3e4–9 mice (Fig. 6E)—as confirmed by quantitative analyses of Homer1b/c and GluA1 puncta (87.2 ± 2.1 % of Shank3+/+ for Homer1b/c, P< 0.01, n = 17 cells/genotype; 80.3 ± 3.4% of Shank3+/+ for GluA1, P< 0.001, n = 27 cells/genotype) (Fig. 6F). The puncta sizes of immunostaining for Homer1b/c and GluA1 were also reduced in Shank3e4–9 mice (Supplementary Material, Fig. S5G). Although GKAP (n = 8/genotype) and NR2A (n= 8/genotype) immunostaining were decreased in Shank3e4–9 mice, this difference was not statistically significant, perhaps reflecting the differing sensitivities of these methods, or differences in synaptic versus non-synaptic pools of these molecules. Together, these findings indicate that specific synaptic scaffolding proteins and receptor subunits are altered by the disruption of major Shank3 isoforms.
Synaptic plasticity is impaired in Shank3e4–9 mice
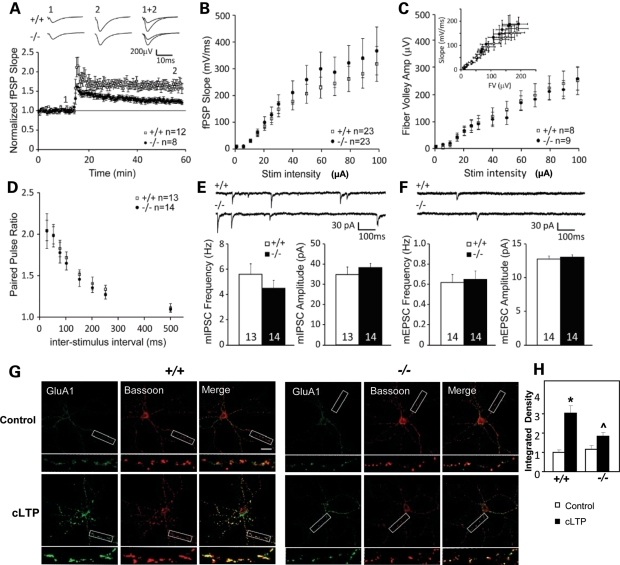
The reductions in GluA1 subunits and other PSD proteins in Shank3e4–9 mice suggest a possible role for Shank3 in synaptic function. To examine this possibility, we analyzed excitatory synaptic transmission and plasticity at the CA1 Shaffer-collateral synapses in 2–4-month-old Shank3e4–9 mice, using field post-synaptic potential (fPSP) recordings. Shank3e4–9 mice showed reduced post-tetanic potentiation compared with Shank3+/+ animals (Shank3+/+ mice: 195 ± 22% baseline fPSP slope, n = 12; Shank3e4–9 mice: 150 ± 10% baseline fPSP slope, n = 8; P< 0.001) (Fig. 7A). Moreover, Shank3e4–9 mice exhibited reduced hippocampal long-term potentiation (LTP) (Shank3+/+ mice: 165 ± 9% baseline fPSP slope, n = 12; Shank3e4–9 mice: 127 ± 6% baseline fPSP slope, n = 8; P< 0.001) (Fig. 7A). No sex differences were observed. Despite the plasticity defect, there were no apparent differences in basal synaptic transmission between Shank3+/+ and Shank3e4–9 mice. The input–output (I/O) curves (Shank3+/+ mice: n = 23; Shank3e4–9 mice: n = 23; P> 0.05) (Fig. 7B), the magnitude of the evoked fiber volley (Shank3+/+ mice: n = 8; Shank3e4–9 mice: n = 9; P> 0.05) (Fig. 7C) and the fiber volley versus fPSP slope (Shank3+/+ mice: n = 8; Shank3e4–9 mice: n = 9; P> 0.05) (Fig. 7C, insert) measurements were similar between genotypes. Finally, the paired-pulse ratio of the synaptically evoked fPSPs was also comparable (Shank3+/+ mice: n = 13; Shank3e4–9 mice: n = 14; P> 0.05) (Fig. 7D). These findings suggest that, when not challenged by high-frequency stimulation, the probability of neurotransmitter release at these synapses is equivalent between adult Shank3+/+ and Shank3e4–9 mice. However, we cannot exclude the possibility that there may be more significant synaptic defects earlier in development that are overcome through age or homeostatic mechanisms.
Figure 7.
Impaired synaptic plasticity and activity-dependent AMPAR GluA1 distribution in Shank3e4–9 mice. (A) Summary graph of LTP experiments and representative fPSP traces in Shank3+/+ (+/+) and Shank3e4–9 (–/–) mice (+/+, n = 12 slices from 8 mice; –/–, n = 8 slices from 8 mice). Tetanic stimulation (100 Hz, 1 s × 2) was applied at 15 min. LTP was significantly impaired in Shank3e4–9 mice. (B) Input–output relationships of fPSPs of Shank3+/+ (+/+, n = 23 slices from 8 mice) and Shank3e4–9 mice (–/–, n = 23 slices from 8 mice) were not significantly different. (C) Summary graph of the fiber volley (FV) amplitude during the input–output test of Shank3+/+ (+/+, n = 8 slices from 8 mice) and Shank3e4–9 mice (–/–, n = 9 slices from 8 mice). Insert depicts FV versus fPSP relationship. (D) Paired-pulse ratio at different inter-stimulus intervals in Shank3+/+ (+/+, n = 13 slices from 8 mice) and Shank3e4–9 mice (–/–, n = 14 slices from 8 mice). (E) Sample traces and summary graphs depicting mIPSC amplitude and frequency in Shank3+/+ (n = 13 cells from 5 mice) and Shank3e4–9 (n = 14 cells from 5 mice) animals. No significant differences were observed between genotypes. (F) Sample traces and summary graphs depicting mEPSC amplitude and frequency in Shank3+/+ (n = 14 cells from 5 mice) and Shank3e4–9 (n = 14 cells from 5 mice) animals. No significant differences were observed between genotypes. (G) Attenuated response of activity-dependent distribution of surface GluA1. Hippocampal neurons in dissociated culture (16–18DIV) from Shank3+/+ and Shank3e4–9 mice were treated with a chemical LTP protocol. Neurons were sequentially stained for surface GluA1 (N-terminus) (green) and bassoon (red) under non-permeant and permeant conditions, respectively. A significant increase of surface GluA1 staining after cLTP was seen in Shank3+/+ neurons; this increase was much attenuated in Shank3e4–9 neurons. (H) The normalized integrated density of surface GluA1. Staining intensity was significantly increased after cLTP in cultured neurons from Shank3+/+, but not from Shank3e4–9 mice. Shank3+/+, n = 17 cells; Shank3e4–9, n = 12 cells. *P <0.001, Shank3+/+ versus Shank3e4–9 mice. ^P< 0.01, cLTP-stimulated Shank3+/+ versus Shank3e4–9 mice.
To further assess possible differences in neurotransmitter release probability or postsynaptic strength, we analyzed miniature excitatory post-synaptic currents (mEPSCs) and miniature inhibitory post-synaptic currents (mIPSCs), using voltage-clamp recordings (Fig. 7E and F). Average mEPSC amplitude (Shank3+/+ mice: 12.7 ± 0.5pA, n = 14; Shank3e4–9 mice: 13.0 ± 0.4pA, n = 14; P> 0.05) and frequency (Shank3+/+ mice: 0.62 ± 0.09 Hz, n = 14; Shank3e4–9 mice: 0.65 ± 0.09 Hz, n = 14; P> 0.05) were similar between Shank3e4–9 and Shank3+/+ mice, confirming that normal basal excitatory synaptic transmission can develop in the absence of expression of major Shank3 isoforms. Inhibitory synaptic transmission, as measured by mIPSC frequency (Shank3+/+ mice: 3.0 ± 0.8 Hz, n = 13; Shank3e4–9 mice: 4.5 ± 0.7 Hz, n = 14; P> 0.05) and amplitude (Shank3+/+ mice: 34.9 ± 3.7pA, n = 13; Shank3e4–9 mice: 38.3 ± 2.2pA, n = 14; P> 0.05), was also similar between genotypes. Together, these results indicate that, despite normal basal synaptic transmission, Shank3 deletion impairs hippocampal CA1 synaptic plasticity.
Impaired activity-dependent GluA1 redistribution in Shank3e4–9 neurons
The reduced GluA1 levels suggest that Shank3 deficiency may alter activity-dependent AMPAR dynamics at the synapse, which might underlie the impaired hippocampal LTP. To address this point, we applied a chemical LTP (cLTP) protocol to cultured neurons. cLTP was associated with a significant increase in surface GluA1 subunits in Shank3+/+ mice (Fig. 7G); this response was significantly attenuated in Shank3e4–9 mice. Consistent with this result, after surface GluA1 immunostaining, the staining intensity of puncta increased in an activity-dependent manner in Shank3+/+ (n = 17, control: 0.99 ± 0.13; cLTP: 3.02 ± 0.41), but not in Shank3e4–9 cells (n = 12, control: 1.16 ± 0.19; cLTP: 1.85 ± 0.16. P< 0.001 for Shank3+/+ versus Shank3e4–9 in control cells and P< 0.01 for Shank3+/+ versus Shank3e4–9 in cLTP stimulated cells) (Fig. 7H). Likewise, the size of stained puncta after surface GluA1 immunostaining was also decreased in Shank3e4–9 mice (Supplementary Material, Fig. S5H). These observations suggest that impaired activity-dependent trafficking of GluA1 subunits in Shank3e4–9 neurons may be a cellular mechanism underlying deficient hippocampal LTP in Shank3e4–9 mice.
DISCUSSION
SHANK3 mutations in PMS and ASD
SHANK3 maps to the critical region of chromosome 22q13.3 deletion syndrome or PMS and this gene is thought to play a key role in the ASD and cognitive dysfunction in this disorder. because many cases have small deletions only disrupting SHANK3 (10–13,22,37). Other genes in the region may also contribute to the condition (23). Microdeletions and de novo missense and frame-shift mutations in SHANK3 have been reported in a small subgroup of ASD cases; these individuals are often comorbid with intellectual disability (16–21). ASD patients with missense and frame-shift mutations in SHANK3 may have loss of some but not all SHANK3 isoforms. At this point, it is unclear which SHANK3 point mutations in ASD patients behave as loss- or gain-of-function in vivo. The Shank3 mutation in our mice includes exons 4–9, resulting in the loss of Shank3a and Shank3b, the two major Shank3 mRNA transcripts in the brain. ASD patients with heterozygous missense mutations in exons 5–9 of SHANK3 have been identified (16,18,19). We have conducted our studies in Shank3 homozygous mutants to identify functional roles of Shank3 in vivo. Our findings support an important role for SHANK3 in PMS- and ASD-like responses.
We found that the combination of five intragenic promoters and alternatively spliced exons results in a complex pattern of mRNA species both in mice and humans (data not shown for human), and these species presumably produce a sizable number of Shank3 protein isoforms with unique protein domain structures, including some that are neuron- or astrocyte-specific (38). Each domain mediates specific protein–protein interactions at synapses, raising the possibility that each Shank3 isoform may play a distinct role in synaptic function and may make distinct contributions to the disease processes of ASD. The deletion of exons 4–9 physically removed the ANK domain but it also disrupted other domains encoded by the Shank3a and Shank3b isoforms because of a frame-shift change. Our data indicate an isoform-specific role of Shank3 in synaptic function. Since SHANK3 mutations in ASD appear isoform-specific, we speculate that different SHANK3 isoforms may lead to different clinical presentations.
Shank3e4–9 mice provide a model for ASD
In humans, the diagnosis of ASD is usually based upon behavioral assessment. ASD patients are typically impaired in social and communication skills and engage in repetitive and stereotyped behaviors (24–26). Cognitive disabilities and movement disorders are common comorbidities in ASD (27–29). Interestingly, de novo missense mutations in exons 5–9 and microdeletions of the entire SHANK3 gene in heterozygotes have been reported in individuals who meet the diagnosis for ASD (17–19). In ASD cases with microdeletions, haploinsufficiency of SHANK3 is apparent, but whether point mutations of SHANK3 behave as loss- or gain-of-functions in vivo is not known. Since our Shank3e4–9 homozygous mutants present with multiple behavioral abnormalities that are reminiscent of key features ASD may support a conclusion of loss-of-function as a mutational mechanism in these cases.
ASD children often fail to develop normal social interactions. Similarly, autistic-like mice fail to demonstrate preference for social novelty (39). In our sociability test, Shank3e4–9 mice showed context-specific sociability. When familiar non-social and novel social stimuli were paired, Shank3+/+ controls showed affiliation for the latter, whereas Shank3e4–9 mutants demonstrated little or no social affiliation. By comparison, when confronted with a novel and a familiar social stimulus, both genotypes showed social preference for the novel animal. These findings indicate that Shank3e4–9 animals can develop a social reference when two social stimuli are paired, but choose not to socially interact given a choice between non-social and social stimuli. This failure to interact is observed also in the dyadic social test, where Shank3e4–9 mice fail to reciprocate social exchanges and engage in non-social self-directed behaviors like self-grooming and sifting. This decrease in sustained social exchanges probably contributes to the reduced social investigations by their C3H partners. Interestingly, a decay in reciprocal exchanges is proposed to be a hallmark of autistic-like responses in mice (39).
Vocal communication is perturbed in Shank3e4–9 mice; ultrasonic calls in Shank3e4–9 mice are abbreviated and lack the complexity of frequency modulation of the Shank3+/+ controls. The increased numbers of calls by Shank3e4–9 males and reduced complexity by both mutant males and females are reminiscent of the ‘echolalia’ and ‘monotone speech’ observed in ASD (27,40). In fact, many of the calls by Shank3e4–9 mice are characterized by single notes or short non-complex whistles of single elements.
Shank3e4–9 mice engage in more stereotyped and repetitive behaviors than Shank3+/+ controls. Self-grooming by mutants is evident during social interactions with an unfamiliar C3H mouse in the dyadic test, and in the home cage in the presence of a novel object. Grooming, however, does not appear to be due to anxiety-like behaviors (41,42), since Shank3e4–9 animals do not display these responses in the light–dark box or zero maze. Enhanced repetitive behaviors are seen also in the hole-board and in the home cage novel object tests. These responses suggest that the behavioral repertoire of Shank3e4–9 mice may be more restricted and less malleable than that of the Shank3+/+ controls. Indeed, decreased plasticity of behavior is evident during reversal testing in the Morris water maze, where Shank3e4–9 mice are unable to learn the new location of the hidden platform. Interestingly, ASD is also associated with enhanced stereotyped responses and impairments in novelty processing, with reductions in the ability to modify ongoing behavior in response to changing environmental conditions (43,44).
The Shank3e4–9 mutants also demonstrate abnormalities in motor performance and learning and memory–comorbidities often accompanying ASD and PMS in humans (13,27–29). The motor deficiencies in the gait and foot-misplacement tests are evident in both sexes, whereas performance on the rotorod is impaired only in Shank3e4–9 males. Spatial memory is slightly perturbed in the Shank3e4–9 mice; acquisition was delayed, although accompanying probe test responses were similar to those of the Shank3+/+ controls. However, mutants failed the task when the hidden platform was moved to a new location, suggesting that behavioral plasticity is restricted. In addition, long-term and remote episodic and social memories are aberrant in Shank3e4–9 mice. These deficiencies in hippocampally based behaviors are concordant with the impaired hippocampal LTP observed in the Shank3 mutant mice (45,46).
It should be emphasized that Shank3e4–9 males display a more severe phenotype than females in motor performance despite the absence of significant sex differences in the biochemical, morphological and electrophysiological studies. In this context, it is noteworthy that human males are four to six times more likely than females to receive a diagnosis of ASD (27,30,31), though possible sex differences have not been analyzed in ASD patients with SHANK3 mutations.
Collectively, the abnormal social behaviors, communication patterns and repetitive behaviors support the Shank3 mutant mouse as a valuable model for further investigations of the neurophysiological basis of ASD. This mouse also provides an important basis to further investigate behavioral profiles in Shank3 heterozygous mutant mice that may have greater relevance for PMS and ASD, which are associated with SHANK3 mutations.
Shank3, AMPAR trafficking and synaptic plasticity
Homer1b/c and GKAP proteins are decreased in the PSD of Shank3e4–9 mice, suggesting that Shank3 may recruit or prevent the removal of these proteins at synapses (1,2,47), thus perturbing postsynaptic signal transduction. In vitro, the ANK domain of Shank3 is known to mediate the interaction between synaptic glutamate receptor complexes and cytoskeletal proteins at the PSD (7,48). Direct interactions between Shank3 and GluA1, GKAP and Homer1b/c have been reported (1,2,8). In Shank3e4–9 mutant mice, all domains in two major Shank3 protein isoforms were disrupted. We find here that activity-dependent redistribution of the GluA1 of the AMPAR is attenuated, LTP in CA1 hippocampus is impaired and spine morphology is subtly altered in Shank3e4–9 mice. We speculate that the deficits in activity-dependent trafficking of GluA1 in Shank3-deficient neurons may be caused by the disrupted interactions between Shank3 and GKAP and Homer1b/c, perhaps leading to impaired exocytic or endocytic recycling of AMPAR trafficking. As expected with deficiencies in GluA1 trafficking and hippocampal LTP, Shank3e4–9 animals are impaired on hippocampus-based behavioral tasks, including the Morris water maze, novel object recognition and STFP. Human patients with deletions or point mutations in SHANK3, who present with moderate-to-severe intellectual disability, may harbor many of the same biochemical and cellular deficiencies found in our mutants.
Shank family proteins and neuropsychiatric disorders
Recent genetic studies indicate that SHANK family proteins play an important role in other neurodevelopmental and neuropsychiatric disorders. For instance, point mutations or microdeletions of SHANK2 occur in cases of ASD and intellectual disability (49,50), whereas mutations in SHANK3 have been identified in individuals with schizophrenia (20,21). Although Shank proteins share similar domain structures, their expression patterns and subcellular localizations within the PSD are different (36,51). Similar to our Shank3e4–9 animals, Shank1–/– mice have reduced levels of Homer1b/c and GKAP proteins, whereas GluA1 and NR2A subunit levels are normal (52). Maturation of dendritic spines is perturbed also in Shank1–/– mice. Nevertheless, basal synaptic transmission is weak, whereas hippocampal CA1 synaptic LTP is intact. Additionally, spatial memory and social behavior are not impaired in Shank1–/– mice (52,53). These biochemical, cellular, morphological and behavioral distinctions indicate that mutations in these Shank family protein genes can produce very different phenotypes. Moreover, since mutations within SHANK3 itself are associated with ASD, intellectual disability and schizophrenia, it remains to be determined how these different mutations produce these varied disorders. Combinations of human genetic studies with murine-targeted exon deletions of Shank family members may provide new insights into basic mechanisms that underlie these and other neuropsychiatric conditions.
MATERIALS AND METHODS
Generation of Shank3 mutant mice, mouse breeding, animal care
The standard protocol for gene targeting in embryonic stem cells and production of knockout mice was previously described (32) (see Supplementary Material, Methods, for details). All experiments were conducted with approved protocols from the Institutional Animal Care and Use Committee at Duke University.
Neuronal culture, cLTP and immunocytochemistry
Primary hippocampal cultures were prepared from brains of newborn mouse pups at postnatal day 0 (P0) to P1 as described previously (54,55) with some modifications. The protocols for cLTP and immunocytochemistry are found in Supplementary Material, Methods.
Spine morphology in hippocampal slice culture and two-photon laser scanning microscopy
Organotypic hippocampal slices were prepared from mouse pups postnatal 6–7 days of age and spines were imaged as described (55) (see Supplementary Material, Methods, for details).
Hippocampal slice preparation and electrophysiology
Hippocampal slices were prepared from 2–4-month-old mice. The recording protocol has been described (56,57) and is included in Supplementary Material, Methods.
Behavioral testing
Shank3 mutant mice and their wild-type littermates were at 3–4 months of age at the time of testing. The estrous cycle in females was not controlled. Descriptions of the many of the behavioral methods have been published (42,58–60); more detailed descriptions of all the methods can be found in Supplementary Material, Methods.
Statistics
All data were analyzed with SPSS 11 (SPSS, Inc., Chicago, IL, USA) and expressed as means ± SEM. Descriptions of the statistical methods can be found in Supplementary Material, Methods. Additionally, all statistical behavioral results can be found in Supplementary Material, Results.
SUPPLEMENTARY MATERIAL
NOTE ADDED IN PROOF
The authors wish to acknowledge that while this paper was under the review and production stages, three papers of different Shank3 mutations in mice have been published online (PMID: 21167025, 21423165, and 21565394).
FUNDING
Some of the experiments were conducted with equipment/software purchased with a grant from the North Carolina Biotechnology Center. Y.-H.J. is a scholar of National Institutes of Health grant (5K12-HD0043494–08) and is also supported by grants from Autism Speaks and the Angelman Syndrome Foundation as well as Duke Institute for Brain Science. These studies were partially supported by National Institute of Health grant (MH019109 to J.S.C., MH082441 to W.C.W., NS039444 to R.J.W. and T32-HD40127 to P.A.M.) and funds from the Duke University Mouse Behavioral and Neuroendocrine Analysis Core Facility. B.D.P. was supported by a grant from the Simons Foundation and J.S.C. was supported by NIH Training Grant MH082441.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Susan Weinberg, Luis Landa, Maguerita Klein, Ting-ting Wang, Matt Kennedy, Yoonji Lee for technical support. We wish to thank also Christopher Means, Susan Cavender, and Erin Moore for help with the behavioral experiments, and Santowana Aryal and Jiechun Zhou for maintaining the mice.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Naisbitt S., Kim E., Tu J.C., Xiao B., Sala C., Valtschanoff J., Weinberg R.J., Worley P.F., Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 2.Tu J.C., Xiao B., Naisbitt S., Yuan J.P., Petralia R.S., Brakeman P., Doan A., Aakalu V.K., Lanahan A.A., Sheng M., et al. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- 3.Boeckers T.M., Winter C., Smalla K.H., Kreutz M.R., Bockmann J., Seidenbecher C., Garner C.C., Gundelfinger E.D. Proline-rich synapse-associated proteins ProSAP1 and ProSAP2 interact with synaptic proteins of the SAPAP/GKAP family. Biochem. Biophys. Res. Commun. 1999;264:247–252. doi: 10.1006/bbrc.1999.1489. [DOI] [PubMed] [Google Scholar]
- 4.Sheng M., Kim E. The Shank family of scaffold proteins. J. Cell. Sci. 2000;113:1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- 5.Gundelfinger E.D., Boeckers T.M., Baron M.K., Bowie J.U. A role for zinc in postsynaptic density asSAMbly and plasticity? Trends Biochem. Sci. 2006;31:366–373. doi: 10.1016/j.tibs.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Kreienkamp H.J. Scaffolding proteins at the postsynaptic density: shank as the architectural framework. Handb. Exp. Pharmacol. 2008;186:365–380. doi: 10.1007/978-3-540-72843-6_15. [DOI] [PubMed] [Google Scholar]
- 7.Bockers T.M., Mameza M.G., Kreutz M.R., Bockmann J., Weise C., Buck F., Richter D., Gundelfinger E.D., Kreienkamp H.J. Synaptic scaffolding proteins in rat brain. Ankyrin repeats of the multidomain Shank protein family interact with the cytoskeletal protein alpha-fodrin. J. Biol. Chem. 2001;276:40104–40112. doi: 10.1074/jbc.M102454200. [DOI] [PubMed] [Google Scholar]
- 8.Uchino S., Wada H., Honda S., Nakamura Y., Ondo Y., Uchiyama T., Tsutsumi M., Suzuki E., Hirasawa T., Kohsaka S. Direct interaction of post-synaptic density-95/Dlg/ZO-1 domain-containing synaptic molecule Shank3 with GluR1 alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor. J. Neurochem. 2006;97:1203–1214. doi: 10.1111/j.1471-4159.2006.03831.x. [DOI] [PubMed] [Google Scholar]
- 9.Roussignol G., Ango F., Romorini S., Tu J.C., Sala C., Worley P.F., Bockaert J., Fagni L. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J. Neurosci. 2005;25:3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhar S.U., del Gaudio D., German J.R., Peters S.U., Ou Z., Bader P.I., Berg J.S., Blazo M., Brown C.W., Graham B.H., et al. 22q13.3 deletion syndrome: clinical and molecular analysis using array CGH. Am. J. Med. Genet. A. 2010;152A:573–581. doi: 10.1002/ajmg.a.33253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson H.L., Wong A.C., Shaw S.R., Tse W.Y., Stapleton G.A., Phelan M.C., Hu S., Marshall J., McDermid H.E. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J. Med. Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderlid B.M., Schoumans J., Anneren G., Tapia-Paez I., Dumanski J., Blennow E., Nordenskjold M. FISH-mapping of a 100-kb terminal 22q13 deletion. Hum. Genet. 2002;110:439–443. doi: 10.1007/s00439-002-0713-7. [DOI] [PubMed] [Google Scholar]
- 13.Phelan K. In: Gene Reviews. Pagon R.A., Bird T.C., Dolan C.R., Stephens K., editors. Seattle WA: University of Washington,; 2007. 22q13.3 Deletion Syndrome. Internet. [Google Scholar]
- 14.Phelan M.C. Deletion 22q13.3 syndrome. Orphanet. J. Rare Dis. 2008;3:14. doi: 10.1186/1750-1172-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philippe A., Boddaert N., Vaivre-Douret L., Robel L., Danon-Boileau L., Malan V., de Blois M.C., Heron D., Colleaux L., Golse B., et al. Neurobehavioral profile and brain imaging study of the 22q13.3 deletion syndrome in childhood. Pediatrics. 2008;122:e376–e382. doi: 10.1542/peds.2007-2584. [DOI] [PubMed] [Google Scholar]
- 16.Durand C.M., Betancur C., Boeckers T.M., Bockmann J., Chaste P., Fauchereau F., Nygren G., Rastam M., Gillberg I.C., Anckarsater H., et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall C.R., Noor A., Vincent J.B., Lionel A.C., Feuk L., Skaug J., Shago M., Moessner R., Pinto D., Ren Y., et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moessner R., Marshall C.R., Sutcliffe J.S., Skaug J., Pinto D., Vincent J., Zwaigenbaum L., Fernandez B., Roberts W., Szatmari P., et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am. J. Hum. Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauthier J., Spiegelman D., Piton A., Lafreniere R.G., Laurent S., St-Onge J., Lapointe L., Hamdan F.F., Cossette P., Mottron L., et al. Novel de novo SHANK3 mutation in autistic patients. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- 20.Awadalla P., Gauthier J., Myers R.A., Casals F., Hamdan F.F., Griffing A.R., Cote M., Henrion E., Spiegelman D., Tarabeux J., et al. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am. J. Hum. Genet. 2010;87:316–324. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gauthier J., Champagne N., Lafreniere R.G., Xiong L., Spiegelman D., Brustein E., Lapointe M., Peng H., Cote M., Noreau A., et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc. Natl Acad. Sci. USA. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonaglia M.C., Giorda R., Mani E., Aceti G., Anderlid B.M., Baroncini A., Pramparo T., Zuffardi O. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J. Med. Genet. 2005;10:822–828. doi: 10.1136/jmg.2005.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson H.L., Crolla J.A., Walker D., Artifoni L., Dallapiccola B., Takano T., Vasudevan P., Huang S., Maloney V., Yobb T., et al. Interstitial 22q13 deletions: genes other than SHANK3 have major effects on cognitive and language development. Eur. J. Hum. Genet. 2008;16:1301–1310. doi: 10.1038/ejhg.2008.107. [DOI] [PubMed] [Google Scholar]
- 24.Lord C., Cook E.H., Leventhal B.L., Amaral D.G. Autism spectrum disorders. Neuron. 2000;28:355–363. doi: 10.1016/s0896-6273(00)00115-x. [DOI] [PubMed] [Google Scholar]
- 25.Volkmar F.R., State M., Klin A. Autism and autism spectrum disorders: diagnostic issues for the coming decade. J. Child Psychol. Psychiatry. 2009;50:108–115. doi: 10.1111/j.1469-7610.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- 26.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. Washington, DC: APA; 1994. [Google Scholar]
- 27.Miles J.H., McCathren R.B., Stichter J.P., Shinawi M. In: Gene Reviews. Pagon R.A., Bird T.C., Dolan C.R., Stephens K, editors. Seattle, WA: University of Washington; 2010. Internet. [Google Scholar]
- 28.Bauman M.L. Medical comorbidities in autism: challenges to diagnosis and treatment. Neurotherapeutics. 2010;7:320–327. doi: 10.1016/j.nurt.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue M., Brimacombe M., Chaaban J., Zimmerman-Bier B., Wagner G.C. Autism spectrum disorders: concurrent clinical disorders. J. Child Neurol. 2008;23:6–13. doi: 10.1177/0883073807307102. [DOI] [PubMed] [Google Scholar]
- 30.Newschaffer C.J., Croen L.A., Daniels J., Giarelli E., Grether J.K., Levy S.E., Mandell D.S., Miller L.A., Pinto-Martin J., Reaven J., et al. The epidemiology of autism spectrum disorders. Annu. Rev. Public Health. 2007;28:235–258. doi: 10.1146/annurev.publhealth.28.021406.144007. [DOI] [PubMed] [Google Scholar]
- 31.Wing L. Asperger's syndrome: a clinical account. Psychol. Med. 1981;11:115–129. doi: 10.1017/s0033291700053332. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y.H., Armstrong D., Albrecht U., Atkins C.M., Noebels J.L., Eichele G., Sweatt J.D., Beaudet A.L. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 33.Clapcote S.J., Roder J.C. Deletion polymorphism of Disc1 is common to all 129 mouse substrains: implications for gene-targeting studies of brain function. Genetics. 2006;173:2407–2410. doi: 10.1534/genetics.106.060749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clapcote S.J., Lipina T.V., Millar J.K., Mackie S., Christie S., Ogawa F., Lerch J.P., Trimble K., Uchiyama M., Sakuraba Y., et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 35.Kim J.Y., Duan X., Liu C.Y., Jang M.H., Guo J.U., Pow-anpongkul N., Kang E., Song H., Ming G.L. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim S., Naisbitt S., Yoon J., Hwang J.I., Suh P.G., Sheng M., Kim E. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J. Biol. Chem. 1999;274:29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- 37.Bonaglia M.C., Giorda R., Borgatti R., Felisari G., Gagliardi C., Selicorni A., Zuffardi O. Disruption of the ProSAP2 gene in a t(12;22)(q24.1;q13.3) is associated with the 22q13.3 deletion syndrome. Am. J. Hum. Genet. 2001;69:261–268. doi: 10.1086/321293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maunakea A.K., Nagarajan R.P., Bilenky M., Ballinger T.J., D'Souza C., Fouse S.D., Johnson B.E., Hong C., Nielsen C., Zhao Y., et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silverman J.L., Yang M., Lord C., Crawley J.N. Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lord C., Shulman C., DiLavore P. Regression and word loss in autistic spectrum disorders. J. Child Psychol. Psychiatry. 2004;45:936–955. doi: 10.1111/j.1469-7610.2004.t01-1-00287.x. [DOI] [PubMed] [Google Scholar]
- 41.Kalueff A.V., Tuohimaa P. Mouse grooming microstructure is a reliable anxiety marker bidirectionally sensitive to GABAergic drugs. Eur. J. Pharmacol. 2005;508:147–153. doi: 10.1016/j.ejphar.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 42.Welch J.M., Lu J., Rodriguiz R.M., Trotta N.C., Peca J., Ding J.D., Feliciano C., Chen M., Adams J.P., Luo J., et al. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature. 2007;448:894–900. doi: 10.1038/nature06104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maes J.H., Eling P.A., Wezenberg E., Vissers C.T., Kan C.C. Attentional set shifting in autism spectrum disorder: differentiating between the role of perseveration, learned irrelevance, and novelty processing. J. Clin. Exp. Neuropsychol. 2011;33:210–217. doi: 10.1080/13803395.2010.501327. [DOI] [PubMed] [Google Scholar]
- 44.Lord C., Risi S., Lambrecht L., Cook E.H., Jr, Leventhal B.L., DiLavore P.C., Pickles A., Rutter M. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 2000;30:205–223. [PubMed] [Google Scholar]
- 45.Bredt D.S., Nicoll R.A. AMPA receptor trafficking at excitatory synapses. Neuron. 2003;40:361–379. doi: 10.1016/s0896-6273(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 46.Kessels H.W., Malinow R. Synaptic AMPA receptor plasticity and behavior. Neuron. 2009;61:340–350. doi: 10.1016/j.neuron.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hung A.Y., Sung C.C., Brito I.L., Sheng M. Degradation of postsynaptic scaffold GKAP and regulation of dendritic spine morphology by the TRIM3 ubiquitin ligase in rat hippocampal neurons. PLoS One. 2010;5:e9842. doi: 10.1371/journal.pone.0009842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qualmann B., Boeckers T.M., Jeromin M., Gundelfinger E.D., Kessels M.M. Linkage of the actin cytoskeleton to the postsynaptic density via direct interactions of Abp1 with the ProSAP/Shank family. J. Neurosci. 2004;24:2481–2495. doi: 10.1523/JNEUROSCI.5479-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berkel S., Marshall C.R., Weiss B., Howe J., Roeth R., Moog U., Endris V., Roberts W., Szatmari P., Pinto D., et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat. Genet. 2010;42:489–491. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- 50.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S., et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tao-Cheng J.H., Dosemeci A., Gallant P.E., Smith C., Reese T. Activity induced changes in the distribution of Shanks at hippocampal synapses. Neuroscience. 2010;168:11–17. doi: 10.1016/j.neuroscience.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hung A.Y., Futai K., Sala C., Valtschanoff J.G., Ryu J., Woodworth M.A., Kidd F.L., Sung C.C., Miyakawa T., Bear M.F., et al. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J. Neurosci. 2008;28:1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silverman J.L., Turner S.M., Barkan C.L., Tolu S.S., Saxena R., Hung A.Y., Sheng M., Crawley J.N. Sociability and motor functions in Shank1 mutant mice. Brain. Res. 2010;1380:120–137. doi: 10.1016/j.brainres.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brewer G.J., Torricelli J.R., Evege E.K., Price P.J. Optimized survival of hippocampal neurons in B27-supplemented neurobasal, a new serum-free medium combination. J. Neurosci. Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- 55.Lu J., Helton T.D., Blanpied T.A., Racz B., Newpher T.M., Weinberg R.J., Ehlers M.D. Postsynaptic positioning of endocytic zones and AMPA receptor cycling by physical coupling of dynamin-3 to Homer. Neuron. 2007;55:874–889. doi: 10.1016/j.neuron.2007.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roberts A.C., Diez-Garcia J., Rodriguiz R.M., Lopez I.P., Lujan R., Martinez-Turrillas R., Pico E., Henson M.A., Bernardo D.R., Jarrett T.M., et al. Downregulation of NR3A-containing NMDARs is required for synapse maturation and memory consolidation. Neuron. 2009;63:342–356. doi: 10.1016/j.neuron.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yashiro K., Riday T.T., Condon K.H., Roberts A.C., Bernardo D.R., Prakash R., Weinberg R.J., Ehlers M.D., Philpot B.D. Ube3a is required for experience-dependent maturation of the neocortex. Nat. Neurosci. 2009;12:777–783. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguiz R.M., Chu R., Caron M.G., Wetsel W.C. Aberrant responses in social interaction of dopamine transporter knockout mice. Behav. Brain Res. 2004;148:185–198. doi: 10.1016/s0166-4328(03)00187-6. [DOI] [PubMed] [Google Scholar]
- 59.Rodriguiz R.M., Gadnidze K., Ragnauth A., Dorr N., Yanagisawa M., Wetsel W.C., Devi L.A. Animals lacking endothelin-converting enzyme-2 are deficient in learning and memory. Genes Brain Behav. 2007;7:418–426. doi: 10.1111/j.1601-183X.2007.00365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pogorelov V.M., Rodriguiz R.M., Insco M.L., Caron M.G., Wetsel W.C. Novelty seeking and stereotypic activation of behavior in mice with disruption of the Dat1 gene. Neuropsychopharmacology. 2005;30:1818–1831. doi: 10.1038/sj.npp.1300724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.