Abstract
Background
V. cholerae is the etiological agent of cholera, a major public health concern in most developing countries. Virulence of V. cholerae relies on the powerful cholera toxin, encoded by the CTX prophage. The emergence of new pathogenic variants in the recent years has been mostly associated with new CTX prophage rearrangements.
Results
In this retrospective study, we show that the epidemic V. cholerae O1 El Tor strain responsible for the 2006 outbreak in Angola is clonally and genetically different from El Tor strains circulating in the 1990s in the same area. Strains from 2006 carry ICEVchAng3 of the SXT/R391 family. This ICE is associated with a narrower multidrug resistance profile compared to the one conferred by plasmid p3iANG to strains of the 1990s. The CTX prophage carried by 2006 El Tor strains is characterized by rstRET and ctxBCla alleles organized in a RS1-RS2-Core array on chromosome I. Interestingly, the newly emerging atypical strain belongs to a clade previously known to comprise only clinical isolates from the Indian subcontinent that also contain the same ICE of the SXT/R391 family.
Conclusions
Our findings remark the appearance of a novel V. cholerae epidemic variant in Africa with a new CTXΦ arrangement previously described only in the Indian Subcontinent.
Background
Vibrio cholerae is the etiological agent of the severe watery diarrhoeal disease known as cholera, a major public health concern in most developing countries.
More than 200 serogroups have been described on the basis of different somatic O antigens [1], but only serogroups O1 and O139 have the ability to cause harsh epidemics. Serogroup O1 is further divided into two main biotypes, Classical and the 7th pandemic El Tor. Beside their phenotypic characteristics, differences in specific genetic markers, such as toxin structure, confer distinct features to these biotypes.
Pathogenic V. cholerae strains carry the genes encoding the cholera toxin (CT) on the CTXΦ prophage. Different CTXΦ arrangements have been described within the O1 serogroup [2]. These arrangements depend on the genotype of the CT gene ctxB and on the organization and chromosomal location of several gene clusters of phage origin, namely the core, RS2, and RS1 [2]. Although the Classical biotype is considered extinct, new El Tor strains holding the Classical ctxB allele, generically labeled as atypical El Tor (including hybrid El Tor, altered El Tor and Mozambique variants) [2], were identified from 1993 to date mostly in Asia [3-8] with few cases in Africa [5,9,10]. The atypical variants are characterized by a new CTXΦ arrangement, holding El Tor and/or Classical alleles of rstR and ctxB genes [2]. As a consequence of these genetic arrangements in CTX prophage, toxigenic V. cholerae O1 El Tor strains have changed in the last 20 years. Initially, atypical variants were only sporadically identified in the Indian Subcontinent along with prototype El Tor. However they are now in the process of replacing it worldwide [2].
Prototype El Tor strains often contain multi-resistant conjugative plasmids [11] whereas O139 and atypical O1 El Tor V. cholerae epidemic strains usually harbor Integrative Conjugative Elements (ICEs) of the SXT/R391 family [12].
SXT/R391 ICEs are self-transmissible mobile elements, ranging in size from 79 to 108 kb, able to integrate into the host bacterial chromosome and to transfer by conjugation. They are recognized for their important role in bacterial genome plasticity [13] and as vectors of antibiotic resistance and alternative metabolic pathways [12]. The name of the SXT/R391 family originates from elements SXTMO10 and R391, respectively discovered in clinical strains of Vibrio cholerae in India [14] and Providencia rettgeri in South Africa [15]. The two elements are associated with different multi-resistance profiles: chloramphenicol, streptomycin, sulfamethoxazole, and trimethoprim for SXTMO10, and kanamycin, and mercury for R391 [12]. They share a highly conserved genetic backbone encoding their integration/excision, conjugative transfer, and regulation, but also contain variable DNA found in five insertion sites of the backbone [12]. Each ICE of the family holds specific genes scattered in the conserved sequence that code for resistance to antibiotics and heavy metals, new toxin/antitoxin systems, restriction/modification systems, and alternative metabolic pathways [12]. To date more than 50 ICEs have been identified and grouped within the SXT/R391 family, most of them discovered in V. cholerae strains.
To date, only a few SXT-related ICEs were identified in Africa, most of them through the characterization of the integrase intSXT. Only ICEVchMoz10 from Mozambique (2004) has been completely sequenced and annotated [12]. This ICE has no close relative in Africa except its sibling ICEVchBan9 isolated in Bangladesh (1994), suggesting the possible spread of SXT-related ICEs between the two continents in recent times. Although the use of horizontally-transferred elements as genetic markers for strain discrimination might appear risky, we recently showed the existence of an ICE/strain association in epidemic V. cholerae strains circulating in the Indian Subcontinent [16]. The association between ICE and V. cholerae reflects the classification proposed by Chun and colleagues to describe homologous intraspecific groups of V. cholerae based on the whole genome alignment of 23 strains isolated over the past 100 years [17].
In this retrospective study, we analysed V. cholerae O1 clinical strains isolated in Luanda (Angola) in 2006. Angola is an endemic area for cholera and was subjected to two major epidemic events in the past three decades. The first outbreak (1987-1993) [18] was followed by a thirteen year remission phase until cholera reemerged in 2006 in one of the most severe epidemic outbreaks of the last decade, counting about 240.000 cases [19].
Here we demonstrate that the V. cholerae O1 El Tor strain responsible for the 2006 Angolan outbreak is an atypical O1 El Tor variant previously detected only in Asia [3]. This variant is significantly different from those isolated during previous cholera outbreaks in the 1990s in the same geographic area. Indeed, it holds a peculiar CTXΦ array and the SXT-like element ICEVchAng3. Ribotype analysis suggests that this strain might have spread to West Africa from the Indian Subcontinent.
Methods
Bacterial strains, susceptibility tests and transfer of drug resistances
We analyzed V. cholerae strains isolated in Angola or India between 1992 and 2006 (Table 1). All strains were isolated from stool samples and/or rectal swabs from patients, and after isolation on thiosulfate citrate bile sucrose agar and biochemical identification, bacterial strains were routinely grown in Luria-Bertani (LB) or agar plates at 37°C and maintained at -80°in LB broth containing 30% (vol/vol) glycerol.
Table 1.
V. cholerae O1 strains analyzed in this study
| Isolation | ||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Place | Year | Antibiotic resistance profile | Antibiotic resistance genes | ICE content | CTXΦ array | Ribotype | Reference |
| VC175 | Angola (Luanda) | 2006 | Ap, Pn, Sm, Su, Tp | floR, strA, strB, dfrA1, sulIIb | ICEVchAng3 | B | R1 | This study |
| VC189 | Angola (Luanda) | 2006 | Ap, Pn, Sm, Su, Tp | floR, strA, strB, dfrA1, sulIIb | ICEVchAng3 | B | R1 | This study |
| VC582 | Angola (Luanda) | 1992 | Ap, Cm, Kn, Pn, Sm, Sp, Su, Tc, Tpa | aph, tetG, cat1, blaP1, dfrA15, aadA8, sul2c | - | A | R2 | [11] |
| VC1383 | Angola (Benguela) | 1994 | Ap, Cm, Kn, Pn, Sm, Sp, Su, Tc, Tpa | aph, tetG, cat1, blaP1, dfrA15, aadA8, sul2c | - | A | R3 | [11] |
| VC547 | Angola (Bengo river) | 1994 | Ap, Cm, Kn, Pn, Sm, Sp, Su, Tc, Tpa | aph, tetG, cat1, blaP1, dfrA15, aadA8, sul2c | - | A | R4 | [11] |
| VC7452 | India (Sevagram) | 1995 | Ap, Nx, Pn, Sm, Sp, Su, Tp | floR, strA, strB, dfrA1, sulIIb | ICEVchInd5d | B | R1 | [16] |
| VC15699 | India (Sevagram) | 1999 | Ap, Nx, Pn, Sm, Sp, Su, Tp | floR, strA, strB, dfrA1, sulIIb | ICEVchInd5 | B | R1 | [16] |
| VC9258 | India (Sevagram) | 1999 | Ap, Nx, Pn, Sm, Sp, Su, Tp | floR, strA, strB, dfrA1, sulIIb | ICEVchInd5 | B | R1 | [16] |
aResistance profile conferred by conjugative plasmid p3iANG [11]; blocated on the ICE; clocated on p3iANG; dICE fully sequenced. Abbreviations: Ap, ampicillin; Cm, chloramphenicol; Kn, kanamycin; Nx, nalidixic acid; Pn, penicillin; Sm, streptomycin; Sp, spectinomycin; Su, sulfamethoxazole; Tc, tetracycline; Tp, trimethoprim.
Antimicrobial susceptibility was tested at the following concentrations: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 20 μg/ml; kanamycin (Km), 50 μg/ml; nalidixic acid (Nx), 40 μg/ml; penicillin (Pn), 20 μg/ml; rifampin (Rf), 100 μg/ml; spectinomycin (Sp), 50 μg/ml; streptomycin (Sm), 50 μg/ml; sulfamethoxazole (Su), 160 μg/ml; tetracycline (Tc), 12 μg/ml; and trimethoprim (Tm), 32 μg/ml. Antibiotic concentrations were defined according to their MIC breakpoints as previously described [18,20] and were included in ISO sensitest (Oxoid) agar plates. Bacterial strains were spotted onto the plates as previously described [11].
Conjugation assays were used to transfer ICEVchAng3 from V. cholerae into rifampin-resistant derivatives of E. coli 803 strain. Mating assays were performed by mixing equal volumes of overnight cultures of donor and recipient strains. Briefly, the cells were harvested by centrifugation and resuspended in a 1/20 volume of LB broth. Cell suspensions were poured onto LB agar plates and incubated at 37°C for 6 h. The cells were then resuspended in 1 ml of LB medium, and serial dilutions were plated onto appropriate selective media to determine the numbers of donors, recipients, and exconjugants. Frequency of transfer was expressed as the number of exconjugant cells per donor cells in the mating mixture at the time of plating.
V. cholerae O139 MO10 [14], V. cholerae E4:ICEVchInd1 [21], V. cholerae O1 VC20 [22], V. cholerae N16961 [23], V. cholerae O1 CO840 [22], V. cholerae O1 VC7452, VC15699, and VC9258 isolated in India (Maharashtra) [16], and E. coli AB1157:R391 [24] were appropriately used as negative or positive controls for class 1 integrons, ICE, tcpA, and rstR detection, CTXΦ array and ribotype analysis.
Molecular biology procedures
Bacterial DNA for PCR analysis was prepared with a Wizard Genomic DNA Purification kit (Promega). Amplicons to be sequenced were directly purified from PCR or extracted from agarose gel by Wizard SV Gel and PCR Clean-up System (Promega) according to the manufacturer's instructions. DNA sequences were determined by BMR Genomics (Padova, Italy).
Class 1 integron detection was performed by PCR amplification with specific primer pairs as previously described [11]. ICEs of the SXT/R391 family were screened by PCR analysis, using 17 specific primer pairs previously described by our group [25,26]. intSXT, prfC/SXTMO10 right junction, floR, strA, strB, sul2, dfrA18, dfrA1, rumAB operon, traI, traC, setR, and Hotspots or Variable Regions s026/traI, s043/traL, traA/s054, s073/traF and traG/eex were screened. A second set of 15 primer pairs designed on the specific sequences of ICEVchInd5 [16] were used to detect ICEVchInd5 and ICEVchAng3 specific Hotspots and Variable Regions.
All PCR reactions were set in a 50-μl volume of reaction buffer containing 1 U of Taq polymerase as directed by the manufacturer (Promega).
Ribotype analysis
Ribotyping of V. cholerae strains was performed by BglI restriction of chromosomal DNA with fluorescent-labeled 16S and 23S DNA (Gene Images 3540 RPn3510, Amersham) generated by reverse transcriptase polymerase chain reaction of ribosomal RNAs, as already described [25].
CTX array analysis and ctxB, tcpA, rstR biotype characterization
The structure of CTX array was determined by multiple PCR analysis (Table 2) and by Southern Blot hybridization. The genetic structure of the two CTX prophage arrays described in Figure 1 was determined using the primers described in Table 2. Briefly, combination of primers tlcF/rstAR, tlcF/rstCR, rstCF/rstAR, ctxAF/rstAR, rstCF/rtxR and ctxAF/rtxR were used to detect the presence of CTXΦ on chromosome 1 and to determine the position of the RS1 element (see Additional file 1 Table S1 for complete amplicon profiles). The absence of CTXΦ or RS1 on chromosome 2 was established using primers chr2F/chr2R. Primers ctxAF/cepR were used to determine the presence of CTX tandem arrays.
Table 2.
Primers used to determine CTX prophage array structure
| Primer | Nucleotide sequence (5' to 3') | Position (GenBank Accession no. NC_002505-6) |
|---|---|---|
| tlcF | CCAAAACAACAGAAGCAACAGAGCAACG | 1574460-1574487 |
| rstCF | GGCGCTTATACAGACGAAATCGCTC | 1564180-1564201 |
| rstCR | AGCGCCTGAACGCAGATATAAA | 1564290-1564311 |
| rstAR | CGACAAAAACAAACGGAGAAGCGT | 1572748-1572771 |
| ctxAF | CTCAGACGGGATTTGTTAGGCACG | 1567895-1567918 |
| rtxR | CAAGCTGCGATCAGCATGGCGTGGTC | 1563652-1563671 |
| cepR | CAGTGTTTTGGTGACTTCCGT | 1571101-1571121 |
| chr2F | CTCACGCTGAACAGCAAGTC | 507564-507583 |
| chr2R | AAACCGGGAGAAGTGATTGC | 509487-509506 |
Figure 1.
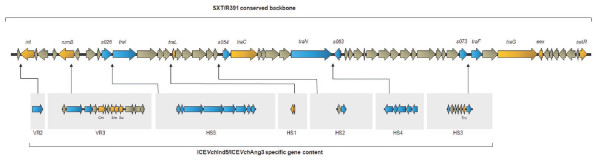
ICEVchAng3 genetic structure. Schematic linear representation (adapted from Wozniak et al., 2009) of the genes amplified by PCR to define the molecular structure of ICEVchAng3. The upper line represents the conserved backbone of the SXT/R391 family members. The black arrows indicate insertion sites for ICEVchInd5/ICEVchAng3 specific gene content. Genes in orange were tested with primer set A. Genes in blue were tested with primer set B. Genes not tested are shown in grey. VR: Variable Region; HS: Hotspot. GenBank accession no. of the full sequence of ICEVchInd5 is GQ463142[12].
Three previously described primer sets were used to detect: (i) Classical, El Tor, or Kolkata type rstR gene [27], (ii) ctxB genotype sequencing [28], (iii) and Classical or El Tor biotypes for tcpA [29].
PCR results on organization and location of CTXΦ on chromosome 1 were further confirmed by Southern Blot hybridization assays. DNA probes were produced by PCR using the chromosomal DNA of V. cholerae strain N16961 as template: ctxA gene (564 bp) with primers CTX-2 (CGGGCAGATTCTAGACCTCCTG) and CTX-3 (CGATGATCTTGGAGCATTCCCAC); rstA gene (789 bp) with primers rstA1F (AAACCTGCAAAATACCCCT) and rstA1R (ACAACTCGATACAAACGCT). Probes for hybridization were labeled with alkaline phosphatase with AlkPhos Direct™ Labelling and Detection System with CDP-Star™ kit (Ge Healthcare), according to manufacturer's instructions.
Strains were cultured in Luria-Bertani medium and 1 ml of culture was used to extract and purify the genomic DNA using the DNeasy Blood & Tissue Kit (Qiagen). Aliquots of the extracted DNA (1,5 μg) were digested with EcoRV for CTXΦ element restriction fragment length polymorphism analysis. The digested fragments were separated by agarose gel electrophoresis (1% gel) and were blotted on nitrocellulose membranes using standard methods [30]. Southern blots were hybridized O/N with ctxA or rstA labeled probes, and washed under stringent conditions, according to manufacturer's instructions. Addition of CDP-Star Detection Reagent was followed by 10 min incubation, and autoradiography (20 min to 1 h) was performed to generate a signal.
Nucleotide sequence accession numbers
Nucleotide sequences of ctxB genes were deposited in GenBank under accession no. HQ599507 (V. cholerae 1383), HQ599508 (V. cholerae 7452), HQ599509 (V. cholerae 547), HQ599510 (V. cholerae 582), and HQ599511 (V. cholerae 175).
Results
V. cholerae strains from 2006 show reduced resistance profile compared to previous epidemic strains
We analyzed two V. cholerae O1 El Tor clinical strains, VC175 and VC189 (Table 1), isolated at the Luanda Central Hospital (Angola). These strains were collected during the peak (May) of the cholera outbreak reported in Angola in 2006.
The two strains were sensitive to tetracycline, chloramphenicol, and kanamycin but showed a multiresistant profile to ampicillin, penicillin, streptomycin, trimethoprim, and sulfamethoxazole (see Table 1 for complete phenotype and genotype). Despite this significant multidrug resistance, these strains showed a narrower resistance profile compared to those isolated in the previous 1987-1993 cholera epidemic, which were also resistant to tetracycline, chloramphenicol, spectinomycin and kanamycin [11]. We found no evidence for the presence of conjugative plasmids or class 1 integrons in the 2006 strains analyzed (data not shown), which might explain their reduced drug resistance profile. Indeed, strains from 1987-1993 were associated with the conjugative plasmid p3iANG that holds genes encoding the resistance to tetracycline, chloramphenicol, kanamycin, and spectinomycin [11].
ICEVchAng3 is a sibling of ICEVchInd5
We assessed the presence of SXT/R391 family ICEs since they are a major cause of antibiotic resistance spread among V. cholerae strains. Both strains were intSXT+, were shown to contain an ICE integrated into the prfC gene, and contained the conserved genes traI, traC and setR, respectively encoding a putative relaxase, a putative conjugation coupling protein, and a transcriptional repressor found in all SXT/R391 family members [31]. Based on these results we included this ICE in the SXT/R391 family and named it ICEVchAng3 according to the accepted nomenclature [32].
SXT/R391 ICEs exhibit significant genetic polymorphisms in hotspot content [12]. We used a first set of primers (primer set A), designed to discriminate between SXTMO10 and R391 specific sequences [25], in order to prove the identity of the ICE circulating in the 2006 Angolan strains. Genes floR, strA, strB, sul2, dfrA18, dfrA1, the rumAB operon, and Hotspots or Variable Regions s026/traI, s043/traL, traA/s054, s073/traF and traG/eex were screened.
The 2006 strains exhibited the same SXTMO10/R391 hybrid ICE pattern. Intergenic regions traG/eex (Variable Region 4) and traA/s054 (Hotspot 2) showed the molecular arrangement described in SXTMO10, whereas region s043/traL (Hotspot 1) was organized as in R391. Variable Region 3, inserted into the rumB locus, contained genes that mediate resistance to chloramphenicol, streptomycin and sulfamethoxazole: floR, strA, strB, sul2. Interestingly, ICEVchAng3 lacks dfr18, the gene conferring resistance to trimethoprim found in SXTMO10, and carries instead dfrA1 in Hotspot 3. This preliminary analysis revealed that ICEVchAng3 exhibits a hybrid genetic content similar to that of the completely sequenced ICEVchInd5, the most widespread ICE circulating in V. cholerae El Tor O1 strains in the Indian Subcontinent [16].
Given these similarities we analyzed ICEVchAng3 using a second set of primers (primer set B) previously designed to assess the hotspot content of ICEVchInd5 [16]. This analysis confirmed that all the peculiar insertions found in ICEVchInd5 were also present in ICEVchAng3: (i) a gene encoding a protein similar to the E. coli dam-directed mismatch repair protein MutL (Variable Region 2); (ii) intI9 integron (Hotspot 3); (iii) a possible transposon of the IS21 family (Hotspot 4); and (iv) a 14.8-kb hypothetical operon of unknown function (Hotspot 5). On account of our results and of the common backbone shared by SXT/R391 ICEs (~65% of the ICE), we are confident that ICEVchAng3 is a sibling of ICEVchInd5 [16]. A map (not to scale) of ICEVchAng3 is shown in Figure 1.
We performed mating experiments to assess the ability of ICEVchAng3 to transfer by conjugation between V. cholerae strain VC 175 or VC 189 and E. coli 803Rif. The frequency of transfer of ICEVchAng3 was 4,4 X 10-5, a frequency of transfer similar to that of most of the ICEs of this family. Ten E. coli exconjugant colonies were tested and proved to be positive for the presence of intSXT, confirming the mobilization of ICEVchAng3.
A new CTXΦ array in Africa
The variability of CTXΦ and the emergence of atypical El Tor variants in the ongoing 7th pandemic [2] les us to analyze the organization of CTXΦ arrays and the presence of different alleles of ctxB, rstR and tcpA genes. The genetic structure of CTX prophage in the genome of the Angolan isolates from both epidemic events was determined by multiple PCR analysis, hybridization, and sequencing, when required.
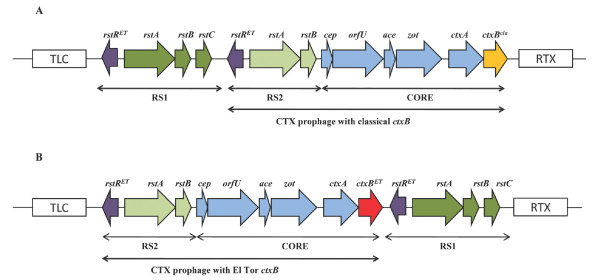
Combining the results obtained by multiple PCR analysis and hybridization we were able to show that the strains analyzed contained two distinct CTXΦ arrays (A and B), both of which were found integrated in the large chromosome (Figure 2, Additional file 1 Table S1). These strains also proved to be negative for any CTXΦ integration on the small chromosome and devoid of CTX tandem arrays as detected by primer pairs chr2F/chr2R and ctxAF/cepR, respectively. The Angolan strains isolated in 2006 (VC 175 and VC 189) belonged to profile A, in which the RS1 element is followed by CTXΦ, both being located between the toxin-linked cryptic (TLC) element and the chromosomal RTX (repeat in toxin) gene cluster (Figure 2a). In contrast, strains from the first outbreak (1987-1993) contained CTXΦ followed by the RS1 element (profile B) (Figure 2b). Both CTXΦ arrays were characterized by El Tor type rstR genes (both in RS1 and RS2) but showed a noteworthy difference in their ctxB genotype (Table 3). CTXΦ arrays belonging to profile A contained a histidine and a threonine at the 39th and 68th amino acid positions, respectively, which are representative of Classical genotype 1 CtxB. The CTXΦ arrays belonging to profile B held a tyrosine, a phenylalanine and an isoleucine at positions 39th, 46th and 68th, respectively, typical of an El Tor genotype 3 CtxB.
Figure 2.
Comparison of the genetic structures of the two CTX prophage arrays identified in the V. cholerae strains under study. Both prophages are integrated into the large chromosome. Arrows indicate the transcription direction of each gene. (A) CTX prophage array profile A: RS1-RS2-CORE; (B) CTX prophage array profile B: RS2-CORE-RS1. Map is not to scale. rstRET (purple arrow): El Tor type rtsR; ctxBET (red arrow): El Tor type ctxB; ctxBcla (yellow arrow): Classical type ctxB; TLC: toxin-linked cryptic plasmid; RTX: RTX (repeat in toxin) gene cluster.
Table 3.
Biotype characterization and ctxB genotype comparison of V. cholerae O1 isolates from Angola and India
| Strain | rstR | tcpA | ctxB | |
|---|---|---|---|---|
| Genotypea | Amino acid positionb | |||
| VC582 | ET | ET | 3 (ET) | 20 (His); 24 (Gln); 28 (Asp); 34 (His); 39 (Tyr); 46 (Phe); 55 (Lys); 68 (Ile) |
| VC547 | ET | ET | 3 (ET) | 20 (His); 24 (Gln); 28 (Asp); 34 (His); 39 (Tyr); 46 (Phe); 55 (Lys); 68 (Ile) |
| VC1383 | ET | ET | 3 (ET) | 20 (His); 24 (Gln); 28 (Asp); 34 (His); 39 (Tyr); 46 (Phe); 55 (Lys); 68 (Ile) |
| VC175 | ET | ET | 1 (Cla) | 20 (His); 24 (Gln); 28 (Asp); 34 (His); 39 (His); 46 (Phe); 55 (Lys); 68 (Thr) |
| VC7452 | ET | ET | 1 (Cla) | 20 (His); 24 (Gln); 28 (Asp); 34 (His); 39 (His); 46 (Phe); 55 (Lys); 68 (Thr) |
Cla, Classical type; ET, El Tor type; aAccording to ctxB genotyping by Safa et al., 2010 [2]; bNucleotide position +1 corresponds to the A of the ATG start codon in ctxB.
Angolan and Indian strains share the same clonal origin
In order to verify their clonal relationship, we analysed by ribotyping the strains from the two Angolan epidemics of the 1990s and of 2006, as well as the Indian strains collected from 1993 to 2005 (Table 1) [16]. Strains from 1987-1993 outbreak (VC582, VC1383 and VC547) were chosen according to their epidemiological role (clinical or environmental isolate) and the presence of plasmid p3iANG [11].
Angolan strains isolated between 1992 and 1994 showed an assorted ribotype profile: clinical strains VC582 and VC1383 were characterized by profiles R2 (2.3,4.2, 4.6, 5.7, 6.0 kb) and R3 (2.3,4.2, 4.6, 5.7, 6.0, 9.6, 18.0 kb), respectively, and environmental isolate VC547 by a third completely different profile R4 (1.0, 1.4, 1.6, 1.8, 2.0, 2.2, 2.4, 3.8, 5.5 kb). This heterogeneity is not surprising if we consider the Angolan clinical strains on a larger sample scale. Indeed, our data showed that there was a clonal shift in Angola from 1992 to 1993/1994 with consequent change of ribotype (D.C personal communication) that can explain the discrepancies observed here. Strains VC175 and VC189 isolated in 2006 were characterized by the same ribotype profile R1 (2.3, 4.2, 5.8, 6.1, 6.3, 8.5, 9.4, 10.8, 22.0 kb) which corresponds to the ribotype profile of the Indian strains carrying ICEVchInd5 [16], suggesting a common clonal origin.
Discussion
2006 was a crucial year for cholera worldwide. The number of reported cases was higher than ever and exceeded the levels of the late 1990s. Major outbreaks affected some of the largest African countries, including Angola, which reported to WHO one of the most exceptional epidemics experienced in Africa in the last decade [19].
This is the first study on the causative agent of this dramatic outbreak and our analysis revealed significant differences between the Angolan strains of 2006 and those isolated in the previous 1987-1993 cholera epidemic. The 1987-1993 epidemic was the longest in Angolan history and the V. cholerae epidemic strains were characterized by the presence of the conjugative plasmid p3iANG that carries three class 1 integrons [11]. Interestingly, the strains from the 2006 outbreak lack p3iANG but harbor an SXT-like ICE sibling of ICEVchInd5, previously described only in Asian V. cholerae strains [16]. The gene content of ICEVchAng3 comprises elements shared with SXTMO10, R391, ICEVchBan9, and ICEPdaSpa1, alongside some unique insertions of unknown function that might provide the strain with increased fitness. In light of its genetic content we included ICEVchAng3 in the subgroup of SXT/R391 ICEs that characterizes V. cholerae O1 El Tor strains circulating in several epidemic areas of the Indian Subcontinent, of which ICEVchInd5 is the reference ICE [12,16].
Beside the analysis of the Mozambican variant, extensive studies of CTXΦ arrangements in V. cholerae strains isolated in Africa lack so far.
Our analysis reports that the strains of the 2006 outbreak contain an RS1-CTX array on the large chromosome with a classical ctxB allele, which classifies them as V. cholerae O1 altered El Tor. This variant was responsible for major epidemics in India in 2004-2006 [3] and in Vietnam in 2007 [8]. It is considered as prevalent in Asia nowadays [33,34] and forms a monophyletic group with other variants of the 7th pandemic clade [17]. This variant arose in the Indian Subcontinent at the beginning of the 90s and slowly diffused to Asian countries [6,7]. The possible spread to Africa was only suggested [3,33] and some authors gave partial evidences supporting this hypothesis by strain ribotyping [22] or ctxB genotyping [5]. With this work we ascertain the presence of this atypical El Tor variant in Africa and demonstrate it holds the responsibility for the 2006 cholera epidemic in Angola.
The Angolan variant is the second example of atypical El Tor variant described in Austral Africa, the first being the Mozambican strain B33 [9]. However, this variant is different from the Angolan one, since it holds a tandem CTXΦ array on the small chromosome [33], contains a different ICE (ICEVchMoz10) [12], and is closely related to the Bangladeshi strain MJ-1236 [7,17].
Unlike B33 whose progenitor was identified as a Kolkata hybrid strain from 1992 [35], we have no clear information on how the variant we found in Angola penetrated Austral Africa. We can speculate that it arrived from the Indian Subcontinent through the same Sub-Saharan corridor used by cholera to enter Africa at the beginning of the 7th pandemic [36]. During the '70s it spread from the Horn of Africa to Senegal, Guinea Bissau and eventually arrived in Angola: the new atypical variant might have disseminated by a similar route. This supposition might find some confirmation in the analysis performed by Sharma and colleagues who proposed the spread of a distinct V. cholerae O1 strain from India to Guinea Bissau, where it was associated with an epidemic of cholera in 1994 [22]. This hypothesis was based on the ribotype analysis of pre- and post- O139 V. cholerae O1 strains circulating in both countries. Our ribotype analysis confirmed these data since the Angolan strain from 2006, the clinical strains isolated in Guinea Bissau in 1994/1995 [37], and clinical post-O139 V. cholerae O1 strains from India [22] share the same profile, suggesting a common clonal origin. Unfortunately, the genetic content of the strains isolated in Guinea Bissau, in terms of ICE structure and CTXΦ array, was never investigated and our speculations cannot go any further.
Whichever route of dissemination used by the new variant to spread from the Indian Subcontinent to Africa, many evidences indicate that atypical V. cholerae strains are in the process of globally replacing the prototype El Tor strains, as observed in Angola.
Conclusions
Cholera remains a global threat to public health and the recent outbreak in Haiti is a distressing example of this situation [38]. In 2006, Angola, which had reported no cholera cases since 1998, was affected by a major outbreak due to an atypical V. cholerae O1 El Tor strain that was analyzed for the first time in our study. This altered El Tor strain holds an RS1-CTX array on the large chromosome and a Classical ctxB allele and likely replaced the previous prototype O1 El Tor strain reported till 1994. The success of the new variant might depend on the combination of the respective predominant features of the El Tor and Classical biotypes: a better survival in the environment [2] and the expression of a more virulent toxin [39].
Authors' contributions
The project was conceived and designed by DC, PC and MMC. All experiments were performed by DC and MS with the help of DB (ribotyping). The paper was written by DC, MS, PC, and MMC. All authors discussed the results, read and approved the final manuscript.
Supplementary Material
Table S1. Amplicon profiles obtained for CTXΦ array A and B. We designed new primer pairs able to discriminate between the different CTXΦ array on the chromosome of V. cholerae. In this table we present the region amplified by each primer pair and the two different arrays obtained for the strains under analysis.
Contributor Information
Daniela Ceccarelli, Email: daniela.ceccarelli@uniroma1.it.
Matteo Spagnoletti, Email: matteo.spagnoletti@uniroma1.it.
Donatella Bacciu, Email: dbacciu@uniss.it.
Piero Cappuccinelli, Email: pcapp@uniss.it.
Mauro M Colombo, Email: mauro.colombo@uniroma1.it.
Acknowledgements
We are grateful to Dr. M. Francisco (Dept. of Microbiology, Faculty of Medicine, University A. Neto, Luanda - Angola) for providing us with Angolan V. cholerae strains from 2006, and to A. Crupi for technical assistance. We are grateful to G. Garriss for manuscript revision.
This work was supported by Ministero Istruzione Università e Ricerca (MIUR) (Grant n. 2007W52X9M to MMC and PC), and Ministero Affari Esteri - Direzione Generale Cooperazione Sviluppo (MAE-DGCS) (Grant n. AID89491 to MMC), Italy. DC was supported by a fellowship from Institute Pasteur - Fondazione Cenci Bolognetti, Italy. MS is the recipient of a PhD fellowship from the Doctorate School in Cellular and Developmental Biology, Sapienza Università di Roma.
References
- Kaper JB, Glenn J, Morris JR, Levine MM. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safa A, Nair GB, Kong RYC. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 2010;18:46–54. doi: 10.1016/j.tim.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Lee JH, Choi SY, Jeon YS, Lee HR, Kim EJ, Nguyen BM, Hien NT, Ansaruzzaman M, Islam MS, Bhuiyan NA, Niyogi SK, Sarkar BL, Nair GB, Kim DS, Lopez AL, Czerkinsky C, Clemens JD, Chun J, Kim DW. Classification of hybrid and altered Vibrio cholerae strains by CTX prophage and RS1 element structure. J Microbiol. 2009;47:783–788. doi: 10.1007/s12275-009-0292-6. [DOI] [PubMed] [Google Scholar]
- Ang GY, Yu CY, Balqis K, Elina HT, Azura H, Hani MH, Yean CY. Molecular evidence of cholera outbreak caused by a toxigenic Vibrio cholerae O1 El Tor variant strain in Kelantan, Malaysia. J Clin Microbiol. 2010. pp. 01086–01010. JCM. [DOI] [PMC free article] [PubMed]
- Safa A, Sultana J, Cam PD, Mwansa JC, Kong RYC. Vibrio cholerae O1 Hybrid El Tor Strains, Asia and Africa. Emerg Infect Dis. 2008;14:987–988. doi: 10.3201/eid1406.080129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair G, Qadri F, Holmgren J, Svennerholm A, Safa A, Bhuiyan N, Ahmad Q, Faruque S, Faruque A, Takeda Y, Sack D. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol. 2006;44:4211–4213. doi: 10.1128/JCM.01304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair GB, Faruque SM, Bhuiyan NA, Kamruzzaman M, Siddique AK, Sack DA. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J Clin Microbiol. 2002;40:3296–3299. doi: 10.1128/JCM.40.9.3296-3299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BM, Lee JH, Cuong NT, Choi SY, Hien NT, Anh DD, Lee HR, Ansaruzzaman M, Endtz HP, Chun J, Lopez AL, Czerkinsky C, Clemens JD, Kim DW. Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J Clin Microbiol. 2009;47:1568–1571. doi: 10.1128/JCM.02040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansaruzzaman M, Bhuiyan N, Nair B, Sack D, Lucas M, Deen J, Ampuero J, Chaignat C. Group MCvDPC. Cholera in Mozambique, variant of Vibrio cholerae. Emerg Infect Dis. 2004;10:2057–2059. doi: 10.3201/eid1011.040682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilici M-L, Massenet D, Gake B, Bwalki B, Olson DM. Vibrio cholerae O1 variant with reduced susceptibility to ciprofloxacin, Western Africa. Emerg Infect Dis. 2010;16:1804–1805. doi: 10.3201/eid1611.100568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli D, Salvia AM, Sami J, Cappuccinelli P, Colombo MM. New cluster of plasmid-located class 1 integrons in Vibrio cholerae O1 and a dfrA15 cassette-containing integron in Vibrio parahaemolyticus isolated in Angola. Antimicrob Agents Chemother. 2006;50:2493–2499. doi: 10.1128/AAC.01310-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RAF, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Déry C, Burrus V, Waldor MK. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 2009;5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith H, Croucher NJ. Genome watch: breaking the ICE. Nat Rev Microbiol. 2009;7:328–329. doi: 10.1038/nrmicro2137. [DOI] [PubMed] [Google Scholar]
- Waldor MK, Tschape H, Mekalanos JJ. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters SE, Hobman JL, Strike P, Ritchie DA. Novel mercury resistance determinants carried by IncJ plasmids pMERPH and R391. Mol Gen Genet. 1991;228:294–299. doi: 10.1007/BF00282479. [DOI] [PubMed] [Google Scholar]
- Ceccarelli D, Spagnoletti M, Bacciu D, Danin-Poleg Y, Mendiratta D, Kashi Y, Cappuccinelli P, Burrus V, Colombo MM. ICEVchInd5 is prevalent in epidemic Vibrio cholerae O1 El Tor strains isolated in India. Int J Med Microbiol. 2011;301:318–324. doi: 10.1016/j.ijmm.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Taviani E, Jeon Y, Kim DW, Lee J, Brettin TS, Bruce DC, Challacombe JF, Detter JC, Han CS, Munk AC, Chertkov O, Meincke L, Saunders E, Walters RA, Huq A, Nair GB, Colwell RR. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA. 2009;106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo MM, Mastrandrea S, Leite F, Santona A, Uzzau S, Rappelli P, Pisano M, Rubino S, Cappuccinelli P. Tracking of clinical and environmental Vibrio cholerae O1 strains by combined analysis of the presence of toxin cassette, plasmid content and ERIC PCR. FEMS Immunol Med Microbiol. 1997;19:33–45. doi: 10.1111/j.1574-695X.1997.tb01070.x. [DOI] [PubMed] [Google Scholar]
- WHO. Cholera 2006. Wkly Epidemiol Rec. 2007;31:273–284. [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; seventeenth informational supplement. CLSI document M100-S17. Wayne, Pennsylvania, USA; 2007. [Google Scholar]
- Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother. 2001;45:2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C, Ghosh A, Dalsgaard A, Forslund A, Ghosh RK, Bhattacharya SK, Nair GB. Molecular evidence that a distinct Vibrio cholerae O1 biotype El Tor strain in Calcutta may have spread to the African continent. J Clin Microbiol. 1998;36:843–844. doi: 10.1128/jcm.36.3.843-844.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath BM, O'Halloran JA, Piterina AV, Pembroke JT. Molecular tools to detect the IncJ elements: a family of integrating, antibiotic resistant mobile genetic elements. J Microbiol Methods. 2006;66:32–42. doi: 10.1016/j.mimet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Bani S, Mastromarino PN, Ceccarelli D, Van AL, Salvia AM, Viet QTN, Hai DH, Bacciu D, Cappuccinelli P, Colombo MM. Molecular characterization of ICEVchVie0 and its disappearance in Vibrio cholerae O1 strains isolated in 2003 in Vietnam. FEMS Microbiol Lett. 2007;266:42–48. doi: 10.1111/j.1574-6968.2006.00518.x. [DOI] [PubMed] [Google Scholar]
- Taviani E, Ceccarelli D, Lazaro N, Bani S, Cappuccinelli P, Colwell RR, Colombo MM. Environmental Vibrio spp., isolated in Mozambique, contain a polymorphic group of integrative conjugative elements and class 1 integrons. FEMS Microbiol Ecol. 2008;64:45–54. doi: 10.1111/j.1574-6941.2008.00455.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharya T, Chatterjee S, Maiti D, Bhadra RK, Takeda Y, Nair GB, Nandy RK. Molecular analysis of the rstR and orfU genes of the CTX prophages integrated in the small chromosomes of environmental Vibrio cholerae non-O1, non-O139 strains. Environ Microbiol. 2006;8:526–634. doi: 10.1111/j.1462-2920.2005.00932.x. [DOI] [PubMed] [Google Scholar]
- Keasler SP, Hall RH. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-F. [DOI] [PubMed] [Google Scholar]
- Olsvik O, Wahlberg J, Petterson B, Uhlen M, Popovic T, Wachsmuth IK, Fields PI. Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J Clin Microbiol. 1993;31:22–25. doi: 10.1128/jcm.31.1.22-25.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: Green Publishing Associates and Wiley; 1990. [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol. 2002;184:4259–4269. doi: 10.1128/JB.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V, Marrero J, Waldor MK. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid. 2006;55:173–183. doi: 10.1016/j.plasmid.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Grim CJ, Hasan NA, Taviani E, Haley B, Chun J, Brettin TS, Bruce DC, Detter JC, Han CS, Chertkov O, Challacombe J, Huq A, Nair GB, Colwell RR. Genome sequence of hybrid V. cholerae O1 MJ-1236, B-33 and CIRS101 and comparative genomics with V. cholerae. J Bacteriol. 2010;192:3524–3533. doi: 10.1128/JB.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YJ, Yi H, Lee JH, Kim DW, Chun J. Genomic evolution of Vibrio cholerae. Curr Opin Microbiol. 2010;13:646–651. doi: 10.1016/j.mib.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Patra T, Ghosh K, Raychoudhuri A, Pazhani GP, Das M, Sarkar B, Bhadra RK, Mukhopadhyay AK, Takeda Y, Nair GB, Ramamurthy T, Nandy RK. Vibrio cholerae O1 clinical strains isolated in 1992 in Kolkata with progenitor traits of the 2004 Mozambique variant. J Med Microbiol. 2009;58:239–247. doi: 10.1099/jmm.0.003780-0. [DOI] [PubMed] [Google Scholar]
- Cappuccinelli P, Colombo MM, Morciano C. Il colera, una malattia da debellare Il colera oggi in America Latina. IILA; 1995. Epidemiologia del colera in Africa. [Google Scholar]
- Dalsgaard A, Mortensen HF, Molbak K, Dias F, Serichantalergs O, Echeverria P. Molecular characterization of Vibrio cholerae O1 strains isolated during cholera outbreaks in Guinea-Bissau. J Clin Microbiol. 1996;34:1189–1192. doi: 10.1128/jcm.34.5.1189-1192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli D, Spagnoletti M, Cappuccinelli P, Burrus V, Colombo MM. Origin of V. cholerae in Haiti. Lancet Infect Dis. 2011;11:260. doi: 10.1016/S1473-3099(11)70037-8. [DOI] [PubMed] [Google Scholar]
- Ghosh-Banerjee J, Senoh M, Takahashi T, Hamabata T, Barman S, Koley H, Mukhopadhyay AK, Ramamurthy T, Chatterjee S, Asakura M, Yamasaki S, Nair GB, Takeda Y. Cholera toxin production by the El Tor variant of Vibrio cholerae O1 compared to prototype El Tor and Classical biotypes. J Clin Microbiol. 2010;48:4283–4286. doi: 10.1128/JCM.00799-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Amplicon profiles obtained for CTXΦ array A and B. We designed new primer pairs able to discriminate between the different CTXΦ array on the chromosome of V. cholerae. In this table we present the region amplified by each primer pair and the two different arrays obtained for the strains under analysis.