Abstract
Hereditary hemochromatosis (HH) is an iron metabolism disorder that leads to excess iron buildup, especially in the heart, liver, and pancreas. Mutations in the HFE gene are the single most common cause of HH, which can be treated effectively if diagnosed early. Patents cover the HFE gene, related proteins, screening methods, and testing kits. Most initial testing for HH is biochemical, but HFE DNA testing or genotyping is used to confirm a diagnosis of inherited hemochromatosis. Concerns over patents covering HFE testing emerged in 2002, when scholars argued that exclusive licensing and the patent-enabled sole provider model then in place led to high prices and limited access. Critics of the sole provider model noted that the test was available at multiple laboratories prior to the enforcement of patents. By 2007, however, Bio-Rad, Limited, acquired the key intellectual property and sub-licensed it widely. In part because of broad, non-exclusive licensing, there are now multiple providers and testing technologies, and research continues. This case study illustrates how both changes in intellectual property ownership and evolving clinical utility of HFE genetic testing in the last decade have effected the licensing of patents and availability of genetic testing.
Keywords: Patents, Intellectual Property, Hemochromatosis, HFE, genetic testing
Introduction
Hereditary hemochromatosis (HH) is an autosomal recessive disorder that results most often from mutations in the HFE gene,1-3 which regulates iron absorption. HH caused by functional mutations in the HFE gene is commonly referred to as HH type 1. Mutations in the HFE gene place the individual at an increased risk for developing symptomatic HH, an iron metabolism disorder that leads to excess iron absorption from the diet, particularly in males. Since the body lacks a natural way to rid itself of the excess iron, it accumulates over time, resulting in organ damage, particularly in the heart, liver, and pancreas. In extreme cases, hemochromatosis can even lead to death, usually due to heart or liver failure.
Early detection of the disorder, and thus earlier treatment by phlebotomy (repeated blood draws), can greatly mitigate its effects and allow HH patients to live normal, healthy lives.4 HFE testing in combination with a patient's family history and physical health record can provide guidance for clinical interventions or lifestyle changes that a patient would not have without genetic testing. Testing for the presence of HFE gene mutations can also help physicians to identify patients experiencing characteristic symptoms of the disorder, clarify their diagnosis, and sometimes prevent irreversible organ damage.
HH is a candidate for genetic screening for many reasons. First, the mutations associated with HH are present at birth, whereas characteristic symptoms of hemochromatosis as a disease usually do not develop until mid-adulthood, beginning in an individual's 40s and 50s. In addition, the variability and non-specific nature of symptoms can make diagnosis difficult, raising the possibility that patients, especially those with no family history, may be diagnosed too late. Therefore, an early, specific diagnosis allows for an effective treatment plan. Secondly, unlike some hereditary disorders, a limited number of genes are associated with HH that can be tested for mutations to determine a patient's risk. Finally, HH is among the most common recessive genetic traits in some populations of Northern European descent, resulting in a relatively high carrier frequency. Between 1 in 200 and 1 in 400 people of Northern European descent, or 0.5% of this population, is homozygous for the HFE mutation and thus at high risk of developing clinical hemochromatosis.5 The estimated carrier frequency of HFE mutation is 1 in every 8 to 10 individuals of Northern European ancestry.6 The reason for higher population frequency in Northern Europe is not known. One intriguing, but still speculative, theory posits a survival advantage among those with HH mutations in resisting infections causing plague and other diseases prevalent in Europe.7 Another hypothesis, which is not incompatible, is co-selection of hemochromatosis and certain major histocompatibility loci involved in immune function.8
Despite this, universal genetic screening has not been recommended for several reasons. First, presence of the mutation does not mean that the individual will develop HH. While testing may assist physicians in diagnosing HH when a patient is presenting characteristic symptoms, presence of the mutation merely indicates one's susceptibility to iron overload and not the certainty of disease for those who are asymptomatic. The symptoms of HH are highly variable among homozygotes (those in whom both chromosomal copies of the HFE gene have hemochromatosis-associated mutations). Some are completely asymptomatic, others are severely affected. Several studies provide evidence that the penetrance of the HFE mutations, or the chance that those with the mutations will have HH symptoms, is lower than first estimated and highly variable.3 The disease is also rarer in non-white populations. Homozygous mutation levels are 0.27 homozygotes per 1,000 Hispanic individuals, less than .0001 homozygotes per 1,000 Asian American individuals, 0.12 homozygotes per 1,000 in Pacific Islanders, and an estimated .14 homozygotes per 1,000 in African–American individuals.9 The American College of Physicians does not recommend genetic or phenotypic (using biochemical tests) screening for HH in the asymptomatic general population.5 The U.S. Preventive Services Task Force (USPTF) similarly found insufficient evidence to support broad population genetic screening.9 Finally, the current price of the genetic diagnostic tests also makes their use as an initial screening procedure for HH prohibitive. Current practice is to identify symptomatic individuals utilizing non-genetic tests that measure iron overload, followed by genetic testing for specific diagnosis and to detect cases in families once an HH proband is identified.
Hereditary hemochromatosis is a natural case study for studying the impact of intellectual property (IP) on patient access to genetic testing. Patents exist on the HFE gene, its related protein, genetic screening test methods, and related testing kits (see Appendix A). Additional genes linked to rarer forms of HH are also patented.
The impact of these patents and their licensing on access to testing for HH type 1 is complicated by the generally subordinate role of clinical genetic testing in hemochromatosis, but also by the complex history of ownership of these patents. Despite an initial controversy about patenting, HFE genetic testing appears to have been adopted in clinical practice and much of the heat may have drained from the public debate. The path to the current state, however, involved transitional periods of turbulence that centered on exclusive licensing of a genetic diagnostic test.
One distinctive feature of this case is how HFE testing has evolved over time. HFE genetic testing illustrates how patent ownership and use by different patent-holders can affect licensing. HFE patent rights were transferred many times, and use and licensing policies changed over time. A 2002 Nature article, written when the licensing schema was based on exclusive licensing and a single-provider model, judged that HFE genetic testing “failed the test” of socially optimal access. In 2007 and 2008, compared to 2002, we found little controversy surrounding HFE genetic testing, and the licensing model has evolved to include several providers and sublicensing for use on different platform technologies. The past licensing practices of SmithKline Beecham Clinical Laboratories (SBCL) (exclusive licensing model) were controversial, but the current owner of patent rights, Bio-Rad Ltd., appears to have adopted a broad sub-licensing model that has resulted in broader clinical and patient access and less public conflict.
HFE genetic testing in the context of HH also shows how genetic testing is part of a larger set of diagnostic tools addressing a clinical syndrome. The clinical utility of those tools, including genetic testing, evolves over time. Growing knowledge about the uncertain penetrance of HFE mutations required additional research to determine the clinical significance of different HFE mutations, and other factors influencing expression of disease. These studies demonstrated a much lower clinical penetrance of HFE mutations than first expected, suggesting that the mutations alone were poor predictors of developing clinically significant hemochromatosis. Population screening was more likely to be pursued, if at all, by chemical or protein assays rather than genetic testing—with genetic tests finding more limited use in confirmatory diagnosis and family risk assessment once an index case is found. This most likely had a significant impact on interest in investing in patent enforcement, since the market for HFE genetic testing became much smaller when general population use seemed highly unlikely.
Lessons Learned
Research
The Mercator Genetics business plan was centered on the identification of candidate genes for a number of complex diseases including asthma, schizophrenia, cardiovascular disease and prostate cancer, all of which presumably had a diagnostic market. The prospects of patents and revenue from diagnostic testing for HH probably stimulated research at Mercator Genetics. However, Dr. Dennis Drayna, co-founder of Mercator Genetics, notes that the company was conceived and initially funded on an agenda much broader than hemochromatosis gene discovery or diagnostic testing alone. Discovery of the HFE gene was nonetheless Mercator's signature success.
The “race” for the HH gene was won by Mercator Genetics with the publication of an August 1996 Nature Genetics article. Two additional groups (one in France and another in Australia, which were both in non-profit institutions) were pursuing similar approaches to candidate gene identification and would likely have been successful in their efforts within months. However, the scale and focus of the positional cloning effort at Mercator, enabled by private R&D investment, probably gave their research group a competitive advantage.
The patent applications filed by Mercator Genetics predated the submission of related manuscripts by nearly a year.10 It is unclear, however, if this delay resulted from scientific issues, patenting activities, corporate strategy, or commercialization efforts by Mercator. It remains possible that such a delay may be the consequence of factors unrelated to patenting, such as the need for additional research or data prior to submission to peer reviewed journals, journal requests for additional data and experiments, delays in peer review, etc. Dr. Dennis Drayna, a senior author of the Nature Genetics paper, indicated that the latter was in fact true, and that Mercator Genetics made every attempt to expedite simultaneous paper submissions and patent filings.
Concerns regarding inhibition of research due to the HFE gene patents do not seem to be supported. Substantial basic research, including identification of genes and mutations associated with other types of hemochromatosis has continued. Similarly, research on improved methods for detection of HFE mutations has also progressed. The adoption of broad sublicensing practices by Bio-Rad, Ltd., has facilitated commercial research and development efforts focused on alternative methods for HFE mutation detection.
Development
Mercator Genetics announced that it was developing a blood test for HFE genotyping within a year of publication of results. It is likely that the prospect of revenues from population wide screening may have served as an incentive for test development. However, no test was marketed before Mercator went out of business and merged with Progenitor.
Intellectual property ownership alone did not provide incentive for test development. As reported by Merz et al., laboratories were able to develop in-house testing and offer it as clinical service soon after information of the gene sequence and its associated mutation had been made public and well before the patents were granted.
Commercialization
HFE patents were potentially valuable assets for Mercator in facilitating its merger with Progenitor. Exclusive licensing of the HFE patents to SmithKline Beecham Clinical Laboratories (SBCL) resulted in significant and guaranteed revenue for Progenitor.
Until it was sold to Quest Diagnostics, SBCL offered the test as part of its commercial diagnostics services. SBCL also undertook enforcement activities, including sending “cease and desist” letters to clinical laboratories.
Similarly, the HFE patents were perceived as valuable assets when Bio-Rad acquired them subject to the exclusive clinical testing license and all pending patents from Progenitor in 1999. Quest transferred the license to Bio-Rad under undisclosed terms.
Acquisition of the HFE patents was integral to Bio-Rad's business plans to develop and market analyte-specific reagents (ASRs) for HFE testing. HFE ASRs became available in 2001.
HFE patents do not appear to have blocked commercial development of additional methods of HFE testing utilizing different platform technologies. For instance, Bio-Rad Ltd. granted a non-exclusive license to Nanogen Ltd for detection of the C282Y and H63D mutations using the NanoChip™ System. We cannot assess whether alternatives were unimpeded in all cases, but at least some alternatives have developed. The patent-associated fees may have discouraged some laboratories from entering the market, but testing is widely available from multiple sources. One external reviewer of an early draft of this case study noted he was aware of at least one potential HFE test developer who decided not to develop a test because of the up-front payments to BioRad.
Several non-profit and for-profit laboratories offer HFE testing for a fee. It is unknown how many providers have acquired a sublicense from Bio-Rad for tests developed in-house or use the Bio-Rad analyte specific reagents (ASRs) (in which case a sublicense is built into the purchase).
It is unclear how much of the price variability among different providers (list price for mutation analysis ranges from approximately $150 to $500) can be attributed to license/royalty fees as opposed to variable overhead costs or costs associated with different testing methodology/platforms.
Communication and Marketing
Patents have had little to no impact on the communication and marketing of HFE testing.
There is no evidence that HFE mutation testing was ever marketed directly to consumers by Mercator Genetics or subsequent holders of HFE patent rights.
Information on promotion of HFE testing by Mercator Genetics among clinicians and other medical professionals is also unavailable. Similarly, it is unclear if SBCL and Bio-Rad Ltd. engaged in specific marketing activities to increase utilization of the test by consumers or health care providers.
Independent campaigns by the Hemochromatosis Foundation, the American Hemochromatosis Society, the American Liver Foundation and the CDC have sought to increase awareness of HH screening and HFE genetic testing among patients and medical professionals.11 The organizations promoting awareness are not the patent-holders, and the motivation appears to be public health awareness.
Direct-to-consumer testing is also available from DNADirect.
Clinical Adoption
Adoption of testing was rapid. As reported by Merz et al., adoption began nearly 17 months before the first patent was issued.10
In a survey of testing providers, Merz et al. reported that 5 of 58 clinical laboratories offering the test in January 1998 elected to stop testing after receiving “cease and desist” letters from SBCL. Out of 31 other laboratories that had not developed the test, 22 indicated patents were the primary reason for not doing so. SBCL began patent enforcement (“cease and desist” letters) approximately two years after the patents were issued, by which time there had been significant adoption of the test.
Although the number of laboratories offering HFE testing decreased, the majority of clinical providers (53) continued HFE genetic testing services. Therefore, it is unclear if the reduction in laboratories offering the test directly reduced clinical access to HFE testing.
As of May 2007, 37 laboratories were listed as providers of HFE testing on the Genetests.org website. In addition, the test is offered directly to consumers by DNADirect.
Adoption by third party payers
Patents do not appear to have had a direct or significant effect on decisions to cover the test by public or private insurance providers. A number of insurance companies cover genetic testing for HH when “medically necessary.”
Consumer utilization
There is little evidence bearing on the impact of patents on consumer utilization.
Patent enforcement activities by SBCL led to the discontinuation of testing in some laboratories. Other laboratories reported being deterred from developing an HFE test by patent enforcement activities. However, most laboratories did continue offering the test as a service. The effects that the reduction in number of laboratories had on patient access or consumer utilization cannot be determined.
HFE testing currently appears to be widely available. A large number of clinical laboratories offer the test in the price range of $160- $500. Consumers can also access testing independent of physicians through DNAdirect. The price offered by DNAdirect ($199) is less than that listed by many clinical laboratories and includes genetic counseling services.
The test is covered by several insurance providers when patients meet the eligibility criteria for testing. In the absence of quantitative data on how many tests are ordered per year and when and how often insurance coverage is denied, it is unclear to what extent third party adoption affects consumer utilization. The effect of patents on such coverage decisions, if any, was not mentioned by those offering tests or seeking reimbursement for them, and was not noted in payer coverage or reimbursement policies.
Background
The clinical syndromes of HH relate to the excessive deposition of iron in various organs. While healthy people usually absorb about 10 percent of the iron contained in their diet to meet their bodies' needs, those with HH absorb more. Chronic iron absorption may lead to a variety of symptoms. The most common symptoms include joint pain, fatigue, lack of energy, abdominal pain, loss of sex drive, and heart problems (including both arrhythmia and cardiomyopathy, or loss of cardiac muscle function). Men are more likely to experience symptoms and experience them earlier in life, between the ages of 30 and 50. Women affected by HH are usually symptomatic after the age of 50. The lower rates of HH in younger women are attributed to the protective effect of physiological blood loss associated with menstruation.12
HH begins as mere iron overload, but over time this overload can result in more serious disease through organ failure. Without early detection, the accumulated iron in various tissues may lead to:
Arthritis (due to joint damage)
Liver failure and cirrhosis (death of liver cells followed by scarring)
Pancreatic damage that can possibly include diabetes (Dr. Paul Adams cautions “that this area remains controversial since screening studies have not shown an increase in prevalence of diabetes. Several metabolic studies have suggested that the diabetes seen in hemochromatosis is more often insulin resistance of cirrhosis” (Personal communication with Paul Adams).
Problems with digestion (due to loss of pancreatic enzymes and paucity of fat-absorbing bile pigments produced by the liver)
Heart abnormalities such as irregular heart rhythms or congestive heart failure
Impotence
Early menopause
Abnormal pigmentation causing the skin to appear gray or bronze
Thyroid deficiency
Damage to the adrenal gland, and infrequently
Liver cancer
There are several known types of HH.3 The most common form, Type 1, affects adults and is usually caused by a defect in the HFE gene. Type 2 or juvenile hemochromatosis, which is not associated with the HFE gene, leads to severe iron overload and liver and heart disease in young adults between the ages of 15 and 30. Unlike adult-onset HH, juvenile HH affects males and females equally. Similarly, Types 3 and 4 of hereditary hemochromatosis are not associated with HFE mutations and they are much rarer.
Since the symptoms of HH can arise from many causes, doctors often focus on treating the individual symptoms and may not identify the underlying HH. Many cases of HH are therefore undiagnosed. This problem of effective diagnosis could be partially solved by genetic screening tests that would easily detect the HFE mutation in symptomatic persons and through a systematic screening process that identifies those presymptomatic individuals with iron overload. Individuals with signs of iron overload could then be evaluated with genetic testing and other means for determining causes of iron overload. In most cases, either an environmental source of overwhelming iron intake (e.g., vitamin overdose, dietary practice, water supply, or environmental exposure) or a known genetic mutation would explain the iron overload.
Genes Associated with Hemochromatosis
The gene most commonly associated with Type1 HH is HFE, located in the region of the gene HLA-A on chromosome 6.9 There are two known mutations of the HFE gene that are most commonly linked to HH. The C282Y mutation is caused by a single base change, resulting in tyrosine replacing the normal cystine at position 282 of the HFE protein. C282Y accounts for almost 90 percent of HH cases.5 Most patients are homozygous for the mutation, which is transmitted in an autosomal recessive manner.2 Environmental factors and other genotypes also contribute to HH.13 Another mutation, H63D, is the result of the substitution of an aspartic acid for a histidine at position 63. It is still unclear exactly how the H63D mutation is associated with HH. When H63D is inherited from one parent, it usually causes little increase in iron absorption and rarely leads to the development of hemochromatosis. Although most patients with a clinical diagnosis of HH are homozygous for the C282Y mutation, approximately 10% are compound heterozygotes carrying a single copy each of the C282Y and H63D mutations.12 S65C is an HFE gene mutation tentatively linked to a mild form of iron overload. Other mutations with less frequency and/or low penetrance have also been described, including V53M, V59M, H63H, Q127H, Q283P, P168X, E168Q, E168X, and W168X.14
Juvenile hemochromatosis, also called HH type 2, (subtypes 2A and 2B), is an autosomal recessive disorder not caused by a defect in the HFE gene. HJV, a gene located on chromosome 1q, was recently identified as the cause of HH type 2A. Juvenile HH type 2B is caused by mutation in the HAMP gene coding for hepcidin, a peptide hormone that has a key role in human iron metabolism.14 The hepcidin protein hormone was initially called “Liver-Expressed Anti-microbial Protein”15, 16 because its function appeared to be related to fighting fungal and bacterial infections (iron is essential to the inflammatory response to certain pathogens). HH type 3 is an autosomal recessive disease caused by mutations in the transferrin receptor 2 gene, TRF2.16 HH type 4, which is an autosomal dominant disease, is caused by mutations in the SLC40A1 gene. SLC40A1 encodes for a protein implicated in iron intestinal export, ferroportin.16
The remainder of this case study focuses on HFE, the gene most commonly associated with Type 1 HH, and for which the patenting and licensing stories are best documented.
Genetic Tests for Hemochromatosis
Several genetic tests are currently available for hemochromatosis. Targeted mutation analysis is the most common form of clinical genetic testing. This process tests for the presence of the two most common known disease-causing alleles in the HFE gene, C282Y and H63D. (Roughly 60 – 90% of the persons tested with an HFE mutation will have two C282Y alleles. While 3 – 8% will have one C282Y mutation and one H63D mutation, the rarest combination, roughly 1 % of those with HFE mutations will have two H63D mutations present. See Appendix D for more information.) Different laboratories use different methods. Several common testing methods for the presence of the C282Y and H63D mutations were used by 90 U.S. laboratories in 2002. These include electrophoresis for restriction fragment length polymorphisms (RFLPs) and size analysis (64% of labs), allele-specific oligonucleotide assay (ASO) (11% of labs), allele-specific polymerase chain reaction and Amplification Refractory Mutation System (PCR/ARMS) (6% of labs), LightCycler (8% of labs), DNA sequencing (3% of labs), and other/unspecified methods (8% of labs).17 Linked linear amplification (LLA) is another means of amplification of DNA to detect HFE mutations.
Some methods are more labor intensive than others, making them suitable only for research rather than diagnostic laboratories. Other methods accommodate the needs of large numbers of specimens requiring short turn-around times. In Canada and Europe, commercial suppliers can provide “kits” to clinical laboratories. However, since such kits used for clinical testing in the United States are regulated by the FDA, increasing the costs associated with development, analyte specific reagents (ASR) rather than test kits are routinely developed and marketed by biotech companies. Four biotechnology companies, Bio-Rad, Nanogen, LightCycler (a subsidiary of Roche), and Orchid Cellmark, provide reagents for the most commonly used methods of large-scale HH gene testing. A full sequence analysis can also be performed, usually to identify mutant alleles associated with HH that are not C282Y or H63D.
Non-Genetic-Based Means of Diagnosis Hemochromatosis
Currently, diagnosis of HH is often based on first-level biochemical tests, followed by second-level genetic testing. Biochemical methods are simple, fast, and inexpensive. The standard test is transferrin saturation (TS). This test determines how much iron is bound to transferrin, the protein that carries iron in the blood. Measuring a morning fasting TS level eliminates 80 percent of false-positive results. Values of 60% or greater in men and 50% or greater in women have an approximate sensitivity of 92%, specificity of 93%, and positive predictive value of 86% for detecting homozygous individuals with HH.12 The above data are primarily from referral studies in which the TS test is embedded in the clinical diagnosis. In general population screening studies, where there is no referral for testing, the sensitivity of TS is much less. There is also a wide biological variability in the test. Fasting TS has also been shown to be of no increased value over random testing.18, 19 The lack of a uniform cutoff percentage for the optimal detection of disease lowers the specificity and positive predictive value of the TS test. Another limitation of TS is that it is a two-step test and therefore more prone to error.
A second possible test is serum ferritin (SF). This test estimates the total body iron stores. Ferritin values greater than 300 μg/L in men and 200 μg/L in women, suggest iron overload. However, ferritin can be falsely elevated as an acute phase reactant and does not become abnormal until iron loading has advanced due to liver involvement.14, 19 Therefore, doctors should consider non-HH causes behind a patient's high serum ferritin levels if transferrin saturation is not elevated.
A more recent biochemical method used to test for HH is unbound iron-binding capacity (UIBC). UIBC is a one-step assay that has high sensitivity and has been suggested as a reliable and potentially inexpensive diagnostic test for HH.20 Prior to the availability of mutation analysis, liver biopsy was the most common second-level diagnostic test for HH. Liver biopsy helps determine the extent of iron accumulation in the liver. However, the biopsy is more often used as a prognostic tool, to review the level of damage in the liver.9, 21 Another non-genetic test used to diagnose HH is quantitative phlebotomy,22 in which specified amounts of blood are drawn. Removing “4 g or more of mobilizable iron stores (16 phlebotomies, each removing 500 mL of blood [250 mg of iron per 500 mL]) before the development of iron-limited erythropoiesis confirms the presence of primary iron overload due to hemochromatosis” (pp. 928-929).22 If any of the tests described above suggest iron overload, HFE genotype testing is strongly suggested.
Treatment of Hemochromatosis
Unlike many other serious genetic disorders, hemochromatosis may be treated simply, safely, and inexpensively. The most common treatment for HH is phlebotomy, a process used to rid the body of excess iron. In phlebotomy, doctors remove a pint of blood once or twice a week for several months or more, depending on the iron levels. Phlebotomy has been widely adopted because it is inexpensive and safe, and has clear face validity as a common-sense treatment for iron overload. Recent studies have demonstrated a reversal of liver fibrosis with phlebotomy treatment.23, 24 Treatment for those who already have organ damage is more complicated. While phlebotomy may stop the progression of liver disease in its early stages, those with more severe cases may need to seek a specialist. Phlebotomy will not cure other conditions associated with hemochromatosis, but it will help most of them, with the exception of arthritis, for which removal of excess iron has little effect.
Current Guidelines for Genetic Testing
Clinical uses of genetic testing include confirmatory diagnostic testing, predictive testing for at-risk relatives, carrier testing to identify heterozygotes, and prenatal diagnosis (technically available but rarely performed).6 The American College of Physicians (ACP) clinical practice guidelines for the screening of HH state evidence is insufficient to recommend for or against screening for HH in the general population.5 They recognize that the C282Y mutation is the most common predictor of whether the patient will develop HH but note that there is still no way of predicting which homozygous patients will develop HH.5 For these reasons, the ACP leaves the decision whether or not to perform tests for HH to clinical judgment, based on: whether patients exhibit symptoms of the associated disorders; whether patients exhibit serum ferritin levels of more than 200 μg/L in women and more than 300 μg/L in men combined with transferrin saturation greater than 55%; or whether the individual has a family history of HH. Each factor increases the risk for developing the disease compared to the general population.5
The ACP also encourages doctors to discuss the risks and benefits of genetic testing with their patients. This should include a discussion of the available treatment and its efficacy, as well as the social impact of disease labeling, insurability, psychological well-being, and as-yet-unknown genotypes associated with HH.5 One observational study found that notification of indeterminate results from screening might pose a potential participant risk. Asymptomatic individuals who underwent HFE genotype testing, or were tested for HH using the SF or FT methods and were found to have elevated levels of uncertain clinical significance, reported diminished general health and mental wellbeing, and more health worries, than normal controls.25 In another study, asymptomatic persons found homozygous or not for the C282Y mutation may develop unnecessary stress or false reassurance.5 The ACP does acknowledge that the lack of information on the natural history of HH makes it difficult to manage patients with the disorder, the effects of which are modified by environmental factors including blood loss from menstruation or donation, alcohol intake, diet, and co-morbid disease including viral hepatitis.5, 9 The ACP considers future technological developments and genetic screening as potential aids in the management of the disease.5 Finally, the ACP recommends more uniform diagnostic criteria.
The American Association for the Study of Liver Disease recommends genetic testing for all patients in whom there is a strong suspicion for iron overload. Such patients should have C282Y and H63D mutation analysis completed (see Appendix C).12
Patents and Licensing
Patenting of Hemochromatosis Genes
Bio-Rad Laboratories, Inc., is the owner and licensee of most of the patents relating to HH genetic testing and the HFE gene. In 1999, Bio-Rad bought many of those rights from Progenitor, which had retained the rights to HH genetic testing following the Mercator-Progenitor merger. Mercator was the initial patent owner and assignee.
Mercator scientists first identified the HFE gene in 1995–96, along with the two mutations, C282Y and H63D, which were present in over 80 percent of people suffering from HH.26 In 1995 and 1996, Mercator applied for patents related to HFE and its mutations. The patents were issued at various times between 1998 and 2000 and covered the whole HFE gene sequence, a method for diagnosing the C282Y and H63D mutations within the HFE sequence, a method of analyzing C282Y and H63D HFE mutations, and a method of analyzing the mutation using a kit. Other patents in the same patent family and with the same group of inventors issued between 2000 and 2006 and were assigned to Bio-Rad. These patents included diagnostic methods for a panel of less prevalent mutations, which did not include C282Y or H63D. They also cover polypeptides related to the HFE gene, and the associated proteins. Another patent covers a method of diagnosis for TRF2, another gene related to HH (see Appendix A).
Some other patents pertinent to HH are not controlled by Bio-Rad, but they are far fewer in number. Billups-Rothenberg, Inc., (BRI), in San Diego, California owns a gene patent, US 6,355,425 “Mutations Associated with Iron Disorders,” which covers a diagnostic method for a panel of HFE mutations including S65C, 193T, G93R, 277C, 105T, 314C but does not include C282Y and H63D. BRI has exclusively licensed this patent to Nanogen. The one HH gene patent owned by a non-profit organization is assigned to Erasmus University in Rotterdam, Netherlands. This patent claims a method of diagnosis for SCL11A3, a mutation of the ferroportin 1 gene. We have been unable to determine if this patent was ever licensed. However, these patents may be less relevant to the case study because the predominant tests related to HH genotyping involve the mutations C282Y and H63D that are covered by the Bio-Rad patents.
We know of no litigation over the DNA sequence patents associated with HFE or other genetic forms of hemochromatosis, although given exclusive licenses to Nanogen for several mutations, this is a conceivable prospect.
Licensing of HH Genes
Merz et al. published a report in 2002 highlighting the patenting of the HFE gene and the licensing practices of the Mercator/Bio-Rad patents. 10 The authors argued that gene patents had a negative impact on clinical practice because of the high prices the patent owners commanded. According to the article, in the late 1990s, Progenitor exclusively licensed the patent rights to perform clinical testing of the HH mutations to SmithKline Beecham Clinical Laboratories (SBCL) for an up-front payment and guaranteed continuing fees worth roughly $3 million. The licensing agreement guaranteed that SBCL's exclusive license and payments to Progenitor would continue until a kit became available for use by clinical laboratories. In June 1998, after SBCL obtained the exclusive licensing for the clinical testing component of HH, it began informing laboratories of their possible infringement activities and offering sublicenses for an up-front fee of $25,000 to academic licensees and for 5 to 10 times that amount to commercial laboratories (Appendix E). It also sought royalties as high as $20 per test. 10 After the sale of SBCL and the patent rights for clinical testing to Quest Diagnostics in 1999, the IP was not enforced again until Bio-Rad began offering analyte-specific reagents (ASRs) in 2001.
When Bio-Rad acquired the portfolio of pending and issued patents covering HFE and its mutations from Progenitor in April 1999, it acquired them subject to the exclusive clinical-testing license held by SBCL. Quest transferred the clinical-testing license it acquired from SBCL to Bio-Rad. 10 The terms and conditions of that license agreement were not made public. Bio-Rad obtained other patents related to HH gene products. It began offering analyte-specific reagents (ASR) for testing of the C282Y and H63D alleles in 2001.
Today, Bio-Rad offers two HH test kits, the mDx Hereditary Hemochromatosis ASR kit and mDx Hereditary Hemochromatosis LLA ASR test kit. Both kits provide for 24 tests at a cost of $2,016, or $84 per test. A purchase of the kit includes the purchase of a sublicense from Bio-Rad to perform the test. According to some providers, the sublicenses attached to Bio-Rad's kits are more cost efficient than the licenses it offers to laboratories that develop and offer their own mutation testing or “in-house” assays. 10 However, Dr Michael Watson at the American College of Medical Genetics indicates that, at least initially, the Bio-Rad test kit's inferior performance essentially forced laboratories to develop their own “in-house” tests, which would require paying the higher fee for a sublicense. Such a sub-license includes up-front payments that are inversely proportional to the testing volume of the laboratory plus a per test fee, which was $20 in 2002.10 It is not known what sublicensing fees are currently paid by laboratories that offer tests they have developed in-house, also known as “home- brews”. The CDC review of analytic validity of HFE testing noted that since Bio-Rad owned the patent for hereditary hemochromatosis, no other commercially available manufactured reagents were available for this test.17 However, ASRs for mutation detection using other platform technologies have become available more recently. For example, ASRs are offered by Nanogen Inc,27 with sublicenses from Bio-Rad and presumably BRI too.
Impact of IP and Licensing on Clinical Genetic Testing for HH
Despite the presence of IP on clinical testing methods, laboratories around the country were performing HH screening on patients before and after the Mercator patents issued.17 In a study of 128 U.S. laboratories identified as capable of offering the HFE test, with 119 of those laboratories responding, 58 laboratories indicated that they were performing HFE testing by 1998.28 Thirty-five of the 58 laboratories were conducting the testing after the Nature Genetics paper published in August 1996 identifying the mutation, but before the patent issued in January, 1998.28 Fifty-four of the 58 laboratories conducting the test received letters from SBCL informing them of the HH IP and offering a sublicense.10 Ninety-one percent of the interviewed laboratories were aware of the HFE patents and 36 revealed that the patents contributed to their decisions not to offer the test.10 Five laboratories out of the initial 128 sample, or 4 percent, chose to stop performing the test. Of these 5 labs, 2 stated that the reason to stop testing was patents. One laboratory stated that patents were one of several reasons for abandonment of the HH test. Two additional laboratories stated that patents were not a reason for their decision to abandon the test. Commercial reasons (e.g., lack of adequate volume to cover fixed costs) appeared to be the predominant reason why these laboratories stopped performing the test (Personal communication with Jon Merz).28
As of May 2007, the GeneTests database (www.genetest.org) listed 37 U.S. laboratories performing targeted mutation analysis for HH. A sampling of 17 of those 37 laboratories revealed a list price for targeted mutation analysis that fell between $125 and $467 indicating a significant range in prices. (The sample was conducted by informal telephone conversations with laboratory staff on April 6, 2007. The providers were surveyed for their laboratory's “list price” for HFE testing. In some situations, staff offered both the individual list price and the insurance list price. See Appendix B.) By way of comparison, a study noted that the cost of HFE-genotyping in Australia cost less than $28.21 The variability in American pricing may be due to several factors, including variability in methods of mutation testing, reagents costs for each method, and potentially licensing fees to perform HH testing. Some laboratories may perform “home-brew” assays with relatively low reagent costs. In these cases, one must consider the cost of the technical time for reagent preparation and the Quality Control/Quality Assurance (QC/QA) costs. The costs of ASR can be relatively high compared to traditional biochemical assays. At the same time, savings in technical staff time for preparation and QC/QA can offset reagent costs. For screening, the relevant figure is the cost per patient tested, not the cost per mutation tested; a diagnostic test may entail running the test case as well as controls, which also consume reagents covered by the reagent kits.17 The exact economics of HFE mutation testing for HH are therefore not completely transparent. The cost of the IP is a minimum of the $20 per test fee and could be higher, depending on how licensing fees are structured into reagent costs that come with associated patent licenses.
Cost Effectiveness of Screening for HFE Mutations
Several studies on the cost-effectiveness and benefits of genotypic screening for the common disease-causing alleles on the HFE gene have been performed. As recently reviewed by Phatak et al., these studies provide evidence that screening would improve health status. However, all the studies reviewed support the use of biochemical tests rather than genetic tests as the initial test.29 In 1999, Adams et al. reported that the genotypic screening of voluntary blood donors and their siblings by genotyping would be less expensive than phenotypic screening with biochemical tests if the genetic test cost less than $28. However, if the genetic test cost $173, then it would cost nearly $110,000 to identify a homozygote with a potentially life-threatening disease. The cost per homozygote identified also increased with decreasing penetrance of the disease. A 10% penetrance (i.e., 10% of those individuals with the relevant mutation actually have HH) resulted in nearly $400,000 in costs per individual identified.30
A literature review and synthesis conducted by Whitlock et al. for the U.S. Preventive Services Task Force provides some outline of the cost effectiveness of HFE screening. However, it could not determine the cost-effectiveness of screening because of uncertainties associated with penetrance of disease in individuals with C282Y mutations, poorly defined natural history of disease progression, and variable prevalence of HFE mutations in different ethnic populations.9 HH testing would not be as effective as the control procedures without evidence establishing that the prevailing symptoms are caused directly by or associated with iron overload. The review outlined several studies suggesting that while members of the general population with symptoms or signs consistent with HH did not have higher levels of C282Y homozygosity, patients in a liver clinic prescreened for higher transferrin saturation levels, hospitalized diabetic patients, and patients referred to specialists for chronic fatigue and arthralgias did.9 Studies have suggested that most individuals with the genetic abnormality do not have shortened life expectancy or progression of disease when compared with control groups.12, 31 Morbidity and mortality in HH are related to the presence of iron overload in the blood, tissue, and organ systems, not the HFE mutation, per se.32 End organ damage is related to the severity of iron overload and reduces life expectancy.4 One study suggests that HFE screening is cost effective if the proportion of C282Y homozygotes that develop end organ damage when left untreated is over twenty percent.4 Allen et al. recently reported that nearly 28 % of men and 1 % of women with C282Y homozygosity will develop iron overload disease.33
To assess the cost-effectiveness of genotype screening for HH, a study would need to address: (1) the prevalence of HH; (2) the probability of developing disease manifestations and cost of managing them; (3) the cost of the screening test; (4) the cost offsets of screening and diagnosis compared to costs avoided by early detection or more effective management; and (5) the discount rate, to accommodate the separation in time from detection to health benefit.
In a recent comprehensive analysis, Gagne et al. evaluated the cost effectiveness of 165 population screening algorithms using biochemical and genetic tests in a simulated virtual population with user defined demographic characteristics including variable HFE mutation frequencies and penetrance. Biochemical penetrance was used as an intermediate phenotype in this study. In the 165 algorithms used in 91 virtual populations of a million individuals, biochemical screening tests were more cost effective than genetic tests when used as the initial test. Genetic testing was once again found to be most cost effective when performed as the final confirmatory step.34
HFE gene testing for the C282Y mutation is a cost-effective method of screening the siblings and children of patients with HH.32 The authors incorporated serum iron studies among persons homozygous for C282Y and compared a no-screening strategy with four screening strategies for HH. All the strategies were developed for treating children and siblings of probands, except for one when the spouse was also given a genetic test. This exception strategy was only applied to children. The study recommended a four step clinical intervention: “(1) serum iron studies; (2) gene testing of the proband. If the proband is [without a C282Y mutation], the spouse undergoes gene testing; if he or she is heterozygous [for the C282Y mutation], the children undergo gene testing; (3) Gene testing of the proband; if he or she is homozygous, relatives undergo gene testing; (4) Direct gene testing of relatives” (p. 261).32 The study concluded that “HFE gene testing of the proband was the most cost-effective strategy for screening one child,” with an incremental cost-effectiveness ratio of $508 per life-year saved (p. 261).32 For screening two or more children, the second most cost effective strategy was “HFE gene testing of the proband followed by testing of the spouse” (p. 261).32 There, the incremental cost-effectiveness ratio was $3665 per life-year saved. The study also concluded that “in siblings, all screening strategies were dominant compared with no screening” and that “strategies using HFE [genetic] testing were less costly than serum iron studies” (p. 261).32 The greater cost-effectiveness of this sequential algorithm, which incorporates genetic testing but does not use genetic testing as the first step, is because the relatively high cost of genetic testing is incurred only in cases where risk is higher than average. The use of clinical genetic testing to confirm a diagnosis of HH among those with iron overload, in this conceptual framework, is an “indicated” preventive intervention targeted at asymptomatic individuals who have evidence of iron overload based on inexpensive biochemical screening tests. Here, we borrow from the terminology of Gordon's classification of preventive strategies, using genetic testing as one step in the prevention strategy.35
Phatak et al. recommend selective or “targeted” screening in groups whose risk is elevated such as adult men greater than 25 years of age of Northern European ancestry and first degree relatives of patients with known HH.29
Lessons Learned
HH was selected for study to assess the impact of patenting and licensing practices on access to genetic testing. Using the conceptual framework developed for a parallel literature synthesis, we now consider what lessons might be learned from this case.
Research
We considered whether the gene patents in question either accelerated or retarded the original discovery that ultimately led to the development of HH mutation analysis and genetic testing. Initially, the discovery of the HH-related genes was characterized as a “race,” which was won by Roger K. Wolff and his colleagues of Mercator Genetics in Mountain View, California. The scientists knew that the gene for HH resided on chromosome 6, but were unable to pinpoint it. They suspected that most people with HH had the same mutations and invested heavily in research to find such mutations. Studying a group of 178 people with iron-overload disease from across the country, the researchers identified a segment of DNA that all patients had in common and used that information to scour that region of chromosome 6 in search of specific mutations. After a long search, they determined that two mutations accounted for 87 percent of iron-overload patients in the study and published their findings in the August 1996 issue of Nature Genetics.26 French and Australian scientists verified these findings a few months later, publishing their findings in the November issue of that same journal.36-38 There is no evidence that the patent retarded the original discovery. On the contrary, the potential of revenues from diagnostic testing may have provided added incentive for basic research linking HFE mutations to HH by drawing Mercator into the race. Of Mercator's four original patents, the first was filed on May 8, 1995 and the last was filed May 23, 1996 (see Appendix A). The May 1995 patent application pre-dates the submission of the Nature Genetics article by over one year. While some speculated that patenting and commercial positioning might account for the delay, Dr. Dennis Drayna, who was a co-founder of Mercator Genetics and a senior author in the 1996 Nature Genetics paper, indicated that “there was no attempt to delay publication for commercial or competitive reasons” (Personal communication with Dr. Dennis Drayna). He said that delay in publication simply resulted from the time taken for scientific review and subsequent efforts to address reviewers' comments and criticisms before resubmitting the manuscript. In fact he believes that the opposite was true and that it was in Mercator Genetics's best interest to publish their results as early as possible. In Dr. Drayna's opinion, “Early scientific discoveries are essential for raising subsequent rounds of funding from additional investors, and publication of scientific discoveries is paramount to the maintenance of an ongoing enterprise. Laboratory discoveries are trumpeted as loudly and quickly as possible, which is basically what Mercator Genetics did” (Personal communication with Dr. Dennis Drayna).
Dr. Margit Krikker, medical director of the Hemochromatosis Foundation, opposed Mercator's approach to patenting in a 1996 AP story published in the New York Times. “[She] complained about the way Mercator was handling the discovery, saying that by filing a patent for the gene, Mercator had limited other scientists' research opportunities” (p. C-7).39 We found no evidence to corroborate this assertion. Substantial basic and clinical research on the genetics of hemochromatosis has continued since the discovery of HFE, including identification of genes and mutations associated with other types of hemochromatosis, suggesting patents have not blocked further research and development. We cannot eliminate the possibility of a “chilling effect” from fear of patent prosecution, but in 2007 and 2008 it did not emerge as a major controversy, as it appears to have been at the time of the patent and again in 2002.
However negotiating licenses for the use of HFE patents may have contributed to a several-month delay in initiating research conducted as part of the Hemochromatosis and Iron Overload Screening Study (HEIRS) sponsored by the NHLBI. The purpose of HEIRS is to determine the prevalence, genetic and environmental determinants, and potential clinical, personal, and societal impact of iron overload and hereditary hemochromatosis, in a multi-center, multiethnic, primary care-based sample of 100,000 adults. Dr. Michael Watson, Executive Director of the American College of Medical Genetics, indicated that “the study was delayed by nearly 6 months” because Third wave Technologies needed a sublicense from Bio-Rad Ltd for the use of patents covering HFE mutations (C282Y and H63D) for the Invader™ assay ASRs (Personal communication with Dr. Michael Watson). Dr. Eckfeldt, another prominent researcher in HEIRS, confirmed that the study was indeed delayed between 4-6 months but indicated that start-up logistics also contributed to this delay. A modified Invader™ assay was used for all HFE genotyping in the study.40 NHLBI paid Bio-Rad a license fee to access HFE patents for genetic testing performed as part of HEIRS, since the study was designed to return test results to the nearly 100,000 patients enrolled and their physicians. Dr John Eckfeldt stated that the royalty fee per test paid to Bio-Rad was reasonable, although the exact amount is confidential and protected by non-disclosure agreements. He also noted that “considering that 100,000 subjects were screened, the overall cost to NHLBI was quite substantial” despite a nominal fee per test. Bio-Rad subsequently granted a general sublicense to Third Wave Technologies. Until recently, Third Wave offered HFE custom ASRs as a service. Following the acquisition of Third Wave by Holologics Inc., in June 2008, custom ASRs for HFE are no longer being marketed.41
Development
Within one year of the Nature Genetics publication, Mercator announced that it was developing a blood test for HH genotype testing. The company pointed to the ultimate goal of population-wide screening for HH, whereby all persons, not just those at higher risk for the mutation, would be tested.38 As demonstrated above, laboratories without IP rights on the HFE gene developed genetic tests for the mutations based on the Nature Genetics paper before the patent issued. This suggests that information on the gene sequence and its associated mutations was sufficient for other clinical providers to develop and offer genetic testing for HFE.28
Commercialization
Mercator Genetics, the company that first patented the HFE gene and its corresponding mutations, was founded by a group of doctors and genetic researchers from Stanford Medical School and the Silicon Valley biotechnology sector. Mercator Genetics described itself as a “gene discovery company” that focused on the identification of genes responsible for major diseases. Mercator's business model consisted of positional cloning to discover genes of interest and then capitalizing on the development of diagnostic tools associated with those genes.42 Financial support was solicited from the pharmaceutical industry and venture capitalists like Robertson Stephens & Co., Interwest Partners, and Oak Investment Partners.42 Investment was possibly tied to the prospect of patents. According to Dr. Dennis Drayna, “Mercator Genetics was conceived and raised funding on the basis of a far broader agenda. Hemochromatosis was never mentioned in any of the discussions that preceded funding of the company. HH was settled upon as a research and commercial target during later discussions with the Scientific Advisory Board. The choice of a diagnostic as a commercial target, as opposed to our competing genomics companies who mostly worked toward therapeutics as commercial targets, generated some discussion at the time, as the investors had already committed their funds” (Personal communication with Dr. Dennis Drayna). Mercator was not only “racing” to clone the HH gene but also to search for genes linked with complex diseases like asthma, schizophrenia, prostate cancer, and cardiovascular disease.38 However, Dr. Drayna said, “While the company did work in a number of other disease areas, these were either small exploratory efforts (such as Werner Syndrome and narcolepsy), or were the subject of primarily business transactions. There was never any work in the laboratory on asthma, schizophrenia, prostate cancer, or cardiovascular disease at Mercator Genetics” (Personal communication). Mercator placed second or later and thus lost to Darwin Molecular Corporation in the “race” to patent the gene for the aging disorder Werner's syndrome.42 Ultimately, the company's only successful entry in a patent race was the search for HFE and its mutations. Mercator Genetics's most valuable IP assets were patent rights to HFE and its mutations. In 1997, after expending $10 million on developing its method of positional cloning and discovering the association between HFE mutations and HH, Mercator went out of business and merged with Progenitor in 1997, which received rights to Mercator's pending and issued patents.10 Dr. Drayna believes that “Mercator Genetics …. was a clear scientific success in the face of exceptionally widespread competition. It was less of a business success largely due to medical, social, and political factors surrounding the adoption of genetic testing on a widespread basis” (Personal communication with Dr. Dennis Drayna).
Progenitor obtained rights to Mercator's HFE patents, and was readying its first initial public offering (IPO) when it was sold to SmithKline Beecham Laboratories, which received assets from both Mercator and Progenitor. Progenitor anticipated an IPO price between $10 and $12 per share and proposed funding its acquisition of Mercator with $22 million of Progenitor Common Stock, based upon an initial public offering price.43 Again, the value of Progenitor was largely based upon the perceived value of its IP more than tangible assets.
Communication/Marketing
There is no evidence that the patented HFE mutation analysis test was ever marketed using direct-to-consumer marketing, although the idea was considered originally. For instance, there has been no ad campaign similar to the one launched by Myriad Genetics during the 2002 Super Bowl and test-marketed in Denver and Atlanta, or Myriad's 2007-2008 BRCA advertising in the Northeast.
Outside of Mercator's promotion activities, organizations committed to HH awareness have led their own marketing campaigns. Following the gene discovery in 1996, Margit Krikker of the Hemochromatosis Foundation bought an advertisement in the New York Times to alert the public to the deadliness of HH. The Foundation was frustrated over the lack of interest in HH displayed by federal officials and wanted to mount an awareness campaign. Another early and active proponent of communicating Mercator's discoveries was the American Liver Foundation (Personal communication with Dr. Dennis Drayna). The American Hemochromatosis Society (AHS) designated May 2007 as “National Hereditary Hemochromatosis Genetic Screening & Awareness Month.” It asked its membership to contact local newspapers, TV and radio stations with AHS press releases that connected screening to saving lives.44
The CDC has also made detailed information available about diagnosis of hemochromatosis for physicians and the use of genetic testing in family based testing for hemochromatosis.45 However, in the absence of family history, CDC recommends genetic testing for HFE mutations only as the confirmatory step of their testing protocol after the appropriate biochemical tests for iron overload (TS and serum ferritin) have been conducted.46 The HH genetic test is currently also available directly to consumers through DNAdirect. Otherwise, HH testing is primarily offered to consumers by healthcare providers.
Adoption
Shortly following the HFE gene discovery, the CDC considered recommending widespread screening for HH and considered advising doctors to order a gene test for all patients 18 years or older. That recommendation has not been made because of inconclusive evidence on the penetrance of HFE mutations and cost-effectiveness of the test. Dr. Dennis Drayna, a Mercator co-founder and NIH molecular geneticist, argued enthusiastically for broad HH genetic screening at a 1997 Ethical, Legal and Social Issues (ELSI) meeting associated with the Human Genome Project.47 Ethical, legal, and social concerns such as fear of genetic discrimination and questions over whether it made sense to “diagnose people based on genotype and not health” were raised as criticisms.47 An account of this meeting suggested that the market would determine whether insurance companies and HMOs adopted the test to save money in HH complications like liver transplants.47 A recent study, which measured the extent of employment and health insurance problems associated with population screening for hereditary hemochromatosis and iron overloads, found that at one year following genotypic and phenotypic screening, only 0.4% of individuals surveyed (3 out of 1154 individuals) reported any problems. Problems primarily involved life insurance and long term care insurance coverage. However, none of the affected individuals reported problems with health insurance coverage or employment. The outcome suggests that genetic discrimination concerns are much lower than originally anticipated.48 It also suggests, however, that they occur in forms of insurance, long-term care and life insurance, that are not covered by the Genetic Information Nondiscrimination Act passed in 2008 (which begins to take effect in 2009 and 2010).
Insurance companies and at least one Medicare carrier have adopted HH genotype testing but not as the broad screening test initially conceived. Rather, HH genotyping is usually a second-level test conducted after less expensive biochemical tests suggest HH or to test family members of identified HH homozygotes. Insurance policies may cover HH testing if it comports with “medical necessity.” To be eligible for testing, the insured individual will likely need to meet defined conditions for testing that in some plans are enforced by preauthorization requirements, such as: (1) prior blood test indicating iron overload; (2) family history of HH; or, (3) member of a family with a known HH mutation. Cost is not cited as an explicit criterion, and patents may not have a direct or significant effect on the decisions to cover the test by insurance providers. However, patents did affect which laboratories offered the test and which laboratories decided to cease testing after patent enforcement by SBCL.28 Yet the majority of laboratories continued to offer the test either with or without a sublicense. As noted earlier, several providers offer these tests currently and presumably interact with a range of carriers for insurance reimbursement.
Consumer Utilization
The HFE test is not available as an initial, universal screening test along the lines originally envisioned. Consumers typically access the tests through clinical laboratories via their physicians. Appendix B provides a sample of some laboratories, their services, and their costs. At least 37 laboratories offered HFE genetic testing as of May 2007. Additional providers not listed on Genetests.org may also offer this test. The test is also easily obtainable without physicians serving as the conduit for HH testing. DNAdirect, a direct-to-consumer genetic testing service, offers HH genetic testing for $199. Consumers using this service can thus choose to avoid involving a doctor or notifying their insurance company.49 DNAdirect sends consumers a test collection kit in the mail that includes cotton swabs for cheek swabbing and a postage-paid envelope to mail the swabs back to the laboratory for DNA analysis. Unlike most direct-to-consumer testing outlets, DNAdirect offers genetic counseling with the test results. DNAdirect provides forms, CPT Codes, and Letters of Medical Necessity for consumers seeking reimbursement from insurance or health plans. The service also offers anonymity and explains why anonymity might be desirable due to the potential of genetic discrimination. Since the $199 price tag is less than several of the clinical laboratories offering the test (see Appendix B), consumers with or without a family history of HH but with some means can easily obtain results, provided that they do not seek insurance reimbursement (insurance coverage would generally be confined to high-risk individuals meeting iron overload or family history criteria). However, DNAdirect and its counterparts are not FDA-regulated, and there is no peer review of the tests' accuracy, although the tests themselves are performed in CLIA-approved laboratories.50
Our study does not provide information regarding the impact of patents on under- or over-utilization of the HFE genetic test. Test utilization would need to be ascertained more systematically by surveying providers about how frequently the test is ordered and matching clinical indication to test use.
We did not uncover evidence about whether consumers are denied coverage for HH genetic tests. Direct assessment of test utilization and the frequency of inability to receive testing due to insurance coverage problems will help address the issue of patient access more comprehensively.
Acknowledgments
John Eckfeldt, Michael Hopkins, Dennis Drayna, Jon Merz, Michael Watson, and Paul Adams kindly reviewed this case study.
Funding: This case study was carried out under grant P50 003391, co-funded by the National Human Genome Research Institute and US Department of Energy, and supplemented by funding from The Duke Endowment. The case study authors have no consultancies, stock ownership, grants, or equity interests that would create financial conflicts of interest. The Center for Genome Ethics, Law & Policy accepts no industry funding. Dr. Robert Cook-Deegan is listed on the British Medical Journal roster of physicians who have pledged to remain independent of industry funding <http://www.tseed.com/pdfs/bmj.pdf>; more details about how the case studies were done are noted in a 29 July 2009 letter to the Secretary's Advisory Committee on Genetics, Health, and Society <http://www.genome.duke.edu/centers/gelp/documents/SACGHSResponsetopubliccomments.pdf>.
Appendix A: US Patents Related to Hemochromatosis Testing
| Patent No. and Title | Date Filed/Issued |
Inventors | Assignee | Claims |
|---|---|---|---|---|
| 5705343, Method to Diagnose Hereditary Hemochromatosis | Feb. 9, 1996 / Jan. 6, 1998 | Drayna et al. | Mercator Genetics Inc., Menlo Park, CA | Mutation analysis of HFE with kit |
| 5712098, Hereditary Hemochromatosis Diagnostic Markers and Diagnostic Methods | Apr. 16, 1996/ Jan. 27, 1998 | Tsuchihashi et al. | Mercator Genetics, Menlo Park, CA | Mutation analysis for HFE |
| 5753438, Method to Diagnose Hereditary Hemochromatosis | May 8, 1995 / May 19, 1998 | Drayna et al. | Mercator Genetics Inc., Menlo Park, CA | Method for diagnosing the mutation of an HFE sequence; mutation sequences, but not the whole gene. |
| 6025130, Hereditary Hemochromatosis Gene | May 23, 1996 / Feb. 15, 2000 | Thomas et al. | Mercator Genetics Inc., Menlo Park, CA | HFE gene and a diagnostic method; whole HFE gene sequence |
| 6140305, Hereditary Hemochromatosis Gene Products | Apr. 4, 1997 / Oct. 31, 2000 | Thomas et al. | Bio-Rad Laboratories, Inc., Hercules, CA | Polypeptides associated with HFE |
| 6228594, Method for Determining the Presence or Absence of Hereditary Hemochromatosis Gene Mutation | Feb. 14, 2000 / May 8, 2001 | Thomas et al. | Bio-Rab Laboratories, Hercules, CA | Diagnostic method for C282Y and H63D detection using DNA and RNA. |
| 6355425, Mutations Associated With Iron Disorders | Mar. 26, 1999 / Mar. 12, 2002 | Rothenberg et al. | Billups-Rothenberg, Inc., San Diego, CA | Diagnostic method for a panel of mutations in HFE, including : S65C, 193T, G93R, 277C, 105T, 314C |
| 6762293, Diagnostics and Therapeutics for Autosomal Dominant Hemochromatosis | Oct. 10, 2001 / Jul, 13, 2004 | van Duijn et al. | Erasmus University Rotterdam, Rotterdam (NL) | Ferroportin (SLC11A3) sequence and method of diagnosis for SLC11A3 |
| 6849399, Methods and Compositions for Diagnosis and Treatment of Iron Misregulation Diseases | Aug. 27, 1997 / Feb. 1, 2005 | Feder et al. | Bio-Rab Laboratories, Hercules, CA | Diagnostic method for transferring receptor (TFR2) and mutation A424G |
| 6955875, Mutations associated with iron disorders | Oct. 16, 2001/ Oct. 18, 2005 | Rothberg et al. | Billups-othberg Inc. | Methods for diagnosing HFE by detecting mutations in nucleotide position 193 |
| 7067255, Hereditary Hemochromatosis Gene | May 2, 2002 / Jun 27, 2006 | Thomas et al. | Bio-Rab Laboratories, Hercules, CA | Method for detecting three mutant alleles (24d1, 2 and 7) |
| 7078513, Plasmids Comprising Nucleic Acids from the Hereditary Hemochromatosis Gene | Feb. 4, 2000 / Jul. 18, 2006 | Thomas et al. | Bio-Rab Laboratories, Hercules, CA | Plasmid containing HFE mutation 24d1 |
| 7026116, Polymorphisms in the Region of the Human Hemochromatosis Gene | May 7, 1997 / Apr. 11, 2006 | Ruddy et al. | Bio-Rad Laboratories, Hercules, CA | Isolated polynucleotide of HFE gene sequence containing SNP variants, and a kit. |
Appendix B: Price Comparison for HFE Testing from A Subset of Providers
| Laboratory | Genetic Testa | List Priceb | CPT Codesc | |
|---|---|---|---|---|
| Arup Laboratory | HFE PCR | $225 | 83890, 83900, 83896 × 4, 83912 | |
| Baylor College of Medicine | $200 | 83914 × 3, 83912, 83898 × 2, 83891 | ||
| Blood Center of Wisconsin | Allele-specific PCR | $175 | 83891, 83900, 83896 × 4, 83912 | |
| Boston University School of Medicine | $250 | |||
| Case Western Reserve Univ. | $275 | 83890, 83892 × 2, 83894 × 2, 83898 × 2, 83912 × 2 | ||
| Cincinnati Children's Hosp. Medical Center | $337 | 83891, 83894, 83898, 83912, 83892 | ||
| Duke Univ. Health System | ARMS | $467.25 | ||
| Greenwood Genetics Center | $250 | 83894, 83898, 83912 | ||
| Kimball Genetics, Inc. | PCR analysis for both the C282Y and the H63D mutations | $190 | ||
| LabCorp | $297 cost w/o insurance | $229.50 with insurance | ||
| Mayo Clinic | PCR-based assay (using LightCycler technology) used to test for 3 mutations in the HFE gene: C282Y, H63D, and S65C. S65C mutation is only reported when it is found with the C282Y mutation. (PCR utilized pursuant to a license agreement with Roche Molecular Systems, Inc.) | $411.20 | 83890, 83898 × 2, 83912 | |
| Michigan State Univ. | Extract DNA from the sample and amplified enzymatically then digested with the following restriction enzymes: Rsa I, Dpn II, and Hinf I. After digestion, the fragments are separated by electrophoresis. Testing can detect the C282Y, H63D, and S65C mutations in the HFE gene. | $227 | 83890, 83898 × 2, 83892 × 2, 83894, 83912 | |
| NorDx | Linked Linear Amplification (LLA) with DNA probes | $372.50 | 83890 83896 × 4 83900 83912 |
|
| SUNY Upstate Medical Univ. | $158 | |||
| Specialty Laboratories | Cleave-based Invader Assay Hemochromatosis GenotypeR | $345 | 83891, 83892×4, 83896 × 10, 83903 × 2, 83908 × 2, 83912 | |
| Spectrum Health | $205.50 | 8 CPT Codes | ||
| University of Alabama @ Birmingham | Detection of C282Y and H63D mutations in the HFE gene using multiplex PCR methods. | $200 | 83890 83898 83892 83894 83912 |
|
| Univ. of Iowa Hospitals & Clinics | $395 | |||
All the tests described are targeted mutation analysis, or allele-specific mutation analysis. The tests are for either (1) a nucleotide repeat expansion, or (2) one or more specific mutations. Some of the labs provided a more specific description of their services. The blank boxes indicate areas for which no information was obtained.
List prices as of May 2007. Prices were obtained either by phone call or from information listed on provider website.
CPT Code Interpretation: 83890 Molecular Isolation and Extraction; 83900 Amplification; 83891 Isolation and extraction of highly purified nucleic acid; 83894; 83896 Nucleic Acid Probes; 38398 Amplification of nucleic acid, each primer pair; 83900 Amplification of nucleic acid, first two sequences; 83912 Interpretation and report; 83914 Mutation identification by enzymatic ligation or primer extension, single segment, each segment (eg, oligonucleotide ligation assay (OLA), single base chain extension (SBCE), or allele-specific primer extension (ASPE)).
Appendix C Diagnostic Algorithm for Hereditary Hemochromatosis
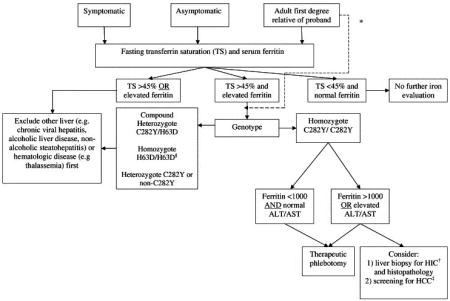
Modified from the American Association for the Study of Liver Disease Diagnostic Algorithm, 2001.
*direct testing of first degree probands is an acceptable alternative
†hepatic iron concentration
‡hepatocellular carcinoma
§Although H63D homozygosity is thought to lead to hemochromatosis in some individuals, this is more the exception, rather than the rule. Since the H63D mutation has a higher prevalence than the C282Y mutation, but accounts for a significantly smaller portion of those with clinically relevant hemochromatosis, abnormal iron studies with H63D homozygosity should prompt further evaluation into other disease processes first, with a diagnosis of hereditary hemochromatosis only after other avenues have been explored.
Reprinted from the American Journal of Medicine, Volume 119, Number 5, Andrew W. Yen, Tonya L. Fancher and Christopher L. Bowlus, “Revisiting Hereditary Hemochromatosis: Current Concepts and Progress,” pp. 391-9, at p. 396, 2006, with permission from Elsevier.
Appendix D: Molecular Genetic Testing: Clinical Methods and Testing Strategy
Appendix D is from http://www.geneclinics.org/profiles/hemochromatosis/details.html [accessed May 3, 2007] and is copyrighted by the University of Washington, Seattle.
There are various ways to detect hemochromatosis:
Targeted mutation analysis: available on a clinical basis for two known disease-causing alleles in the HFE gene (C282Y and H63D). About 87% of individuals of European origin with HFE-HH are either homozygotes for the C282Y mutation or compound heterozygotes for the C282Y and H63D mutations. Most clinical laboratories do not routinely test for the S65C allele because it appears to account for only 1% of individuals affected clinically and its clinical significance is unclear.
Sequence analysis: available in a limited number of clinical and research laboratories to identify other mutant alleles associated with HFE-HH laboratories
The table below summarizes molecular genetic testing for this disorder.51
Table 1. Molecular Genetic Testing Used In HFE-HHC.
| Test Method | Mutations Detected | Mutation Detection Rate | |
|---|---|---|---|
| % of Individuals [in populations of European origin] with HHC52 | Genotype | ||
| Targeted mutation analysis | HFE mutations: p.C282Y, p.H63D | ∼60%-90% | p.C282Y/p.C282Y |
| 3%-8% | p.C282Y/p.H63D | ||
| ∼1% | p.H63D/p.H63Da | ||
| Sequence analysis | HFE sequence alterations | Unknown | Unknownb |
There is no evidence that p.H63D/p.H63D is associated with a hemochromatosis phenotype in the absence of another cause of iron overload.
A few individuals who are compound heterozygyotes for the p.C282Y allele, and one of a small number of rare HFE mutations, have the hemochromatosis phenotype.
Testing Strategy for a Proband
Adults with transferrin-iron saturation higher than 45% warrant targeted mutation analysis. Individuals homozygous for the C282Y mutation or compound heterozygous for the C282Y and H63D mutations can be diagnosed as having the genetic make-up to develop HFE-HHC.
Individuals who are not C282Y homozygotes generally represent a heterogeneous group who may suffer from liver disease unrelated to HFE or have other metabolic syndromes. These individuals should undergo liver biopsy with assessment of histology and measurement of hepatic iron concentration as a next diagnostic step.
The figure below represents the testing strategy to establish the diagnosis of HFE-HH for the two groups listed above.

Appendix E: Sample Letter of Patent Enforcement For HFE Testing from SBCL

Letter reproduced with permission from Dr. Debra Leonard.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
HUMAN STUDIES: All interviews were conducted under Duke University IRB-approved protocol 1277 and usually conducted by phone and recorded. Researchers obtained informed consent from subjects.
Contributor Information
Subhashini Chandrasekharan, Center for Public Genomics, Center for Genome Ethics, Law & Policy, Institute for Genome Sciences and Policy, Duke University.
Emily Pitlick, Center for Public Genomics, Center for Genome Ethics, Law & Policy, Institute for Genome Sciences and Policy, and Duke School of Law, Duke University.
Christopher Heaney, Center for Public Genomics, Center for Genome Ethics, Law & Policy, Institute for Genome Sciences and Policy, Duke University.
Robert Cook-Deegan, Center for Public Genomics, Center for Genome Ethics, Law & Policy, Institute for Genome Sciences and Policy, Duke University.
References
- 1.National Digestive Diseases Information Clearinghouse (NDDIC) Hemochromatosis. [November 10, 2008]; http://digestive.niddk.nih.gov/ddiseases/pubs/hemochromatosis/index.htm.
- 2.Schmitt B, Aronson M, Fitterman N, Snow V, Weiss K, Owens D. Screening primary care patients for hereditary hemochromatosis with trasnferrin saturation and serum ferritim level: systematic review for the American College of Physicians. Ann Intern Med. 2005;143:522–536. doi: 10.7326/0003-4819-143-7-200510040-00011. [DOI] [PubMed] [Google Scholar]
- 3.Olynyk J, Trinder D, Ramm G, Britton R, Bacon B. Hereditary hemocromatosis in the post-HFE era. Hepatology. 2008;48(3):991–1001. doi: 10.1002/hep.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford D, Hickman P. Screening for hemochromatosis. Hepatology. 2000;31(5):1192–1193. doi: 10.1053/he.2000.7354. [DOI] [PubMed] [Google Scholar]
- 5.Qaseem A, Aronson M, Fitterman N, et al. Screening for hereditary hemochromatosis: a clinical practice guideline from the American College of Physicians. Ann Intern Med. 2005;143:517–521. doi: 10.7326/0003-4819-143-7-200510040-00010. [DOI] [PubMed] [Google Scholar]
- 6.Kowdley KV, Tait JF, Robin BL, Motulsky AG. HFE-Associated Hemochromatosis. [November 10, 2008]; http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=hemochromatosis.
- 7.Moalen S, Weinberg E, Percy M. Hemochromatosis and the enigma of misplaced iron: implications for infectious disease and survival. Biometals. 2004;17(2):135–139. doi: 10.1023/b:biom.0000018375.20026.b3. [DOI] [PubMed] [Google Scholar]
- 8.Cardozo C, Alves H, Mascarenhas M, et al. Co-selection of the C64D mutation and HLA-A29 allele: a new paradigm of linkage disequilibrium? Immunogenetics. 2002;53:1002–1008. doi: 10.1007/s00251-001-0414-8. [DOI] [PubMed] [Google Scholar]
- 9.Whitlock E, Garlitz B, Harris E, Beil T, Smith P. Screening for hereditary hemochromatosis: a systematic review for the U.S. Preventative Services Task Force. Ann Intern Med. 2006;145:209–223. doi: 10.7326/0003-4819-145-3-200608010-00009. [DOI] [PubMed] [Google Scholar]
- 10.Merz J, Kriss A, Leonard D, Cho M. Diagnostic testing fails the test. Nature. 2002;415:577–579. doi: 10.1038/415577a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonnell SW, DL, Cogswell M, McIntyre R. Strategies to increase detection of hemochromatosis. Ann Intern Med. 1998;129:987–992. doi: 10.7326/0003-4819-129-11_part_2-199812011-00010. [DOI] [PubMed] [Google Scholar]
- 12.Yen A, Fancher T, Bowlus C. Revisiting hereditary hemochromatosis: current concepts and progress. Am J Med. 2006;119(5):391–399. doi: 10.1016/j.amjmed.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 13.Wood M, Powell L, Ramm G. Environmental and genetic modifiers of the progression to fibrosis and cirrhosis in hemochromatosis. Blood. 2008;111:4456–4462. doi: 10.1182/blood-2007-11-122374. [DOI] [PubMed] [Google Scholar]
- 14.Franchini M, Veneri D. Hereditary hemochromatosis. Hematology. 2005;10(2):145–149. doi: 10.1080/10245330500065771. [DOI] [PubMed] [Google Scholar]
- 15.Park C, Valore E, Waring A, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 16.Krause A, Neitz S, Mägert H. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000 September 1;480(2-3):147–150. doi: 10.1016/s0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 17.ACC Review of HHC/General Adult Population Analytic Validity. Centers for Disease Control [January 12, 2009];2003 http://www.cdc.gov/genomics/gtesting/ACCE/FBR/HH/HHAnaVal_17.htm.
- 18.Adams P, Reboussin D, Press R. Biological variability of transferrin saturation and unsaturated iron binding capacity. Am J Med. 2007;120:999.e991–e997. doi: 10.1016/j.amjmed.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams P, Reboussin D, Eckfeldt J. A comparison of the unsaturated iron binding capacity to transferrin saturation as a screening test to detect C282Y homozygotes for hemochromatosis in 101,168 participants in the HEIRS study. Clin Chem. 2005;51:1048–1052. doi: 10.1373/clinchem.2005.048371. [DOI] [PubMed] [Google Scholar]
- 20.Murtagh L, Whiley M, Wilson S, Tran H, Bassett M. Unsaturated iron binding capacity and transferrin saturation are equally reliable in detection of HFE hemochromatosis. Am J Gastroenterol. 2002;97(8):2093–2099. doi: 10.1111/j.1572-0241.2002.05927.x. [DOI] [PubMed] [Google Scholar]
- 21.Adams P, Kertesz A, McLaren C, Barr R, Bamford A, Chakrabarti S. Population screening for hemochromatosis: a comparison of unbound iron-binding capacity, transferrin saturation, and C282Y genotyping in 5,211 voluntary blood donors. Ann Intern Med. 2000;31(5):1160–1164. doi: 10.1053/he.2000.6984. [DOI] [PubMed] [Google Scholar]
- 22.Powell L, George D, McDonnell S, Kowdley K. Diagnosis of hemochromatosis. Ann Intern Med. 1998;129:925–931. doi: 10.7326/0003-4819-129-11_part_2-199812011-00002. [DOI] [PubMed] [Google Scholar]
- 23.Falize L, Guillygomarch A, Perrin M. Reversibility of hepatic fibrosis in treated genetic hemochromatosis: A study of 36 cases. Hepatology. 2006;44:472–477. doi: 10.1002/hep.21260. [DOI] [PubMed] [Google Scholar]
- 24.Powell L, Dixon J, Ramm G. Screening for hemochromatosis in asymptomatic subjects with or without a family history. Arch Intern Med. 2006;166:294–301. doi: 10.1001/archinte.166.3.294. [DOI] [PubMed] [Google Scholar]
- 25.Anderson R, Wenzel L, Walker A, et al. Impact of hemochromatosis screening in patients with indeterminate results: the hemochromatosis and iron overload screening study. Genet Med. 2006;8(11):681–687. doi: 10.1097/01.gim.0000245631.07117.ac. [DOI] [PubMed] [Google Scholar]
- 26.Feder J, Gnirke A, Thomas W, et al. A novel MHC class-I like gene is mutated in patients with hereditary hemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 27.Nanogen and Bio-Rad sign licensing agreement. Biotech Patent News. 2002 June 1; http://www.encyclopedia.com/doc/1G1-89013216.html.
- 28.Cho M. Effects of gene patents and licenses on clinical genetic testing. Secretary's Advisory Committee on Genetics, Health, and Society. 2008 [Google Scholar]
- 29.Phatak P, Bonkovsky H, Kowdley K. Hereditary hemochromatosis: time for targeted screening. Ann Intern Med. 2008;149(4):270–272. doi: 10.7326/0003-4819-149-4-200808190-00009. [DOI] [PubMed] [Google Scholar]
- 30.Adams P, Valberg L. Screening blood donors for hereditary hemochromatosis: decision analysis model comparing genotyping to phenotyping. Am J Gastroenterol. 1999;94:1593–1600. doi: 10.1111/j.1572-0241.1999.1120_f.x. [DOI] [PubMed] [Google Scholar]
- 31.Waalen J, Nordestgaard B, Beutler E. The penetrance of hereditary hemochromatosis. Best Pract Res Clin Haematol. 2005;18(2):203–220. doi: 10.1016/j.beha.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 32.El-Serag H, Inadomi J, Kowdley K. Screening for hereditary hemochromatosis in siblings and children of affected patients, a cost-effectiveness analysis. Ann Intern Med. 2000;2000(132):261–269. doi: 10.7326/0003-4819-132-4-200002150-00003. [DOI] [PubMed] [Google Scholar]
- 33.Allen KJ, Gurrin LC, Constantine CC. Iron-overload–related disease in HFE hereditary hemochromatosis. N Engl J Med. 2008;358:221–230. doi: 10.1056/NEJMoa073286. [DOI] [PubMed] [Google Scholar]
- 34.Gagne G, Reinharz D, Laflamme N, Adams P, Rousseau F. Hereditary hemochromatosis: effect of mutation penetrance and prevalence on cost-effectiveness of screening modalities. Clin Genet. 2007;71:46–58. doi: 10.1111/j.1399-0004.2006.00727.x. [DOI] [PubMed] [Google Scholar]
- 35.Gordon R. An operational definition of disease prevention. Public Health Reports. 1983;98:107–109. [PMC free article] [PubMed] [Google Scholar]
- 36.Jazwinska EC, Cullen LM, Busfield F, et al. Haemochromatosis and HLA–H. Nat Genet. 1996;14:249–251. doi: 10.1038/ng1196-249. [DOI] [PubMed] [Google Scholar]
- 37.Jouanolle A, Gandon G, Jézéquel P. Hemochromatosis and HLA-H (Letter) Nat Genet. 1996;14:251–252. doi: 10.1038/ng1196-251. [DOI] [PubMed] [Google Scholar]
- 38.Fackelmann K. Rusty origins: researchers identify the gene for iron-overload disease. [January 12, 2009];Science News. 1997 January 18; http://findarticles.com/p/articles/mi_m1200/is_n3_v151/ai_19056180/pg_1?tag=artBody;col1.
- 39.Associated Press Gene found for iron buildup. New York Times. 1996 July 31;:C-7. [Google Scholar]
- 40.Adams P, Reboussin D, Barton J. Hemochromatosis and iron-overload screening in a racially diverse population. N Engl J Med. 2005;352(17):1769–1778. doi: 10.1056/NEJMoa041534. [DOI] [PubMed] [Google Scholar]
- 41.Transcript of a conference call held by Hologic, Inc on June 9, 2008. SEC Accession No 0001193125-08-130599 SEC. 2008 [Google Scholar]
- 42.Capitalizing on the genome (Editorial) Nat Genet. 1996;13(1):1. doi: 10.1038/ng0596-1. [DOI] [PubMed] [Google Scholar]
- 43.Progenitor files registration statements for initial public offering of 2,750,000 shares and for acquisition of Mercator Genetics Inc. Business Wire. 1997 March 14; [Google Scholar]
- 44.American Hemochromatosis Society home page [May 3, 2007]; http://www.americanhs.org.
- 45.Hemochromatosis for health care professionals: genetic testing and basic counseling. [November 12, 2008]; http://www.cdc.gov/ncbddd/hemochromatosis/training/family_detection/testing_and_counseling.htm.
- 46.Hemochromatosis for health care professionals: diagnostic testing. [November 12, 2008]; http://www.cdc.gov/ncbddd/hemochromatosis/training/diagnostic_testing/testing_protocol.htm.
- 47.Allen A. Policing the gene machine. [May 2, 2007];Lingua Franca. 1997 http://linguafranca.mirror.theinfo.org/9703/Policing6.html.
- 48.Hall M, Barton J, Adams P, et al. Genetic screening for iron overload: no evidence of discrimination at one year. J Fam Practice. 2007;56:829–833. [PubMed] [Google Scholar]
- 49.Who should consider testing for hemochromatosis? [January 12, 2009]; http://www.dnadirect.com/web/article/testing-for-genetic-disorders/hemochromatosis/36/who-shouldconsider-testing.
- 50.Shute N. Unraveling your DNA's secret. US News & World Report. 2007:50–54. 57–58. [PubMed] [Google Scholar]
- 51.Ramrakhiani S, Bacon B. Hemochromatosis: advances in molecular genetics and clinical diagnosis. J Clin Gastroenterol. 1998;27(1):41–46. doi: 10.1097/00004836-199807000-00008. [DOI] [PubMed] [Google Scholar]
- 52.Morrison E, Brandhagen D, Phatak P, et al. Serum ferritin level predicts advanced hepatic fibrosis among U.S. patients with phenotypic hemochromatosis. Ann Intern Med. 2003;138(8):627–633. doi: 10.7326/0003-4819-138-8-200304150-00008. [DOI] [PubMed] [Google Scholar]


