Abstract
Ubiquitination is one of the most conserved post-translational modifications of proteins, and is involved in essential eukaryotic cellular processes. These include protein degradation, transcriptional regulation, cell-cycle progression, and signaling. Microbial pathogens have evolved sophisticated systems to hijack host cellular functions for their own benefit. Central to these systems are protein transport machineries; many pathogenic bacteria inject “effector proteins” to modulate host cellular processes including the ubiquitin pathway. Numerous bacterial pathogens have been found to modulate the host ubiquitin system in various ways. In this review, we focus on three examples of temporal and spatial regulation of bacterial effectors, which are mediated by the host ubiquitin system. Subversion of the host ubiquitin system must be a widespread strategy among pathogenic bacteria to accomplish successful infection.
Keywords: bacterial pathogen, effector, secretion system, ubiquitin, proteasome, degradation, temporal regulation, metaeffector
Introduction
Pathogenic bacteria have evolved specialized protein secretion systems to transfer a wide array of bacterial proteins into eukaryotic cells, exhibiting an intimate relationship between bacteria and host cells. Historically referred to as type III, IV, and VI secretion systems, distinct types of “nanomachines” are used by pathogenic bacteria to directly inject bacterial proteins, so called “effector proteins,” into the host cell cytosol (Galan, 2001; Christie et al., 2005; Backert and Meyer, 2006; Galan and Wolf-Watz, 2006; Filloux et al., 2008). These effector proteins modulate many cellular processes essential for the establishment of infection, such as actin cytoskeleton rearrangement, vesicular trafficking manipulation, interference of numerous cellular signaling pathways, and escape of host immune systems (Galan, 2001; Waterman and Holden, 2003; Backert and Meyer, 2006; Pukatzki et al., 2009). Therefore, it can be said that effector proteins play a key role in the hijacking of host cellular systems by bacteria.
In some cases, effector proteins translocated into host cells by a bacterium display opposing functions on a single host protein. For instance, Legionella pneumophila DrrA/SidM and LepB are effector proteins with opposing effects on the host Rab1 GTPase; DrrA functions as a guanine nucleotide exchange factor (GEF), and LepB has GTPase-activating protein (GAP) activity (Machner and Isberg, 2006, 2007; Murata et al., 2006; Ingmundson et al., 2007). As discussed later in this article, the Salmonella enterica serovar Typhimurium effectors SopE and SptP also display GEF and GAP activities, respectively, for the Rho family of GTPases. To conduct opposing functions according to the progressive stages of infection, effector expression, translocation, and activity within host cells must be precisely regulated in a temporal and/or spatial manner.
Ubiquitin is a small protein of 76 amino acids and is conserved among all eukaryotic organisms (Goldstein et al., 1975). Ubiquitination is a process that involves the covalent addition of one or several ubiquitins onto a target protein, and is one of the essential post-translational modifications of eukaryotic proteins (Hershko and Ciechanover, 1998). The process involves at least three distinct types of enzymes: E1 activating enzymes, E2 conjugation enzymes, and E3 ubiquitin ligases (Hershko et al., 1983). E3 ubiquitin ligases are responsible for substrate specificity by directly interacting with target proteins. They are a large and diverse group of proteins and are subclassified into two major types depending on the presence of distinct motifs: the really interesting new gene (RING)/U-box domain and the homologous to E6-associated protein C terminus (HECT) domain (Pavletich, 2002; Ardley and Robinson, 2005).
The conjugation of a single ubiquitin, known as monoubiquitination, can trigger an alteration in the activity and/or localization of target proteins, and is involved in many cellular processes including membrane trafficking, endocytosis, DNA repair, and transcription regulation (Haglund and Dikic, 2005). The fate of proteins possessing four or more chains of ubiquitin is determined by the linkage pattern of the polyubiquitin chains. Polyubiquitinated proteins with Lys-48-linked chains are commonly targeted for 26S proteasome-dependent degradation, whereas Lys-63-linked polyubiquitination is involved in a wide range of cellular processes (Thrower et al., 2000; Haglund and Dikic, 2005).
Previous studies have established that many pathogenic bacteria use the host ubiquitin system to establish infection and to evade the host immune system (Angot et al., 2007; Rytkonen and Holden, 2007; Hicks and Galan, 2010). Furthermore, evidence indicates that ubiquitination of effector proteins plays a critical role in the temporal or spatial regulation of proteins within the host cytoplasm. Here, we review three examples of ubiquitin-mediated spatial or temporal regulation of bacterial effectors: (1) Legionella metaeffector LubX, which functions as an E3 ubiquitin ligase on another effector SidH, leading to proteasomal degradation; (2) Salmonella effectors SopE and SptP, both of which are subjected to proteasomal degradation with distinct half-lives; and (3) Salmonella effector SopB, the cellular localization of which is regulated by ubiquitination of the protein.
Bacterial Effector Proteins Mimicking Eukaryotic E3 Ligases
Recent studies have revealed that many bacterial effectors have E3 ubiquitin ligase activity. SopA, a Salmonella Typhimurium effector, is an example of a bacterial HECT-type E3 ubiquitin ligase and is involved in the host inflammatory response following Salmonella infection (Zhang et al., 2006; Diao et al., 2008). SopA itself is reported to be ubiquitinated by the host E3 ubiquitin ligase HsRMA1 (Zhang et al., 2005). A homolog of SopA, the enterohemorrhagic Escherichia coli (EHEC) O157 effector protein NleL, was recently characterized by the same group (Lin et al., 2011), showing that it has E3 ligase activity and shares structural similarity with SopA.
There are some examples of bacterial RING/U-box-type E3 ligases. The Pseudomonas syringae effector AvrPtoB has no detectable sequence similarity to eukaryotic E3 ligases, but structural studies demonstrated that AvrPtoB has higher-order structural similarity to the U-box domain (Janjusevic et al., 2006). AvrPtoB suppresses programmed cell death in infected plants depending on E3 ligase activity (Abramovitch et al., 2003, 2006). The host targets of AvrPtoB have been identified. Several kinases important in plant immunity, including tomato Fen kinase (Rosebrock et al., 2007), plant pattern-recognition receptor FLS2 (Gohre et al., 2008), and LysM receptor kinase CERK1 (Gimenez-Ibanez et al., 2009), were subjected to AvrPtoB-mediated polyubiquitination, resulting in proteasomal degradation. Recent studies also revealed that E. coli O157 NleG effectors have higher-order, but not primary, structural similarity to the RING/U-box domain (Wu et al., 2010).
SKP1/CUL1/F-box protein (SCF) complexes are multi-subunit RING-type E3 ubiquitin ligases (Schulman et al., 2000). The F-box proteins provide specificity for ubiquitination by directly binding the target proteins. Pathogenic bacteria encode numerous F-box proteins; examples include Agrobacterium tumefaciens VirF, Ralstonia solanacearum GALA proteins, and proteins from other plant pathogens and human pathogens, including Coxiella burnetii and L. pneumophila (Chen et al., 2010; Price and Abu Kwaik, 2010).
Recent structural studies revealed a new family of E3 ligases, termed novel E3 ligases (NELs), which are conserved in bacteria and are distinct from both the RING and the HECT-type ligases (Hicks and Galan, 2010). The NEL E3 ubiquitin ligases include IpaH proteins from Shigella spp. (Singer et al., 2008; Zhu et al., 2008), and SspH1 and SspH2 from Salmonella spp. (Rohde et al., 2007; Quezada et al., 2009; Levin et al., 2010). IpaH9.8, a Shigella flexneri effector, is a NEL-type E3 ubiquitin ligase (Rohde et al., 2007) that promotes polyubiquitination and the subsequent proteasome-dependent degradation of the NFκB essential modifier (NEMO), which influences NFκB activation (Ashida et al., 2010).
The P. syringe effector HopM1 is thought to belong to an E3 ubiquitin ligase complex as an adaptor protein. Even without classical E3 ligase features, it mediates ubiquitination and the proteasome-dependent elimination of host AtMIN7, a plant protein involved in host defense (Nomura et al., 2006).
Temporal Regulation by a Metaeffector: Legionella LubX and SidH
Legionella effector proteins
Legionella pneumophila is a Gram-negative bacterium ubiquitously found in freshwater environments (Fields, 1996). L. pneumophila is able to replicate in a wide range of eukaryotic host cells, from free-living amoeba to human macrophages. Human infection can result in a severe pneumonia called Legionnaires’ disease (Fields et al., 2002). The pathogenesis of Legionella is associated with intracellular replication of the bacteria. To replicate within phagocytic cells and to establish infection, L. pneumophila delivers numerous effector proteins (Ninio and Roy, 2007; Ensminger and Isberg, 2009; Franco et al., 2009; Isberg et al., 2009; Hubber and Roy, 2010; Zhu et al., 2011) via specialized protein transport machinery, the Dot/Icm type IV secretion system (T4SS; Segal et al., 1998; Vogel et al., 1998). Through the function of the effector proteins, L. pneumophila remodels the Legionella-containing phagosome to form a replicative compartment (Horwitz, 1983; Swanson and Isberg, 1995; Tilney et al., 2001). Effector proteins also modulate various host cellular functions for bacterial benefit and host cell survival, avoiding host immune responses, and bacterial egress (Shin and Roy, 2008).
It has been demonstrated experimentally that L. pneumophila translocates approximately 150 effector proteins via the Dot/Icm T4SS. The growing list of potential L. pneumophila effectors includes a significant number of proteins that share regions of similarity to eukaryotic protein domains or motifs (Franco et al., 2009; Isberg et al., 2009). One example is LubX, which possesses two U-box domains, conserved among U-box-type E3 ubiquitin ligases (Kubori et al., 2008). The U-box1 of LubX has E2 binding activity critical to the ligase function, while U-box2 serves as a substrate binding site. U-box2 binds to host Cdc2-like kinase 1 (Clk1; Kubori et al., 2008), as well as to another bacterial effector, SidH (Kubori et al., 2010), resulting in the polyubiquitination of these proteins in infected cells. Although the consequence of Clk1 ubiquitination is still being determined, polyubiquitinated SidH was shown to be subjected to proteasome-mediated protein degradation as described below.
Metaeffector: an effector regulating another effector
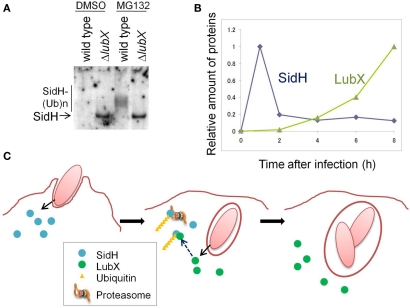
The product of sidH, located just downstream of lubX, was translocated into the host cell cytosol and the cellular level of SidH was increased by the disruption of lubX (Kubori et al., 2010; Figure 1A). SidH was reported to be a Legionella Dot/Icm T4SS substrate (Luo and Isberg, 2004; Laguna et al., 2006), but its cellular function has not been clarified. LubX showed the ability to mediate polyubiquitination of SidH in vitro and in vivo (Kubori et al., 2010). In host cells polyubiquitinated SidH was shown to be subjected to proteasome-mediated protein degradation, as higher molecular weight species of SidH were observed in cells treated with the proteasome inhibitor MG132, but not in untreated cells (Figure 1A).
Figure 1.
The Legionella “metaeffector” LubX temporally regulates SidH. (A) LubX is responsible for polyubiquitination and proteasomal degradation of SidH in infected cells. CHO-FcR cells were infected with a wild-type L. pneumophila strain and a lubX deletion mutant for 8 h and the cell lysate was analyzed by immunoprecipitation, followed by western immunoblotting to detect SidH. The cells infected with the lubX deletion mutant, but not the wild-type strain, possessed an intact SidH protein. In the presence of a proteasome inhibitor, MG132, polyubiquitinated species of SidH were detected only after infection with the wild-type strain. This panel is reproduced from Kubori et al. (2010) under the Creative Common Attribution License. (B) Time course of the relative amounts of SidH and LubX in the cells infected with the wild-type L. pneumophila strain. (C) A model of the temporal regulation of SidH. SidH is delivered via Legionella T4SS just after infection (left panel). Compared with SidH, LubX synthesis, and delivery is delayed. SidH is polyubiquitinated by the LubX ubiquitin ligase (middle panel), which leads to degradation of SidH by the host proteasome (middle and right panels). This panel is adopted from Kubori et al. (2010).
The time course of SidH and LubX levels in infected CHO-FcR cells was analyzed (Figure 1B). SidH is expressed in culture-grown bacteria and is ready for delivery upon infection, while LubX expression is induced upon infection of host cells (Kubori et al., 2008, 2010). SidH was detected in host cells immediately after infection and the level gradually decreased over time. LubX was not detected initially, but became detectable in a few hours with level peaking at 8 h after infection. The distinct time frames for protein expression/delivery lead to the temporal accumulation of SidH, which is eventually degraded by the host proteasome that is induced by polyubiquitination mediated by LubX (Figure 1C).
SidH is an intrinsically stable effector protein within the host cytosol and its function is deactivated at later stage of infection by LubX-mediated proteasomal degradation. LubX is a “metaeffector,” a designation for an effector protein which regulates another effector protein (Kubori et al., 2010). In a fly infection model, an L. pneumophila strain lacking lubX showed hyper lethality to flies, suggesting that the persistent presence of SidH is harmful to the model host. In summary, the regulatory circuit involving the metaeffector LubX eliminates excess SidH at a later stage of infection, which would be beneficial for Legionella.
Temporal Regulation by Differential Stability: Salmonella SopE and SptP
Bacterial internalization requires the function of SopE and SptP
Salmonella Typhimurium has the ability to invade non-phagocytic cells. The process of internalization of Salmonella into the intestinal epithelium was carefully described more than 40 years ago (Takeuchi, 1967). Electron microscopic observations revealed that Salmonella induced pronounced changes in the plasma membrane, disrupting microvilli, and entering intestinal epithelial cells by converting the bacterium-containing cavity into a membrane-bound vacuole. Importantly, the disrupted brush border was reconstituted even in the presence of internalized bacteria, showing the reversibility of the morphology of the plasma membrane according to the infection stage.
Internalization of Salmonella into non-phagocytic cells is the result of marked actin cytoskeleton rearrangements that cause drastic changes in the plasma membrane of infected cells, which resemble “membrane ruffles” induced by growth factors, hormones, and activated oncogenes (Finlay et al., 1991; Ginocchio et al., 1992; Francis et al., 1993). The function of the Rho family GTPases is central to the cellular responses leading to actin cytoskeleton rearrangements, as well as to the nuclear responses upon infection with Salmonella (Chen et al., 1996). The formation of membrane ruffles upon Salmonella infection results from the coordinated action of Salmonella effectors delivered by the type III secretion system (T3SS; Patel et al., 2005). SopE and its homolog SopE2 are responsible for activating the Rho family GTPases Cdc42 and Rac1 via the function of GEF (Hardt et al., 1998; Rudolph et al., 1999; Bakshi et al., 2000; Stender et al., 2000). Another effector, SopB (also known as SigD), a phosphoinositide phosphatase, is also involved in the process of actin remodeling and bacterial uptake into cells, but has no guanine nucleotide exchange activity (Zhou et al., 2001). Other bacterial proteins, SipA and SipC, are known to be involved in efficient Salmonella entry by directly binding actin to modulate actin dynamics. SipC, an integral component of the T3SS translocon, is inserted into host plasma membranes (Scherer et al., 2000) and confers nucleation of actin polymerization and actin bundling (Hayward and Koronakis, 1999). SipA preferentially associates with peripheral cortical actin filaments (Higashide et al., 2002) and induces actin bundling, as well as preventing F-actin depolymerization (Zhou et al., 1999; Lilic et al., 2003; Dai et al., 2004). SipA substantially stimulates SipC-mediated nucleation of actin polymerization and actin bundling, demonstrating the cooperative action of these two effectors (McGhie et al., 2004). A Salmonella strain defective in SopE, SopE2, and SopB is, however, unable to induce actin rearrangement and fails to be internalized into host cells (Zhou et al., 2001), indicating that these three proteins are primary effectors responsible for Salmonella entry.
The process of restoring cellular morphology at a later stage of infection is dependent on the action of another effector, SptP (Fu and Galan, 1999). The amino-terminal domain of SptP possesses GAP activity toward Cdc42 and Rac1. SptP switches the regulatory molecules to an inactive GDP-bound state, resulting in disruption of the actin cytoskeleton, thus shutting off membrane ruffling (Fu and Galan, 1998, 1999). Salmonella uptake is established by morphological changes in host cell membranes, induced by the reversible temporal activation of the Rho family GTPases.
Differential stability of SopE and SptP accounts for transient induction of membrane ruffling
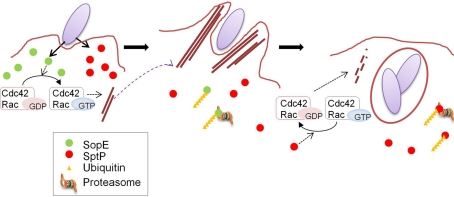
SopE and SptP represent a remarkable “yin and yang,” displaying opposing functions in the same molecules. The mechanism determining the balance between these two effectors was examined in sopE2- and sopB-defective strain backgrounds in which SopE was the sole inducer for actin remodeling (Kubori and Galan, 2003). SopE and SptP are co-delivered into host cells shortly after infection, but SopE is predominant at inducing membrane ruffling, leading to bacterial internalization (Figure 2). In host cells, both SopE and SptP are subjected to polyubiquitination and proteasomal degradation, but display different half-lives; SopE is rapidly degraded, while SptP exhibits much slower degradation kinetics (Figure 2). This difference in stability disrupts the balance between the functions of these two proteins. SptP dominates during the later stages of infection, allowing for the recovery of the normal cellular architecture (Figure 2). By chimeric analysis, the signals for ubiquitin modification to SopE and SptP were found to be located within the amino-terminal region of the two proteins (Kubori and Galan, 2003), which possibly includes the signaling domain for secretion/translocation, as suggested by structural and functional analyses (Fu and Galan, 1998; Hardt et al., 1998; Stebbins and Galan, 2000, 2001; Buchwald et al., 2002). The fact that the degradation signal is encoded in the proteins means that bacteria can determine the fate of their own proteins, the process of which is mediated by host cellular systems.
Figure 2.
Coordination of the functions of SopE and SptP. A model for the temporal regulation of SopE and SptP functions. SopE and SptP are delivered in equivalent amounts via the Salmonella T3SS upon infection, and SopE dominantly activates Rho GTPases Cdc42 and Rac as a GEF (left panel), resulting in bacterial uptake by profuse membrane ruffling (middle panel). Shortly after infection, SopE is polyubiquitinated in the cell and subjected to host proteasomal degradation (middle panel), making SptP dominant in the later stages of infection. SptP functions to shut off membrane ruffling by its GAP activity against Cdc42 and Rac, and normal cellular architecture is regained (right panel). This figure is adopted from Kubori and Galan (2003).
Salmonella SopB: Spatial Regulation Mediated by Monoubiquitination
Multifunctional effector SopB
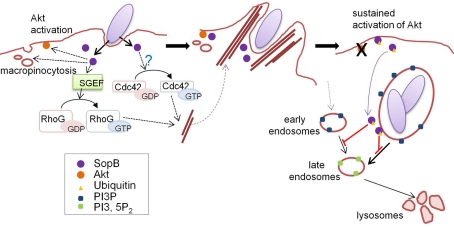
Salmonella phosphoinositide phosphatase SopB mediates actin-dependent bacterial internalization, as described above (Figure 3; Zhou et al., 2001; Patel and Galan, 2006). SopB activates SH-3-containing guanine nucleotide exchange factor (SGEF), an exchange factor for RhoG, which leads to specific cellular responses including actin remodeling (Patel and Galan, 2006). SopB phosphatase function also contributes to actin remodeling via Cdc42 signaling, but the mechanism remains unclear (Patel and Galan, 2006). SopB is also required for the efficient intracellular replication of bacteria by modulating Salmonella-containing vacuole (SCV) trafficking by inducing macropinocytosis upon bacterial uptake (Hernandez et al., 2004). The inositol phosphatase activity of SopB is necessary to generate PtdIns(3)P-decorated SCVs, which serve as a spacious niche where Salmonella can reside and replicate (Figure 3). Furthermore, SopB activates the serine protein kinase Akt (Steele-Mortimer et al., 2000), although the underlying mechanism behind this is poorly understood.
Figure 3.
Ubiquitin-mediated regulation of SopB localization. Just after contact with host cells, Salmonella delivers SopB via the T3SS. SopB inositol phosphatase activity plays key roles in bacterial internalization and SCV maturation by inducing macropinocytosis accompanied by bacterial uptake. At the early stages of infection, SopB localizes to the plasma membrane and activates the protein kinase Akt. SopB also activates SGEF, a RhoG GEF, and indirectly activates Cdc42 (left panel), resulting in actin cytoskeleton reorganization to form “membrane ruffles” which enable bacteria to be internalized into host cells (middle panel). SopB is monoubiquitinated by the function of an unknown host ubiquitin ligase, and ubiquitinated SopB is then removed from the plasma membrane and recruited to the SCV, where SopB functions to modulate vesicular trafficking by altering phosphoinositide metabolism (right panel). This figure is adopted from Patel et al. (2009).
Spatio-temporal regulation of SopB mediated monoubiquitination
Recent studies have shown that SopB is an effector protein whose function is spatio-temporally regulated in an ubiquitin-dependent manner (Knodler et al., 2009; Patel et al., 2009). Multiple lysine residues in SopB are monoubiquitinated and linked via the Lys-63 of ubiquitin in the cells, and the ubiquitinated SopB is persistently present for a long time after bacterial internalization (Knodler et al., 2009; Patel et al., 2009). Analyses using a ubiquitin-deficient SopB mutant (in which all lysine residues were substituted with glutamate or arginine) revealed that ubiquitination is not required for bacterial uptake, but plays a significant role in cellular localization of SopB (Knodler et al., 2009; Patel et al., 2009). Early in infection, SopB localizes to the plasma membrane to mediate bacterial entry and Akt activation. In the later stages of infection, SopB is translocated to SCVs depending on its ubiquitination (Figure 3). Ubiquitin-deficient SopB was found to persist at the plasma membrane and failed to shut off Akt activation (Knodler et al., 2009; Patel et al., 2009).
Conclusion and Perspectives
In this article, we have focused on Legionella and Salmonella effector proteins, which are subjected to ubiquitin-mediated temporal or spatial regulation, highlighting the sophisticated strategies adopted by pathogenic bacteria to precisely regulate cellular processes during infection using the host ubiquitin system. Of particular interest, Legionella LubX functions as a metaeffector that targets another effector protein, SidH, and temporally regulates its proteasome-mediated degradation. Recent evidence indicates that pathogenic bacteria possess numerous effector proteins involved in ubiquitination reactions. Such effector proteins may function as metaeffectors like Legionella LubX. Comprehensive elucidation of how bacteria use the host ubiquitin system may lead to important insights into the molecular basis of bacterial pathogenesis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the authors of related works that were not cited here because of space constraints. Research in the Nagai laboratory was supported in part by the Targeted Proteins Research Program (TPRP) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Mitsubishi Foundation (Japan).
References
- Abramovitch R. B., Janjusevic R., Stebbins C. E., Martin G. B. (2006). Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. U.S.A. 103, 2851–2856 10.1073/pnas.0507892103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitch R. B., Kim Y. J., Chen S., Dickman M. B., Martin G. B. (2003). Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22, 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angot A., Vergunst A., Genin S., Peeters N. (2007). Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 3, e3. 10.1371/journal.ppat.0030003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardley H. C., Robinson P. A. (2005). E3 ubiquitin ligases. Essays Biochem. 41, 15–30 10.1042/EB0410015 [DOI] [PubMed] [Google Scholar]
- Ashida H., Kim M., Schmidt-Supprian M., Ma A., Ogawa M., Sasakawa C. (2010). A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat. Cell Biol. 12, 66–73; sup. 61–69. 10.1038/ncb2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S., Meyer T. F. (2006). Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9, 207–217 10.1016/j.mib.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Bakshi C. S., Singh V. P., Wood M. W., Jones P. W., Wallis T. S., Galyov E. E. (2000). Identification of SopE2, a Salmonella secreted protein which is highly homologous to SopE and involved in bacterial invasion of epithelial cells. J. Bacteriol. 182, 2341–2344 10.1128/JB.182.8.2341-2344.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G., Friebel A., Galan J. E., Hardt W. D., Wittinghofer A., Scheffzek K. (2002). Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J. 21, 3286–3295 10.1093/emboj/cdf329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Banga S., Mertens K., Weber M. M., Gorbaslieva I., Tan Y., Luo Z. Q., Samuel J. E. (2010). Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc. Natl. Acad. Sci. U.S.A. 107, 21755–21760 10.1073/pnas.1004060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. M., Hobbie S., Galan J. E. (1996). Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science 274, 2115–2118 10.1126/science.274.5290.1176 [DOI] [PubMed] [Google Scholar]
- Christie P. J., Atmakuri K., Krishnamoorthy V., Jakubowski S., Cascales E. (2005). Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59, 451–485 10.1146/annurev.micro.58.030603.123630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S., Sarmiere P. D., Wiggan O., Bamburg J. R., Zhou D. (2004). Efficient Salmonella entry requires activity cycles of host ADF and cofilin. Cell. Microbiol. 6, 459–471 10.1111/j.1462-5822.2004.00375.x [DOI] [PubMed] [Google Scholar]
- Diao J., Zhang Y., Huibregtse J. M., Zhou D., Chen J. (2008). Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat. Struct. Mol. Biol. 15, 65–70 10.1038/nsmb1346 [DOI] [PubMed] [Google Scholar]
- Ensminger A. W., Isberg R. R. (2009). Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr. Opin. Microbiol. 12, 67–73 10.1016/j.mib.2008.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B. S. (1996). The molecular ecology of Legionellae. Trends Microbiol. 4, 286–290 10.1016/0966-842X(96)10041-X [DOI] [PubMed] [Google Scholar]
- Fields B. S., Benson R. F., Besser R. E. (2002). Legionella and Legionnaires’ disease: 25 years of investigation. Clin. Microbiol. Rev. 15, 506–526 10.1128/CMR.15.3.506-526.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A., Hachani A., Bleves S. (2008). The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154, 1570–1583 10.1099/mic.0.2008/016840-0 [DOI] [PubMed] [Google Scholar]
- Finlay B. B., Ruschkowski S., Dedhar S. (1991). Cytoskeletal rearrangements accompanying Salmonella entry into epithelial cells. J. Cell Sci. 99(Pt 2), 283–296 [DOI] [PubMed] [Google Scholar]
- Francis C. L., Ryan T. A., Jones B. D., Smith S. J., Falkow S. (1993). Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature 364, 639–642 10.1038/364639a0 [DOI] [PubMed] [Google Scholar]
- Franco I. S., Shuman H. A., Charpentier X. (2009). The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell. Microbiol. 11, 1435–1443 10.1111/j.1462-5822.2009.01351.x [DOI] [PubMed] [Google Scholar]
- Fu Y., Galan J. E.(1998). The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol. Microbiol. 27, 359–368 10.1046/j.1365-2958.1998.00684.x [DOI] [PubMed] [Google Scholar]
- Fu Y., Galan J. E. (1999). A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 401, 293–297 10.1046/j.1365-2958.1998.00684.x [DOI] [PubMed] [Google Scholar]
- Galan J. E. (2001). Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17, 53–86 10.1038/45829 [DOI] [PubMed] [Google Scholar]
- Galan J. E., Wolf-Watz H. (2006). Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573 10.1146/annurev.cellbio.17.1.53 [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Hann D. R., Ntoukakis V., Petutschnig E., Lipka V., Rathjen J. P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429 10.1038/nature05272 [DOI] [PubMed] [Google Scholar]
- Ginocchio C., Pace J., Galan J. E. (1992). Identification and molecular characterization of a Salmonella typhimurium gene involved in triggering the internalization of Salmonellae into cultured epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 89, 5976–5980 10.1016/j.cub.2009.01.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohre V., Spallek T., Haweker H., Mersmann S., Mentzel T., Boller T., de Torres M., Mansfield J. W., Robatzek S. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18, 1824–1832 10.1073/pnas.89.13.5976 [DOI] [PubMed] [Google Scholar]
- Goldstein G., Scheid M., Hammerling U., Schlesinger D. H., Niall H. D., Boyse E. A. (1975). Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl. Acad. Sci. U.S.A. 72, 11–15 10.1016/j.cub.2008.10.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K., Dikic I. (2005). Ubiquitylation and cell signaling. EMBO J. 24, 3353–3359 10.1073/pnas.72.6.2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt W. D., Chen L. M., Schuebel K. E., Bustelo X. R., Galan J. E. (1998). S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93, 815–826 10.1038/sj.emboj.7600808 [DOI] [PubMed] [Google Scholar]
- Hayward R. D., Koronakis V. (1999). Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 18, 4926–4934 10.1016/S0092-8674(00)81442-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez L. D., Hueffer K., Wenk M. R., Galan J. E. (2004). Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science 304, 1805–1807 10.1093/emboj/18.18.4926 [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 10.1126/science.1098188 [DOI] [PubMed] [Google Scholar]
- Hershko A., Heller H., Elias S., Ciechanover A. (1983). Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258, 8206–8214 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Hicks S. W., Galan J. E. (2010). Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr. Opin. Microbiol. 13, 41–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashide W., Dai S., Hombs V. P., Zhou D. (2002). Involvement of SipA in modulating actin dynamics during Salmonella invasion into cultured epithelial cells. Cell. Microbiol. 4, 357–365 10.1016/j.mib.2009.11.008 [DOI] [PubMed] [Google Scholar]
- Horwitz M. A. (1983). Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158, 1319–1331 10.1046/j.1462-5822.2002.00196.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber A., Roy C. R. (2010). Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26, 261–283 10.1084/jem.158.6.2108 [DOI] [PubMed] [Google Scholar]
- Ingmundson A., Delprato A., Lambright D. G., Roy C. R. (2007). Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450, 365–369 10.1146/annurev-cellbio-100109-104034 [DOI] [PubMed] [Google Scholar]
- Isberg R. R., O’Connor T. J., Heidtman M. (2009). The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24 10.1038/nature06336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janjusevic R., Abramovitch R. B., Martin G. B., Stebbins C. E. (2006). A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311, 222–226 10.1038/nrmicro1967 [DOI] [PubMed] [Google Scholar]
- Knodler L. A., Winfree S., Drecktrah D., Ireland R., Steele-Mortimer O. (2009). Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cell. Microbiol. 11, 1652–1670 10.1126/science.1120131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T., Galan J. E. (2003). Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. Cell 115, 333–342 10.1111/j.1462-5822.2009.01356.x [DOI] [PubMed] [Google Scholar]
- Kubori T., Hyakutake A., Nagai H. (2008). Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol. Microbiol. 67, 1307–1319 10.1016/S0092-8674(03)00849-3 [DOI] [PubMed] [Google Scholar]
- Kubori T., Shinzawa N., Kanuka H., Nagai H. (2010). Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 6, e1001216. 10.1371/journal.ppat.1001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguna R. K., Creasey E. A., Li Z., Valtz N., Isberg R. R. (2006). A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc. Natl. Acad. Sci. U.S.A. 103, 18745–18750 10.1371/journal.ppat.1001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin I., Eakin C., Blanc M. P., Klevit R. E., Miller S. I., Brzovic P. S. (2010). Identification of an unconventional E3 binding surface on the UbcH5 ~ Ub conjugate recognized by a pathogenic bacterial E3 ligase. Proc. Natl. Acad. Sci. U.S.A. 107, 2848–2853 10.1073/pnas.0609012103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilic M., Galkin V. E., Orlova A., VanLoock M. S., Egelman E. H., Stebbins C. E. (2003). Salmonella SipA polymerizes actin by stapling filaments with nonglobular protein arms. Science 301, 1918–1921 10.1073/pnas.0914821107 [DOI] [PubMed] [Google Scholar]
- Lin D. Y., Diao J., Zhou D., Chen J. (2011). Biochemical and structural studies of a HECT-like ubiquitin ligase from Escherichia coli O157:H7. J. Biol. Chem. 286, 441–449 10.1126/science.1088433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z. Q., Isberg R. R. (2004). Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. U.S.A. 101, 841–846 10.1074/jbc.M110.204537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machner M. P., Isberg R. R. (2006). Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 11, 47–56 10.1073/pnas.0304482101 [DOI] [PubMed] [Google Scholar]
- Machner M. P., Isberg R. R. (2007). A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science 318, 974–977 10.1016/j.devcel.2006.05.013 [DOI] [PubMed] [Google Scholar]
- McGhie E. J., Hayward R. D., Koronakis V. (2004). Control of actin turnover by a Salmonella invasion protein. Mol. Cell 13, 497–510 10.1126/science.1149121 [DOI] [PubMed] [Google Scholar]
- Murata T., Delprato A., Ingmundson A., Toomre D. K., Lambright D. G., Roy C. R. (2006). The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8, 971–977 10.1016/S1097-2765(04)00053-X [DOI] [PubMed] [Google Scholar]
- Ninio S., Roy C. R. (2007). Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15, 372–380 10.1038/ncb1463 [DOI] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y. H., Pumplin N., Jones J., He S. Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313, 220–223 10.1016/j.tim.2007.06.006 [DOI] [PubMed] [Google Scholar]
- Patel J. C., Galan J. E. (2006). Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J. Cell Biol. 175, 453–463 10.1126/science.1129523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J. C., Hueffer K., Lam T. T., Galan J. E. (2009). Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell 137, 283–294 10.1083/jcb.200605144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J. C., Rossanese O. W., Galan J. E. (2005). The functional interface between Salmonella and its host cell: opportunities for therapeutic intervention. Trends Pharmacol. Sci. 26, 564–570 10.1016/j.cell.2009.01.056 [DOI] [PubMed] [Google Scholar]
- Pavletich N. P. (2002). Structural biology of ubiquitin-protein ligases. Harvey Lect. 98, 65–102 10.1016/j.tips.2005.09.005 [DOI] [PubMed] [Google Scholar]
- Price C. T., Abu Kwaik Y. (2010). Exploitation of host polyubiquitination machinery through molecular mimicry by eukaryotic-like bacterial F-box effectors. Front. Microbio. 1:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S., McAuley S. B., Miyata S. T. (2009). The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12, 11–17 10.3389/fmicb.2010.00131 [DOI] [PubMed] [Google Scholar]
- Quezada C. M., Hicks S. W., Galan J. E., Stebbins C. E. (2009). A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proc. Natl. Acad. Sci. U.S.A. 106, 4864–4869 10.1016/j.mib.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde J. R., Breitkreutz A., Chenal A., Sansonetti P. J., Parsot C. (2007). Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 1, 77–83 10.1073/pnas.0811058106 [DOI] [PubMed] [Google Scholar]
- Rosebrock T. R., Zeng L., Brady J. J., Abramovitch R. B., Xiao F., Martin G. B. (2007). A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448, 370–374 10.1016/j.chom.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph M. G., Weise C., Mirold S., Hillenbrand B., Bader B., Wittinghofer A., Hardt W. D. (1999). Biochemical analysis of SopE from Salmonella typhimurium, a highly efficient guanosine nucleotide exchange factor for RhoGTPases. J. Biol. Chem. 274, 30501–30509 10.1038/nature05966 [DOI] [PubMed] [Google Scholar]
- Rytkonen A., Holden D. W. (2007). Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe 1, 13–22 10.1074/jbc.274.17.11824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer C. A., Cooper E., Miller S. I. (2000). The Salmonella type III secretion translocon protein SspC is inserted into the epithelial cell plasma membrane upon infection. Mol. Microbiol. 37, 1133–1145 10.1016/j.chom.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Schulman B. A., Carrano A. C., Jeffrey P. D., Bowen Z., Kinnucan E. R., Finnin M. S., Elledge S. J., Harper J. W., Pagano M., Pavletich N. P. (2000). Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386 10.1046/j.1365-2958.2000.02066.x [DOI] [PubMed] [Google Scholar]
- Segal G., Purcell M., Shuman H. A. (1998). Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U.S.A. 95, 1669–1674 10.1038/35042620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Roy C. R. (2008). Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell. Microbiol. 10, 1209–1220 10.1073/pnas.95.9.4804 [DOI] [PubMed] [Google Scholar]
- Singer A. U., Rohde J. R., Lam R., Skarina T., Kagan O., Dileo R., Chirgadze N. Y., Cuff M. E., Joachimiak A., Tyers M., Sansonetti P. J., Parsot C., Savchenko A. (2008). Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat. Struct. Mol. Biol. 15, 1293–1301 10.1111/j.1462-5822.2008.01151.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins C. E., Galan J. E. (2000). Modulation of host signaling by a bacterial mimic: structure of the Salmonella effector SptP bound to Rac1. Mol. Cell 6, 1449–1460 10.1038/nsmb.1511 [DOI] [PubMed] [Google Scholar]
- Stebbins C. E., Galan J. E. (2001). Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414, 77–81 10.1016/S1097-2765(00)00141-6 [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O., Knodler L. A., Marcus S. L., Scheid M. P., Goh B., Pfeifer C. G., Duronio V., Finlay B. B. (2000). Activation of Akt/protein kinase B in epithelial cells by the Salmonella typhimurium effector sigD. J. Biol. Chem. 275, 37718–37724 10.1038/35102073 [DOI] [PubMed] [Google Scholar]
- Stender S., Friebel A., Linder S., Rohde M., Mirold S., Hardt W. D. (2000). Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 36, 1206–1221 10.1074/jbc.M008187200 [DOI] [PubMed] [Google Scholar]
- Swanson M. S., Isberg R. R. (1995). Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63, 3609–3620 10.1046/j.1365-2958.2000.01933.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi A. (1967). Electron microscope studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am. J. Pathol. 50, 109–136 [PMC free article] [PubMed] [Google Scholar]
- Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000). Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L. G., Harb O. S., Connelly P. S., Robinson C. G., Roy C. R. (2001). How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell Sci. 114, 4637–4650 10.1093/emboj/19.1.94 [DOI] [PubMed] [Google Scholar]
- Vogel J. P., Andrews H. L., Wong S. K., Isberg R. R. (1998). Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876 [DOI] [PubMed] [Google Scholar]
- Waterman S. R., Holden D. W. (2003). Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5, 501–511 10.1126/science.279.5351.648 [DOI] [PubMed] [Google Scholar]
- Wu B., Skarina T., Yee A., Jobin M. C., Dileo R., Semesi A., Fares C., Lemak A., Coombes B. K., Arrowsmith C. H., Singer A. U., Savchenko A. (2010). NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-box E3 ubiquitin ligases. PLoS Pathog. 6, e1000960. 10.1371/journal.ppat. 1000960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C., Pearl L. H. (2005). Chaperoned ubiquitylation – crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20, 525–538 10.1371/journal.ppat.1000960 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Higashide W. M., McCormick B. A., Chen J., Zhou D. (2006). The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol. Microbiol. 62, 786–793 10.1016/j.molcel.2005.09.023 [DOI] [PubMed] [Google Scholar]
- Zhou D., Chen L. M., Hernandez L., Shears S. B., Galan J. E. (2001). A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39, 248–259 10.1111/j.1365-2958.2006.05407.x [DOI] [PubMed] [Google Scholar]
- Zhou D., Mooseker M. S., Galan J. E. (1999). Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science 283, 2092–2095 10.1046/j.1365-2958.2001.02224.x [DOI] [PubMed] [Google Scholar]
- Zhu W., Banga S., Tan Y., Zheng C., Stephenson R., Gately J., Luo Z. Q. (2011). Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE 6, e17638. 10.1371/journal.pone.0017638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Li H., Hu L., Wang J., Zhou Y., Pang Z., Liu L., Shao F. (2008). Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat. Struct. Mol. Biol. 15, 1302–1308 10.1371/journal.pone.0017783 [DOI] [PubMed] [Google Scholar]