Abstract
Peroxiredoxins (Prxs), thioredoxins (Trxs), and NADPH-thioredoxin reductases (NTRs) constitute central elements of the thiol-disulfide redox regulatory network of plant cells. This study provides a comprehensive survey of this network in the model legume Lotus japonicus. The aims were to identify and characterize these gene families and to assess whether the NTR-Trx systems are operative in nodules. Quantitative reverse transcription-polymerase chain reaction and immunological and proteomic approaches were used for expression profiling. We identified seven Prx, 14 Trx, and three NTR functional genes. The PrxQ1 gene was found to be transcribed in two alternative spliced variants and to be expressed at high levels in leaves, stems, petals, pods, and seeds and at low levels in roots and nodules. The 1CPrx gene showed very high expression in the seed embryos and low expression in vegetative tissues and was induced by nitric oxide and cytokinins. In sharp contrast, cytokinins down-regulated all other Prx genes, except PrxQ1, in roots and nodules, but only 2CPrxA and PrxQ1 in leaves. Gene-specific changes in Prx expression were also observed in response to ethylene, abscisic acid, and auxins. Nodules contain significant mRNA and protein amounts of cytosolic PrxIIB, Trxh1, and NTRA and of plastidic NTRC. Likewise, they express cytosolic Trxh3, Trxh4, Trxh8, and Trxh9, mitochondrial PrxIIF and Trxo, and plastidic Trxm2, Trxm4, and ferredoxin-Trx reductase. These findings reveal a complex regulation of Prxs that is dependent on the isoform, tissue, and signaling molecule and support that redox NTR-Trx systems are functional in the cytosol, mitochondria, and plastids of nodules.
In plants, reactive oxygen species (ROS), such as the superoxide radical and hydrogen peroxide (H2O2), are mainly formed in the chloroplasts, mitochondria, peroxisomes, and apoplast during photosynthesis, respiration, and other processes involving electron transfer (del Río et al., 2002; Mittler, 2002; Foyer and Noctor, 2005). Plant cells also produce reactive nitrogen species, such as nitric oxide (NO), S-nitrosoglutathione (GSNO), and peroxynitrite, under physiological conditions (Lamattina et al., 2003; Valderrama et al., 2007; Neill et al., 2008). Overproduction of both types of reactive species is potentially deleterious, but, at tightly controlled concentrations, they fulfill essential functions in plant development, defense response, and redox signaling (Foyer and Noctor, 2005; Besson-Bard et al., 2008). Thus, antioxidant defenses are linked to cellular regulation through a complex network involving redox input elements, transmitters, targets, and sensory proteins, such as peroxiredoxins (Prxs), thioredoxins (Trxs), and glutaredoxins (Grxs; Meyer et al., 2009; Dietz and Pfannschmidt, 2011).
Prxs constitute a ubiquitous family of nonheme thiol peroxidases that catalyze the reduction of H2O2, alkylhydroperoxides, and peroxynitrite to water, alcohols, or nitrite, respectively (Rouhier and Jacquot, 2005; Tripathi et al., 2009). These enzymes contain one or two Cys residues at the active site and usually function as monomers or dimers. Their common catalytic mechanism involves the catalytic Cys (peroxidatic) thiol, which is oxidized by peroxides to sulfenic acid. In most Prxs, the sulfenic acid is then reduced by a second Cys (resolving) thiol forming an intra or intermolecular disulfide bond. A new catalytic cycle is allowed after the reduction of the disulfide bond using electron donors, such as Trxs, Grxs, or cyclophilins (Dietz et al., 2006). There are four types of Prxs in plants (1CPrx, 2CPrx, PrxII, and PrxQ), which play specific roles according to their spatio-temporal expression patterns and subcellular localizations. Plant Prxs protect the nuclei (1CPrx), plastids (2CPrxA, 2CPrxB, PrxQ, and PrxIIE), cytosol (PrxIIB, PrxIIC, and PrxIID), and mitochondria (PrxIIF) against excess ROS in stressful conditions but are also implicated in redox signaling (Romero-Puertas et al., 2007; Tripathi et al., 2009).
Unlike most other organisms, plants have a large number of Trx genes, at least 20 in the fully sequenced genomes of Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), which are classified into seven types (for review, see Vieira Dos Santos and Rey, 2006; Meyer et al., 2009). The Trxf, Trxm, Trxx, Trxy, and Trxz are localized in the chloroplasts, the Trxh isoforms in the cytosol, and the Trxo in the mitochondria. However, some Trxh isoforms have been found also in the mitochondria, nuclei, phloem, and apoplast (Gelhaye et al., 2004). Oxidized Trxs produced as a result of reactions with Prxs and other substrates are reduced back to the functional reduced state by NADPH-thioredoxin reductases (NTRA and NTRB) in the cytosol and mitochondria (Schürmann and Jacquot, 2000; Reichheld et al., 2005) or by ferredoxin-thioredoxin reductase (FTR) in the chloroplasts (Dai et al., 2004). Another NADPH-thioredoxin reductase (NTRC) has been recently found in green tissues (Serrato et al., 2004). This peculiar enzyme contains both NTR and Trx domains in the same polypeptide and may act as a complete NTR-Trx system, reactivating plastidic 2CPrx without the assistance of classical Trxs (Moon et al., 2006; Pérez-Ruiz et al., 2006; Alkhalfioui et al., 2007).
Legume root nodules are formed as a result of the molecular interaction between the roots and soil rhizobia. The bacteroids inside the nodules fix atmospheric N2 into ammonia and in return host cells supply the bacteroids with carbon metabolites. Two model legumes, Medicago truncatula and Lotus japonicus, have been proposed for genetic analyses of indeterminate and determinate nodulation, respectively. The two types of nodules differ in some structural and biochemical features (Hirsch, 1992). The antioxidants of nodules, in particular the superoxide dismutase, catalase, and ascorbate-glutathione pathway enzymes, have been studied in some detail (for review, see Puppo et al., 2005; Becana et al., 2010), whereas there is a dearth of information concerning other antioxidant and redox sensor enzymes, such as Prxs, Trxs, and NTRs. The study of these enzymes in legumes, and particularly in nodules, is important because N2 fixation requires a strict regulation of the redox state in the host cells and bacteroids. Thus, nodules contain abundant metalloproteins that are prone to oxidation, such as nitrogenase, ferredoxin, hydrogenase, and leghemoglobin, with a high potential for ROS generation (Dalton et al., 1998; Becana et al., 2010). Knowledge of the redox regulatory network of nodules is only slowly emerging and is still in a fragmentary state. In pea (Pisum sativum), the content of PrxIIF in nodules is similar to that in roots and remains constant during nodule development (Groten et al., 2006), whereas in soybean (Glycine max), a Trxh isoform is essential for nodulation (Lee et al., 2005), and in M. truncatula, two isoforms of a new type of Trx, designated Trxs (“s” for symbiosis), are highly expressed in nodules (Alkhalfioui et al., 2008).
This study, designed to gain insights into the Prx, Trx, and NTR gene families of L. japonicus, is organized in two parts. First, we identified the Prx genes and determined their expression profiles in nodulated plants and in response to signaling compounds to better understand their functional diversity and regulation. Second, we focused on the expression of the Trx and NTR isoforms in nodules to identify possible Prx regenerating systems in these symbiotic organs.
RESULTS
Identification and Characterization of LjPrx Genes
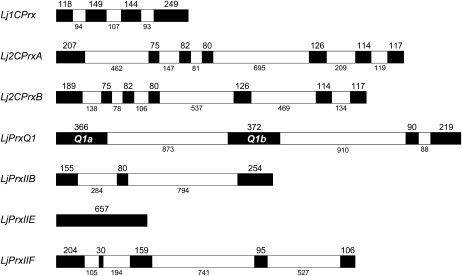
The L. japonicus Prx (LjPrx) genes were identified by searching genomic and EST databases using the Arabidopsis Prx protein sequences as BLAST queries. The open reading frames of seven LjPrx genes were found to be complete based on their tentative consensus (TC) sequences (Table I), and the exon-intron structures were elucidated by comparison between the gene and TC sequences (Fig. 1). An additional gene, here termed LjPrxQ2, was detected in the selected genome assembly contig (Sato et al., 2008), but it is not transcribed or its expression is below detection limits, in agreement with the absence of ESTs for this gene. All the LjPrx genes, except LjPrxQ2, could be mapped (Table I). The two Lj2CPrx genes are highly homologous, with 93% (nucleotide) and 84% (amino acid) identities in their sequences and with 81% to 82% (nucleotide) and 90% to 93% (amino acid) identities with respect to the 2CPrxA (At3g11630) and 2CPrxB (At5g06290) genes of Arabidopsis. The Lj2CPrx genes were designated A and B based on the higher expression of the Lj2CPrxA gene in the leaves, as occurs for Arabidopsis 2CPrxA.
Table I. Prx genes and proteins of L. japonicus.
| Gene | Clonea | Chra | TCb | No. of ESTsb | Lengthc | Mol. Massc | Localizationd | Arabidopsis Orthologe | Medicago Orthologe |
| Lj1CPrx | LjT20M01 | 4 | TC57452 | 4 | 219 | 24.7 | Nucleus/cytosol | At1g48130 | TC176842 |
| LjPrxQ1a | LjT31N02 | 4 | TC62358 | 26 | 224 | 24.4 | Chloroplast | At3g26060 | TC174754 |
| LjPrxQ1b | LjT31N02 | 4 | TC60736 | 6 | 226 | 24.6 | Chloroplast | At3g26060 | TC174754 |
| LjPrxQ2 | LjSGA_149250 | ND | – | – | – | – | Chloroplast | At3g26060 | TC174754 |
| Lj2CPrxA | LjT18K22 | 1 | TC75376 | 80 | 266 | 29.2 | Chloroplast | At3g11630 | TC179904 |
| Lj2CPrxB | LjT04E07 | 5 | TC76501 | 54 | 260 | 28.6 | Chloroplast | At5g06290 | TC174211 |
| LjPrxIIB | LjT33L17 | 2 | TC64422 | 22 | 162 | 17.5 | Cytosol | At1g65980 | TC182619 |
| LjPrxIIE | LjT19I13 | 1 | TC76090 | 20 | 218 | 23.2 | Chloroplast | At3g52960 | TC174129 |
| LjPrxIIF | LjT27H06 | 6 | TC60826 | 16 | 197 | 21.2 | Mitochondrion | At3g06050 | TC176989 |
Designation of genomic clones and chromosome location (ND, not determined).
Designation of TC sequences and number of ESTs according to the DFCI Lotus Gene Index (6.0).
Predicted number of amino acid residues and molecular mass (kD) of precursor proteins.
Predicted subcellular localizations of mature proteins.
Ortholog genes of Arabidopsis and M. truncatula according to The Arabidopsis Information Resource and DFCI Medicago Gene Index (11.0), respectively.
Figure 1.
Exon-intron organization of LjPrx genes. Exons are depicted in black boxes and intron in white boxes. Exon and intron sizes are indicated in numbers of base pairs and are drawn to scale.
The number of exons and introns of the LjPrx genes (Fig. 1) is identical to that of the Arabidopsis Prx genes (Rouhier and Jacquot, 2005), with the exception of the LjPrxQ1 gene. This single gene locus is transcribed in two mRNAs, LjPrxQ1a and LjPrxQ1b, by alternative splicing, using a different first exon but the same second and third exons (Fig. 1). The first exons display high homology, with identities of 92% (nucleotide) and 83% (amino acid). Although this high overall sequence identity precluded a separate analysis of each alternative spliced form, two sets of primers were designed that allowed us to quantify, respectively, the LjPrxQ1b mRNA and the sum of the LjPrxQ1a and LjPrxQ1b mRNAs. This quantitative reverse transcription (RT)-PCR experiment revealed that the LjPrxQ1b mRNA accounted for only <10% of the total LjPrxQ1 mRNAs in leaves, stems, petals, and pods and that it was undetectable in the other tissues examined (data not shown).
The LjPrx family includes at least one member of each Prx type, as confirmed by phylogenetic analysis (Supplemental Fig. S1). Furthermore, in silico analyses predicted that Lj2CPrxs, LjPrxQs, and LjPrxIIE are targeted to plastids, LjPrxIIB to the cytosol, and LjPrxIIF to the mitochondria (Table I). The Lj1CPrx sequence contains two highly conserved motifs: PVCTTE, which is thought to be the catalytic site of the enzyme, and KE(X13)KK(X2)LRFT, which is a putative nuclear localization signal (Mowla et al., 2002, and refs. therein). In addition, sequence analysis of Lj1CPrx with TargetP predicted that it is a cytosolic enzyme, consistent with the dual localization in the nucleus and cytosol of the 1CPrx proteins of barley (Hordeum vulgare), wheat (Triticum aestivum), and Arabidopsis (Stacy et al., 1999; Haslekås et al., 2003; Pulido et al., 2009).
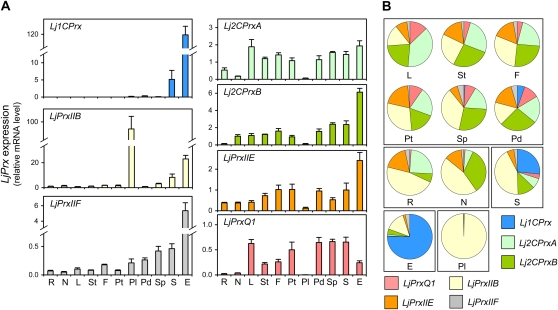
To gain insights into the functional diversification of LjPrxs, their expression profiles were determined in plant tissues. Two genes, Lj1CPrx and LjPrxIIB, are highly expressed in specific tissues, as can be noted by the different scales used to represent their mRNA levels compared to those of the other genes (Fig. 2A). Thus, expression of Lj1CPrx is almost confined to the embryo and hence is also relatively high in whole seeds. Only low levels of Lj1CPrx mRNA could be detected in vegetative organs such as roots, nodules, and leaves. Likewise, LjPrxIIB shows very high expression in pollen, moderate expression in embryos and seeds, and low expression in other organs. By contrast, LjPrxIIF is expressed in all organs but at maximal levels in the embryo. The genes encoding the plastidic Prx isoforms, namely, Lj2CPrxA, Lj2CPrxB, LjPrxIIE, and LjPrxQ1, are also expressed throughout the plant, albeit for some of these genes the mRNA levels were close to detection limits in pollen and roots (Fig. 2A). The relative abundance of all LjPrx mRNAs within each plant tissue was determined (Fig. 2B). The leaves, stems, flowers, petals, and pods exhibit similar expression profiles, with 68% to 82% of the transcripts encoding plastidic LjPrxs. The pollen and embryos show unique expression profiles. Thus, in pollen >99% of the transcripts correspond to LjPrxIIB mRNA, and in the embryo, the Lj1CPrx and LjPrxIIB mRNAs account for 75% and 15%, respectively, of the total transcripts (Fig. 2B).
Figure 2.
Expression profiles of the LjPrx gene family. A, Relative LjPrx mRNA levels in roots (R), nodules (N), leaves (L), stems (St), flowers (F), petals (Pt), pollen (Pl), pods (Pd), seedless pods (Sp), seeds (S), and embryos (E). The mRNA levels were normalized with respect to ubiquitin, and the LjPrxIIB mRNA level in roots was arbitrarily assigned a value of 1. All data are means ± se of four to eight biological replicates. B, Relative LjPrx mRNA level within each plant organ, calculated from data in A.
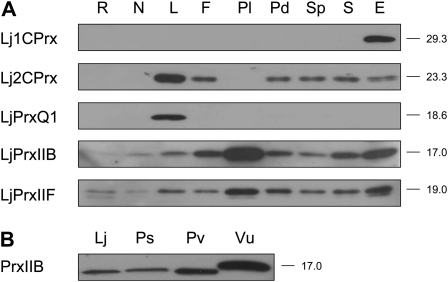
The content of the LjPrx proteins in plant tissues was examined using immunoblots (Fig. 3A). The Lj1CPrx protein was found specifically in the embryo and was undetectable in any other tissues or even in seed extracts (Fig. 2A). Because seeds contain significant Lj1CPrx mRNA levels and the protein is present in the embryos, it may be below detection limits in the whole seed extracts due to a dilution effect. Similarly, the LjPrxQ1 protein was found exclusively in leaves. The Lj2CPrx proteins accumulated in leaves and to a lower extent in flowers, pods, seeds, and embryos, whereas they were undetectable in roots, nodules, and pollen. In contrast, LjPrxIIB and LjPrxIIF were found in all organs, although the amount of LjPrxIIB was very low in roots. Also, it was necessary to load 5-fold more protein on immunoblots to unambiguously detect PrxIIB in nodules of L. japonicus and other legumes, such as pea, common bean (Phaseolus vulgaris), and cowpea (Vigna unguiculata; Fig. 3B).
Figure 3.
Immunoblot analyses of LjPrx proteins in different organs. A, Relative abundance of LjPrx proteins in roots (R), nodules (N), leaves (L), flowers (F), pollen (Pl), pods (Pd), seedless pods (Sp), seeds (S), and embryos (E). B, Detection of cytosolic PrxIIB proteins in nodules of L. japonicus (Lj), P. sativum (Ps), P. vulgaris (Pv), and V. unguiculata (Vu). Gels were loaded with 10 μg (A) or 50 μg (B) of protein per lane, and the apparent molecular masses (kD) of the proteins are indicated on the right. Blots are representative of two independent protein extractions.
Regulation of LjPrx Expression by Hormones and NO
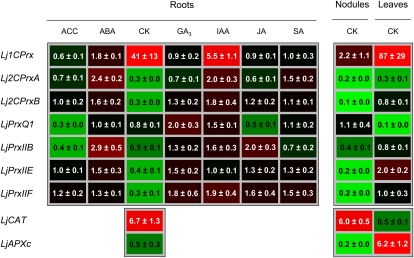
The effect of several hormones and stress signaling molecules was also studied to gain information about their role in developmental and acclimatory regulation of LjPrx gene expression. To this purpose, nodulated plants were grown in hydroponic medium supplemented with hormones for 48 h, and the expression levels of LjPrx genes were determined in the roots (Fig. 4). Treatment of plants with GA3, jasmonic acid (JA), or salicylic acid did not cause substantial changes in the LjPrx mRNA levels, whereas 1-aminocyclopropane-1-carboxylic acid (the immediate ethylene precursor) down-regulated LjPrxQ1 and LjPrxIIB, abscisic acid (ABA) up-regulated Lj2CPrxA and LjPrxIIB, and indole-3-acetic acid up-regulated Lj1CPrx. However, the most marked effects were observed with cytokinins (CKs), which decreased the expression of all LjPrx genes, except Lj1CPrx and LjPrxQ1, in roots (Fig. 4). This finding led us to investigate the effects of CK on the expression of LjPrx genes also in nodules and leaves. The response of most LjPrx genes to CK in nodules was similar to that observed in roots, whereas in leaves, CK caused down-regulation of LjPrxQ1 but had no effect on the LjPrxII genes. Although the Lj1CPrx mRNA levels were very low in vegetative organs, a strong induction of this gene in roots and leaves and a much weaker induction in nodules were detected (Fig. 4). Additional experiments showed that the increase of Lj1CPrx mRNA level in roots occurred after only 3 h and persisted for at least 48 h. This gene was induced with only 5 μm CK and had maximal expression with 100 μm CK.
Figure 4.
Heat map of the hormone response of the expression of LjPrx genes in roots. Plants grown in hydroponic cultures were treated for 48 h with 50 μm of each hormone. The effects of CK on LjPrx expression in leaves and nodules, and on LjCAT and LjAPXc genes, are also shown. Transcript levels were normalized with ubiquitin and expressed relative to those found in control plants, which were arbitrarily given a value of 1. Values are means ± se of four to 10 biological replicates from at least two independent treatments. ACC, 1-Aminocyclopropane-1-carboxylic acid; IAA, indole-3-acetic acid; SA, salicylic acid.
The effects of CK on LjPrx expression suggest an important function of this hormone in the control of the cellular level of H2O2. To test this possibility, the mRNA levels of important genes implicated in H2O2 metabolism were quantified. In roots, CK did not cause any significant change in the contents of transcripts encoding mitochondrial Mn-superoxide dismutase, cytosolic and plastidic Fe- and CuZn-superoxide dismutases, or thylakoidal, stromal, and peroxisomal ascorbate peroxidases (data not shown). However, in response to CK, the expression of catalase (LjCAT) increased in roots and nodules but not in leaves, whereas the expression of cytosolic ascorbate peroxidase (LjAPXc) decreased in nodules and increased in leaves (Fig. 4). The effect of NO on Lj1CPrx expression was also examined because this signal molecule has been implicated in the response of plant cells to CK (Tun et al., 2001; Carimi et al., 2005). Plants were supplied with two NO donors and Lj1CPrx expression levels were determined in the roots. Both S-nitroso-N-acetyl-dl-penicillamine (SNAP) and GSNO increased gene expression, although maximal induction was achieved after 24 h with SNAP and after only 3 h with GSNO (Supplemental Fig. S2).
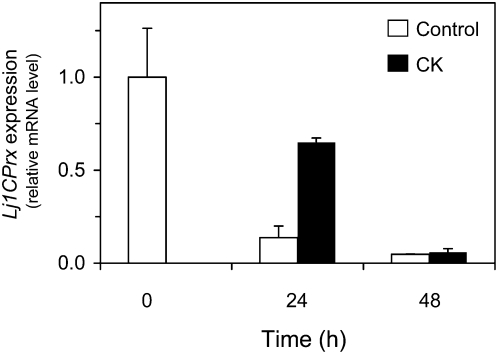
The stimulatory effect of CK on Lj1CPrx mRNA accumulation and the fact that this gene is almost exclusively expressed in the embryo (Fig. 2A) prompted us to investigate the effects of this hormone on Lj1CPrx expression in germinating seeds. In the absence of CK, the content of Lj1CPrx mRNA progressively decreased following germination and was hardly detectable after 48 h of imbibition (Fig. 5). In the presence of CK, the mRNA level was also reduced but, after 24 h, it was 65% of the initial value compared to 14% for the control seeds. These results suggest that the CK treatment induced Lj1CPrx expression in seeds and that CK was unable to completely overcome the down-regulation of the gene that takes place during germination, as can be seen at 48 h (Fig. 5).
Figure 5.
Effect of CK on expression of the Lj1CPrx gene during seed germination. Seeds were stratified for 24 h at 4°C and then germinated in agar plates for up to 48 h in the absence (control) or presence of 50 μm CK. Data are expressed with respect to the mRNA levels at time zero, which were arbitrarily given a value of 1, and are means ± se of two biological replicates, each of them corresponding to the total RNA from 10 germinating seeds.
Identification and Characterization of LjTrx and LjNTR Genes
Most Prx isoforms are reduced efficiently by Trxs, and, in turn, the nonplastidic Trxs are regenerated by NTRA and NTRB. To complete our study, we pursued the identification of the Trx and NTR proteins of L. japonicus. The search focused on the isoforms expressed in nodules to determine if the NTR-Trx system might be operative in these unique plant organs. Because of the complexity of the Trx gene family, we combined mRNA expression with immunoblot and proteomic analyses of nodules.
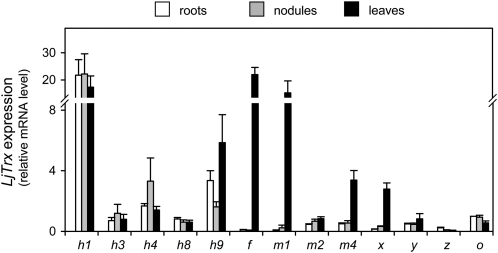
The L. japonicus EST and genomic databases were screened to identify Trxs using the Arabidopsis and M. truncatula protein sequences as BLAST queries. This analysis identified 14 LjTrx genes coding for six isoforms of Trxh, three isoforms of Trxm, and one isoform each of Trxf, Trxx, Trxy, Trxz, and Trxo, but failed to detect any homologs of Trxs (Table II). All these gene sequences were already deposited in the data banks except the LjTrxh1 clone (LjT45J20), which was isolated using TC65928 sequence information. This clone was completely sequenced for this study (accession no. AP012058). The LjTrx genes were designated according to the similarities of their derived proteins with respect to those of M. truncatula (Alkhalfioui et al., 2008; Renard et al., 2011). An alignment of the Trxh sequences (Supplemental Fig. S3) and a phylogenetic analysis of the Trxs (Supplemental Fig. S4) of L. japonicus and other model plants were performed to verify protein assignments to the Trx types and the three Trxh subgroups. Thus, LjTrxh1 and LjTrxh3 belong to subgroup I, LjTrxh4 and LjTrxh6 to subgroup II, and LjTrxh8 and LjTrxh9 to subgroup III. LjTrxh9 exhibits a very peculiar active site and may rather possess a protein disulfide isomerase activity that depends on glutathione instead of NTR (Gelhaye et al., 2004, Serrato et al., 2008). Only one isoform of subgroup I, LjTrxh1, contains the N-terminal motif MAAEE (Supplemental Fig. S3) found in the Trxh1 and Trxh2 of M. truncatula, which might allow these proteins to be secreted to the phloem or apoplast in addition to being localized to the cytosol (Supplemental Fig. S3; Renard et al., 2011). The LjTrxh3 gene was found to be transcribed (Table II), whereas no ESTs are available to date for the orthologous Trxh3 gene of M. truncatula (Renard et al., 2011). The expression profiles of the LjTrx genes were determined in roots, nodules, and leaves (Fig. 6). Notably, LjTrxh1 showed by far the greatest expression levels, whereas the LjTrxh6 mRNA was virtually undetectable in the three plant organs. As expected, the LjTrxf, LjTrxm1, LjTrxm4, and LjTrxx genes, which encode plastidic isoforms, were highly expressed in leaves compared to roots or nodules. In fact, the amounts of LjTrxf mRNA in roots and nodules, or of LjTrxm1 and LjTrxx mRNAs in roots, were near detection limit. Almost no expression of LjTrxz was observed in roots, nodules, and leaves. By contrast, in these three plant organs, the LjTrxh3, LjTrxh8, LjTrxy, and LjTrxo genes had low but significant expression, whereas LjTrxh4 and LjTrxh9 showed moderate expression (Fig. 6). Proteomic analyses allowed the unambiguous identification of the cytosolic Trxh1 isoform in nodules of L. japonicus, M. truncatula, and common bean (Table III). These analyses also identified in nodules several Prxs (PrxIIB, PrxIIE, and PrxIIF) as well as two Grxs (GrxC2 and GrxC4) that may act as putative electron donors of Prxs (Table III).
Table II. Trx and NTR genes and proteins of L. japonicus.
| Gene | Clonea | Chra | TCb | No. of ESTsb | Lengthc | Mol. Massc | Localizationd | Arabidopsis Orthologe | Medicago Orthologe |
| LjTrxh1 | LjT45J20 | 5 | TC65928 | 34 | 121 | 13.3 | Cytosol | At3g51030 | TC197256 |
| LjTrxh3 | LjT17E09 | 2 | TC68183 | 12 | 121 | 13.3 | Cytosol | At3g51030 | CR955005f |
| LjTrxh4 | LjT40C04 | 1 | TC65406 | 6 | 131 | 14.4 | Cytosol | At5g39950 | TC177162 |
| LjTrxh6 | LjT58M11 | 2 | TC65208 | 2 | 126 | 14.3 | Cytosol | At1g69880 | TC188623 |
| LjTrxh8 | LjSGA_031277 | ND | TC58009 | 16 | 138 | 15.5 | Cytosol | At3g08710 | TC176865 |
| LjTrxh9 | LjSGA_132520 | ND | TC63066 | 33 | 123 | 13.8 | Cytosol | At1g11530 | TC177045 |
| LjTrxf | LjSGA_082631 | ND | TC59402 | 23 | 179 | 19.3 | Chloroplast | At5g16400 | TC174294 |
| LjTrxm1 | LjSGA_017977 | ND | TC60229 | 8 | 181 | 19.8 | Chloroplast | At4g03520 | TC180914 |
| LjTrxm2 | LjSGA_126827 | ND | TC71331 | 15 | 183 | 19.8 | Chloroplast | At3g15360 | TC178205 |
| LjTrxm4 | LjSGA_126077 | ND | TC67299 | 20 | 182 | 19.8 | Chloroplast | At3g15360 | TC193088 |
| LjTrxx | LjT02A04 | 5 | TC61897 | 9 | 185 | 20.2 | Chloroplast | At1g50320 | TC173902 |
| LjTrxy | LjT08O18 | 5 | TC61826 | 6 | 168 | 18.7 | Chloroplast | At1g76760 | TC175512 |
| LjTrxz | LjSGA_025025 | ND | TC62611 | 9 | 188 | 21.3 | Chloroplast | At3g06730 | NP7258770 |
| LjTrxo | LjT11M06 | 4 | TC61209 | 19 | 179 | 20.0 | Mitochondrion | At2g35010 | TC184686 |
| LjNTRA | LjB04N17 | ND | TC63269/73407 | 49 | 369 | 39.5 (35.6) | Mitochondrion (cytosol) | At2g17420 | TC177239 |
| LjNTRB | LjT16K13 | ND | TC73044/80146 | 6 | 387 | 40.4 (30.7) | Mitochondrion (cytosol) | At2g17420 | TC177239 |
| LjNTRC | LjB24C14 | ND | TC57567/68679 | 9 | 518 | 56.6 | Chloroplast | At2g41680 | TC189527 |
Designation of genomic clones and chromosome location (ND, not determined).
TC sequences and number of ESTs according to the DFCI Lotus Gene Index (6.0).
Predicted number of amino acid residues and molecular mass (kD) of precursor proteins. The molecular mass in parentheses corresponds to the protein encoded by the putative alternative mRNA.
Predicted subcellular localizations of mature proteins. The localization in parentheses corresponds to the protein encoded by the putative alternative mRNA.
Ortholog genes of Arabidopsis and M. truncatula according to The Arabidopsis Information Resource and DFCI Medicago Gene Index (11.0), respectively.
Genomic clone (BAC number) is given because no ESTs are available (Renard et al., 2011).
Figure 6.
Expression profiles of LjTrx genes in roots, nodules, and leaves. The mRNA levels were normalized with respect to ubiquitin, and the LjTrxo mRNA level in roots was arbitrarily assigned a value of 1. All data are means ± se of four biological replicates.
Table III. Identification of Prxs and their putative physiological reductants by proteomic analyses of legume nodules.
| Protein | Legume | TCa | UniProtb | Peptidesb |
| 2CPrxB | P. vulgaris | TC32275 | Q9FE12 | ASSELPLVGNTAPDFEAEAVFDQEFIK, SGGLGDLNYPLISDVTK, SYDVLIPDQGIALR |
| PrxIIB | M. truncatula | TC182619 | B7FH22 | YTHALGLELDLSDK, FALLVEDLK |
| PrxIIE | M. truncatula | TC174129 | C6TFM7c | AIGVELDLSDKPVGLGVR, LFNLEEGGAFTFSGADDILK |
| PrxIIF | M. truncatula | TC176989 | B7FGM0 | VATGSDIISAASNVSLQK, SLELTTDLSGALLGTR |
| P. vulgaris | TC33229 | Q6KBB1c | VATGTDIVSAAPNVSLQK, SLELVTDLSGALLGTR | |
| Trxh1 | L. japonicus | TC65928 | Q6RJZ7c | FIAPILAEIAK, TVAEEWNVEAMPTFLFLK |
| Trxh1 | M. truncatula | TC197256 | A1BLP6 | FIAPILAEIAK, TVAEEWAVDAMPTFLFLK |
| Trxh1 | P. vulgaris | TC38652 | A1BLP7c | FIAPILAEIAK, WSIEAMPTFLFLK |
| GrxC4 | M. truncatula | TC181572 | C6TCR3c | IQDVLVNIVGK, HLGGSDETVEAYESGLLAK |
| GrxC2 | P. vulgaris | TC40141 | B3F8F4c | LIEMDVEPDGADIQAALLEWTGQR, LVPLLTSAGAITK |
| NTRA | M. truncatula | TC177239 | A6XJ26 | VSGLFFAIGHEPATK, TSVEGVFAAGDVQDKK |
TC sequences according to the DFCI Lotus Gene Index (6.0), Medicago Gene Index (11.0), or Bean Gene Index (4.0). Data of M. truncatula nodules were taken from Larrainzar et al. (2007), stored in the ProMEX spectral library (http://promex.pph.univie.ac.at/promex/), and updated to the current DFCI version.
UniProt accessions (UniRef100) and peptides detected.
UniProt accessions of ortholog proteins showing best match hit.
Similarly, a search of L. japonicus databases and genomic libraries allowed the identification of three LjNTR genes with their complete open reading frames (Table II). The sequence of one genomic clone (LjT16K13) was available from public databases, whereas two other clones (LjB04N17 and LjB24C14) were isolated from bacterial artificial chromosome (BAC) libraries using sequence information on TC63269 (accession no. AP012059) and TC57567 (accession no. AP012060), respectively. Phylogenetic analysis confirmed protein assignments to the NTRA/B and NTRC types (Supplemental Fig. S5). Designation of LjNTRA and LjNTRB was based on sequence identity (86% and 80%, respectively) to the single NTR isoform (NTRA) of M. truncatula (Alkhalfioui et al., 2007) rather than to the Arabidopsis NTRA and NTRB genes (approximately 80%). The motifs characteristic of NTRs, namely, catalytic Cys residues and FAD- and NADPH-binding sites, were fully conserved in the L. japonicus NTRA/B enzymes (Supplemental Fig. S6).
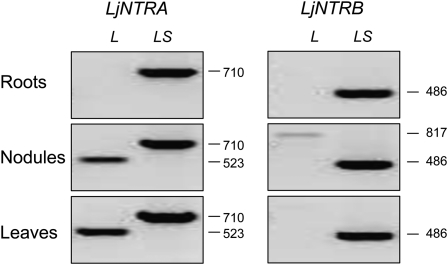
Because each of the Arabidopsis NTRA and NTRB genes can generate two types of transcripts, encoding cytosolic and mitochondrial NTR isoforms (Laloi et al., 2001; Reichheld et al., 2005), we investigated if this also occurred for the LjNTR genes by performing a semiquantitative RT-PCR analysis (Fig. 7). Specific primers were designed so that one pair of primers amplified solely the long cDNA, whereas the second pair amplified both long and short cDNAs. The long LjNTRA mRNA was found in nodules and leaves but not in roots, whereas the long LjNTRB mRNA was only detected in nodules at very low levels (Fig. 7). Consistent with this, two ESTs are available for the long LjNTRB mRNA for nodules (accession no. CB829112) and nodulating roots (accession no. DC595411). Together, the data indicate that, under our plant growth conditions, the mitochondrial isoform of LjNTRA is produced only in nodules and leaves. On the other hand, LjNTRC is predicted to be localized in the chloroplasts, as occurs for rice NTRC (Serrato et al., 2004).
Figure 7.
Steady-state levels of alternative transcripts for the LjNTRA and LjNTRB genes in roots, nodules, and leaves. Semiquantitative RT-PCR analysis was carried out using gene-specific primers (Supplemental Table S1). One pair of primers amplified exclusively the long cDNA (L), and the second pair amplified both long and short cDNAs (LS). Numbers of base pairs are given on the right.
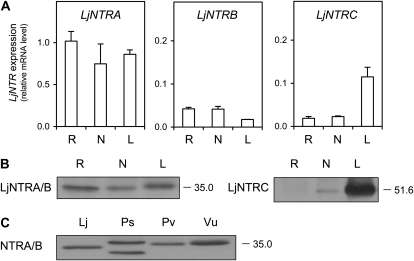
The expression of the LjNTR genes was investigated in roots, nodules, and leaves. Although the two alternative mRNAs for LjNTRA and LjNTRB were probably present, they could not be quantified separately. Instead, the total mRNA levels of LjNTRA, LjNTRB, and LjNTRC were determined (Fig. 8A). All these genes were expressed in the three plant organs, but the LjNTRA mRNA levels were considerably higher. As expected for a gene coding for a chloroplastic enzyme, the expression of LjNTRC was enhanced in leaves relative to roots or nodules. Additional experiments with other plant tissues indicated that expression levels of LjNTRA in pollen and LjNTRB in embryos were approximately 12- and 0.5-fold, respectively, those found in roots. The abundance of the LjNTRA/B and LjNTRC proteins in roots, nodules, and leaves was compared using immunoblots (Fig. 8B). A single LjNTRA/B protein (35 kD) was observed in all three organs, whereas the LjNTRC protein (51.6 kD) was very abundant in leaves, detectable in nodules, and undetectable in roots. The NTRA/B protein was also present in other legume nodules (Fig. 8C), and its identity was confirmed by proteomic analyses of M. truncatula nodules (Table III).
Figure 8.
Expression of LjNTR genes in plant organs. A, Relative LjNTR mRNA levels in roots (R), nodules (N), and leaves (L). The mRNA levels were normalized with respect to ubiquitin, and the LjNTRA mRNA level in roots was arbitrarily assigned a value of 1. All data are means ± se of four biological replicates from two independent treatments. B, Immunoblots of LjNTR proteins in roots, nodules, and leaves. C, Immunoblots of NTRA/B proteins in nodules of L. japonicus (Lj), P. sativum (Ps), P. vulgaris (Pv), and V. unguiculata (Vu). Gels were loaded with 10 μg (B) or 50 μg (C) of protein per lane, and the apparent molecular mass (kD) of the proteins is indicated on the right. Blots are representative of at least two independent protein extractions.
It is also worth noting that we detected significant mRNA levels not only of some plastidic Trxs but also of the FTRB gene (TC64844) in nodules of L. japonicus. This gene encodes the catalytic subunit of FTR, an essential component of the redox FTR-Trx system in the chloroplasts; hence, the FTRB mRNA levels were expected to be high in the leaves. However, the roots and nodules also contained significant amounts of transcript, approximately 7-fold lower than in the leaves (Supplemental Fig. S7).
DISCUSSION
In this work, we identified the Prx, Trx, and NTR multigenic families of L. japonicus and determined their expression profiles in plant tissues and, for Prxs, also in response to signaling compounds. Prxs play major roles in preventing oxidative damage and in maintaining redox homeostasis. These essential functions are consistent with the presence of Prx isoforms in most, if not all, cellular compartments. Transcriptional regulation of the Prx genes depends on developmental and environmental factors (Dietz et al., 2006). In L. japonicus, the genes encoding plastidic Prx isoforms show high (Lj2CPrxA and LjPrxQ1) or moderate (Lj2CPrxB and LjPrxIIE) expression in the leaves. The Lj2CPrx proteins, indistinguishable in immunoblots, were found to accumulate in leaves and, less abundantly, in flowers, pods, seeds, and embryos, which suggests that this type of Prxs is also present in nonphotosynthetic plastids. This was confirmed by the finding of Lj2CPrxB in the proteome of common bean nodules. A recent study linked the redox state of 2CPrx in animal and plant cells to the circadian clock and described it as a mechanism that functions independently of transcription (O’Neill et al., 2011). Such a function in timing metabolism could be also important in nonphotosynthetic plant cells.
Interestingly, two alternative spliced variants of the LjPrxQ1 gene and a putative LjPrxQ2 pseudogene could be identified. The high similarity of the deduced LjPrxQ1a and LjPrxQ1b proteins suggests that they do not differ, at least substantially, in their catalytic properties. Rather, the reason for the occurrence of two LjPrxQ1 spliced variants may reside on a different regulation because the LjPrxQ1b mRNA levels were 10-fold lower than those of LjPrxQ1a. The two PrxQ1 isoforms might be expressed in different types of leaf cells or under different environmental conditions. In Arabidopsis, the PrxQ protein is attached to thylakoids (Lamkemeyer et al., 2006), and its transcript is highly responsive to light, ascorbate, and compounds inducing oxidative stress (Horling et al., 2003). In poplar (Populus spp.), the PrxQ mRNA level increases following pathogen infection (Rouhier et al., 2004). Although the LjPrxQ1 mRNA was present in leaves, flowers, and embryos, the protein was only detectable in the leaves, suggesting posttranscriptional regulation of the gene. This organ specificity was also observed for poplar PrxQ (Rouhier et al., 2004) and suggests that the protein may be exclusively implicated in chloroplast protection. Interestingly, the expression of the LjPrxQ1 gene was down-regulated by two hormones, ethylene and JA, that play a major role in stress signaling. Unlike PrxQ, the chloroplastic PrxIIE of Arabidopsis is largely present as a soluble protein in the stroma (Dietz et al., 2006) and the gene is constitutively expressed (Bréhélin et al., 2003), which suggests that PrxIIE also plays a role in other types of plastids. Our finding of a high level of LjPrxIIE mRNA in seeds and particularly in the embryo is consistent with a function of its protein product in seed development and/or germination.
The Arabidopsis genome contains three genes encoding cytosolic PrxIIs: PrxIIB is ubiquitously expressed in plant tissues, whereas PrxIIC and PrxIID are expressed at high levels in pollen (Bréhélin et al., 2003) and at low levels in other tissues (Pena-Ahumada et al., 2006). We could identify only one homolog of such genes, PrxIIB, in the L. japonicus databases. The LjPrxIIB mRNA and protein were found in all organs, although at much higher levels in pollen, seeds, and embryos than in roots, nodules, and leaves. These observations point to a role of LjPrxIIB in the antioxidative protection of pollen grains in order to cope with oxidative stress during dessication (Bréhélin et al., 2003). They also suggest that PrxIIB is important in seeds and maturing fruits (Matamoros et al., 2010) and that, in legumes, this single protein could fulfill the functions of the three cytosolic PrxIIs of Arabidopsis. Previous work failed to detect a typical cytosolic PrxII in pea nodules (Groten et al., 2006; Matamoros et al., 2010), although the presence of a putative PrxIIA homolog (68 kD) was reported (Groten et al., 2006). This was probably due to the low abundance of PrxIIB in nodules because a genuine cytosolic PrxII (17 kD) was found here using higher protein loadings and its identity was verified by proteomic analyses of various legume nodules.
In contrast, the LjPrxIIF mRNA and protein were readily detected in all tissues, consistent with the hypothesis that PrxIIF is important in redox homeostasis and antioxidant defense of mitochondria (Finkemeier et al., 2005). This enzyme is widely distributed in all plant tissues and probably has a housekeeping function in mitochondria (Gama et al., 2007). In pea leaves, PrxIIF is induced by salt, cadmium, and cold stress (Barranco-Medina et al., 2007), whereas the poplar enzyme is relatively unresponsive (Gama et al., 2007). The high levels of LjPrxIIF mRNA and protein in the embryo may reflect an increased need for protection against ROS generated when respiration is resumed during imbibition, as proposed for cereal seeds (Stacy et al., 1999; Pulido et al., 2009). In fact, the production of superoxide radicals and H2O2 is enhanced in mitochondria from soybean embryonic axes during imbibition (Puntarulo et al., 1988), and PrxIIF could thus play a role in protecting mitochondrial DNA in seed cells. This protein was also found in nodules using immunoblots and proteomics. In pea nodules, the PrxIIF content remained unaffected with aging or after exogenous supply of ascorbate (Groten et al., 2006). In contrast, exogenous CK caused down-regulation of LjPrxIIF in roots and nodules, although this effect was not specific because the hormone also decreased the expression of most other LjPrx genes in both plant organs.
The strong effect of CK on expression of Prx genes has not been described so far and could be of physiological relevance by either linking Prxs to CK-dependent signal transduction or by adjusting the cellular redox milieu in plants. The cell cycle is under control of CK and redox state (den Boer and Murray, 2000), and glutathione is recruited to the nucleus in proliferating cells (Díaz Vivancos et al., 2010). Thus, the up-regulation by CK of Lj1CPrx, which encodes a nuclear protein, supports the hypothesis that CK and Prx collaborate in tuning the proper redox state of the dividing cell. To understand whether the effects of CK are related to ROS metabolism, the expression of several other genes encoding H2O2-scavenging enzymes was examined. Plant treatment with CK resulted in up-regulation of LjCAT and down-regulation of LjAPXc in roots and nodules and had the opposite effects in leaves. Also, external application of CK increases antioxidant enzyme activities and delays leaf senescence (Zavaleta-Mancera et al., 2007). Taken all these results together, we conclude that CK may affect H2O2 homeostasis in plant cells through changes in the regulation of critical antioxidant enzymes, such as Prxs, catalase, and ascorbate peroxidase. In this regard, a novel finding in this study is that Lj1CPrx is induced by CK in roots and leaves. This hormone promotes cell division (Romanov, 2009), which requires enhanced protection of DNA against ROS; therefore, the induction of 1CPrx would favor such a role, as has been proposed to occur during the dessication and early imbibition of seeds (Aalen et al., 1994). The protective and regulatory functions proposed for 1CPrx (Pulido et al., 2009) would explain the presence of low levels of Lj1CPrx mRNA in vegetative tissues. The localization of 1CPrx is considered to be highly restricted to the nuclei and cytosol of the developing embryo and aleurone cells of seeds (Stacy et al., 1999; Haslekås et al., 2003; Pulido et al., 2009). The protein has nevertheless been recently detected in vegetative tissues of the resurrection plant (Xerophyta viscosa) under abiotic stress or following ABA application (Mowla et al., 2002). These results suggest that 1CPrx is also expressed, although at low levels, in some plant tissues or species, where the protein may exert an antioxidant and/or signaling function in the nuclei. It is also worth mentioning that Lj1CPrx is induced by NO. This induction has not been reported to date for plant Prxs, but it was recently described for PrxI and PrxVI in murine macrophages and proposed to play a protective role against nitrosative stress and, indirectly, in H2O2 signaling (Diet et al., 2007). In any case, the NO-mediated induction of Lj1CPrx, should this occur also in seeds, would be consistent with the stimulating effect of NO on germination (Lamattina et al., 2003).
The presence of Prxs in nodules of L. japonicus shown in this work raised the question of whether the system most commonly used for Prx regeneration, consisting of Trx and NTR, is operative in these specialized organs. Consequently, the expression of LjTrxs and LjNTRs, particularly in nodules, was investigated. The genes encoding all Trx types, except the s-type, were identified in the L. japonicus genome and found to be transcribed. The Trxs genes were reported to be functional in M. truncatula (Alkhalfioui et al., 2008) but could not be found in the genomes of L. japonicus or soybean, which suggests that they are restricted to specific tribes or genera of legumes. In contrast, we could clearly detect a Trxh1 isoform in nodules of L. japonicus, M. truncatula, and common bean. However, LjTrxh4 rather than LjTrxh1 is probably the ortholog of a soybean Trxh previously reported as being essential for ROS scavenging in nodules (Lee et al., 2005). This soybean Trxh isoform has higher amino acid identity with LjTrxh4 (73%) than with LjTrxh1 (53%), LjTrxh3 (55%), or LjTrxh8 (42%). Interestingly, LjTrxh4 shows greater expression in nodules than in roots or leaves. Also, we could identify three functional LjNTR genes that are expressed in nodules. Of these, LjNTRA and LjNTRB produce long and short mRNAs presumably encoding the mitochondrial and cytosolic isoforms, as described for their Arabidopsis counterparts (Laloi et al., 2001; Reichheld et al., 2005).
The presence of NTR enzymes in nodules had not been previously reported and provides strong support to the functioning of redox NTR-Trx systems, in conjunction with Prxs, in the symbiotic tissue (Fig. 9). This is most evident by the finding of the PrxIIB, Trxh1, and NTRA proteins in the nodule cytosol. Such an NTR-Trx system requires a steady supply of NADPH, which is mainly produced by the enzymes Glc-6-P dehydrogenase, 6-phosphogluconate dehydrogenase, and isocitrate dehydrogenase, all of them very active in nodules (Marino et al., 2007). These dehydrogenases also may provide the NADPH needed for glutathione reductase and the ascorbate-glutathione pathway (Dalton et al., 1998), or for Grxs, which were also identified in nodules and are efficient reductants of PrxII (Rouhier et al., 2002). In this regard, a recent study has shown that a NTRA-Trxh3 system can intervene as a functional backup for cytosolic glutathione reductase in Arabidopsis leaves (Marty et al., 2009).
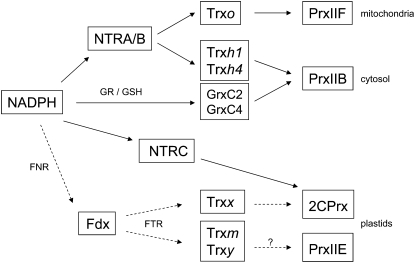
Figure 9.
Model of the redox NTR-Trx systems and their putative Prx targets, which may be operating in the mitochondria, cytosol, and plastids of nodule host cells. A putative FTR-Trx system of plastids is also indicated in dashed lines. This model essentially has been built based on the known biochemical specificities observed in vitro. The NTRA/B, NTRC, FNR, Grx, Trxh1, PrxIIB, PrxIIE, PrxIIF, and 2CPrx isoforms were detected by immunoblots and/or proteomic analysis, whereas expression of FTR, Trxo, Trxh4, Trxm, Trxx, and Trxy was detected at the mRNA level. FNR, Ferredoxin-NADP reductase; Fdx, ferredoxin; GR, glutathione reductase; GSH, glutathione.
Two other NTR-Trx pathways, localized in mitochondria and plastids, may be also functional in nodules (Fig. 9). First, the expression of mitochondrial NTRA/B and Trxo described here, in addition to PrxIIF, points to the functioning of an NTR-Trx system in nodule mitochondria. This is supported by in vitro reconstitution systems with recombinant Trxo and PrxIIF from pea leaves, which have shown that Trxo strongly interacts with, and can act as an electron donor to, PrxIIF (Finkemeier et al., 2005; Barranco-Medina et al., 2008). Second, we could detect LjNTRC in nodules, albeit at the low levels expected for a nonphotosynthetic tissue. The identification of low levels of 2CPrxB and PrxIIE in nodules using proteomics would suggest that NTRC could act as an electron donor to those Prxs, lending support to the operation of such an antioxidant system in nodule proplastids or amyloplasts. Moreover, it is known that NTRC exerts functions that are independent of Prx reduction (Pulido et al., 2010), such as redox regulation of starch synthesis in photosynthetic and nonphotosynthetic tissues (Michalska et al., 2009). This also might be the case in nodules. Thus, it will be important to define the relative contribution of the NTR-Trx and ascorbate-glutathione pathways in peroxide removal and redox signaling, as both of them are likely to be operative in the cytosol, mitochondria, and plastids of legume nodules (Dalton et al., 1998; Iturbe-Ormaetxe et al., 2001). A comparison of the two pathways will need to consider the differences in abundance of ascorbate peroxidase (up to 0.9% of the total nodule soluble proteins; Dalton et al., 1998) and PrxIIB (low levels found here) in the nodule cytosol or in the responses of these enzymes to developmental or environmental cues. Besides the components of the NTR-Trx systems, we detected significant expression of the FTRB gene in nodules of L. japonicus. Considering that FTR is an electron donor of some Trxs in the chloroplasts, an FTR-Trx system, comprising ferredoxin, FTR, and plastidic Trxs, might be also functional in nodules (Fig. 9). Although this system is beyond the scope of this work, it can be anticipated that FTR activity provides a means by which plastidic Trxs are regenerated in nodules because available information suggests that the NTRC enzyme is unable to reduce Trx in the chloroplasts and presumably in nonphotosynthetic plastids (Serrato et al., 2004; Traverso et al., 2008). The functionality of an FTR-Trx system in nodules is further supported by the presence of such a redox system in wheat endosperm amyloplasts (Balmer et al., 2006) and of ferredoxin-NADP reductase in M. truncatula nodules (Larrainzar et al., 2007).
MATERIALS AND METHODS
Plant Growth and Treatments
One-week-old seedlings of Lotus japonicus ‘MG20’ were inoculated with Mesorhizobium loti strain R7A, transferred to aerated hydroponic cultures lacking combined nitrogen (1:4 strength B&D nutrient solution), and grown under controlled environment conditions (Bustos-Sanmamed et al., 2011). Roots, leaves, nodules, and stems were harvested from 45-d-old plants (late vegetative stage), and flowers, pollen, pods, seeds, and embryos were collected from 60-d-old plants (pods of approximately 3.5 cm; late flowering-fruiting stage). Plant material was immediately flash-frozen in liquid nitrogen and stored at −85°C until use.
Plants grown in hydroponics were treated for 48 h with 50 μm ABA, GA3, JA, indole-3-acetic acid, 1-aminocyclopropane-1-carboxylic acid, or CK (an equimolar mixture of kinetin and 6-benzyl-aminopurine) as described previously (Bustos-Sanmamed et al., 2011). To study the effects of CK on germination, seeds were surface-disinfected, stratified for 24 h in 0.5% agar plates at 4°C, and treated with the hormone for up to 48 h during germination. Control seeds were germinated on plates in the presence of 0.1 mm NaOH, which was also used to dissolve CK. The pH value was kept at 6.6 for both control and CK-treated seeds with 5 mm MES. The effect of NO on gene expression in roots of 15-d-old nonnodulated seedlings was studied by application of two NO-releasing compounds, SNAP (500 μm; Sigma-Aldrich) and GSNO (250 μm; Calbiochem), for up to 24 h. In the case of GSNO, a control treatment with 250 μm of glutathione was included because this physiological NO donor releases both NO and glutathione during the incubation period.
Gene Identification and Expression Profiling
Transformation-competent artificial chromosome and BAC genomic libraries of L. japonicus were screened with probes based on the cDNA sequences. The partial or full nucleotide sequences of the isolated transformation-competent artificial chromosome/BAC clones were determined according to the bridging shotgun method (Sato et al., 2008).
Total RNA was extracted from plant material with the RNAqueous isolation kit (Ambion), and cDNA was synthesized from 2 μg DNase-treated RNA with (dT)17 and Moloney murine leukemia virus reverse transcriptase (Promega). Quantitative RT-PCR analysis was performed with an iCycler iQ instrument using iQ SYBR-Green Supermix reagents (Bio-Rad) and gene-specific primers (Supplemental Table S1). The PCR program and other details were already described (Bustos-Sanmamed et al., 2011). The amplification efficiency of primers, calculated by serial dilutions of cDNAs, was >80%. Gene expression levels were normalized with ubiquitin. The PP2A gene, encoding a subunit of the Ser/Thr protein phosphatase 2A (Czechowski et al., 2005), was used as an additional reference gene to verify that ubiquitin expression was not affected by any of the treatments.
Phylogenetic Analyses and Prediction of Subcellular Localization of Proteins
Multiple alignment of amino acid sequences was performed using the MegAlign-DNASTAR program (Lasergene) by the neighbor-joining ClustalW2 method, and the phylogenetic trees were built with 1,000 bootstrap replicates. Predictions of subcellular localizations were carried out using the MitoProt (http://ihg.gsf.de/ihg/mitoprot.html), TargetP v1.1 (http://www.cbs.dtu.dk/services/TargetP/), and PSORT (http://psort.hgc.jp/) programs. The nuclear localization of 1CPrx was predicted according to the presence of a bipartite nuclear localization signal in the C-terminal region of the amino acid sequence (Stacy et al., 1999).
Immunoblots
For protein extraction, frozen plant organs were pulverized with liquid nitrogen and homogenized in ice-cold extraction buffer (1 mL per 0.2 g fresh weight) containing 50 mm potassium phosphate (pH 7), 5 mm dithiothreitol, 1% Triton X-100, and complete protease inhibitor cocktail (Roche). The extracts were cleared by centrifugation and stored at −20°C if necessary. Total proteins were separated by 12% SDS-PAGE and blotted onto polyvinylidene difluoride membranes, and immunoblots were carried out as described elsewhere (Matamoros et al., 2010). The sources of the polyclonal antibodies were as follows: 2CPrx, PrxQ, PrxIIC, and PrxIIF of Arabidopsis (Arabidopsis thaliana; Horling et al., 2003); 1CPrx of barley (Hordeum vulgare; Stacy et al., 1999); Trxh1 to Trxh5 of poplar (Populus spp.; Gelhaye et al., 2004); NTRA/B of wheat (Triticum aestivum; Serrato et al., 2002); and NTRC of rice (Oryza sativa; Serrato et al., 2004). The PrxIIC antibody was used to detect LjPrxIIB as this antibody recognizes the two cytosolic PrxII isoforms but does not cross-react with plastidic PrxIIE (Horling et al., 2003) or mitochondrial PrxIIF (Finkemeier et al., 2005).
Proteomic Analyses of Nodules
All proteomic analyses were performed at the University of Vienna using a gel-free protocol based on liquid chromatography and tandem mass spectrometry as outlined in detail by Larrainzar et al. (2007) and Hoehenwarter and Wienkoop (2010). After mass spectrometry analysis, raw files were searched against the Dana-Farber Cancer Institute (DFCI) Lotus Gene Index (6.0), Medicago Gene Index (11.0), or Bean Gene Index (4.0) databases using the Sequest algorithm. For identification and spectral count based data, the matrix generation Proteome Discoverer (version 1.1; Thermo Fisher Scientific) was used. A decoy database enabled false positive rate analysis. Only high confidence peptides (false positive rate < 0.1%) better than 5 ppm precursor mass accuracy per protein passed criteria.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic analysis of Prx proteins from higher plants.
Supplemental Figure S2. Effect of NO donors on Lj1CPrx gene expression in roots.
Supplemental Figure S3. Sequence alignment of Trxh proteins of model legumes.
Supplemental Figure S4. Phylogenetic analysis of Trx proteins of model plants.
Supplemental Figure S5. Phylogenetic analysis of NTR proteins of higher plants.
Supplemental Figure S6. Sequence alignment of NTRA/B proteins of plants.
Supplemental Figure S7. Expression analysis of the LjFTRB gene.
Supplemental Table S1. Primers used for RT-PCR analyses.
Supplementary Material
Acknowledgments
We thank Stefanie Wienkoop and Christiana Staudinger (University of Vienna) for proteomic analyses of nodules and two anonymous reviewers for helpful comments. A.T-M. was the recipient of a postdoctoral contract (JAE-CSIC program).
References
- Aalen RB, Opsahl-Ferstad HG, Linnestad C, Olsen OA. (1994) Transcripts encoding an oleosin and a dormancy related protein are present in both the aleurone layer and the embryo of developing barley (Hordeum vulgare L.) seeds. Plant J 5: 385–396 [DOI] [PubMed] [Google Scholar]
- Alkhalfioui F, Renard M, Frendo P, Keichinger C, Meyer Y, Gelhaye E, Hirasawa M, Knaff DB, Ritzenthaler C, Montrichard F. (2008) A novel type of thioredoxin dedicated to symbiosis in legumes. Plant Physiol 148: 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhalfioui F, Renard M, Montrichard F. (2007) Unique properties of NADP-thioredoxin reductase C in legumes. J Exp Bot 58: 969–978 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Cai N, Manieri W, Schürmann P, Hurkman WJ, Buchanan BB. (2006) A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc Natl Acad Sci USA 103: 2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barranco-Medina S, Krell T, Bernier-Villamor L, Sevilla F, Lázaro JJ, Dietz KJ. (2008) Hexameric oligomerization of mitochondrial peroxiredoxin PrxIIF and formation of an ultrahigh affinity complex with its electron donor thioredoxin Trx-o. J Exp Bot 59: 3259–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barranco-Medina S, Krell T, Finkemeier I, Sevilla F, Lázaro JJ, Dietz KJ. (2007) Biochemical and molecular characterization of the mitochondrial peroxiredoxin PsPrxII F from Pisum sativum. Plant Physiol Biochem 45: 729–739 [DOI] [PubMed] [Google Scholar]
- Becana M, Matamoros MA, Udvardi M, Dalton DA. (2010) Recent insights into antioxidant defenses of legume root nodules. New Phytol 188: 960–976 [DOI] [PubMed] [Google Scholar]
- Besson-Bard A, Pugin A, Wendehenne D. (2008) New insights into nitric oxide signaling in plants. Annu Rev Plant Biol 59: 21–39 [DOI] [PubMed] [Google Scholar]
- Bréhélin C, Meyer EH, de Souris JP, Bonnard G, Meyer Y. (2003) Resemblance and dissemblance of Arabidopsis type II peroxiredoxins: similar sequences for divergent gene expression, protein localization, and activity. Plant Physiol 132: 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos-Sanmamed P, Tovar-Méndez A, Crespi M, Sato S, Tabata S, Becana M. (2011) Regulation of nonsymbiotic and truncated hemoglobin genes of Lotus japonicus in plant organs and in response to nitric oxide and hormones. New Phytol 189: 765–776 [DOI] [PubMed] [Google Scholar]
- Carimi F, Zottini M, Costa A, Cattelan I, De Michele R, Terzi M, Lo Schiavo F. (2005) NO signalling in cytokinin-induced programmed cell death. Plant Cell Environ 28: 1171–1178 [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Johansson K, Miginiac-Maslow M, Schürmann P, Eklund H. (2004) Structural basis of redox signaling in photosynthesis: structure and function of ferredoxin:thioredoxin reductase and target enzymes. Photosynth Res 79: 233–248 [DOI] [PubMed] [Google Scholar]
- Dalton DA, Joyner SL, Becana M, Iturbe-Ormaetxe I, Chatfield JM. (1998) Antioxidant defenses in the peripheral cell layers of legume root nodules. Plant Physiol 116: 37–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río LA, Corpas FJ, Sandalio LM, Palma JM, Gómez M, Barroso JB. (2002) Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J Exp Bot 53: 1255–1272 [PubMed] [Google Scholar]
- den Boer BGW, Murray JAH. (2000) Triggering the cell cycle in plants. Trends Cell Biol 10: 245–250 [DOI] [PubMed] [Google Scholar]
- Díaz Vivancos P, Dong Y, Ziegler K, Markovic J, Pallardó FV, Pellny TK, Verrier PJ, Foyer CH. (2010) Recruitment of glutathione into the nucleus during cell proliferation adjusts whole-cell redox homeostasis in Arabidopsis thaliana and lowers the oxidative defence shield. Plant J 64: 825–838 [DOI] [PubMed] [Google Scholar]
- Diet A, Abbas K, Bouton C, Guillon B, Tomasello F, Fourquet S, Toledano MB, Drapier JC. (2007) Regulation of peroxiredoxins by nitric oxide in immunostimulated macrophages. J Biol Chem 282: 36199–36205 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, de Miranda SM, Baier M, Finkemeier I. (2006) The function of peroxiredoxins in plant organelle redox metabolism. J Exp Bot 57: 1697–1709 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Pfannschmidt T. (2011) Novel regulators in photosynthetic redox control of plant metabolism and gene expression. Plant Physiol 155: 1477–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I, Goodman M, Lamkemeyer P, Kandlbinder A, Sweetlove LJ, Dietz KJ. (2005) The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J Biol Chem 280: 12168–12180 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. (2005) Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ 28: 1056–1071 [Google Scholar]
- Gama F, Keech O, Eymery F, Finkemeier I, Gelhaye E, Gardeström P, Dietz KJ, Rey P, Jacquot JP, Rouhier N. (2007) The mitochondrial type II peroxiredoxin from poplar. Physiol Plant 129: 196–206 [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP. (2004) The thioredoxin h system of higher plants. Plant Physiol Biochem 42: 265–271 [DOI] [PubMed] [Google Scholar]
- Groten K, Dutilleul C, van Heerden PDR, Vanacker H, Bernard S, Finkemeier I, Dietz KJ, Foyer CH. (2006) Redox regulation of peroxiredoxin and proteinases by ascorbate and thiols during pea root nodule senescence. FEBS Lett 580: 1269–1276 [DOI] [PubMed] [Google Scholar]
- Haslekås C, Viken MK, Grini PE, Nygaard V, Nordgard SH, Meza TJ, Aalen RB. (2003) Seed 1-cysteine peroxiredoxin antioxidants are not involved in dormancy, but contribute to inhibition of germination during stress. Plant Physiol 133: 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch AM. (1992) Developmental biology of legume nodulation. New Phytol 122: 211–237 [DOI] [PubMed] [Google Scholar]
- Hoehenwarter W, Wienkoop S. (2010) Spectral counting robust on high mass accuracy mass spectrometers. Rapid Commun Mass Spectrom 24: 3609–3614 [DOI] [PubMed] [Google Scholar]
- Horling F, Lamkemeyer P, König J, Finkemeier I, Kandlbinder A, Baier M, Dietz KJ. (2003) Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol 131: 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I, Matamoros MA, Rubio MC, Dalton DA, Becana M. (2001) The antioxidants of legume nodule mitochondria. Mol Plant Microbe Interact 14: 1189–1196 [DOI] [PubMed] [Google Scholar]
- Laloi C, Rayapuram N, Chartier Y, Grienenberger JM, Bonnard G, Meyer Y. (2001) Identification and characterization of a mitochondrial thioredoxin system in plants. Proc Natl Acad Sci USA 98: 14144–14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamattina L, García-Mata C, Graziano M, Pagnussat G. (2003) Nitric oxide: the versatility of an extensive signal molecule. Annu Rev Plant Biol 54: 109–136 [DOI] [PubMed] [Google Scholar]
- Lamkemeyer P, Laxa M, Collin V, Li W, Finkemeier I, Schöttler MA, Holtkamp V, Tognetti VB, Issakidis-Bourguet E, Kandlbinder A, et al. (2006) Peroxiredoxin Q of Arabidopsis thaliana is attached to the thylakoids and functions in context of photosynthesis. Plant J 45: 968–981 [DOI] [PubMed] [Google Scholar]
- Larrainzar E, Wienkoop S, Weckwerth W, Ladrera R, Arrese-Igor C, González EM. (2007) Medicago truncatula root nodule proteome analysis reveals differential plant and bacteroid responses to drought stress. Plant Physiol 144: 1495–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Shin KH, Kim YK, Suh JY, Gu YY, Kim MR, Hur YS, Son O, Kim JS, Song E, et al. (2005) Induction of thioredoxin is required for nodule development to reduce reactive oxygen species levels in soybean roots. Plant Physiol 139: 1881–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D, González EM, Frendo P, Puppo A, Arrese-Igor C. (2007) NADPH recycling systems in oxidative stressed pea nodules: a key role for the NADP+-dependent isocitrate dehydrogenase. Planta 225: 413–421 [DOI] [PubMed] [Google Scholar]
- Marty L, Siala W, Schwarzländer M, Fricker MD, Wirtz M, Sweetlove LJ, Meyer Y, Meyer AJ, Reichheld JP, Hell R. (2009) The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc Natl Acad Sci USA 106: 9109–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros MA, Loscos J, Dietz KJ, Aparicio-Tejo PM, Becana M. (2010) Function of antioxidant enzymes and metabolites during maturation of pea fruits. J Exp Bot 61: 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Buchanan BB, Vignols F, Reichheld JP. (2009) Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet 43: 335–367 [DOI] [PubMed] [Google Scholar]
- Michalska J, Zauber H, Buchanan BB, Cejudo FJ, Geigenberger P. (2009) NTRC links built in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc Natl Acad Sci USA 106: 9908–9913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Moon JC, Jang HH, Chae HB, Lee JR, Lee SY, Jung YJ, Shin MR, Lim HS, Chung WS, Yun DJ, et al. (2006) The C-type Arabidopsis thioredoxin reductase ANTR-C acts as an electron donor to 2-Cys peroxiredoxins in chloroplasts. Biochem Biophys Res Commun 348: 478–484 [DOI] [PubMed] [Google Scholar]
- Mowla SB, Thomson JA, Farrant JM, Mundree SG. (2002) A novel stress-inducible antioxidant enzyme identified from the resurrection plant Xerophyta viscosa Baker. Planta 215: 716–726 [DOI] [PubMed] [Google Scholar]
- Neill S, Barros R, Bright J, Desikan R, Hancock J, Harrison J, Morris P, Ribeiro D, Wilson I. (2008) Nitric oxide, stomatal closure, and abiotic stress. J Exp Bot 59: 165–176 [DOI] [PubMed] [Google Scholar]
- O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. (2011) Circadian rhythms persist without transcription in a eukaryote. Nature 469: 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Ahumada A, Kahmann U, Dietz KJ, Baier M. (2006) Regulation of peroxiredoxin expression versus expression of Halliwell-Asada-Cycle enzymes during early seedling development of Arabidopsis thaliana. Photosynth Res 89: 99–112 [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Spínola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ. (2006) Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18: 2356–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P, Cazalis R, Cejudo FJ. (2009) An antioxidant redox system in the nucleus of wheat seed cells suffering oxidative stress. Plant J 57: 132–145 [DOI] [PubMed] [Google Scholar]
- Pulido P, Spínola MC, Kirchsteiger K, Guinea M, Pascual MB, Sahrawy M, Sandalio LM, Dietz KJ, González M, Cejudo FJ. (2010) Functional analysis of the pathways for 2-Cys peroxiredoxin reduction in Arabidopsis thaliana chloroplasts. J Exp Bot 61: 4043–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntarulo S, Sánchez RA, Boveris A. (1988) Hydrogen peroxide metabolism in soybean embryonic axes at the onset of germination. Plant Physiol 86: 626–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppo A, Groten K, Bastian F, Carzaniga R, Soussi M, Lucas MM, de Felipe MR, Harrison J, Vanacker K, Foyer CH. (2005) Legume nodule senescence: roles for redox and hormone signalling in the orchestration of the natural aging process. New Phytol 165: 683–701 [DOI] [PubMed] [Google Scholar]
- Reichheld JP, Meyer E, Khafif M, Bonnard G, Meyer Y. (2005) AtNTRB is the major mitochondrial thioredoxin reductase in Arabidopsis thaliana. FEBS Lett 579: 337–342 [DOI] [PubMed] [Google Scholar]
- Renard M, Alkhalfioui F, Schmitt-Keichinger C, Ritzenthaler C, Montrichard F. (2011) Identification and characterization of thioredoxin h isoforms differentially expressed in germinating seeds of the model legume Medicago truncatula. Plant Physiol 155: 1113–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov GA. (2009) How do cytokinins affect the cell? Russ J Plant Physiol 56: 268–290 [Google Scholar]
- Romero-Puertas MC, Laxa M, Mattè A, Zaninotto F, Finkemeier I, Jones AME, Perazzolli M, Vandelle E, Dietz KJ, Delledonne M. (2007) S-Nitrosylation of peroxiredoxin IIE promotes peroxynitrite-mediated tyrosine nitration. Plant Cell 19: 4120–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Jacquot JP. (2002) Glutaredoxin-dependent peroxiredoxin from poplar: protein-protein interaction and catalytic mechanism. J Biol Chem 277: 13609–13614 [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Gualberto JM, Jordy MN, De Fay E, Hirasawa M, Duplessis S, Lemaire SD, Frey P, Martin F, et al. (2004) Poplar peroxiredoxin Q. A thioredoxin-linked chloroplast antoxidant functional in pathogen defense. Plant Physiol 134: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Jacquot JP. (2005) The plant multigenic family of thiol peroxidases. Free Radic Biol Med 38: 1413–1421 [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. (2008) Genome structure of the legume Lotus japonicus. DNA Res 15: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann P, Jacquot JP. (2000) Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol 51: 371–400 [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Pérez-Ruiz JM, Cejudo FJ. (2002) Cloning of thioredoxin h reductase and characterization of the thioredoxin reductase-thioredoxin h system from wheat. Biochem J 367: 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrato AJ, Pérez-Ruiz JM, Spínola MC, Cejudo FJ. (2004) A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem 279: 43821–43827 [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Guilleminot J, Meyer Y, Vignols F. (2008) AtCXXS: atypical members of the Arabidopsis thaliana thioredoxin h family with a remarkably high disulfide isomerase activity. Physiol Plant 133: 611–622 [DOI] [PubMed] [Google Scholar]
- Stacy RAP, Nordeng TW, Culiáñez-Macià FA, Aalen RB. (1999) The dormancy-related peroxiredoxin anti-oxidant, PER1, is localized to the nucleus of barley embryo and aleurone cells. Plant J 19: 1–8 [DOI] [PubMed] [Google Scholar]
- Traverso JA, Vignols F, Cazalis R, Serrato AJ, Pulido P, Sahrawy M, Meyer Y, Cejudo FJ, Chueca A. (2008) Immunocytochemical localization of Pisum sativum TRXs f and m in non-photosynthetic tissues. J Exp Bot 59: 1267–1277 [DOI] [PubMed] [Google Scholar]
- Tripathi BN, Bhatt I, Dietz KJ. (2009) Peroxiredoxins: a less studied component of hydrogen peroxide detoxification in photosynthetic organisms. Protoplasma 235: 3–15 [DOI] [PubMed] [Google Scholar]
- Tun NN, Holk A, Scherer GFE. (2001) Rapid increase of NO release in plant cell cultures induced by cytokinin. FEBS Lett 509: 174–176 [DOI] [PubMed] [Google Scholar]
- Valderrama R, Corpas FJ, Carreras A, Fernández-Ocaña A, Chaki M, Luque F, Gómez-Rodríguez MV, Colmenero-Varea P, del Río LA, Barroso JB. (2007) Nitrosative stress in plants. FEBS Lett 581: 453–461 [DOI] [PubMed] [Google Scholar]
- Vieira Dos Santos C, Rey P. (2006) Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci 11: 329–334 [DOI] [PubMed] [Google Scholar]
- Zavaleta-Mancera HA, López-Delgado H, Loza-Tavera H, Mora-Herrera M, Trevilla-García C, Vargas-Suárez M, Ougham H. (2007) Cytokinin promotes catalase and ascorbate peroxidase activities and preserves the chloroplast integrity during dark-senescence. J Plant Physiol 164: 1572–1582 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.