Abstract
Rac1, a protein of the Rho GTPase subfamily, has been implicated in neuronal and spine development as well as the formation of synapses with appropriate partners. Dendrite and spine abnormalities have been implicated in several psychiatric disorders such as Fragile-X syndrome, where neurons show a high density of long, thin, and immature dendritic spines. Although abnormalities in dendrites and spines have been correlated with impaired cognitive abilities in mental retardation, the causes of these malformations are not yet well understood. Fragile X syndrome is the most common type of inherited mental retardation caused by the absence of FMRP protein, a RNA-binding protein implicated in the regulation of mRNA translation and transport, leading to protein synthesis. We suggest that FMRP might act as a negative regulator on the synthesis of Rac1. Maintaining an optimal level of Rac1 and facilitating the reorganization of the cytoskeleton likely leads to normal neuronal morphology during activity-dependent plasticity. In our study, we first demonstrated that Rac1 is not only associated but necessary for normal spine development and long-term synaptic plasticity. We further showed that, in Fmr1 knockout mice, lack of FMRP induces an overactivation of Rac1 in the mouse brain and other organs that have been shown to be altered in Fragile X syndrome. In those animals, pharmacological manipulation of Rac1 partially reverses their altered long-term plasticity. Thus, regulation of Rac1 may provide a functional link among deficient neuronal morphology, aberrant synaptic plasticity and cognition impairment in Fragile X syndrome.
Keywords: Rac1, Fragile X syndrome, plasticity, LTP
1. INTRODUCTION
Fragile X Syndrome (FXS) is the most common form of mental retardation after Down’s syndrome. FXS is due to the absence of FMRP (Verkerk et al., 1991), an RNA-binding protein implicated in the regulation of mRNA transport and translation during protein synthesis (O’Donnell and Warren, 2002). FMRP is encoded by the Fmr1 gene and in normal individuals is expressed in many tissues being particularly abundant in brain neurons and gonad (Devys et al., 1993; Bakker et al., 2000). FMRP is expressed in the hippocampus (a brain area associated with learning and memory), as well as in Purkinje cells of the cerebellum (Abitbol et al., 1993; Devys et al., 1993). In neurons, this protein is involved in regulation of translation and transport of mRNAs (Brown et al., 2001; Jin and Warren, 2003; Miyashiro et al., 2003). A large body of data suggests that FMRP is involved in synaptic plasticity and dendrite morphogenesis by regulating protein synthesis of specific mRNAs in response to synaptic stimulation (Weiler et al., 1997; Antar et al., 2005). FMRP organizes the translation of messages important for synaptic structural changes (Weiler et al., 1997; Weiler et al., 2004; Willemsen et al., 2004). Failure to regulate protein synthesis can profoundly affect synapse morphology, as seen in neocortex of FXS patients (Irwin et al., 2001) and in neocortex and hippocampus of adult Fmr1 knockout mice, where an increased density of abnormally long dendritic spines has been observed (Comery et al., 1997; Braun and Segal, 2000; Greenough et al., 2001; Galvez & Greenough, 2005; Grossman et al., 2006). Moreover, abnormal spine morphogenesis has been associated with cognitive impairment and mental retardation (Purpura, 1974). Thus, FMRP can have a twofold role in synaptic plasticity, delivering selected mRNAs to synaptic locations, and releasing mRNAs for activity-dependent translation (Davidovic et al., 2005).
Neuronal plasticity requires architectural changes in dendrites and spines (Yuste and Bonhoeffer, 2001) through modification of the actin cytoskeleton (Lin et al., 2005). It has been reported that these changes in spine morphogenesis involve actin-regulatory proteins with a direct role on the stabilization of individual filaments (Lynch et al., 2007; Rex et al., 2009; Rex et al., 2010). However, the upstream signaling molecules that trigger these actin dynamics remain unidentified. Time lapse imaging studies have suggested that changes in spine morphology are controlled by glutamatergic activity (Engert and Bonhoeffer, 1999; Maletic-Savatic et al., 1999; Chen et al., 2000) and modifications of the actin cytoskeleton regulated by small GTPases (Linseman et al., 2000; Luo et al., 1996; Nakayama et al., 2000; Tashiro et al., 2000).
Rac1 is a member of the Rho-family of small GTPases and it is best known for its role in regulating actin polymerization. Rac1 also has a very well known role in superoxide generation, gene transcription (Van Aelst and D’Souza-Schorey, 1997), and G1 cell cycle progression (Minden et al., 1995). Rac1 cycles between an inactive (GDP-bound) and an active (GTP-bound) conformation through the catalytic action of guanine exchange factors (GEF) and GTPase-activating proteins (GAP). Rac1 also cycles between cytosolic and membrane-associated forms through the action of guanine dissociation inhibitor (GDI) proteins. How these two cycles are coupled is critical for the proper interaction of Rac1 with their targets (Harwood and Braga, 2003). Recent studies have proposed a model for Rho GTPase-mediated dendritic arbor growth (Li et al., 2000; Li et al., 2002). Neural activity, through glutamatergic receptors activates Rac1 (Tejada-Simon et al., 2006) and decreases RhoA activation. This tight coordination leads to elaboration of dendritic arbors (Li et al. 2002).
Rac1 functions in the development and structural plasticity of dendrites and dendritic spines (Luo et al., 1996; Threadgill et al., 1997; Ruchhoeft et al., 1999; Lee et al., 2000; Nakayama et al., 2000; Tashiro et al., 2000; Wong et al., 2002). Additionally, Rac1 signaling pathway has been implicated in learning (Diana et al., 2007; Martinez et al., 2007; Haditsch et al., 2009), as well as some forms of cognitive disorders and X-linked mental retardation syndromes (Chelly and Mandel, 2001; Chen et al., 2010) that are characterized with abnormalities in dendritic spine structure. A consistent abnormality observed in brain of FXS patients and of Fmr1 knockout mice is abnormal dendritic spine morphology (Comery et al., 1997; Irwin et al., 2000), a phenomenon that requires actin cytoskeleton remodeling and local protein translation in response to synaptic activity. FMRP and Rac1 have both been observed in dendritic spines, and they are believed to be involved in neuronal plasticity. Several lines of evidence suggest that FMRP may modulate spine morphogenesis via proteins related to Rac1 signaling (Luo et al., 1996; Kobayashi et al., 1998; Nakayama et al., 2000; Schenck et al., 2001; el Bekay et al., 2007; de Diego-Otero et al., 2008; Chen et al., 2010), suggesting that FMRP and Rac1 could control actin cytoskeleton remodeling in neuronal plasticity through a common pathway. Thus, dysregulation of local translation affecting FMRP, Rac1 and other associated factors at synapses may impair normal neuronal plasticity contributing to the cognitive deficits associated with FXS.
We hypothesize that FMRP regulates synthesis of Rac1 and that in FXS, lack of FMRP results in hyperactivation of Rac1, leading to defects in spine morphology and synaptic plasticity. This study was undertaken to first verify the importance of Rac1 in dendritic spine development and synaptic plasticity, and to further examine a possible link for Rac1 in FXS. Herein, we are reporting that Rac1 is indeed associated and necessary for normal spine development and long-term synaptic plasticity, both abnormal in FMRP-deficient mice. We found that Rac1 synthesis and activation state are elevated in the Fmr1 knockout mouse. Thus, regulation of Rac1 signaling may provide a functional link among altered dendritic spines, aberrant synaptic plasticity and cognition impairment in FXS, pointing to Rac1 as a novel target for development of therapeutic strategies in Fragile X syndrome.
2. RESULTS
2.1 Abnormal Rac1-dependent dendritic spine morphology
Rac1 is involved in the structural changes that take place in dendrites and spines upon activity (Luo et al., 1996; Threadgill et al., 1997; Ruchhoeft et al., 1999; Lee et al., 2000; Nakayama et al., 2000; Tashiro et al., 2000; Wong et al., 2002). It has been reported that the shape and density of actin-rich dendritic spines is altered in patients with FXS as well as in Fmr1 knockout mice (Comery et al., 1997; Irwin et al., 2000). We hypothesize that this likely reflects a problem in the regulation of Rac1. In this study, we first wanted to assess whether in healthy animals shutting down Rac1 activity in the hippocampus was indeed critical for dendritic spine morphology and long-term plasticity. Mice carrying a floxed allele of Rac1 (Rac1flox) were crossed with mice expressing the Cre recombinase. By using the Cre-LoxP system we removed Rac1 function in mature hippocampus creating transgenic mice that express Cre recombinase under the control of the CamKII promoter (Tsien et al., 1996). Using this approach we generated a mouse with a brain specific deletion of the Rac1 gene (Rac1 mutant) in the hippocampus. For all animals the genotype was determined by PCR (Figure 1A), using the primers described. Rac1 mutant mice were grossly indistinguishable from wild-type and heterozygous littermates and showed a similar survival rate. During development we detected no gross abnormalities in the size or histological structure of the brain based on Nissl-stained sections (Figure 1B), technique commonly used to identify basic neuronal structure.
Figure 1. Development of a forebrain-specific Rac1 mutant mouse model.
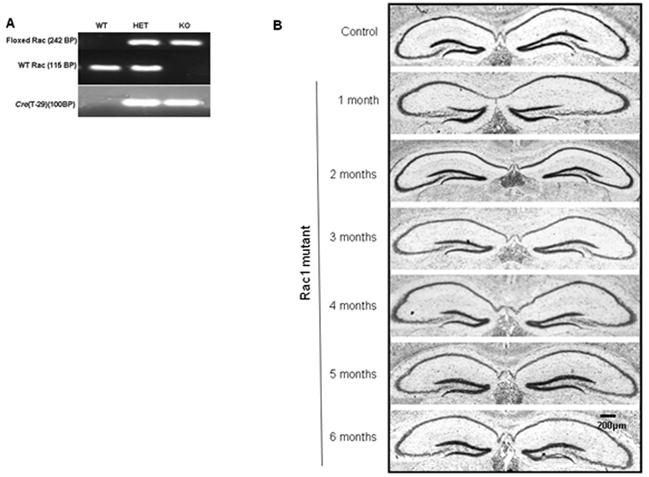
A) Genotype determination by PCR of control mice (WT, HET) and a forebrain-specific Rac1 mutant mouse (KO) at 21 days, B) Nissl staining of control (only 3 month old shown for simplicity) and Rac1 mutant mice (1-6 months) used to compare the gross morphology of the hippocampus.
To confirm loss of Rac1, its location and to determine the time at which this event happened, we used immunoblotting with a Rac1 specific antibody. At different ages (1-6 months) brain extracts of Rac1 mutant mice showed reduction of Rac1 in the CA1 region as compared to wild-type and heterozygous littermates (Figure 2A), demonstrating that Cre-mediated removal of Rac1 was effective. Since the Cre recombinase mouse line used in this study can work over the entire forebrain, we also analyzed other brain regions for ablation of Rac1. Reduction of Rac1 was also apparent in the CA3/dentate gyrus region as well as cortex at 5 months of age, but as expected no loss of Rac1 was detected in any other organ besides the brain (Figure 2A). Heterozygous brains showed no difference as compared to the wild-type brains.
Figure 2. Loss of Rac1 protein and mRNA expression in a newly developed forebrain specific Rac1 mutant mouse model.
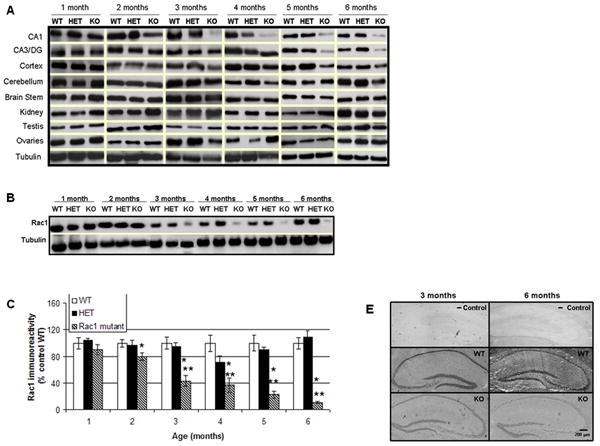

A) Representative immunoblots depicting levels of Rac1 protein in the brain and other tissues of 1-6 months old Rac1 mutant (KO) and control mice (wild type, WT and heterozygotes, HET), B) Representative E-PAGE® depicting gradual loss of Rac1 protein in the CA1 area of 1-6 months old Rac1 mutant mice (KO) relative to controls (wild type, WT and heterozygotes, HET), C) Group data (mean ± SEM, n=6) of Rac1 immunoreactivity in CA1 area of Rac1 mutant mice relative to control mice. Asterisks denote statistically significant difference respect to * wild type (WT) or ** heterozygote (HET) (p<0.05, ANOVA), D) Double staining showing localization and expression of Rac1 in hippocampal slices. Hippocampal slices were stained with anti-Rac1 (green) and DAPI (blue). Images show immunoreactive Rac1 not colocalized with DAPI (nuclear marker) and disappearing in the Rac1 mutant mice, E) In situ hybridization showing expression of Rac1 mRNA in Rac1 mutant (KO) and control animals (WT) at 3 and 6 months of age. A negative control (- Control) indicates no unspecific binding.
Quantification of protein levels in the CA1 area revealed that loss of Rac1 is statistically significant at 2 months (20 % reduction) and continues decreasing as the Rac1 mutant mice aged (~50% at 3 months, ~60% at 4 months, ~75% at 5 months and 85% at 6 months; Figure 2B, 2C). Following our previous observations that Rac1 is localized in the dendritic area at the synaptic site (Tejada-Simon et al., 2006), we sought to confirm the cellular reduction of Rac1 in hippocampal slices using immunohistochemistry. Using hippocampal slices derived from Rac1 mutant animals (3 months old), we probed for Rac1 alongside the nuclear marker DAPI (neuronal body). Control mice showed Rac1 localized in dendritic areas. However, and in line with our biochemical results, Rac1 was dramatically reduced in dendrites derived from the mutant animals (Figure 2D).
To further verify that the brain specific deletion of Rac1 was effective at the mRNA level, in situ hybridization on 3 and 6 months old mice was performed. In situ hybridization confirmed loss of Rac1 mRNA in the CA1 hippocampal region (3 and 6 months) as well as the CA3 and dentate gyrus (6 months old, Figure 2E), while the distribution of Rac1 mRNA was normal in wild-type (WT) and heterozygous animals. To demonstrate that these outcomes are not due to unspecific binding by the Rac1 biotin oligomer probe used or the detection enzyme, negative controls were run in parallel. No hybridization was observed when either probe or enzyme was omitted.
It is worth to note that some residual expression of Rac1 mRNA and protein was always detected, what it is likely attributable to expression in glial cells.
Loss of Rac1 might induce compensation of other related members of the Rho GTPase family. Thus, we estimated biochemically the levels of Cdc42, RhoA and RhoB. Loss of Rac1 did not affect the levels of related Rho GTPases in the brain of Rac1 mutant mice, that is Cdc42 (Figure 3A and 3F), Rho A (Figure 3B and 3G) and Rho B (Figure 3C and 3H).
Figure 3. Effect of Rac1 gene ablation on other related Rho GTPases and signaling pathways.
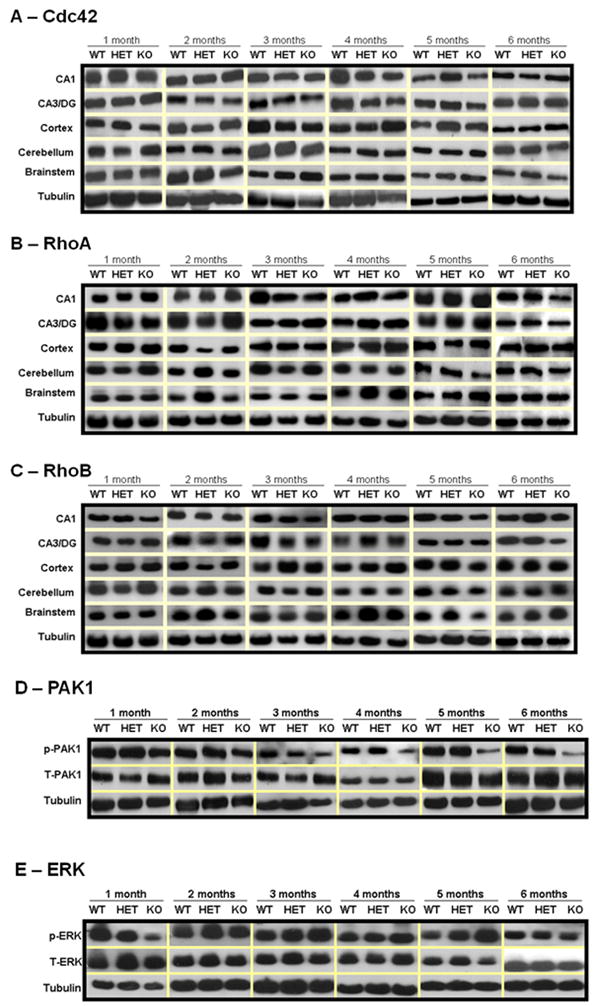
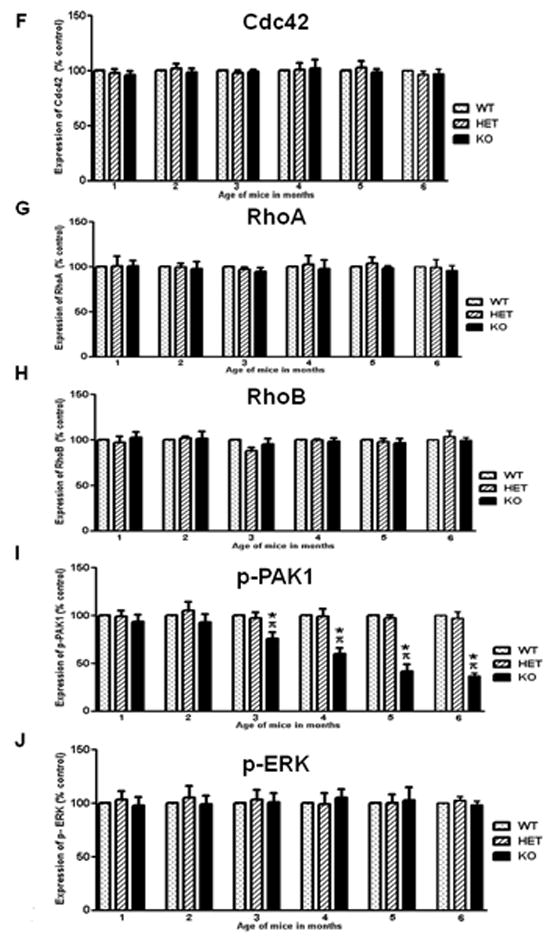
Representative immunoblots depicting levels of A) Cdc42, B) RhoA, C) RhoB, D) p-PAK1 and E) p-ERK, in wild type (WT), heterozygous (HET) and Rac1 mutant (KO) mice (n=6). Group data (mean ± SEM, n=6) of F) Cdc42, G) RhoA, H) RhoB, I) p-PAK1 and J) p-ERK immunoreactivity in CA1 area of Rac1 mutant mice relative to control mice. Asterisks denote statistically significant difference respect to control mice (WT, HET) (p<0.05, ANOVA). Where appropriate, data have been normalized to total PAK1 or total ERK.
Following our observations that Rac1 is activated via NMDA receptors (Tejada-Simon et al., 2006), during hippocampus-dependent learning through the activation of PAK1 (Martinez et al., 2007), we sought to investigate whether PAK1, a Rac1 effector protein associated with actin cytoskeleton remodeling and spine morphology was affected in the Rac1 mutant mouse. Rac1 mutant mice displayed reduced levels of p-PAK1 as compared to control mice (Figure 3D and 3I). This is consistent with the idea that loss of Rac1 alters actin dynamics in neurons by modulating PAK1 activity. Rac1 participates also in transcription through activation of ERK, however we did not find any alteration on p-ERK in the Rac1 mutant mice as compared to controls (Figure 3E and 3J).
We next wanted to corroborate that neuronal morphology is indeed affected by the genetic deletion of Rac1 in the Rac1 mutant as reported by others (Hadistch et al., 2009). Golgi-Cox staining of both Rac1 mutant and littermate control mouse brains was used to investigate dendritic structure and spine density in pyramidal cells from the hippocampus (Figure 4A-F). Alterations were detected at the hippocampal level, where especially spine density appears to be significantly reduced in the Rac1 mutant compared with littermate controls (Figure 4G) following ablation of Rac1 (3 and 6 months).
Figure 4. Golgi-Cox impregnated neurons in the hippocampus of control (WT) and Rac1 mutant mice (KO).

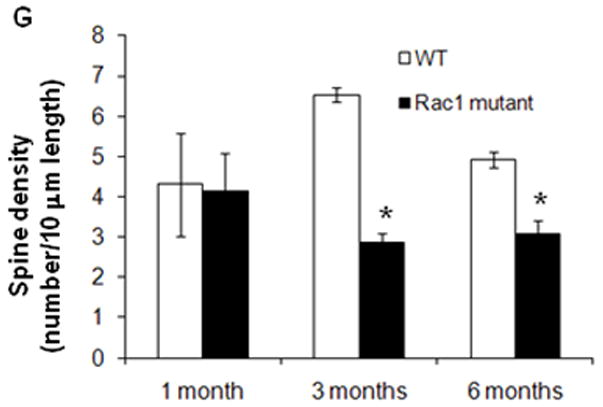
Panels A1-2, B1-2, C1-2, D1-2, E1-2, F1-2 illustrate the morphology of neurons in the CA1 (10 X, scale bar = 60μm). Panels A3-4, B3-4, C3-4, D3-4, E3-4, F3-4 show a close up of a dendritic branch and spines (100 X, scale bar = 5μm). G) Quantification of spine densities in the dendrites of CA1 neurons of Rac1 mutant mice showed a significant decrease in spine number starting at 3 months of age respect to control mice (WT) (* p<0.01, t-test)
2.2 Rac1 association with long-term synaptic plasticity
Besides dendritic spine morphology, long-term plasticity has been shown to be also altered in FXS. We and others have suggested that Rac1 might be critical for these two phenomena (Hadistch et al., 2009; Martinez and Tejada-Simon, 2011). Thus, searching for a connection between Rac1 and FXS, we next studied whether Rac1 is involved not only in LTP but also in LTD, and whether Rac1 inhibition alters this type of plasticity. This is very relevant since an exaggerated LTD is one of the strongest phenotypes observed in Fmr1 knockout mice. Towards this end, LTD was induced in hippocampal slices from wild-type mice treated with a Rac1 inhibitor, NSC23766 (Gao et al., 2004). LTD was induced either with PP-LFS delivered at 1Hz for 15 min (Huber et al., 2001), or by treating the hippocampal slices with 100μM DHPG for 5 min in the presence of the N-methyl-D-aspartate (NMDA) receptor antagonist D,L-2-amino-5-phosphonovalerate (D,L-AP5, 100 μM; Nosyreva and Huber, 2006). The slope of the fEPSP was expressed as percent of the baseline averaged and expressed as the mean ± SEM and compared among responses from all mice before and after LTD induction. Our results indicate that application of NSC23766 to hippocampal slices of wild-type mice inhibits LTD regardless of the induction protocol (Figure 5A, 5B). To confirm the involvement of Rac1, LTD was also induced in hippocampal slices from Rac1 mutant mice in the same manner as described above. Control slices derived from wild-type mice showed a significant lasting decrease in the fEPSP slope. However, slices derived from the Rac1 mutant mice were unable to sustain this response (Figure 5C). These results further suggest that Rac1 is required and also important for LTD.
Figure 5. Rac1 is also associated with long-term depression.
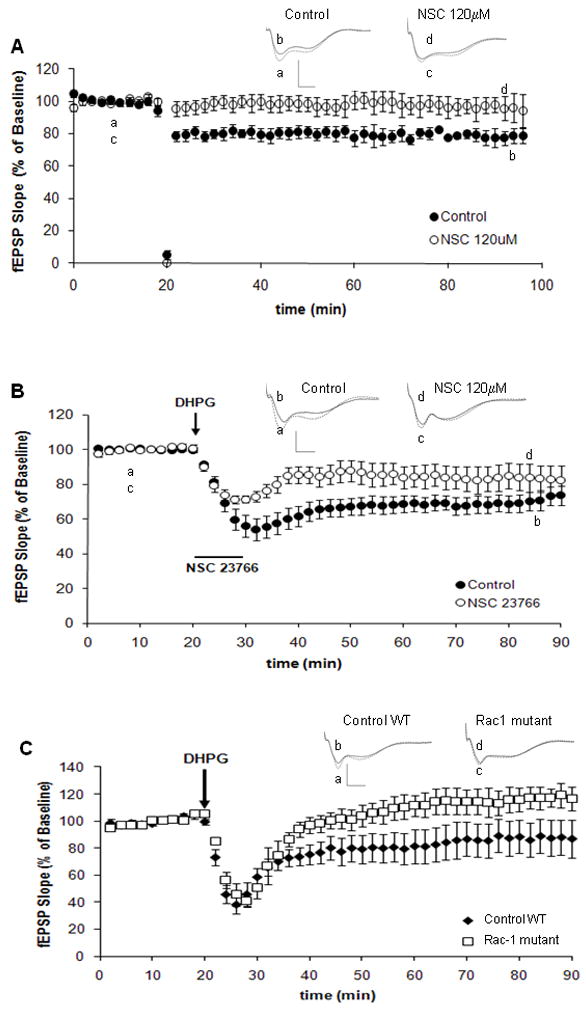
A) LTD-inducing stimulation is blocked by NSC23766. LTD was induced in hippocampal slices with LFS delivered at 1 Hz for 15 min. Perfusion of slices with NSC23766 (120μM) altered LTD induction (n=8). Control slices (closed circles), NSC23766 treated slices (open circles). Inset shows fEPSP sample traces recorded during baseline (a, c) and 1hr after LFS (b, d). Scale: 15mV, 5ms; p < 0.05, two-way ANOVA, B) LTD was induced in hippocampal slices with 100 μM DHPG for 5 min. All experiments were performed in the presence of the N-methyl-D-aspartate inhibitor APV (100 μM). Perfusion of slices with NSC23766 (120μM) altered LTD induction (n=8). Control slices (closed circles), NSC23766 treated slices (open circles). Inset shows fEPSP sample traces recorded during baseline (a, c) and 1hr after DHPG (b, d). Scale: 15mV, 5ms; p < 0.05, two-way ANOVA, C) LTD was induced in hippocampal slices from wild type and Rac 1 mutant mice with 100 μM DHPG for 5 min. Rac1 mutant mice presented altered LTD induction (n=8). Slices from control wild type (closed circles) or Rac1 mutant (open squares). Inset shows fEPSP sample traces recorded during baseline (a, c) and 1hr after DHPG (b, d). Scale: 15mV, 5ms; p < 0.05, two-way ANOVA.
2.3 Elevated levels of Rac1 in the Fmr1 knockout mouse
After verifying that Rac1 modulates spine density and synaptic plasticity, we wanted to examine whether Rac1 has a role in the alteration of spines and plasticity reported in FXS. Rac1 signaling is important for NMDA receptor-dependent learning and memory (Diana et al., 2007; Martinez et al., 2007; Haditsch et al., 2009). Furthermore, Rac1 has been implicated in some forms of cognitive disorders and X-linked mental retardation syndromes that present abnormalities in dendritic spine structure. One of these syndromes bearing these abnormalities is FXS (Chelly and Mandel, 2001). Thus, we aimed to examine the levels of Rac1 in the brain of Fmr1 knockout mice at different stages of development. Relative to age-matched wild type littermates, Rac1 was elevated in brain either at 1 or 3 months of age (Figure 6A). It is worth to note that at 3 months the brainstem showed unusual high levels of Rac1 compared to the other brain regions tested. Rac1-GTP, which represents the active form of this protein, was also elevated, with high levels in the brainstem, as well as the hippocampus and cortex (both associated to learning and memory, Figure 6B). While expression of Rac1 was upregulated at 3 months in Fmr1 knockout mice, Rac1-GTP appeared elevated but no longer significant in the hippocampus.
Figure 6. Rac1 is upregulated in Fmr1 knockout mice.
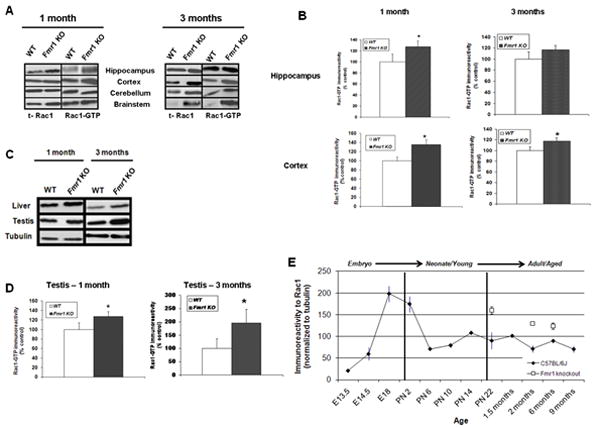
A) Representative Western Blots depicting relative upregulation of Rac1 and active Rac1 (Rac1-GTP) in brain lysates from different regions of the Fmr1 knockout mice as compared to littermate controls (1 and 3 months old, n=8), B) Group data (mean ± SEM, n=8) of Rac1-GTP immunoreactivity in the hippocampus and cortex of wild type (WT,white bars) and Fmr1 knockout (filled bars) mice (1 and 3 months old). Data have been normalized to tubulin as control. * Statistically significant difference respect to control, p<0.05, t-test. C) Representative immunoblots depicting up-regulation of Rac1-GTP in the liver and testis of control (WT) and Fmr1 knockout mice (1 and 3 months old, n=8), D) Group data (mean ± SEM, n=8) of Rac1-GTP immunoreactivity in testis of wild type (WT, white bars) and Fmr1 knockout (filled bars) mice (1 and 3 months old). Data have been normalized to tubulin as control. * Statistically significant difference respect to wild type control, p<0.05, t-test. E) Group data (mean ± SEM, n=6) of Rac1 immunoreactivity in the hippocampal CA1 area of wild type mice from embryonic stage to adulthood (E13.5-9 months) and from Fmr1 knockout mice (1-6 months of age).
Besides cognitive impairment, FXS patients present a variety of other phenotypic characteristics that affect specific organs and are related to cell growth, cell motility, surface remodeling and organization of the actin cytoskeleton, where Rac1 is a key regulator. Thus, we also examined the levels of Rac1-GTP in different organs of the Fmr1 knockout mouse. We found that levels of Rac1-GTP were higher not only in the brain of Fmr1 knockout mice, but also in the other organs examined such as liver (tumorigenic in FXS; Wirojanan et al., 2008) and testis (enlarged in FXS and known as macroordism; Pourbagher et al., 2005) (Figure 6C). Analysis of the levels of Rac1-GTP in testis of Fmr1 knockout mice revealed that this protein is significantly upregulated in these organs (Figure 6D, p<0.05).
To more directly address Rac1 hyperactivity in the brain of Fmr1 knockout mice as related to cognition, we first determined the expression pattern of Rac1 in control mice from embryonic stage (E13) to aged animals (9 months old) in the CA1 area of the hippocampus. We then compared to Fmr1 knockout mice from 1 to 6 months of age. Toward this end, we prepared lysates from hippocampal area CA1 of all these animals, and assessed Rac1 levels by immunoblotting. Hippocampal CA1 lysates from Fmr1 knockout mice exhibited increased Rac1 levels at young age (1 month, 75% increase relative to control group; Figure 6E). Rac1 protein decreased in older Fmr1 knockout mice (3 and 6 months), but still showed higher levels of Rac1 as compared to controls (66% and 48% respectively; Figure 6E). Together, our results suggest that Rac1 is overexpressed in the Fmr1 knockout mice, pointing to FMRP as a regulator of Rac1 synthesis and activation, regulation that is altered in Fmr1 knockout animals due to the ablation of this protein.
2.4 Synaptic transmission of Fmr1 knockout mice after pharmacological reduction of Rac1
Since Rac1 is upregulated in the Fmr1 knockout mouse, a promising way to manipulate these observed high levels of Rac1 is using a pharmacological approach, using specific Rac1 inhibitors. We have shown that Rac1 is involved in long-term plasticity and that Rac1 inhibition alters plasticity (Martinez and Tejada-Simon, 2011). Previously it has been reported that Fmr1 knockout mice present exaggerated LTD. To test whether Rac1 inhibition by pharmacological manipulation affects LTD in Fmr1 knockout mice, we applied NSC23766 to hippocampal slices from Fmr1 knockout mice during electrophysiology experiments. LTD was induced as described before. Our results indicate that application of a Rac1 inhibitor (NSC23766) to hippocampal slices from Fmr1 knockout mice decreases LTD expression in these animals as compared to Frm1 knockout slices used as control (Figure 7).
Figure 7. Pharmacological inhibition of Rac1 decreases LTD expression in Fmr1 knockout mice.

LTD was induced in hippocampal slices derived from either wild type C57BL6/J mice (WT control, solid line) or Fmr1 knockout mice (Fmr1 KO control, closed circles) with 100 μM DHPG for 5 min (n=12). Hippocampal slices derived from Fmr1 knockout mice were perfused with 120μM NSC23766 (Fmr1 KO + NSC, open squares; n=8). Inset shows fEPSP sample traces recorded during baseline (a, c) and 1hr after DHPG (b, d). Scale: 15mV, 5ms; p > 0.05, two-way ANOVA.
Together our results suggest that Rac1 is altered in Fmr1 knockout mice and likely affects both synapse structure and function in the mature brain, pointing to this protein as a potential candidate for the synaptic alterations in structure and function reported in Fmr1 knockout mice as a model of FXS.
3. DISCUSSION
FXS is a common form of mental retardation. One of the main tools used to study FXS is the Fmr1 knockout mouse. These animals, which are missing the FMRP protein, present altered behavior and learning (Galvez and Greenough, 2005), altered cortical LTP (Li et al., 2002), and enhanced hippocampal LTD (Huber et al., 2002) together with an aberrant dendritic spine density and morphology (Comery et al., 1997; Irwin et al., 2000). Interestingly, overexpression of the small GTPase Rac1 results also in similar abnormal structure of dendritic spines (Luo et al., 1996). Overexpression of FMRP over the Fmr1 knockout background produces opposing responses in this animal model (Peier et al., 2000). In this study we focused on establishing a possible connection between the small GTPase Rac1, a protein that regulates actin cytoskeleton remodeling, and the abnormal spine morphogenesis and altered plasticity associated with FXS. FMRP and Rac1 have both been observed in dendritic spines and, in drosophila, FMRP binds to the mRNA encoding Rac1 (Lee et al., 2003). Thus, it is possible that both proteins control actin cytoskeleton remodeling in neuronal plasticity through a common pathway, and that lack of FMRP leads to a dysregulation of neuronal morphology through modification of Rac1.
Previous to this study, other groups have suggested that neuronal plasticity requires proteins involved in cytoskeleton reorganization and actin polymerization (Lin et al., 2005; Rex et al., 2007). Most of these studies however have centered on actin-binding proteins that directly regulate branching and stabilization of actin filaments (Lynch et al., 2007; Rex et al., 2009; Rex et al., 2010). Herein, we focused on identifying the role of upstream molecules that trigger these cytoskeletal rearrangements by having a role closer to NMDA receptor activation and induction of long-term plasticity. By using pharmacological and genetic approaches, we illustrated that the small GTPase Rac1 is associated and necessary for normal dendritic morphology and long-term synaptic plasticity, both abnormal in FMRP-deficient mice. First, by generating a viable mice with a post-developmental deletion of Rac1 in the CA1 region of the hippocampus (as young animals) as well as the CA3/dentate gyrus region (as aged mice), we have provided an exceptional system for addressing the contribution of Rac1 to dendritic spine morphology and long-term plasticity in the adult mice. As Rac1 activity is important for the regulation of actin dynamics and receptor clustering (Hall, 1994; Wiens et al., 2005), it is reasonable to hypothesize that loss of Rac1 in excitatory neurons in the hippocampus will result in impaired long-term plasticity. Conversely, excess Rac1 expression will result in exaggerated long-term plasticity, as reported in FXS. In our study, a tissue-specific Rac1 deficient mouse with Rac1 deletion in the CA1 area of the hippocampus presented an alteration on the dendritic spine density suggesting that Rac1 participates in a mechanism to expand spines. This process might be compromised in brains lacking Rac1 and, conversely, Rac1 overexpression might be associated to increased spine density or other cytoskeleton-dependent alterations leading to impaired plasticity, as reported in FXS.
Recent work from our group (Martinez and Tejada-Simon, 2011) together with the research presented herein indicates that Rac1 is critical for long-term plasticity in the CA1 region of the hippocampus. Altering Rac1 impairs LTP and LTD induction. Because there is no change in paired-pulse facilitation in slices derived from Rac1 deficient animals (data not shown), the reported decrease in synaptic strength associated with lack of Rac1 points to a postsynaptic effect most likely produced by a loss of functional synapses and a reduction in dendritic spine number as shown here. Regulation of synaptic plasticity can involved several signaling pathways that lead to activation of transcription and changes in synaptic morphology. Rac1 regulates actin polymerization through PAK phosphorylation. Rac1 also plays a role in the regulation of transcription mediated through Erk1/2 signaling cascades. In agreement with our previous published data (Martinez et al., 2007), our results showed that lack of Rac1 also alters the effector proteins involved in plasticity. Mice with genetic deletion of Rac1 showed reduced PAK1 phosphorylation indicating that Rac1/PAK1 signaling is involved in changes in spine morphology.
Because this study uses genetically altered mouse mutants with arrested expression of our gene of interest, it is possible that compensatory processes may take place. Other small GTPases from the Rho family have been implicated in the development and maintenance of dendritic spines (Luo et al., 1996; Nakayama et al., 2000; Tashiro et al., 2000) and small GTPases such as RhoB have been seen to have a role in synaptic plasticity and the regulation of neuronal morphology (O’Kane et al., 2003; McNair et al., 2010). However, in our study alterations in plasticity were seen despite having normal levels of related GTPases RhoB, RhoA and Cdc42. After studying levels of related small GTPases, we consider the option of compensatory mechanisms by these GTPases to be of minor importance.
Revising these observations, it is tempting to speculate that accumulation of Rac1 might contribute to the spine alteration as well as the decline in behavioral plasticity, raising the possibility that some of the effects reported in this study may reflect compromised development of hippocampal neurons. Aberrant development of neurons has been implicated in a number of diseases such as FXS, William’s syndrome and other autistic disorders. The present study shows that indeed Rac1 is aberrant in the Fmr1 knockout mouse, suggesting that FMRP might act as a regulator of Rac1. Lack of FMRP in Fmr1 knockout mice translates into an excessive synthesis and activation of Rac1, leading to aberrant Rac1-induced actin remodeling. These anomalies in spine structure and possibly in function will result in cognitive deficiencies. Consistent with these observations, in other systems such as fibroblasts, it has been reported that absence of FMRP induces an atypical development of actin remodeling, which is associated with upregulation of Rac1 downstream effectors PAK, CYFIP1 and also PP2A (Castets et al., 2005). Moreover, it has been reported that overproduction of reactive oxygen species (ROS) occurs in the FXS mouse model as a result of an enhancement on the NADPH-oxidase activity (el Bekay et al., 2007), which is critical in memory consolidation and synaptic plasticity (Serrano & Klann, 2004). Overexpression of this enzyme in the cortex, hippocampus and amygdale was associated with up-regulation of the NADPH-oxidase components, including Rac1. We want to note that while expression of Rac1 is upregulated in 3 months Fmr1 knockout mice, Rac1-GTP appears elevated but no longer significant at this age. FMRP regulates the translation of hundreds of mRNAs in brain tissue, affecting possibly other proteins that participate in the GDP-GTP exchange (Brown et al., 2001). Changes in the levels of these proteins could affect basal levels of Rac1-GTP. However, the identity of these proteins and their effect on Rac1 activation is still under investigation.
The involvement of actin cytoskeleton regulatory proteins in synaptic plasticity is not a new concept. It is well known that LTP is associated with changes in dendrites and creation of new spines (Yuste and Bonhoeffer, 2001; Fukazawa et al., 2003). These modifications are mediated by proteins that increase actin polymerization (Lin et al., 2005), such as PAK, cofilin (Rex et al., 2007) and myosin IIb (Rex et al., 2010). In agreement with our findings, Chen et al. (2010) have recently proposed that activation of Rac/PAK signaling is defective in FXS animal model, and further proved a deficient Rac-GTP in the hippocampal CA1 area following LTP-inducing stimulation. We did not investigate the activated status of Rac1 during LTP in the Fmr1 knockout mouse since previous reports indicate that hippocampal LTP is normal in these animals. Although our work shows that basal levels of Rac1/Rac1-GTP are higher in the Fmr1 knockout mouse, it is possible that the activity-mediated activation of Rac1 (e.g. upon LTP induction) does not move in the same direction.
Our study also found that overactivation of Rac1 is not limited to the brain, but also to other organs affected in FXS patients. This fact is likely due to the role of Rac1 in cell migration and invasion processes related to cancer (Zanca et al., 2010) as well as cell growth, leading to organ enlargement.
Rac1 in the CNS is highly expressed in the hippocampus (Tejada-Simon et al., 2006). Levels of Rac1 in the mouse CNS are high in the embryo and decrease steadily after birth. This fact correlates with morphological maturation at birth and dendritic pruning during adolescence. Thus, it is tempting to speculate that fluctuation of Rac1 might contribute to the spine alteration and decline in behavioral plasticity, as detected in the Fmr1 knockout mouse, where levels of Rac1 are nearly 2 fold up. It has recently been proposed that in the Fmr1 knockout mouse these alterations are due to a developmental delay during the perinatal periods (Harlow et al., 2010). While this theory could also explain our findings, we cannot confirm that Fmr1 knockout mice are developmentally delayed in their Rac1 expression pattern because we did not determine Rac1 levels during embryonic/perinatal states.
It is well known that spine morphology is important for learning and aging (Bailey and Kandel, 1993; Harris and Kater, 1994). Other studies have shown that Rac1 knockout mice have a specific memory deficit (Hadish et al., 2009). This fact, together with our results suggests that Rac1 might be responsible for the cognitive deficit observed in FXS and that correction of its levels might be a way to ameliorate these deficiencies, as indicated by the rescued LTD in our experiments.
In conclusion, we propose that Rac1 modulates spine number and synaptic function in the brain, and that overexpression of Rac1 may be responsible for the aberrant hippocampal plasticity in the FXS animal model. Our findings suggest that Rac1 signaling may provide a functional link among altered dendritic spines, aberrant synaptic plasticity and cognition impairment in FXS. These conclusions point to Rac1 inhibition as a novel therapeutic strategy in FXS.
4. EXPERIMENTAL PROCEDURES
4.1 Animals
All animal use was in accordance with the principles and procedures of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Protocols for experimental tests were approved by the Institutional Animal Care and Use committee (IACUC) at the University of Houston. Fmr1 knockout mouse: The Fmr1 knockout animals (Fmr1tm1Cgr/Fmr1tm1Cgr) were kindly donated by Dr. Richard Paylor (Baylor College of Medicine). Genotyping of mouse tail digests was performed by PCR using the following primers: Fmr1_S1 (5_-GTGGTTAGCTAAAGTGAGGATGAT-3_) and Fmr2_S2 (5_-CAGGTTTGTTGGGATTAACAGATC-3_) for the Fmr1 WT allele (527bp band); and Fmr1_S1 primer and Fmr1_N2 (5_-TGGGCTCTATGGC TTCTGA-3_) for the Fmr1 knockout allele (501 bp band). PCR conditions were as follows: 2min 94°C, (30s 90°C, 30s 55°C, 60s 72°C) cycle 34x and 10min 72°C (Spencer et al., 2008). Rac1-deficient mice (Rac1 mutant): To obtain a Rac1 deficient mouse, two transgenic mice were used as founders: a Rac1flox line (Rac1tm1Djk/J; Walmsley et al. 2003) and Cre T29-1 line (Tsien et al. 1996). Mice were purchased from The Jackson Laboratory (Bar Harbor, Maine). In the T29-1 Cre recombinase transgenic mice, Cre expression is restricted to the forebrain region by the CaMKII-α promoter (Mayford et al., 1995; Mayford et al., 1996; Dragatsis and Zeitlin, 2000). To generate mice with a conditional deletion of rac1 gene in the area CA1 of the hippocampus, Rac1flox mice were out crossed with Cre transgenic mice. Subsequently, the F1 generation was backcrossed with Rac1flox. Mice generated from this cross were genotyped by PCR using primers specific to the Cre recombinase and to floxed alleles of the Rac1 locus. DNA was obtained from mouse tails and processed using the DNeasy Tissue Kit (QIAGEN Inc., Valencia, California) according to manufacturer’s instructions. The extracted DNA was subjected to PCR for amplification. PCR was run using cre specific primers (Cre forward: gccaccagccagctatcaac, Cre reverse: gctaatcgccatcttccagc) and primers that identify floxed alleles of the Rac1 locus (5’tccaatctgtgctgcccatc3’ and 5’gatgcttctaggggtgagcc3’). A PCR mix of 12μl was prepared according to the number of reactions. Ten (10) μl of PCR mix was aliquoted into small PCR tubes and 2 μl of DNA was added. Two separate PCRs were run using the following thermal cycles: Racflox: 1) 94°C for 3 min, 2) 94°C for 30 sec, 3) 59.9°C for 1 min, 4) 72°C for 1 min (repeat 2-4 for 35 cycles), 5) 72°C for 2 min, and hold at 10°C; Cre: 1) 94°C for 3min, 2) 94°C for 30 sec 3) 65°C for 1 min, 4) 72°C for 1 min (repeat 2-4 for 35 cycles), 5) 72°C for 2 min, and hold at 10°C. A 1.5% agarose gel (1.5 g agarose in 100ml of 1X TBE [Tris Borate Ethylenediaminetetraacetic acid: 54g Tris, 20 ml 0.5M EDTA, 27.6g Boric acid]) and 60μl of 2mg/ml ethidium bromide) was run in 1X TBE using the amplified DNA, and visualization was done using UV light and ethidium bromide staining in an AlphaEaseFC-FluorChem 8900-version 4.0 Gel Imager (Alpha Innotech Corporation, San Leandro, California). Mice genotypes were classified as wild type (WT) and heterozygous littermates (HET) or Rac1 deficient (KO or Rac1 mutant) by comparing the bands expressed on the gel to a standard with specific number of base pairs run in parallel. Wild type littermates showed only the Rac1 band (115 bp), heterozygous littermates showed the Cre band (100 bp), Rac1 band and floxed Rac1 band (242 bp), while Rac1 mutant mice showed the Cre band and the floxed Rac1 band. Thus, F2 generation was classified as Rac1 mutants, heterozygotes or wild type littermates according to the genotype results.
Transgenic and wild type C57BL/6J animals used for experiments were obtained by breeding and maintained in a controlled environment (14/10h light/dark cycle, constant temperature).
4.2 Preparation of brain samples and hippocampal slices
Animals were sacrificed by decapitation after cervical disarticulation. Brains were rapidly removed and briefly submerged in ice-cold cutting saline solution (5mM glucose, 110mM sucrose, 60mM NaCl, 28mM NaHCO3, 3mM KCl, 1.25mM NaH2PO4, 7mM MgCl2, and 0.5mM CaCl2, 0.6mM ascorbate). All solutions were saturated with 95% O2/5% CO2. Whole brains were then dissected into different brain regions on cutting solution-soaked filter paper and mounted on a glass platform resting on ice.
For hippocampal slices, 400 μm thick hippocampal slices were prepared using a 1000Plus Vibratome sectioning system (Vibratome Co., St. Louis, Missouri). Slices were allowed to equilibrate in a 50% cutting saline solution and 50% artificial cerebrospinal fluid solution (aCSF, 25mM glucose, 125mM NaCl, 2.5mM KCl, 1.25mM NaH2PO4, 25 NaHCO3,1mM MgCl2, and 2mM CaCl2) saturated with 95% O2/5% CO2 at room temperature for 30 minutes. Hippocampi were carefully removed from brain slices and, when appropriate, CA1, CA3 and dentate gyrus regions were sectioned with the help of a microscope.
4.3 Gross morphology of the hippocampus
Nissl staining was used to compare the gross morphology of the hippocampus of Rac1 mutant mice to that of their wild type littermates. Mice were sacrificed by cervical dislocation. Brains were quickly removed and placed in -80°C for rapid freezing until ready for use. Thin coronal brain sections of 10-15μm were prepared on a cryostat at -20 °C and arranged on cold labeled slides. Sections were fixed in 4% formaldehyde/PBS for 30 minutes followed by soaking in alcohol (95% ethanol for 15 min, 70% ethanol for 1 min, and 50% ethanol for 1 min) and rinsing twice in distilled water (once for 2 min and once for 1 min). Brain sections were then submerged in Cresyl Violet Stain for 2 min followed by rinsing in distilled water for 1 min and soaking in alcohol (50% ethanol for 1 min, 70% acid ethanol [2 ml glacial acetic acid in 200 ml 70% ethanol] for 2 min, 95% ethanol for 2 min, few dips in 95% ethanol, and 100% ethanol for 1 min). Finally sections were submerged in Histoclear (clearing agent) for 5 min to clear unstained parts of the tissue. Mounting medium was applied on every section and a cover slip was placed on the slide to hold the section in place for visualization on a confocal microscope (Olympus FluoView Version 1.7C, FV 1000).
4.4 Affinity purification assay
To determine expression levels of Rac1-GTP 10 μg of GST tagged PAK-PBD protein beads (Millipore, Temecula, CA) were mixed with 200 μg protein of fresh hippocampal homogenates containing phosphatase and protease inhibitor cocktail (EMD, San Diego, CA). Fifty μl of Homogenizing Buffer Complete (HBC; 200mM HEPES, 2.5 mM NaCl, 100mM EDTA, 100mM EGTA, 200mM Na4P2O7, 50mM NaF, 1 mM sodium orthovanadate, 10ug/ml leupeptin, 2ug/ml aprotinin, 1 μM microcystin-LR, and 200 nM calyculin A) were added to increase the reaction volume. Samples were placed on a rocker for 2 hours at 4°C. PAK-PBD-GST beads then were pelleted by centrifugation at 14,000g for 10 minutes at 4°C. The supernatant was carefully removed without disturbing the pelleted beads, followed by three more washes and centrifugations. The final pellet was resuspended in 50 μl Laemmli buffer. Samples were placed in a 100°C water bath for 3 minutes to remove Rac1-GTP from the beads. Amount of Rac1 protein was then analyzed by SDS-PAGE and immunobloting analysis.
4.5 Immunocytochemistry
Brains were quickly removed and placed in -80°C for rapid freezing. Thin coronal brain sections of 10-15μm were prepared on a cryostat at -20 °C and arranged on cold labeled slides. Sections were fixed in 4% formaldehyde/PBS for one hour followed by washing three times in PBS for 5 minutes. Sections were then incubated in 0.2% Triton X-100/PBS for 20 minutes at room temperature, washed 3 times for 10 minutes each in PBS, rinsed in distilled water and allowed to air dry. Rac1 (1:100 Millipore) primary antibody solution was applied on the sections and incubated for 24 hours in a humidified container at 4°C. The antibody solution was gently shaken off and slides were washed five times for 5 minutes each at room temperature in PBS. Dylight 488 anti-mouse florescent dye (1:200, Jackson ImmunoLabs) was applied to the sections and allowed to incubate for 3 hours at room temperature in a humidified container. The antibody solution was gently shaken off and slides were washed 3 times for 10 minutes each at room temperature in PBS. Slides were allowed to air dry and incubated in DAPI stain (DAPI dihydrochloride; CALBIOCHEM, La Jolla, California) for 5 minutes followed by washing three times in PBS for ten minutes. An aqueous mounting medium suitable for fluorescence was applied and slides were coverslipped and imaged using and Olympus BX51WI Microscope with Spinning Disk Confocal (DSU) and Hamamatsu EM-CCD Digital Camera C9100.
4.6 In situ hybridization
In situ hybridization was used to detect Rac1 mRNA expression in the Rac1 mutant and wild type littermates using a biotin-labeled Rac1 oligoprobe as described previously (Braissant and Wahli, 1998; Lazarov et al., 1998) with minor modifications. Mice were sacrificed by cervical dislocation and brains were quickly removed and put in -80°C for rapid freezing. Thin coronal brain sections of 10-15μm were prepared on a cryostat at -20 °C and arranged on cold labeled slides. Sections were fixed in 4% formaldehyde/PBS (Thermo Scientific, Rockford, IL) for 1 hour at 4°C and then washed three times in phosphate-buffered saline solution (0.01 M PBS, pH 7.4, Sigma, St. Louis, MO). Tissue was then permeabilized with 0.25% acetic anhydride in 0.1M triethanolamine-HCL (pH 8.0) in PBS for 10 minutes at 4°C followed by 0.5% Triton-X in PBS for 5 minutes at 4°C. Tissue samples were then incubated in 70% ethanol for 10 minutes, then in 95% ethanol at 4°C until ready for the hybridization step. Next, tissue was incubated in 100 μl of hybridization buffer (50% deionized formamide, 4x SSC [Saline Sodium Citrate], 1% Dernhardts solution, 2% dextran sulfate, 250 μg/ml salmon sperm DNA, 250 μg/ml yeast tRNA with a Rac1 biotin double labeled oligomer probe [sequence ACAACAGCAGGCATTTTCTCTTCCTCTTCTTGACAGGAGGGGGAC; 10μg/ml, Invitrogen, Carlsbad, CA]) overnight in humid chamber with tissue wetted from a solution of 50% formamide and 4x SSC. Tissue was then washed two times to remove unbound probe with a 2x SSC solution in nuclease free water at 40°C for 15 minutes. The tissue was washed in 1x SSC solution two times for 30 minutes at 40°C and further washed in a 0.5x SSC solution for 45 minutes at 40°C, then transferred to a 0.5x SSC solution at room temperature for 5 minutes. Afterwards, tissue samples were rinsed in TBS (Tris-HCl 50mM pH 7.5 and NaCl 150mM) twice for 15 minutes each, and blocked in blocking solution (0.3% Triton X-100 and 1% I-Block) for 30 minutes at room temperature. A 1:1000 solution of AP-Streptavidin (Sigma, St. Louis, MO) was made in blocking solution, and 50 μl was added to each slice and incubated at 37°C for 1 hour. The slides were rinsed 2 times in TBS, and allowed to equilibrate in detection buffer (pH 9.5, 100mM Tris-HCl, 100mM NaCl, 50mM MgCl2). Alkaline phosphatase activity was detected by incubating the tissue in pre-mixed BCIP/NBT liquid substrate/B1911 (Sigma, St. Louis, MO) overnight at room temperature in humid chamber. The reaction was stopped by placing tissue in TE buffer (10mM Tris-HCl pH 8.0, 0.1mM EDTA) for ten minutes. Slides were briefly rinsed with distilled water and coverslipped. To image mRNA staining, an Olympus BX51WI Microscope with Spinning Disk Confocal (DSU) and Hamamatsu EM-CCD Digital Camera C9100 was used.
4.7 Analysis of neuronal morphology
The FD Rapid GolgiStain™ Kit (FD Neurotechnologies, Ellicot City, MD) was used for Golgi staining of neurons according to manufacturer’s instructions. Briefly, to prepare the tissue, the impregnation solution was prepared by mixing equal volumes of Solutions A and B, 24 hours prior to use and stored in the dark. Mice brains were quickly and carefully removed from the skull and rinsed in 0.1 M Phosphate Buffer (pH 7.4). Brains were immersed in the impregnation solution (fixative) and stored at room temperature for 2 weeks in the dark. The impregnation solution was replaced after 12 hours of immersion. Brains were transferred into Solution C and stored at 4°C for one week in the dark. Solution C was also replaced after 12 hours of brain immersion. From solution C, brains were snapped frozen in cold isopentene on dry ice for 30 minutes after which they were stored in -80°C for three days before staining. To proceed to stain the tissue, sections of 60-100 μm were obtained from fixed brains imbedded in Tissue-Tek Optimal Cutting Temperature (O.C.T.) embedding medium for frozen tissue (Sakura Finetek U.S.A., Inc. Torrance, California). Brain sectioning was done in a cryostat set at -32°C. Slices were arranged on cold already labeled microscope slides. A drop of solution C was added to each slice upon removal from the cryostat and allowed to air dry. Staining was done in staining jars by first rinsing brain slices twice in distilled water for 2 minutes each, and then incubating in a staining solution composed of 1 part Solution D, 1 part Solution E and 2 parts distilled water (1D:1E:2DH2O) for 10 minutes followed by rinsing twice in distilled water for 4 minutes. Slices were then dehydrated in 50%, 75%, and 95% ethanol for 4 minutes each and finally in absolute ethanol 4 times for 4 minutes each. Slices were cleared in xylene 3 times, for 4 minutes each followed by mounting using a resinous mounting medium (Permount) added to each slice. A cover slip was placed on the slide to hold sections in place for visualization in confocal microscope and nail hardener was used to seal the slides to avoid drying.
To image cell morphology, confocal microscopy was used. For fixed samples, high-resolution confocal images were obtained using an Olympus BX51WI Microscope with Spinning Disk Confocal (DSU) and Hamamatsu ORCA-ER Deep Cooled Camera. The microscope is completely configured and compatible with Microbrightfield Stereology Software including StereoInvestigator and Neurolucida (MBF, Williston, Vermont). A z series projection of ~60 images at a resolution of 0.37 microns per pixel was used to cover the entire z dimension of the labeled neurons. A 20x lens [0.75 numerical aperture (NA)] was used to image whole-cell and dendritic morphology. A 60x (1.35 NA) and 100x (1.4) oil lenses was used to image fine structures such as dendritic spines. To determine spine density (number of spines per length of dendrite) ten randomly selected fully impregnated neurons were examined and measured. For comparisons, Student’s t-tests were performed using the mean spine density per animal averaged across all neurons.
4.8 Immunoblot analysis
Brain and organs of interest were obtained and sonicated in Homogenizing buffer complete (HBC) (10mM Hepes, 1mM EDTA, 1mM EGTA, 150mM NaCl, 50mM NaF, 10 mM sodium pyrophosphate, 1mM sodium orthovanadate, 10 μg/ml leupeptin, 2 μg/ml aprotinin, 1 μM microcystin-LR, and 200nM calyculin A). Protein concentrations in samples were determined using the Bradford assay (Bradford, 1976). Equal amounts of protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 12%) followed by electrophoretic transfer to a polyvinylidene difluoride (PVDF) membrane, and incubated in Tris-buffered saline with Tween 20 (TTBS, 50 mM Tris-HCl [pH 7.5-8.0], 150 mM NaCl, and 0.1% Tween 20) containing 5% non-fat milk for 1 hour at room temperature. Membranes were incubated with the desired antibody (Rac1, 1:10.0000 from Millipore; Cdc42 1:1000 from Santa Cruz; RhoA and RhoB 1:1000, total PAK1 and phosphorylated PAK1 1:1000, total and phosphorylated ERK, 1:1000, from Cell Signaling) overnight at 4°C. Blots were washed three times for 15 min in TTBS, followed by incubation in the corresponding secondary antibody (horseradish peroxidase-conjugated goat anti-mouse IgG [1:5,000] from Promega, Madison, WI) for Rac1 and Cdc42; anti-rabbit for RhoA, RhoB, total PAK1 and phosphorylated PAK1, total and phosphorylated ERK. This was followed by washing three times in 1X TTBS and visualized using enhanced chemiluminescence (ECL, Amersham Biosciences, Piscataway, NJ) and by exposing the blots to X-Omat Blue film (Kodak, Rochester, NY) for ~ 15 s to 1 min. Densitometric analysis of all samples were performed using NIH Image J software. Differences in the amount of antibody binding between control and treated samples were evaluated statistically.
Immunoblotting was also performed using the E-PAGE™ 48 Protein Electrophoresis System (Invitrogen™, Version A). Samples were prepared in a total volume of 10μl composed of 2.5 μl E-PAGE™ 4X Loading buffer, 1μl Nupage® 10 Sample Reducing Agent (Invitrogen™, Carlsbad, California), protein sample and deionized water (as needed to bring volume to 10μl. Samples were incubated at 70°C for 10 minutes and loaded on the 8% 48 well E-PAGE™ gels inserted into the E-PAGE™ mother Base. Loading of samples was preceded by loading of 5μl of deionized water into each well to make a total volume of 15μl (plus sample) per well. Gels were run for 30 minutes followed by a seven minute quick transfer onto a PVDF membrane on an IBlot™ Dry Blotting System (Transfer Stack Invitrogen™ Version C). Membranes were activated with methanol for few seconds to 1 minute, incubated in Tris-buffered saline with Tween 20 (TTBS, 50 mM Tris-HCl [pH 7.5-8.0], 150 mM NaCl, and 0.1% Tween 20) containing 5% non-fat milk for 1 hour at room temperature, and probed for desired antibody as described before in the conventional method.
4.9 Electrophysiology
Transverse hippocampal slices (400 μm) were prepared from age-matched animals as described before. Slices were transferred to an interface chamber (Harvard Apparatus, Holliston, MS) at 30-32°C, perfused with oxygenated aCSF (perfusion rate: 1-2ml/min) and allowed to recover for a minimum of 1 hr prior to recording. For drug treatment, a Rac1 inhibitor, NSC23766 (120μM) was perfused during induction. Extracellular field recordings were obtained from area CA1. A bipolar enamel-coated platinum stimulating electrode was placed in CA3 Schaffer collateral/commissural fibers and a glass recording electrode (resistance 1-4 MΩ) filled with aCSF was placed into stratum radiatum of area CA1. Synaptic responses to electrical stimulation were collected every 20 sec and averaged over 2min using a stimulus intensity that produces 30-50% of the maximum initial slope of the extracellular field excitatory postsynaptic potential (fEPSP). Baseline fEPSPs were monitored for at least 20 min before the application of any drug. NMDAR-dependent LTD was induced with low-frequency stimulation (LFS, 900 pulses at 1Hz for 15min). mGluR-LTD was induced by 100μM (S)-3,5-dihydroxyphenylglycine, DHPG (Tocris, Ellisville, MO) in the presence of 100μM ((2R)-amino-5-phosphonovaleric acid, AP-5 (Tocris, Ellisville, MO) in aCSF applied for 5min after >30min of stable baseline. Data points were normalized to the baseline mean prior to delivery of LFS or drug and collected using pClamp version 10 (Molecular Devices, Sunnyvale, CA). Data for the electrophysiological experiments comparing the initial slope of the fEPSP at various time points before, during and after LTD were evaluated statistically as described below. Basal synaptic transmission was assessed by the Input/Output relationship between the stimulus strength (0-15mV) and the corresponding fEPSP amplitude. This response range was also used to determine the stimulus intensity (30-50% of the max response) applied during baseline recordings. Baseline stability was tested by recording signals for ≥1.5hrs using the same stimulus value. Paired-pulse facilitation (PPF), a transient form of plasticity induced by presenting two closely spaced pulses of equal intensities, was measured using 50-300ms intervals (data not shown).
4.10 Data analysis
Antibody binding to the antigen of interest in homogenates was expressed as a percent of antibody binding to the respective antigens in control samples. Statistical analyses for two sample comparisons were made using the paired Student’s t test for normally distributed variables to determine differences between control wild type and knockout /Rac1 mutant. Statistical analyses for multiple comparisons were made using a ANOVA followed by Dunnett’s comparison to control or Tukey multiple comparison post tests. A p value of less than 0.05 was considered statistically significant. Two-way ANOVA was used for electrophysiological data analysis with a p value of less than 0.05 as significance criteria.
Research Highlights.
Rac1 has a critical role in dendritic morphology and synaptic function
Inactivation of Rac1 impairs long-term plasticity
Rac1 is upregulated in Fmr1 knockout mice
Inactivation of Rac1 in Fmr1 knockout mice rescues altered LTD
Acknowledgments
This work was supported by generous grants from the FRAXA Research Foundation, the Jérôme LeJeune Foundation and the National Institutes of Health Grant NS48037 (M.V.T.S). We thank Dr. Lindsay Schwarz for her thoughtful commentaries and help editing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abitbol M, Menini C, Delezoide A-L, Rhyner T, Vekemans M, Mallet J. Nucleus basalis magnocellularis and hippocampus are the major sites of FMR-1 expression in the human fetal brain. Nat Genet. 1993;4:147–153. doi: 10.1038/ng0693-147. [DOI] [PubMed] [Google Scholar]
- Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Gen Brain Beh. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Bailey CH, Kandel ER. Structural changes accompanying memory storage. Annu Rev Physiol. 1993;55:397–426. doi: 10.1146/annurev.ph.55.030193.002145. [DOI] [PubMed] [Google Scholar]
- Bakker CE, de Diego Otero Y, Bontekoe C, Raghoe P, Lutejin T, Hoogeveen AT, Oostra BA, Willemsen R. Immunocytochemical and biochemical characterization of FMRP, FXR1P, and FXR2P in the mouse. Exp Cell Res. 2000;258:162–170. doi: 10.1006/excr.2000.4932. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Braissant O, Wahli W. Differential expression of peroxisome proliferator-activated receptor-alpha, -beta, and -gamma during rat embryonic development. Endocrinol. 1998;139:2748–2754. doi: 10.1210/endo.139.6.6049. [DOI] [PubMed] [Google Scholar]
- Braun K, Segal M. FMRP involvement in formation of synapses among cultured hippocampal neurons. Cereb Cortex. 2000;10:1045–1052. doi: 10.1093/cercor/10.10.1045. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in Fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Castets M, Schaeffer C, Bechara E, Schenck A, Khandjian EW, Luche S, Moine H, Rabilloud T, Mandel JL, Bardoni B. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum Mol Genet. 2005;14:835–844. doi: 10.1093/hmg/ddi077. [DOI] [PubMed] [Google Scholar]
- Chelly J, Mandel JL. Monogenic causes of X-linked mental retardation. Nat Rev Genet. 2001;2:669–680. doi: 10.1038/35088558. [DOI] [PubMed] [Google Scholar]
- Chen BE, Lendvai B, Nimchinsky EA, Burbach B, Fox K, Svoboda K. Imaging high-resolution structure of GFP-expressing neurons in neocortex in vivo. Learn Mem. 2000;7:433–441. doi: 10.1101/lm.32700. [DOI] [PubMed] [Google Scholar]
- Chen LY, Rex CS, Babyan AH, Kramar EA, Lynch G, Gall CM, Lauterborn J. Physiological Activation of Synaptic Rac>PAK (p-21 Activated Kinase) Signaling Is Defective in a Mouse Model of Fragile X Syndrome. J Neurosci. 2010;30:10977–10984. doi: 10.1523/JNEUROSCI.1077-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT. Abnormal dendritic spines in fragile X knockout mice: Maturation and pruning deficits. Proc Natl Acad Sci USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovic L, Huot ME, Khandjian EW. Lost once, the Fragile X Mental Retardation protein is now back onto brain polyribosomes. RNA Biol. 2005;2:1–3. doi: 10.4161/rna.2.1.1368. [DOI] [PubMed] [Google Scholar]
- de Diego-Otero Y, Romero-Zerbo Y, el Bekay R, Decara J, Sanchez L, Rodriguez de Fonseca F, del Arco-Herrera I. Alpha-tocopherol protects against oxidative stress in the Fragile X knockout mouse: an experimental therapeutic approach for the Fmr1 deficiency. Neuropsychopharmacol. 2008;34:1–16. doi: 10.1038/npp.2008.152. [DOI] [PubMed] [Google Scholar]
- Devys D, Lutz T, Rouyer N, Bellocq JP, Mandel JL. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Diana G, Valentini G, Travaglione S, Falzano L, Pieri M, Zona C, Meschini S, Fabbri A, Fiorentini C. Enhancement of learning and memory after activation of cerebral Rho GTPases. Proc Natl Acad Sci USA. 2007;104:636–641. doi: 10.1073/pnas.0610059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragatsis I, Zeitlin S. CaMKIIalpha-Cre transgene expression and recombination patterns in the mouse brain. Genesis. 2000;6:133–135. doi: 10.1002/(sici)1526-968x(200002)26:2<133::aid-gene10>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- El Bekay R, Romero-Zerbo Y, Decara J, Sanchez-Salido L, Del Arco-Herrera I, Rodriguez-de Fonseca F, de Diego-Otero Y. Enhanced markers of oxidative stress, altered antioxidants and NADPH-oxidase activation in brains from Fragile X mental retardation 1-deficient mice, a pathological model for Fragile X syndrome. Eur J Neurosci. 2007;26:3169–3180. doi: 10.1111/j.1460-9568.2007.05939.x. [DOI] [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- Fukazawa Y, Sitoh Y, Ozawa F, Ohta Y, Mizuno K, Inokuchi K. Hippocampal LTP is accompanied by enhanced F-Actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron. 2003;38:447–460. doi: 10.1016/s0896-6273(03)00206-x. [DOI] [PubMed] [Google Scholar]
- Galvez R, Greenough WT. Sequence of abnormal dendritic spine development in primary somatosensory cortex of a mouse model of the fragile X mental retardation syndrome. Am J Med Genet. 2005;135:155–160. doi: 10.1002/ajmg.a.30709. [DOI] [PubMed] [Google Scholar]
- Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenough WT, Klintsove AY, Irwin SA, Galvez R, Bates KE, Weiler IJ. Synaptic regulation of protein synthesis and the fragile X protein. Proc Natl Acad Sci USA. 2001;98:7101–7106. doi: 10.1073/pnas.141145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AW, Aldridge GM, Weiler IJ, Greenough WT. Local Protein Synthesis and Spine Morphogenesis: Fragile X Syndrome and Beyond. J Neurosci. 2006;26:7151–7155. doi: 10.1523/JNEUROSCI.1790-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haditsch U, Leone DP, Farinelli M, Chrostek-Grashoff A, Brakebusch C, Mansuy IM, McConnell SK, Palmer TD. A central role for the small GTPase Rac1 in hippocampal plasticity and spatial learning and memory. Mol Cell Neurosci. 2009;41:409–419. doi: 10.1016/j.mcn.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Ann Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- Harlow EG, Till SM, Russell TA, Wijetunge LS, Kind P, Contractor A. Critical period plasticity is disrupted in the barrel cortex of Fmr1 knockout mice. Neuron. 2010;65:385–398. doi: 10.1016/j.neuron.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Kater SB. Dendritic spines: cellular specializations imparting both stability and flexibility to synaptic function. Annu Rev Neurosci. 1994;17:341–371. doi: 10.1146/annurev.ne.17.030194.002013. [DOI] [PubMed] [Google Scholar]
- Harwood A, Braga VMM. Cdc42 and GSK-3: signals at the crossroads. Nat Cell Biol. 2003;5:275–277. doi: 10.1038/ncb0403-275. [DOI] [PubMed] [Google Scholar]
- Huber KM, Roderm JC, Bear MF. Chemical induction of mGluR5-and protein synthesis-dependent long-term depression in hippocampal area CA1. J Neurophysiol. 2001;86:321–325. doi: 10.1152/jn.2001.86.1.321. [DOI] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci USA. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin SA, Galvez R, Greenough WT. Dendritic Spine Structural Anomalies in Fragile-X Mental Retardation Syndrome. Cereb Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Patel B, Idulpulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, et al. Abnormal dendritic spine characteristics in temporal and visual cortices of patients with fragile-X-syndrome: a quantitative examination. Am J Med Genet. 2001;8:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Jin P, Warren ST. New insights into fragile X syndrome: from molecules to neurobehaviors. Trends Biochem Sci. 2003;28:52–158. doi: 10.1016/S0968-0004(03)00033-1. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, Matsuura Y, Yoshida-Kubomura Y, Iwamatsu A, Kaibuchi K. p140Sra-1 (Specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem. 1998;273:291–295. doi: 10.1074/jbc.273.1.291. [DOI] [PubMed] [Google Scholar]
- Lazarov NE, Schmidt U, Wanner I, Pilgrim C. Mapping of D1 dopamine receptor mRNA by non-radioactive in situ hybridization. Histochem Cell Biol. 1998;109:271–279. doi: 10.1007/s004180050227. [DOI] [PubMed] [Google Scholar]
- Lee T, Winter C, Marticke SS, Lee A, Luo L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000;25:307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- Li Z, Van Aelst L, Cline HT. Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat Neurosci. 2000;3:217–225. doi: 10.1038/72920. [DOI] [PubMed] [Google Scholar]
- Li J, Pelletier MR, Perez Velasquez JL, Carlen PL. Reduced cortical synaptic plasticity and GluR1 expression associated with fragile X mental retardation protein deficiency. Mol Cell Neurosci. 2002;19:138–151. doi: 10.1006/mcne.2001.1085. [DOI] [PubMed] [Google Scholar]
- Lin B, Kramar EA, Bi X, Brucher FA, Gall CM, Lynch G. Theta stimulation polymerizes actin in dendritic spines of hippocampus. J Neurosci. 2005;25:2062–2069. doi: 10.1523/JNEUROSCI.4283-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linseman DA, Hofmann F, Fisher SK. A role for the small molecular weight GTPases, Rho and Cdc42, in muscarinic receptor signaling to focal adhesion kinase. J Neurochem. 2000;74:2010–2020. doi: 10.1046/j.1471-4159.2000.0742010.x. [DOI] [PubMed] [Google Scholar]
- Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN. Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature. 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- Lynch G, Rex CS, Gall CM. LTP consolidation: substrates, explanatory power, and functional significance. Neuropharmacology. 2007;52(1):12–23. doi: 10.1016/j.neuropharm.2006.07.027. [DOI] [PubMed] [Google Scholar]
- McNair K, Spike R, Guilding C, Prendergast GC, Stone TW, Cobb SR, Morris BJ. A role for RhoB in synaptic plasticity and the regulation of neuronal morphology. J Neurosci. 2010;30:3508–3517. doi: 10.1523/JNEUROSCI.5386-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K. Rapid Dendritic Morphogenesis in CA1 Hippocampal Dendrites Induced by Synaptic Activity. Science. 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- Martinez LA, Klann E, Tejada-Simon MV. Translocation and activation of Rac in the hippocampus during associative contextual fear learning. Neurobiol Learn Mem. 2007;88:104–113. doi: 10.1016/j.nlm.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Martinez LA, Tejada-Simon MV. Pharmacological inactivation of the small GTPase Rac1 impairs long-term plasticity in the mouse hippocampus. Neuropharmacol. 2011;61:305–312. doi: 10.1016/j.neuropharm.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Wang J, Kandel ER, O’Dell TJ. CaMKII regulates the frequency-response function of hippocampal synapses for the production of both LTD and LTP. Cell. 1995;81:891–904. doi: 10.1016/0092-8674(95)90009-8. [DOI] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- Minden A, Lin A, Claret FX, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the Maintenance of Dendritic Spines and Branches in Hippocampal Pyramidal Neurons. J Neurosci. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosyreva E, Huber K. Metabotropic receptor-dependent long-lerm depression persists in the absence of protein synthesis in the mouse model of Fragile X Syndrome. J Neurophysiol. 2006;95:3291–3295. doi: 10.1152/jn.01316.2005. [DOI] [PubMed] [Google Scholar]
- O’Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- O’Kane EM, Stone TW, Morris BJ. Activation of Rho GTPases by synaptic transmission in the hippocampus. Mol Brain Res. 2003;114:1309–1312. doi: 10.1046/j.1471-4159.2003.02102.x. [DOI] [PubMed] [Google Scholar]
- Peier AM, McIlwain KL, Kenneson A, Warren ST, Paylor R, Nelson DL. (Over)correction of FMR1 deficiency with YAC transgenics: behavioral and physical features. Hum Mol Genet. 2000;9:1145–1159. doi: 10.1093/hmg/9.8.1145. [DOI] [PubMed] [Google Scholar]
- Pourbagher MA, Pourbagher A, Erol I. Fragile X syndrome associated with testicular microlithiassis in siblings. J Ultrasound Med. 2005;24:1727–1729. doi: 10.7863/jum.2005.24.12.1727. [DOI] [PubMed] [Google Scholar]
- Purpura DP. Dendritic spine “Dysgenesis” and mental retardation. Science. 1974;186:1126–1128. doi: 10.1126/science.186.4169.1126. [DOI] [PubMed] [Google Scholar]
- Rex CS, Lin C-Y, Kramar EA, Chen LY, Gall CM, Lynch G. Brain-derived neurotrophic factor promotes long-term potentiation-related cytoskeletal changes in adult hippocampus. J Neurosci. 2007;27:3017–3029. doi: 10.1523/JNEUROSCI.4037-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex C, Chen KY, Sharma A, Liu J, Babayan AH, Gall CM, Lynch G. Different Rho GTPase-dependent signaling pathways initiate sequential steps in the consolidation of long-term potentiation. J Cell Biol. 2009;186:85–97. doi: 10.1083/jcb.200901084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex CS, Gavin CF, Rubio MD, Kramar EA, Chen LY, Jia Y, Huganir RL, Muzyczka N, Gall CM, Miller CA, Lynch G, Rumbaugh G. Myosin IIb regulates actin dynamics during synaptic plasticity and memory formation. Neuron. 2010;67:603–617. doi: 10.1016/j.neuron.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchhoeft ML, Ohnuma S, McNeill L, Holt CE, Harris WA. The neuronal architecture of Xenopus retinal ganglion cells is sculpted by rho-family GTPases in vivo. J Neurosci. 1999;19:8454–8463. doi: 10.1523/JNEUROSCI.19-19-08454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FX1P and FX2P. Proc Natl Acad Sci USA. 2001;98:8844–8849. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano F, Klann E. Reactive oxygen species and synaptic plasticity in the aging hippocampus. Short survey. Age Res Rev. 2004;3:431–443. doi: 10.1016/j.arr.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Spencer CM, Graham DF, Yuva-Paylor LA, Nelson DL, Paylor R. Social Behavior in Fmr1 Knockout Mice Carrying a Human FMR1Transgene. Beh Neurosci. 2008;122:710–715. doi: 10.1037/0735-7044.122.3.710. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Minden A, Yuste R. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex. 2000;10:927–938. doi: 10.1093/cercor/10.10.927. [DOI] [PubMed] [Google Scholar]
- Tejada-Simon MV, Villasana LE, Serrano F, Klann E. NMDA receptor activation induces translocation and activation of Rac in mouse hippocampal area CA1. Biochem Bioph Res Comm. 2006;343:504–512. doi: 10.1016/j.bbrc.2006.02.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill R, Bobb K, Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER. Tonegawa S. Subregion- and Cell Type-Restricted Gene Knockout in Mouse Brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes & Develop. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DPA, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Walmsley MJ, Ooi SKT, Reynolds LF, Smith SH, Ruf S, Mathiot A, Vanes L, Williams DA, Cancro MP, Tybulewicz VLJ. Critical Roles for Rac1 and Rac2 GTPases in B Cell Development and Signaling. Science. 2003;302:459–462. doi: 10.1126/science.1089709. [DOI] [PubMed] [Google Scholar]
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Everwine J, Greenough WT. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler IJ, Spangler CC, Klintsova AY, et al. Fragile X mental retardation protein is necessary for neurotransmitter-activated protein translation at synapses. Proc Natl Acad Sci USA. 2004;101:17504–17509. doi: 10.1073/pnas.0407533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen R, Oostra BA, Bassell GJ, et al. The fragile X syndrome: from molecular genetics to neurobiology. Ment retard Dev Disabil Res Rev. 2004;10:60–67. doi: 10.1002/mrdd.20010. [DOI] [PubMed] [Google Scholar]
- Wiens KM, Lin H, Liao D. Rac1 induces the clustering of AMPA receptors during spinogenesis. J Neurosci. 2005;25:10627–10636. doi: 10.1523/JNEUROSCI.1947-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirojanan J, Kraff J, Hawkins D, Laird C, Gane L, Angkustsiri K, Tassone F, Hagerman R. Two boys with Fragile X syndrome and hepatic tumors. J Ped Hematol/Oncol. 2008;30:239–241. doi: 10.1097/MPH.0b013e31815f88c9. [DOI] [PubMed] [Google Scholar]
- Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:810–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- Zanca C, Cozzolino F, Quntavalle C, Di Costanzo S, Ricci-Vitani L, Santoriello M, Monti M, Pucci P, Condorelli G. PED interacts with Rac1 and regulates cell migration/invasion processes in human non-small cell lung cancer cells. J Cell Physiol. 2010;225:63–72. doi: 10.1002/jcp.22197. [DOI] [PubMed] [Google Scholar]


