Abstract
Elevated reactive oxidative species (ROS) are cytotoxic, and chronic elevated levels of ROS have been implicated in multiple diseases as well as cellular transformation and tumor progression. However, the potential for a transient and minimally toxic episode of ROS exposure, or a minimal threshold dose of ROS, to initiate disease or cellular transformation is unclear. We examined both transcriptional and phospho-proteomic responses of murine embryonic stem (ES) cells to a single brief exposure of minimally toxic hydrogen peroxide (H2O2). The cellular response was distinct from those induced by either an acute exposure to H2O2 or the topoisomerase II poison etoposide. Analysis of tumorigenesis-related transcripts revealed a significant up-regulation of oncogenes and down-regulation of tumor suppressors. Analysis of the phospho-proteomic response demonstrated insulin-signaling induction, including insulin receptor Y972 hypophosphorylation, similar to insulin-resistance mouse models and observed in diabetic patients. In addition, ES cells were more resistant to ROS than differentiated cells, and retained their transcriptional self-renewal signature, suggesting stem cells have a higher potential for ROS-mediated mutagenesis and proliferation in vivo. These results are a direct demonstration that even brief and non-toxic exposures to ROS may induce transduction of insulin resistance and transformation signaling in stem cells leading to diabetes and cancer.
Keywords: reactive oxygen species, oxidative stress, DNA damage, stem cells, gene expression, microarray, proteomics, insulin, diabetes, cancer
Introduction
Reactive oxygen species (ROS) are produced by metabolizing molecular oxygen to produce hydroxyl free radicals (OH), superoxide anions (O2−•), singlet oxygens (102), and hydrogen peroxide (H2O2). ROS are generated by endogenous reduction of oxygen, by the mitochondrial respiratory pathway, as well as by exogenous exposure to UV or environmental damaging agents [1]. Superoxides produced by NADPH oxidase activity are quickly dismutated by superoxide dismutases (SODs) to the more stable H2O2. ROS levels in cells are highly regulated. Increases in ROS above basal cellular concentrations lead to oxidative stress (OS) [2]. OS is thought to damage 20,000 bases per day per human cell and be one of the major causes of DNA damage and mutation [3,4].
At submicromolar concentrations, ROS act as proliferation and growth signaling molecules. Elevated levels of ROS induced by mutations of metabolic enzyme genes, ischemia/reperfusion, chemotherapy, or chronic exposure to 10–100μM H2O2 induce multiple effects ranging from cell cycle arrest to death, depending on the cell type [5,6]. High levels of ROS have been implicated in human diseases including cancer, diabetes, cardiac disease, neurodegeneration, and aging [4,7,8]. In support of this, high levels of OS induced by metabolic enzyme deficiency are associated with head and neck cancers as well as child T-cell leukemia [9,10]. High concentrations of H2O2 (3mM) can induce multiple insulin-like effects in rat adipose tissue including phosphorylation of the insulin receptor (INSR) subunit at E80 and Y20 [11]. However, the potential for a transient and minimally toxic episode of ROS exposure or what minimal threshold dose of ROS to initiate disease or cellular transformation is unclear.
Upon differentiation of embryonic stem (ES) cells, superoxide production, cellular levels of intracellular ROS, and DNA damage levels increase. At the same time, expression of major antioxidant genes and genes involved in multiple DNA repair pathways is downregulated [12], and DNA repair by homologous recombination is reduced [13]. Thus, elevated ROS may promote tumorigenesis in more differentiated somatic cells indirectly through increased illegitimate repair of the ensuing DNA damage. It is not clear how susceptible stem cells are to a single brief exposure of ROS, particularly at minimally toxic doses not expected to induce apoptosis.
A significant body of literature exists on the transcriptional response of multiple cell types to high toxic or low chronic doses of ROS [14–23] but not the impact of a single minimally toxic ROS episode. In addition, the immediate coordination of both transcriptional and post-translational responses in response to ROS is not understood. In this study we measured the immediate cellular response of mouse ES cells to a single minimally toxic episode of hydrogen peroxide (H2O2). The cellular response was distinct from those induced by either an acute exposure to H2O2 or by the topoisomerase II poison etoposide. Parallel examination of transcriptional profiles with the post-translational modifications of a significant though limited number of signaling molecules demonstrated that a single minimally toxic exposure to ROS is sufficient to induce significant increases in oncogenic and metastatic pathways and specifically induce insulin signaling, similar to insulin-resistance mouse models and observed in diabetic patients. Despite the significant signaling changes induced by ROS, cells maintained their stem cell signatures suggesting a mechanism for maintenance, survival, and transformation in early stem cell pools.
Results
Growth arrest, cytotoxicity, and DNA fragmentation
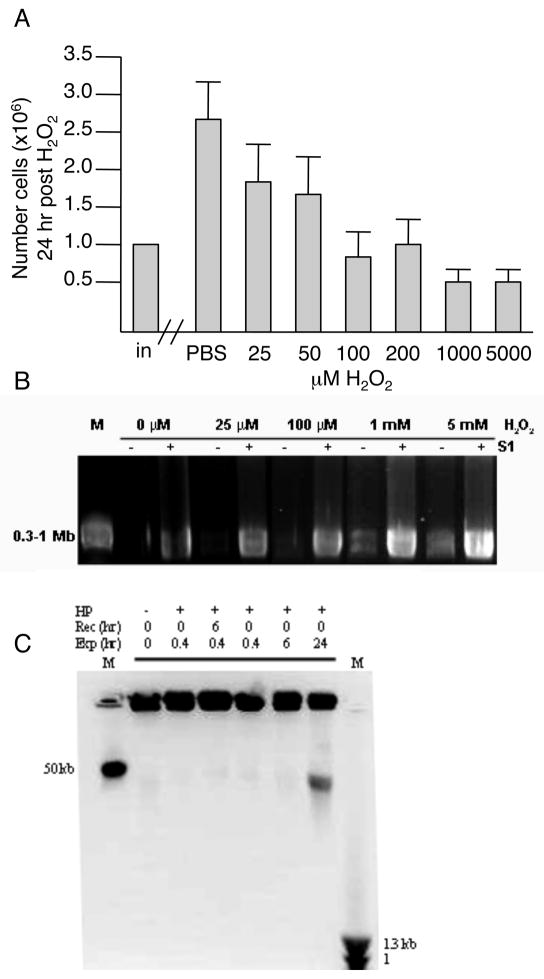
ES cells were exposed to a minimally toxic 100 M hydrogen peroxide (H2O2) for 15 min. Dose was chosen as H2O2 concentrations up to 50μM have been reported in human plasma and 100 M H2O2 induces ROS levels similar to those observed in ischemia/reperfusion or respiratory burst conditions [24]. Alternatively, cells were exposed to 5mM hydrogen peroxide (H2O2) or 20 M etoposide for 30 min. 5mM H2O2 induces ROS levels similar to those observed in vivo during acute inflammatory reactions. Etoposide is an inhibitor of the topoisomerase II religation reaction and a known inducer of DNA double-strand breaks (DSBs). 20 M etoposide is in close agreement with pharmacokinetic studies demonstrating peak patient plasma levels [25].
As expected, ROS and DSBs induced a dose-dependent response of cell cycle arrest and cell death. Cell cycle profiles by BrdU analysis of cells exposed to all three conditions demonstrated G2 cell cycle arrest through 6 hrs post-exposure and release by 24 hrs post-exposure (data not shown). Cells exposed to etoposide demonstrated an intra-S phase arrest at early times and release by 24 hrs post-exposure. The lack of a significant G1/S arrest was expected since ES cells have a defective p53-mediated stress response [26–28]. 100μM H2O2 induced minimal cell death; however, doses beyond 200μM induced significant cell death by 24 hrs post-exposure (IC50=5mM), relative to controls, Fig. (1A).
Fig. 1.
Response of ES cells in culture 24 hrs following exposure to minimally toxic ROS. A. Cells were exposed to H2O2 for 15 min, recovered in normal medium for 20 hrs, then the number of viable cells in culture scored (dead cells were excluded by trypan blue). Data are the average and standard deviation of at least 4 independent experiments. B. Dose-dependent accumulation of single-strand breaks (SSBs) following exposure to H2O2 for 15 min. Pulse field gel electrophoresis of agarose embedded DNA without (−; DSB) or with (+; SSB and DSB) S1 nuclease digestion. DNA fragmentation was not evident in samples exposed to H2O2 at lower doses. Fragmentation was observed at higher doses (1mM and 5mM). As expected, visible fragmentation was detected in all samples, including untreated, following S1 nuclease digestion. Visible fragments were within the 0.3-1 Mb range. C. Time-dependent accumulation of 50 kb fragments following exposure (Exp) to 100 M H2O2 with or without recovery (Rec). 50 kb fragments observed only after chronic 24 hour continuous exposure and no recovery. M -- size standard marker.
Pulse field gel electrophoresis along with S1 nuclease digestion confirmed ES cells had a dose-dependent ssDNA digestion of chromatin DNA that led to 0.3–1 Mb fragments, capable of inefficient religation and cell survival [29]. There was minimal ssDNA fragmentation following 100 M H2O2 and more significant fragmentation at higher toxic exposures, Fig. (1B). 50 kb DNA fragments were not detectable following acute 30 min exposure up to 10mM of H2O2. Only when ES cells were exposed to 100 M of H2O2 continuously for 24 hrs were 50kb DNA fragments produced, but without oligonucleosomal fragmentation, Fig. (1C).
Transcriptional response
Immediate cellular transcriptional response following a transient minimally toxic exposure to ROS was determined using Affymetrix MG-U74VerA chips and MS 5.0 and GeneSpring software comparing four samples exposed to the minimally toxic H2O2, five samples exposed to acute H2O2, and three replicate control samples. Among all conditions, 2742 transcripts were found significantly altered (p≤0.05), and, of these, 501 (433 known, 68 unknown) transcripts had a two-fold or greater change. Real-time RT-PCR results of 19 selected transcripts were concordant with microarray results. Correlation coefficient values for 16 of 19 transcripts were r(100)=0.71 and r(5)=0.78 for the two exposures to ROS, Table (1).
Table 1.
Real-time qPCR validation of transcriptional mbroarray results
| Microarray | Real Time qPCR | ||||
|---|---|---|---|---|---|
| 100 μM-15 min | 5 mM-30 min | 100 μM-15 min | 5 mM-30 min | ||
| Mito (EST) | 2.1 ± 0.3 | 2.5 ± 0.3 | 1.7 ± 0.3 | 1.9 ± 0.2 | Upregulated |
| Ysg2 | 2 ± 0.2 | 2.1 ± 0.5 | 1.8 ± 0.4 | 2.3 ± 0.5 | |
| Zfp143 | 3.7 ± 0.5 | 3.5 ±0.8 | 2.7 ± 0.7 | 2.7 ±1.1 | |
| Sgpp1 | 2 ± 0.8 | 2 ± 0.7 | 2.2 ± 0.6 | 1.5 ±0.4 | |
| Phldb2 | 12.6 ± 3 | 2.2 ± 1.3 | 4.8 ± 1.4 | 1.4 ± 0.5 | |
| Slc23a3 | 2.7 ± 0.7 | 2.1 ± 1 | 13.5 ± 4.5 | 2.4 ± 1.1 | |
| Ub-Lig (EST) | 3 ± 0.6 | 4.7 ± 1.4 | 4.7 ± 1.4 | 6.3 ± 0.8 | |
| Id2 | 1.9 ± 0.3 | 1 ± 0.2 | 3.5 ± 0.9 | 1.7 ± 0.6 | |
| Id4 | 2.7 ± 0.3 | 0.7 ± 0.2 | 4 ± 1.2 | 1.1 ± 0.1 | |
| nab1 | 2 ± 0.6 | 3 ± 0.3 | 2.1 ± 0.5 | 2.8 ± 0.9 | Upregulated |
| nab2 | 2.4 ± 0.7 | 2.7 ± 0.6 | 2.3 ± 0.4 | 3.7 ± 0.8 | |
| Gbx2 | 10.5 ± 1.6 | 3.4 ± 0.7 | 6.2 ± 0.7 | 3.2 ± 0.5 | |
| Bmp4 | 5.3 ± 0.6 | 2.8 ± 0.4 | 4.9 ± 0.7 | 2.1 ± 0.2 | |
| c-myc | 5.44 ± 0.9 | 2.2 ± 0.5 | 2.7 ± 0.5 | 1.9 ± 0.2 | |
| Atp11a | 2.2 ± 0.2 | 2 ± 0.4 | 2.6 ± 1 | 1.9 ± 0.6 | |
| Gtl2 | 4.6 ± 4 | 3 ± 5 | 2.5 ± 0.5 | 1.2 ± 0.3 | |
| Trp53 | 1.8 ± 0.5 | 1 ± 0.1 | 1 ± 0.3 | 1 ± 0.2 | |
| fgfbp1 | 3.6 ± 1.5 | 13.4 ± 8 | 1.8 ± 0.4 | 4.1 ± 0.8 | |
| Id1 | 2.5 ± 0.3 | 1 ± 0.4 | 1.9 ± 0.7 | 1.5 ± 0.6 | |
| n=4 | n=5 | n=8 | n=10 | ||
Correlation without 3 outliers highlighted in bold: r(100)=0.71; r(5)=0.78. Correlation with outliers: r(100)=0.28; r(5)=0.57
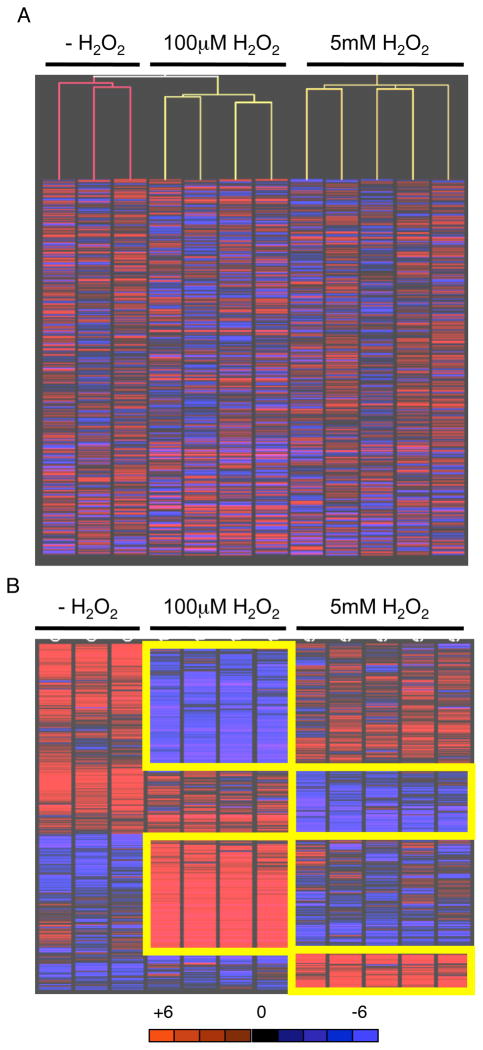
Transient exposure to a minimally toxic dose of H2O2 was sufficient to induce down-regulation of 185 transcripts and up-regulation of 175 transcripts. The response signature was distinct from a transient exposure to an acute dose that induced down-regulation of 114 transcripts and up-regulation of 62 transcripts. Overall, only 19 transcripts were commonly down-regulated and 12 transcripts commonly up-regulated by the two exposures (31 total; Table (2), Fig. (2B)) suggesting major differences, rather than common mechanisms, of immediate cellular response between minimally toxic and acute ROS exposure. A distinct cellular response to ROS exposures was further supported by an inverse dose effect in 11 transcripts, similar to studies in mammalian cells and yeast exposed to low or acute doses of irradiation [30,31] (Out of 360 transcripts altered following minimally toxic ROS and 176 following acute ROS, the expected false discovery rate would be 0.25%, or 2 transcripts). Four transcripts (calml4, cdkn1c, S100a6, gap43) were significantly decreased after minimally toxic ROS but increased after acute ROS, as compared to untreated samples. Conversely, seven transcripts (phtr1, dab2, bhmt2, prg1, sox17, gata6 and col4a1) were significantly increased after the minimally toxic ROS but decreased after acute ROS, as compared to untreated samples.
Table 2.
Commonly regulated transcripts following minimally toxic and acute exposures to H2O2
| Gene Name | Fold change relative to no treatment | |||
|---|---|---|---|---|
| Symbol | GenBank | 100 μM 15 min | 5 mM 30 min | |
| upregulated | ||||
| fibroblast growth factor binding protein 1 | Fgfbpl | AF065441 | 3.6 | 13.4 |
| sarco(endo)plasmic reticulum calcium ATPase | Atp2a2; SERCA2 | AF029982 | 2 | 6.3 |
| Taglin; SM22 | Tagln; Sm22a | Z68618 | 2.8 | 3.5 |
| gastrulation brain homeobox 2 | Gbx2 | Z48800 | 10.5 | 3.4 |
| Ngfi-A binding protein 1 | Nab1 | U47008 | 2 | 3 |
| GTL2, imprinted maternally expressed untranslated mRNA | Gtl2 | Y13832 | 4.6 | 3 |
| BMP-4 gene | Bmp4 | L47480 | 5.3 | 2.8 |
| Ngfi-A binding protein 2 | Nab2 | U47543 | 2.4 | 2.7 |
| EST00652 | AA407332 | 2.3 | 2.7 | |
| solute carrier family 25 (adenine nucleotide translocator), member 5 | Slc25a5 | U10404 | 2.8 | 2.6 |
| Dusp6 | Dusp6 | AI845584 | 2.8 | 2.4 |
| cytochrome c oxidase, subunit VIIc | Cox7c | AI648091 | 3.2 | 2.4 |
| myeloid-associated differentiation marker | Myadm | AJ001616 | 2.1 | 2.3 |
| c-myc exon 3 | c-myc | L00039 | 5.44 | 2.2 |
| stratifin | Sfn | AF058798 | 2.9 | 2.1 |
| urokinase plasminogen activator receptor | Plaur | X62700 | 2.6 | 2.1 |
| CCR4 carbon catabolite repression 4-like | Ccrn4l | AW047630 | 3.3 | 2 |
| Nes | Nes | AW061260 | 3 | 2 |
| ATPase, class VI, type 11A | Atp11a | AA690863 | 2.2 | 2 |
| downregulated | ||||
| pleckstrin homology-like domain, family B, member 2 | Phldb2 | AW125043 | 12.6 | 2.2 |
| receptor (calcitonin) modifying protein 2 | Ramp2 | AJ250490 | 6 | 2.7 |
| zinc finger protein 143 | Zfp143 | U29513 | 3.7 | 3.5 |
| protein kinase, lysine deficient 1 | Prkwnk1 | AV319920 | 3.2 | 2.2 |
| RIKEN cDNA 2310014L17, ubiquitin ligase | 2310014L17Rik | AA794189 | 3 | 4.7 |
| solute carrier family 23 (nucleobase transporters), member 3 | Slc23a3 | AV222871 | 2.7 | 2.1 |
| (clone lambda-MG5.3) acid phosphatase type 5 gene | Acp5; TRAP | M99054 | 2.4 | 2.1 |
| DnaJ (Hsp40) homolog, subfamily C, member 3 | Dnajc3; hsp40 | U28423 | 2.3 | 2 |
| FBJ osteosarcoma oncogene B | Fosb | X14897 | 2.3 | 2.1 |
| RIKEN cDNA 2310005014 gene, mitochondrion | 2310005014Rik | AW124582 | 2.1 | 2.5 |
| sphingosine-1-phosphate phosphatase 1 | Sgpp1 | AI835784 | 2 | 2 |
| sialic acid acetylesterase; yolk sac gene 2 | Ysg2 | U61183 | 2 | 2.1 |
Fig. 2.
A. Relatedness of sample groups confirmed by unsupervised Pearson correlation clustering of transcripts deregulated by two-fold between controls and replicate samples. Analysis performed by GeneSpring data analysis software. B. Distinct gene expression signatures induced by low and high doses of H2O2. Parametric t-test with unequal variance (Welch t-test) analysis was performed by GeneSpring data analysis software.
Maintenance of stem cell markers
Transient minimally toxic exposure to ROS revealed no significant change in 14 of 19 common transcripts associated with stem cell populations and pluripotency including signature transcripts oct3/4, nanog and sox2, Table (3) [32]. Markers specific for differentiation of ES cells upon LIF withdrawal were unchanged [33]. Three stem cell transcripts with known roles in DNA or stress response (Mdr1/abcb1 and ercc5 and gpx3) were up-regulated following a minimally toxic dose of H2O2, Table (3) [12,34]. Increases in SOD activity can reduce growth and malignant phenotypes of tumor cells in culture although its mechanism of action is not well understood and can be inconsistent [23,35,36]. However, in this study, ROS led to down-regulation of sod2, a stem cell marker identified by Sartzki et al [12], suggesting the promotion of growth and survival of mouse ES cells following a single minimally toxic exposure. Two transcripts (gbx2 [37], c-myc [38]) were down-regulated. Gbx2 is within the wnt pathway and not a known myc target gene ([39]; http://www.myccancergene.org/index.asp). Overall, these results suggest that pluripotent stem cell populations have the capacity to respond to ROS while retaining their major self-renewal signatures.
Table 3.
Transcriptional state of pluripotency genes following minimally toxic and acute exposures to H2O2
| Description | Symbol | Effect of OS does on Transcription |
|---|---|---|
| Embryonic stem cell specific gene 1 | esgl | No change |
| Octamer binding transcription factor | oct3/4 | No change |
| Zinc finger protein 42 | zfp42 (rexl) | No change |
| Fibroblast growth factor 4 | fgf4 | No change |
| SRY-box 2 | sox2 | No change |
| Nanog homeobox | nanog | No change |
| X-ray repair complementing 5 | xrcc5 | No change |
| Radiation repair 23 homolog | rad23b | No change |
| MutS homolog 2 (E. coli) | msh2 | No change |
| Zinc finger protein 42 | zfp42 | No change |
| Teratocarcinoma derived growth factor, CRIPTO | tdgf1 | No change |
| Integrin alpha 6 | itga6 | No change |
| Signal transducer and activator of transcription 3 | stat3 | No change |
| Multidrug resistance /ATP binding cassette | Mdr1/abcb1 | 2× up low dose (2 isoforms) |
| Excision repair complementing 5 | ercc5 | 2× up low dose |
| Gastrulation Brain Homeobox 2 | gbx2 | 10× down low dose; 3.4× down high dose |
| Forkhead homeobox D3 | foxod3 | 5× down high dose |
| Undifferentiated embryonic transcription factor | utf1 | 2× down low dose |
| myelocytomatosis viral oncogene homolog | c-myc | 5.4 down low dose; 2.2 down high dose |
Oncogenic immediate transcriptional signature response
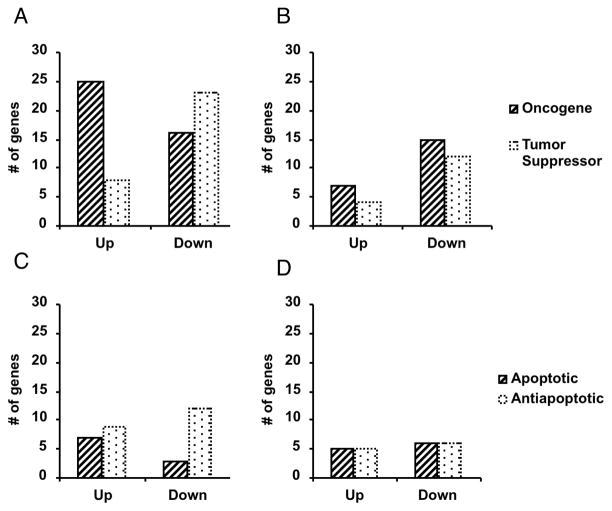
Transcriptional profiles were assessed by GO, Entrez, Locuslink, and Ingenuity Pathway Analysis (IPA) (Ingenuity® Systems, www.ingenuity.com). Pathway analysis demonstrated the minimally toxic and acute ROS stratified top significantly altered biological functions, Table (4). IPA functionally classified all altered transcripts according to tumorigenesis, DNA repair, cell growth/maintenance, development, cell cycle, and pro- or anti-apoptosis (Table 3). IPA noted a significant number of altered transcripts classified in tumorigenesis (139/360; 39%) following the single minimally toxic dose of H2O2, specifically a significant up-regulation of oncogenes and down-regulation of tumor suppressors, Table (4), Fig. (3). Consistent with this, IPA noted a significant up-regulation of anti-apoptotic transcripts, Fig. (3). The additional IPA classifications did not reveal significant clustering. These data indicate a nonlinear relationship and strong association between ROS dose and oncogene activation or tumor suppressor silencing, along with cell survival.
Table 4.
Top significant functions and pathways affected by minimally toxic and acute exposures to H2O2.
| Low Dose Regulated Genes
| ||
|---|---|---|
| Biological Function. | # transcripts | p-valuea |
| Tumorigenesis | 139 | 4.7×10−9-3.8×10−2 |
| Cell Death | 102 | 1.0×10−4 – 3.8×10−2 |
| Cellular Development | 81 | 1.0×10−6 – 3.8×10−2 |
| Gene Expression | 74 | 2.3×10−5 – 3.8×10−2 |
| Gastrointestinal Disease | 55 | 1.8×10−5 – 3.8×10−2 |
|
| ||
| Canonical Pathway
| ||
| Wnt/ -Catenin Signaling | 11 | 7.1×10−3 |
| Biosynthesis of Steroids | 6 | 7.5×10−5 |
|
| ||
|
High Dose Regulated Genes
| ||
| Biological Function | # transcripts | p-value |
|
| ||
| Tumorigenesis | 49 | 1.9×10−4 – 3.2×10−2 |
| Cellular Development | 39 | 1.7×10−5 – 3.3×10−2 |
| Cell Cycle | 36 | 2.6×10−5 – 3.4×10−2 |
| Embryonic Development | 19 | 1.7×10−5 – 2.7×10−2 |
| Respiratory Disease | 7 | 1.9×10−4 – 2.9×10−2 |
|
| ||
| Canonical Pathway
| ||
| Coagulation system | 6 | 2.2×10−2 |
| VEGF Signaling | 5 | 4.9×10−2 |
| Riboflavin Metabolism | 4 | 1.8×10−2 |
| Cell Cycle: G1/S Checkpoint | 3 | 3.9×10−2 |
|
| ||
|
Commonly Regulated Genes
| ||
| Biological Function | # transcripts | p-value |
|
| ||
| Tumorigenesis | 13 | 1.7×10−5 – 3.3×10−2 |
| Cellular Development | 7 | 1.9×10−5 – 3.2×10−2 |
| Neurological Development | 7 | 1.9×10−5 – 2.9×10−2 |
| Cell Cycle | 4 | 1.7×10−5 – 2.7×10−2 |
| Neurological Disease | 2 | 2.6×10−5 – 3.4×10−2 |
|
| ||
| Canonical Pathway
| ||
| Coagulation | 6 | 2.2×10−2 |
| VEGF Signaling | 5 | 4.9×10−2 |
| Riboflavin Metabolism | 4 | 1.8×10−2 |
| Cell Cycle: G1/S Checkpoint | 3 | 3.9×10−2 |
Fisher’s exact test
Fig. 3.
IPA clustering shows distinct response of ES cells to minimally toxic ROS. IPA clustering was performed on statistically significant altered transcripts from four samples exposed to 100uM H2O2, five samples exposed to 5mM H2O2, and three replicate control samples analyzed by Affymetrix MG-U74VerA chips and GeneSpring software. A. Altered oncogene and tumor suppressor transcripts following 100 M H2O2. B. Altered oncogene and tumor suppressor transcripts following 5mM H2O2. C. Altered apoptotic and anti-apoptotic transcripts following 100 M H2O2. D. Altered apoptotic and anti-apoptotic transcripts following 5mM H2O2.
Oncogenic immediate post-translational signature response
To more completely assess the signaling pathways induced in stem cells by a transient and minimally toxic exposure to ROS, we used Kinexus® phospho-proteomic screening technology to analyze 92 phosphorylation sites covering 53 proteins in p53, Rb, p38/MAPK/ERK, NFκB, PI3K/Akt and insulin-signaling pathways, Table (5). Reproducibility was validated by detecting concordant normalized CPM values for common epitopes (Mapk1-T185+Y187, Mapk3-T202+Y204, p38Mapk-T180+Y182). Samples were exposed to 100 M H2O2 for 15 min, recovered in normal media for 1 hr, then harvested for analysis. Specificity of the response was determined by comparison against a 15 min exposure to 5mM H2O2 or 30 min exposure to 20 M etoposide. Based on an absolute cut-off of 300 normalized CPM, nearly two-thirds (52 of 92) of the epitopes had no detectable phosphorylation in either control samples or following ROS or etoposide exposure. The remaining third of epitopes (34 of 92) had a detectable baseline phosphorylation in control samples. We focused on sites that were found differentially phosphorylated by at least 25% with respect to controls. 85% (29 of 34) of these were altered one hour following exposure to either ROS or etoposide, Table (5).
Table 5.
Phosphorylation alterations in response to minimally toxb and acute exposures to H2O2 and etoposide
| Phosphorylation detected in control and significantly altered following stress. | |||||
|---|---|---|---|---|---|
| fold phos. relative to control | |||||
| Mouse (epitope) | Human (epitope) | Controla | Etoposide | 100 μM | 5mM |
| INSR (Y972) | INSR (Y972) | 500 | 1 | 0.45 | 1 |
| PKN1 (PRK1)(T778) | PRK1 (T774) | 435 | 1 | 0.6 | 1 |
| PRKCD (T505) | Prkcd (T505) | 544 | 0.6 | 0.6 | 1 |
| RAF1 (S259) (60 kD) | RAF1 (S259) | 471 | 1 | 0.6 | 1 |
| PKN2 (N/A) | PRK2 (T816) | 397 | 0.7 | 0.6 | 1 |
| PPP1CA (T320) | PP1a (T320) | 422 | 0.56 | 0.61 | 0.5 |
| RAF1 (S259) (70 kD) | RAF1 (S259) | 999 | 1 | 0.63 | 0.68 |
| MAP2K1 (T292) | MEK1 (T291) | 1700 | 0.75 | 0.67 | 0.49 |
| MAP2K1 (T386) | MEK1 (T385) | 475 | 1 | 1 | 0 |
| RB1 (S773)b | Rb1 (S780) | 296 | 1 | 1 | 0 |
| SRC (Y423) | SRC (Y418) | 400 | 0.33 | 1 | 0.17 |
| PTK2 (S722) | FAK (S722) | 700 | 1 | 1 | 0.27 |
| PRKCZ (T40/T402)c | Prkcz/I(T40/T403) | 498 | 1 | 1 | 0.6 |
| SRC (Y534) | SRC (Y529) | 2800 | 1 | 1 | 1.3 |
| MAP2K1 (S298) | MEK1 (S297) | 400 | 1 | 1 | 1.5 |
| CDC2 (Y15) | CDK1 (Y15) | 2530 | 1 | 1 | 1.6 |
| MAPK8 (JNK/SAPK) (T183/Y185) (47kD) | JNK (SAPK) (T183/Y185) | 800 | 0.7 | 1 | 1 |
| PTK2 (S910) | FAK (S910) | 700 | 0.47 | 1.25 | 1 |
| NPM1 (S4) | NPM1 (S4) | 2719 | 1 | 1.3 | 1 |
| PRKCM (S916) | Prkcm (S910) | 305 | 1 | 1.3 | 1.3 |
| AKT1 (S473) | PKBa (Akt1)(S473) | 400 | 0.65 | 1.3 | 3.76 |
| EIF2B5 (S539) | eIF2Be (S540) | 300 | 1.43 | 1.34 | 1.48 |
| CDC2 (T161/T160) | CDK1 (T161/T160) | 386 | 1.65 | 1.74 | 2.6 |
| MAPK14 (T180+Y182) | p38aMAPK (T180/Y182) | 450 | 3.92 | 1.9 | 3.66 |
| Phosphorylation detected in control and not altered following stress. | |||||
| fold phos. relative to control | |||||
| LYN (Y507) (50kD) | Lyn (Y507) (50kD) | 721 | 1 | 1 | 1 |
| LYN (Y507) (47kD) | Lyn (Y507) (47kD) | 895 | 1 | 1 | 1 |
| PDPK1 (S241) | PDK1 (S244) | 1066 | 1 | 1 | 1 |
| MAP2K6 (S207) | MKK6 (S207) | 1306 | 1 | 1 | 1 |
| CDC2 (T14+Y15) | CDK1 (T14/Y15) | 3500 | 1 | 1 | 1 |
| phorylation below 300 (normalized intensities) in control and significantly altered to near 300 following stress. | |||||
| normalized intensities | |||||
| MAPK1 (T185+Y187) | ERK2 (T185/Y187) | 36 | 52 | 53 | 376 |
| MAP2K1 (S217/S221) | MEK1 (S217/S221) | 91 | 104 | 175 | 380 |
| AKT1 (T308) | PKBa/Akt1 (T308) | 99 | 64 | 49 | 296 |
| GSK3a (S21) | GSK3a (S21) | 144 | 255 | 206 | 166 |
Green shading in control column indicates that the normal effect of protein phosphorylation is activation. Red shading in control indicates that the normal effect of protein phosphorylation is inhibition.
PRKCZ was significantly up-regulated following 5 mM H2O2 in parallel transcriptional microarray analysis.
Rb1 was significantly down-regulated following 5 mM H2O2 in parallel transcriptional microarray analysis.
Overall, stem cell response to ROS was distinct from etoposide exposure and could be stratified by dose, with a specific phosphorylation pattern induced by each. Only 5 of the 34 epitopes (15%) with baseline phosphorylation were post-translationally altered by both H2O2 and etoposide treatment--activating phoshorylation of CDC2 (CDK1) at T160/161 and of p38a MAPK at T180+Y182, activating dephosphorylation of the catalytic subunit of protein phosphatase 1 (PPP1CA) at T320 and of MAP2K1 (MEK1) at T291, and inhibitory phosphorylation of eIF2B5 at S539. Decreased phosphorylation of eIF2B5 would be expected as a consequence of slowed translation in response to DNA damage and stress. These results suggest that the immediate response to DNA damage involves changes to homeostatically phosphorylated proteins in ES cells rather than phosphorylation of new ones, at least within the signaling pathways examined.
Eight epitopes were hypophosphorylated and 7 epitopes were hyperphosphorylated in response to 100 M H2O2. Consistent with the IPA analysis of transcriptional response, exposure to 100 M H2O2 uniquely induced hypophosphorylation of protein kinase C related kinase (Prk1) at T778 associated with cell migration and tumor metastasis and hyperphosphorylation of Npm1 at S4 associated with cell survival and growth, Table (5).
Transient exposure to minimally toxic 100 M H2O2 uniquely led to dysregulated insulin signaling activation observed by hypophosphorylation of insulin receptor (INSR) at Y972, Table (5). This response was supported by increased phosphorylation of GSK3 at S21 and increased phosphorylation of FAK/Ptk2 at S910, Table (5). The INSR juxtamembrane autophosphorylation site Y972 promotes interaction and stability between INSR and intracellular substrates [40] while FAK1 acts as an intracellular positive downstream regulator of signaling. Hypophosphorylation of INSR at Y972, phosphorylation of FAK1, and inhibitory phosphorylation of GSK3 at S21 have all been associated with insulin-resistant signaling. Further, protein kinase B/Akt1 plays a role in multiple signaling pathways including insulin-stimulated GLUT4 membrane localization[41]. Partial activation of Akt by S473 phosphorylation, essential for Akt activation, was observed in a dose-dependent manner.
Discussion
Defining the cellular response of stem cells to ROS is critical to understanding the unique sensitivity of stem cells to minimally toxic episodes of ROS exposure, or a minimal threshold dose of ROS, to initiate disease or cellular transformation. In this study we directly examined the response of ES cells to a single transient and minimally toxic exposure of ROS and correlated the immediate transcriptional and post-translational modifications that resulted.
ES cells demonstrated a high resistance to ROS consistent with previous work showing ES cells withstanding extreme hyperoxic conditions (40% 02) compared with cells grown under normoxic culture conditions [12]. ES cells were more resistant to OS-induced high molecular weight DNA fragmentation than differentiated cell types [29,42]. ES cells may have an increased DNA repair capacity or endonuclease-protected higher order chromatin similar to some cancer cell lines [43,44]. Consistent with this, the dose-dependent resistance of ES cells to ROS-mediated cell death was similar to Caco2 colon cancer cells and elevated compared to terminally differentiated primary glial cells [44,45]. Low and high dose ROS-inducing therapies have different cytotoxicities and short- and long-term efficacies [46,47] that are likely defined by the immediate cell-specific response to these treatments, similar to the observations made here. Consistent with increased resistance of ES cells to ROS, transcripts of the majority of canonical self-renewing genes were similarly unaffected by 100 M H2O2. Following this minimally toxic episode of ROS, we did not observe a significant change in transcripts known to be affected following LIF withdrawal-induced differentiation of ES cells [33]. However, a mild transcriptional signal of differentiation could be discerned following acute levels.
Binning of significant biological functions determined that a minimally toxic ROS exposure primarily affected transcription of tumorigenesis-related genes. We curated oncogenes and tumor suppressors and found a significant proportion of oncogenes were up-regulated and tumor suppressors were down-regulated uniquely following 100 M H2O2. This is direct evidence of an oncogenic transcriptional signature induced specifically following a transient and minimally toxic ROS exposure. We did not observe any significant up-regulation in transcripts of classical antioxidant or DNA repair genes following either minimally toxic or acute dose, similar to studies in yeast models [14].
Results showed that early post-translational response to ROS affects mainly homeostatically phosphorylated proteins rather than phosphorylation of new moieties. Focusing on regulated resting state phosphorylated sites, we discerned no apparent paradigm of stem cell response to multiple genotoxic exposures indicating that unique responses might be specific to each compound. Consistent with the transcriptional data, transient exposure to a minimally toxic dose of of H2O2 specifically increased oncogenic (e.g. hypophosphosphorylation of RAF1 at S259, hyperphosphosphorylation of NPM1 at S4) and metastatic (hypophosphosphorylation of PTK2 at S910) signals common to pathways leading to survival, growth and proliferation, G2/M transition and migration.
We previously demonstrated that etoposide induces similar cytotoxic and genotoxic effects as the minimally toxic ROS used here [48]; however, post-translational response to the two agents was distinct. Early signaling induced by etoposide could be distinguished from that of ROS through activation of β-catenin survival pathway and reduction of integrin and migration signaling as well as an inhibition of the 47kD isoform of JNK. These data provide evidence that a single exposure to mild ROS is sufficient to promote a distinct cellular response marked by significant oncogenic signals that may intitiate cell transformation in a surviving stem cell population.
We were surprised to observe activating marks of survival (e.g. hyperphosphorylated Akt at S473), growth (e.g. hypophosphorylated 70kD RAF1 isoform at S910), and proliferation (e.g. hyperphosphorylated Prkcm at S916 and hypophosphorylation of Ptk2 at S722) in ES cells one hour following a transient exposure to ROS suggesting that ES cells’ initial response to stress is to maintain a rapid growth rate and bypass DNA repair. This initial rapid growth is then temporally followed by the well characterized induction of cell cycle checkpoints and reduced E2F-dependent transcription, manifested by almost complete dephosphorylation of Rb at S773, hyperphosphorylation of cdc2 at Y15, and supported by transcriptional microarray data showing down-regulation of E2F targets such as foxd3.
This study demonstrated that a single minimally toxic exposure to ROS uniquely led to dysregulated insulin signaling. The tyrosine kinase insulin receptor (INSR) is required to mediate insulin signaling, and the early steps of INSR activation are well understood. INSR is a heterotetrameric membrane glycoprotein composed of two and two subunits, linked together by disulfide bonds with activation cascade initiated by binding of insulin to the receptor’s extracellular β-subunit [49,50]. The INSR tyrosine kinase is activated upon binding of insulin binds to the receptor’s extracellular β-subunit, initiating subunit colocalization, conformational changes, autophoshorylation, and activation of the receptor’s kinase activity on intracellular protein substrates [49,50]. Mutations in the INSR gene can reduce receptor autophosphorylation and tyrosine kinase activity toward an exogenous substrate, resulting in both in vivo and in vitro insulin resistance and diabetes mellitus [51–56]. In our study, a transient minimally toxic exposure to ROS mediated by H2O2 was sufficient to induce immediate hypophosphorylation of Y972 providing a direct link between ROS and insulin resistance. Y972F mutation has been shown to cause severe impairment of downstream effector IRS-1 adaptor tyrosine phosphorylation and, thus, downstream signaling of the insulin pathway [57]. Further, Y972 hypophosphorylation in HEK cells was shown to be dependent on Grb14 which is over-expressed in insulin resistance mouse models and human Type II diabetic patients [58]. In support of the suggestion that appropriate insulin signaling is altered by ROS, we also observed an almost 2-fold increase in the inhibitory S21 phosphorylation of GSK3. GSK3 is active in a cell’s resting state and inhibited by insulin, and complete inhibition of GSK3 by acute insulin exposure occurs through phosphorylation of Ser21. It has been shown that over-expression of GSK3 impairs insulin responsiveness while knockdown of GSK3 improves insulin action [59]. GSK3 is elevated in patients with poorly controlled type 2 diabetes and animal models of insulin resistance [60,61]. Taken together, our data shed new light on the possible mechanism of even transient mild ROS exposure on hypophosphorylation of INSR and insulin resistance [58,62,63].
Overall, this screen demonstrated that ES cell early response to ROS is dose dependent and a single transient minimally toxic exposure is sufficient to promote an early post-translational response with significant oncogenic signals supporting the transcriptional data. In addition to new data related to stem cell signaling, this work supports the hypothesis that cancer emanates from a transformed stem cell and underscores the potential role of even a single exposure to ROS to promote this transformation. These data underscore the importance of deciphering methods to either spare wild type stem cells from transformation after ischemia/reperfusion or chemotherapy approaches or to target cancer stem cells.
Materials and Methods
DNA damage and oxidative stress
E14TG2a-derived mouse embryonic stem (ES) cells were cultured as previously described [64,65]. 2 × 107 cells in suspension were exposed to one of the following: 2 ml PBS containing 100μM or 5mM hydrogen peroxide for 15 and 30 min, 20μM etoposide (Sigma-Aldrich; 20mM stock solution prepared in dimethylosulphoxide (DMSO)) for 30 min, or PBS alone. Cells were replated and recovered for 1 hr at 37°C in 5% CO2 before harvest.
Cell cycle analysis and growth arrest
5 × 106 cells were plated in 10 cm dishes and allowed to recover for 24 hr. Adherent cells were harvested and viable cells determined by hemocytometer and trypan blue exclusion or by BrdU labeling.
Pulse Field Gel Electrophoresis
Treated cells were suspended in embedding buffer (15mM Tris—HCl, pH 7.4, 1mM EGTA, 60mM KCl, 15mM NaCl, 2mM EDTA, 0.5mM spermidine, 0.15mM spermine), embedded in 0.8% low melting agarose at 40°C, casted in BioRad plugs (cat# 170-3622) (3 × 105 cells per 50μl) then cooled for 1 min at −20°C. Lipid and protein extraction was performed by two overnight incubations in extraction buffer (10mM Tris-HCl, pH 9.5, 10mM NaCl, 25mM EDTA, 1mM EGTA, 1.5% SDS, 0.1% mercaptoethanol) at room temperature and gentle rocking. Plugs were washed three times in TE pH 7.6 for two hours each followed by RNA digestion with RNase for one hr at 37°C. Proteinase K digestion for 6 hours at 50°C was followed by washing in TE pH 7.6 three times for two hours each. For single strand break analysis, DNA plugs were digested with 3 units S1 nuclease for one hour at 37°C in 200 μl S1 nuclease buffer (30mM NaAc pH 4.6,100mM NaCl, 0.5mM ZnCl2). DNA breaks were analyzed by field inversion gel electrophoresis (FIGE). Plugs containing purified DNA were inserted in wells of 1% 0.5X TBE pulse field-certified agarose gel and resolved by BioRad CHEF Mapper (cat# 170-3670) at 14°C 0.5X TBE buffer circulated by a pump, 20 min of forward voltage (6 v/cm) without field reversion. Resolution of DNA was programmed as: forward voltage: 5 V/cm, forward initial switch time: 0.3s, forward final switch time: 30s, reverse voltage: 5 V/cm, reverse initial switch time: 0.1s, reverse final switch time: 10s, A= linear, Run time: 16 hours. Following electrophoresis, gel was stained with ethidium bromide and DNA visualized by UV.
Microarray hybridization and analysis
Samples were exposed to H2O2 for 15 min, recovered for 1 hr, then harvested for analysis. cRNA derived from 10 μg of total RNA from treated cell samples was prepared and hybridized to MG_U74Av2 oligonucleotide chip according to Affymetrix’s protocol. Prior to filtering, unsupervised hierarchical clustering using standard correlation as a similarity measure algorithm confirmed the relatedness of samples within treatment groups (Supplemental Fig. S1A). “Absent” calls in at least 10 out of 12 samples were filtered, leaving 6970 transcripts (out of 12,488) and consistent with previous data that ES cells express approximately 30% of potential transcripts [66]. Analysis of variance in gene expression between control group of replicates and one of the treated groups was performed using the Welch t-test with a 2 fold or greater change and a p-value of 0.05 or lower. Per gene and chip normalization was used as well as the Cross Gene Error Model. Gene expression normalized values were analyzed using GeneSpring GX software (Agilent Technologies). Data is available at http://www.ncbi.nlm.nih.gov/geo/ Accession number #GSE18708 and http://biology.uncc.edu/Faculty/Richardson/index.htm. SYBR® green and LightCycler® real-time RT-PCR was used to validate data.
Phosphoprotein Analysis
Samples were exposed to H2O2 for 15 min, recovered for 1 hr, then harvested for analysis. Cells were washed in ice cold PBS, lysed in 500 μl lysis buffer (150 mM NaCl, 20 mM Tris pH 8.0, 0.5% (w/v) Nonidet P-40, 1 mM dithiothreitol (DTT), 20 mM (β-glycerophosphate, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin and 1 μg/ml pepstatin A) and sonicated for 30 sec pulsing 1 sec ON 1 sec OFF. Cell debris was removed by centrifugation at 13,000 rpm for 15 min at 4°C. Protein concentration was determined by the Bradford assay. Samples were performed in triplicates. For phosphoscreening the Kinetworks™ platform was used (Kinexus--KPSS-2 and KPSS-4). Images available at http://biology.uncc.edu/Faculty/Richardson/index.htm. 300 μg of total protein were resolved on a 13% single lane SDS-polyacrylamide gel and transferred to nitrocellulose membrane. The membrane was incubated with mixtures of up to three antibodies per lane that react with a distinct subset of at least 95 known phosphorylated sites on 53 cell signaling proteins of distinct molecular masses, then horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology). Blots were developed using ECL Plus reagent (Amersham Biosciences) and signals were quantified using Quantity One software (Bio-Rad). The overall early response to OS or etoposide is similar with a Spearman correlation value of 0.94 between etoposide and low OS dose, 0.89 between etoposide and high dose, and 0.90 between low dose and high dose. However, the correlation of response with respect to treatment drops significantly to 0.3–0.4 when only the affected epitopes are analyzed indicating that each treatment can be correlated with a unique identifier.
Acknowledgments
We gratefully acknowledge the assistance of Vladin M. of the Columbia University Microarray Core Facility in microarray sample preparation. CR is supported by NCI/NIH (2R01-CA100159).
References
- 1.Jackson AL, Loeb LA. Mutation Res. 2001;477:7–21. doi: 10.1016/s0027-5107(01)00091-4. [DOI] [PubMed] [Google Scholar]
- 2.Davies KJ. Biochem Soc Symp. 1995;61:1–31. doi: 10.1042/bss0610001. [DOI] [PubMed] [Google Scholar]
- 3.Beckman KB, Ames BN. J Biol Chem. 1997;272:19633–6. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 4.Waris G, Ahsan H. J Carcinog. 2006;5:14. doi: 10.1186/1477-3163-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burdon RH. Free Radio Biol Med. 1995;18:775–94. doi: 10.1016/0891-5849(94)00198-s. [DOI] [PubMed] [Google Scholar]
- 6.Burdon RH, Alliangana D, Gill V. Free Radio Res. 1995;23:471–86. doi: 10.3109/10715769509065268. [DOI] [PubMed] [Google Scholar]
- 7.Olinski R, Gackowski D, Foksinski M, Rozalski R, Roszkowski K, Jaruga P. Free Radio Biol Med. 2002;33:192–200. doi: 10.1016/s0891-5849(02)00878-x. [DOI] [PubMed] [Google Scholar]
- 8.Wang MC, Bohmann D, Jasper H. Dev Cell. 2003;5:811–6. doi: 10.1016/s1534-5807(03)00323-x. [DOI] [PubMed] [Google Scholar]
- 9.Baysal BE. Trends Endocrinol Metab. 2003;14:453–9. doi: 10.1016/j.tem.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Baysal BE. PLoS ONE. 2007;2:e436. doi: 10.1371/journal.pone.0000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes GR, Lockwood DH. Proc Natl Acad Sci U S A. 1987;84:8115–9. doi: 10.1073/pnas.84.22.8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saretzki G, Armstrong L, Leake A, Lako M, von Zglinicki T. Stem Cells. 2004;22:962–71. doi: 10.1634/stemcells.22-6-962. [DOI] [PubMed] [Google Scholar]
- 13.Francis R, Richardson C. Genes Dev. 2007;21:1064–74. doi: 10.1101/gad.1522807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birrell GW, Brown JA, Wu HI, Giaever G, Chu AM, Davis RW, Brown JM. Proc Natl Acad Sci USA. 2002;99:8778–83. doi: 10.1073/pnas.132275199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weigel AL, Handa JT, Hjelmeland LM. Free Radio Biol Med. 2002;33:1419–32. doi: 10.1016/s0891-5849(02)01082-1. [DOI] [PubMed] [Google Scholar]
- 16.Anantharam V, Lehrmann E, Kanthasamy A, Yang Y, Banerjee P, Becker KG, Freed WJ, Kanthasamy AG. Neurochem Int. 2007;50:834–47. doi: 10.1016/j.neuint.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purdom-Dickinson SE, Lin Y, Dedek M, Morrissy S, Johnson J, Chen QM. J Mol Cell Cardiol. 2007;42:159–76. doi: 10.1016/j.yjmcc.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang YY, et al. Cancer Res. 2002;62:6246–54. [PubMed] [Google Scholar]
- 19.Yu Q, He M, Lee NH, Liu ET. J Biol Chem. 2002;277:13059–66. doi: 10.1074/jbc.M111403200. [DOI] [PubMed] [Google Scholar]
- 20.Amundson SA, Do KT, Vinikoor L, Koch-Paiz CA, Bittner ML, Trent JM, Meltzer P, Fornace AJ., Jr Oncogene. 2005;24:4572–9. doi: 10.1038/sj.onc.1208653. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Fong CC, Wong MS, Tzang CH, Lai WP, Fong WF, Sui SF, Yang M. Apoptosis. 2005;10:545–56. doi: 10.1007/s10495-005-1885-0. [DOI] [PubMed] [Google Scholar]
- 22.Islaih M, et al. Mutation Res. 2005;578:100–16. doi: 10.1016/j.mrfmmm.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Allen RG, Tresini M. Free Radio Biol Med. 2000;28:463–99. doi: 10.1016/s0891-5849(99)00242-7. [DOI] [PubMed] [Google Scholar]
- 24.Halliwell B, Clement MV, Long LH. FEBS Letters. 2000;486:10–3. doi: 10.1016/s0014-5793(00)02197-9. [DOI] [PubMed] [Google Scholar]
- 25.Hardman J, Limbird L, Goodman Gilman A. A pharmacological basis of therapeutbs. Mc-Graw Hill; New York: 2001. [Google Scholar]
- 26.Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, Wahl GM. Curr Biol. 1998;8:145–55. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- 27.Corbet SW, Clarke AR, Gledhill S, Wyllie AH. Oncogene. 1999;18:1537–44. doi: 10.1038/sj.onc.1202436. [DOI] [PubMed] [Google Scholar]
- 28.Sabapathy K, Klemm M, Jaenisch R, Wagner EF. EMBO J. 1997;16:6217–29. doi: 10.1093/emboj/16.20.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mouzannar R, Miric SJ, Wiggins RC, Konat GW. Neurochem Int. 2001;38:9–15. doi: 10.1016/s0197-0186(00)00066-8. [DOI] [PubMed] [Google Scholar]
- 30.Furuno-Fukushi I, Tatsumi K, Takahagi M, Tachibana A. Int J Radiat Biol. 1996;70:209–17. doi: 10.1080/095530096145201. [DOI] [PubMed] [Google Scholar]
- 31.Vilenchik MM, Knudson AG. Proc Natl Acad Sci U S A. 2003;100:12871–6. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckfeldt CE, Mendenhall EM, Verfaillie CM. Nat Rev Mol Cell Biol. 2005;6:726–37. doi: 10.1038/nrm1713. [DOI] [PubMed] [Google Scholar]
- 33.Duval D, et al. Cell Death Differ. 2006;13:564–75. doi: 10.1038/sj.cdd.4401789. [DOI] [PubMed] [Google Scholar]
- 34.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 35.Yan T, Oberley LW, Zhong W, St Clair DK. Cancer Res. 1996;56:2864–71. [PubMed] [Google Scholar]
- 36.Zhong W, Oberley LW, Oberley TD, St Clair DK. Oncogene. 1997;14:481–90. doi: 10.1038/sj.onc.1200852. [DOI] [PubMed] [Google Scholar]
- 37.Rathjen J, Lake JA, Bettess MD, Washington JM, Chapman G, Rathjen PD. J Cell Sci. 1999;112 ( Pt 5):601–12. doi: 10.1242/jcs.112.5.601. [DOI] [PubMed] [Google Scholar]
- 38.Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. Development. 2005;132:885–96. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 39.Katoh Y, Katoh M. Int J Oncol. 2005;27:581–5. [PubMed] [Google Scholar]
- 40.Kido Y, Nakae J, Accili D. J Clin Endocrinol Metab. 2001;86:972–9. doi: 10.1210/jcem.86.3.7306. [DOI] [PubMed] [Google Scholar]
- 41.Coffer PJ, Jin J, Woodgett JR. Biochem, J. 1998;335 (Pt 1):1–13. doi: 10.1042/bj3350001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaneko S, et al. J Pharmacol Sci. 2006;101:66–76. doi: 10.1254/jphs.fp0060128. [DOI] [PubMed] [Google Scholar]
- 43.Bai H, Konat GW. Neurochem Int. 2003;42:123–9. doi: 10.1016/s0197-0186(02)00072-4. [DOI] [PubMed] [Google Scholar]
- 44.Wijeratne SS, Cuppett SI, Schlegel V. J Agric Food Chem. 2005;53:8768–74. doi: 10.1021/jf0512003. [DOI] [PubMed] [Google Scholar]
- 45.Konat GW, Mouzannar R, Bai H. Neurochem Int. 2001;39:179–86. doi: 10.1016/s0197-0186(01)00030-4. [DOI] [PubMed] [Google Scholar]
- 46.Han W, Takano T, He J, Ding J, Gao S, Noda C, Yanagi S, Yamamura H. Antioxid Redox Signal. 2001;3:1065–73. doi: 10.1089/152308601317203576. [DOI] [PubMed] [Google Scholar]
- 47.Raffaghello L, Lee C, Safdie FM, Wei M, Madia F, Bianchi G, Longo VD. Proc Natl Acad Sci USA. 2008;105:8215–20. doi: 10.1073/pnas.0708100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Libura J, Slater DJ, Felix CA, Richardson C. Blood. 2005;105:2124–31. doi: 10.1182/blood-2004-07-2683. [DOI] [PubMed] [Google Scholar]
- 49.Hubbard SR. EMBO J. 1997;16:5572–81. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hubbard SR, Wei L, Ellis L, Hendrickson WA. Nature. 1994;372:746–54. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
- 51.Moller DE, Flier JS. N Engl J Med. 1988;319:1526–9. doi: 10.1056/NEJM198812083192306. [DOI] [PubMed] [Google Scholar]
- 52.Odawara M, et al. Science. 1989;245:66–8. doi: 10.1126/science.2544998. [DOI] [PubMed] [Google Scholar]
- 53.Taira M, et al. Science. 1989;245:63–6. doi: 10.1126/science.2544997. [DOI] [PubMed] [Google Scholar]
- 54.Grunberger G, Zick Y, Gorden P. Science. 1984;223:932–4. doi: 10.1126/science.6141638. [DOI] [PubMed] [Google Scholar]
- 55.Le Marchand-Brustel Y, Gremeaux T, Ballotti R, Van Obberghen E. Nature. 1985;315:676–9. doi: 10.1038/315676a0. [DOI] [PubMed] [Google Scholar]
- 56.Freidenberg GR, Henry RR, Klein HH, Reichart DR, Olefsky JM. J Clin Invest. 1987;79:240–50. doi: 10.1172/JCI112789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaburagi Y, et al. J Biol Chem. 1993;268:16610–22. [PubMed] [Google Scholar]
- 58.Nouaille S, Blanquart C, Zilberfarb V, Boute N, Perdereau D, Burnol AF, Issad T. Biochem Pharmacol. 2006;72:1355–66. doi: 10.1016/j.bcp.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 59.Ciaraldi TP, Nikoulina SE, Bandukwala RA, Carter L, Henry RR. Endocrinology. 2007;148:4393–9. doi: 10.1210/en.2006-0932. [DOI] [PubMed] [Google Scholar]
- 60.Eldar-Finkelman H, Schreyer SA, Shinohara MM, LeBoeuf RC, Krebs EG. Diabetes. 1999;48:1662–6. doi: 10.2337/diabetes.48.8.1662. [DOI] [PubMed] [Google Scholar]
- 61.Nikoulina SE, Ciaraldi TP, Mudaliar S, Mohideen P, Carter L, Henry RR. Diabetes. 2000;49:263–71. doi: 10.2337/diabetes.49.2.263. [DOI] [PubMed] [Google Scholar]
- 62.Dokken BB, Saengsirisuwan V, Kim JS, Teachey MK, Henriksen EJ. Endocrinol Metab. 2008;294:E615–21. doi: 10.1152/ajpendo.00578.2007. [DOI] [PubMed] [Google Scholar]
- 63.Goldstein BJ, Mahadev K, Wu X, Zhu L, Motoshima H. Antioxid Redox Signal. 2005;7:1021–31. doi: 10.1089/ars.2005.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. Nature. 1987;326:292–5. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 65.Richardson C, Jasin M. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 66.Eckfeldt CE, Mendenhall EM, Flynn CM, Wang TF, Pickart MA, Grindle SM, Ekker SC, Verfaillie CM. PLoS Biol. 2005;3:e254. doi: 10.1371/journal.pbio.0030254. [DOI] [PMC free article] [PubMed] [Google Scholar]