Abstract
We present an intuitive strategy for predicting the effect of sequence variation on splicing. In contrast to transcriptional elements, splicing elements appear to be strongly position dependent. We demonstrated that exonic binding of the normally intronic splicing factor, U2AF65, inhibits splicing. Reasoning that the positional distribution of a splicing element is a signature of its function, we developed a method for organizing all possible sequence motifs into clusters based on the genomic profile of their positional distribution around splice sites. Binding sites for serine/arginine rich (SR) proteins tended to be exonic whereas heterogeneous ribonucleoprotein (hnRNP) recognition elements were mostly intronic. In addition to the known elements, novel motifs were returned and validated. This method was also predictive of splicing mutations. A mutation in a motif creates a new motif that sometimes has a similar distribution shape to the original motif and sometimes has a different distribution. We created an intraallelic distance measure to capture this property and found that mutations that created large intraallelic distances disrupted splicing in vivo whereas mutations with small distances did not alter splicing. Analyzing the dataset of human disease alleles revealed known splicing mutants to have high intraallelic distances and suggested that 22% of disease alleles that were originally classified as missense mutations may also affect splicing. This category together with mutations in the canonical splicing signals suggest that approximately one third of all disease-causing mutations alter pre-mRNA splicing.
Keywords: cis-element, computational biology, RNA, SNPs
Splicing is catalyzed by the spliceosome, a riboprotein complex that rivals the ribosome in size and complexity. The ribosome has a large and small subunit whose assembly on the mRNA substrate corresponds to a functional switch from initiation to elongation. The spliceosome is composed of five subunits that appear to exist in at least four different stable configurations and, like the ribosomal subunits, transition between different assembled states corresponding to different stages of function (1–3). Mass spectroscopy has identified at least 300 RNA and protein components in this catalytic complex and studies have demonstrated heterogeneity in spliceosomal complexes isolated from different splicing substrates (4–6). The spliceosomal components that recognize the basic cis-elements of the splicing process are known. How the spliceosome assembles and reorganizes on these elements is also fairly well understood. However, several computational analyses estimate that these basic splicing elements contain at most half the information necessary for splice site recognition (7, 8). The remaining information lies outside these splice sites presumably as enhancers or silencers.
This information required to specify splicing presents a considerable mutational target—estimates of the fraction of disease mutations that affect splicing range from 15% (9) to 62% (10). Transcript analysis of genotyped cell lines has discovered numerous cases of allelic splicing demonstrating that polymorphisms also disrupt splicing (11, 12). These types of functional variants likely account for a similarly large fraction of the detected genetic risk for complex disease and could eventually be a target for molecular intervention. As physical methods for the detection of alternative splicing require large panels of genotyped accessible tissue, these studies will probably continue to be limited to samples harvested from human blood. An alternate approach is the prediction of causative variations from single-nucleo polymorphism (SNPs) that fall within splicing elements. The key to this approach is being able to identify what the splicing elements are and whether a variation is disruptive.
Recently, a variety of experimental and computational methods have emerged to identify sequence elements capable of functioning as enhancers and silencers (13–14). Considerable data has been gathered on the proteins that recognize these elements. The prototypical splicing activator that recognizes exonic splicing enhancers (ESEs) is one of the serine/arginine rich (SR) proteins (15). The heterogeneous ribonucleoprotein (hnRNP) family of proteins has generally been regarded as repressors as they inhibit splicing when bound to exons in pre-mRNA. However, hnRNP A, B, C, F, and H stimulate splicing when bound at intronic positions (16, 17). Conversely, SR proteins do not always promote splicing; SR proteins bound at intronic positions tend to function negatively in splice site recognition, a fact exploited by several viral alternative splicing systems (18–21). Experiments that relocated these intronic silencers into exons converted them into enhancers (19), and the reverse experiment of moving a natural ESE into an intronic location resulted in splicing repression (22). Positional effects on function appear at a finer scale than binning sequence into intron versus exon. Indeed an element’s location within an exon can also affect its function (23). This notion that an element’s activity is a function of its position has led to the routine use of “RNA maps” in cross-linking immunoprecipitation (CLIP) studies. An RNA map separates immunoprecipitated tags that fall around positively regulated exons from tags that fall around negatively regulated exons and plots the location of each tag set relative to the regulated splice site. In the genome-wide CLIP studies of hnRNP C, nova, and Fox1/2 specificity, the RNA maps illustrate that function differs according to positional distribution (24–26).
In this work, we exploit the relationship between location and function as a discovery tool. We show that splicing elements have signature positional distributions around constitutively spliced exons—they are abundant where they function positively and rare where they are inhibitory. Thus in a dataset of successful splicing events an element’s positional distribution is a proxy measure for where it enhances splicing. As different types of elements will have different positional distributions, we hypothesize that different positional distributions will define different splicing elements. Here, we describe the development of this discovery tool. All possible hexamers are mapped around splice sites. We discover 51 types of positional distributions (splicing elements) and demonstrate that these are predictive of function in vivo. We find that mutations that create new hexamers with radically different positional distributions are more likely to cause striking differences in splicing in vivo. We use this tool to analyze disease alleles within the human population.
Results
The Splicing Activator, U2AF65, Inhibits Splicing when Bound at an Exonic Site.
To test the relationship between the function of a splicing factor and the location of its predicted binding element, we initially focused on one well-characterized factor-ligand binding event, U2AF65’s recognition of the polypyrimidine tract. The binding motif consists of a Poly U-rich tract that typically contains runs of four or five uridines followed by cytosine frequently initiated with a G (Fig. 1A). Mapping U2AF65’s binding motif across all exons revealed the largest peak occurring immediately upstream of the 3′ splice site (3′ss). This location was consistent with its role as the principal recognizer of the polypyrimidine tract. The U2AF motif was overrepresented in the regions where it was known to function positively (i.e., in 3′ss recognition) and depleted in the exon (where U2AF binding has not been shown to support the normal spliceosomal complex). This suggested that the positional distribution pattern of an element around the splice sites was indicative of the transacting factor’s function in splicing.
Fig. 1.
Exonic binding of the intronic activator, U2AF65, inhibits splicing. (A) SELEX motifs were mapped to a dataset of 312,275 human splice site regions and plotted on an amalgamated exon. (B) The synthetic polypyrimidine tract returned by the SELEX consensus U2AF65 motifs and a genomic polypyrimidine tract were ligated into an exon and tested for U2AF65 binding by UV cross-linking in extract without antibody (lane 1, 3, and 5) or in extract that was blocked by an anti-U2AF65 antibody (lane 2 and 4). The radiolabel transferred to several products of differing mobility—a 65 kD interaction that was sensitive to preincubation with antiU2AF65 antibody is indicated with an arrow. (C) The sizes of RT-PCR products reflecting varying degrees of splicing are shown by the arrows. The disruptive effects of ligating the synthetic and natural PPT into the test exon of pZW4 is shown by RT-PCR in lane 7 and 8.
To experimentally test the role of the binding location of a particular factor in splicing function, we relocated the normally positive-acting intronic U2AF65 binding site into an exonic location and assayed splicing. For this study we utilized two polypyrimidine tracts. One tract was a synthetic consensus U2AF65 binding site derived from a Systematic Evolution of Ligands by Exponential Enrichment (SELEX) study and another was a natural polypyrimidine tract located upstream of the 3′ss of exon 5 of the KCNN1 gene (27). UV cross-linking indicated that numerous cellular proteins contacted both probes after incubation. The 65 kD interaction was blocked by preincubation with anti-U2AF65 antibodies thereby establishing specific U2AF65 contacts with the polypyrimidine tract with both of these inserts (Fig. 1B lanes 2 and 4 compared to no antibody control lanes 3 and 5) but not in the “no insert” control (Fig. 1B, lane 1).
The sequences used to probe binding were then assayed for function in the test exon of pZW4, an in vivo splicing reporter. The splicing phenotype was assayed by RT-PCR from total RNA following transfection into 293 cells. Whereas the no insert control spliced normally (Fig. 1C, black arrow in lane 6), both reporters containing U2AF65 binding elements exhibited evidence of disrupted splice site recognition by skipping exon 2 in some fraction of the transcripts observed. The polypyrimidine tract from the KCNN1 gene also generated an intron inclusion product and several other aberrant species that were not characterized. This result demonstrated that U2AF65, a factor with a well-characterized role of activating splicing when bound in the intron, disrupts splicing when bound in the exon.
To determine if the relationship observed between U2AF65 binding and its function was general, we expanded our analysis to some members of the SR and hnRNP protein family. As SR proteins are generally regarded as activators that function by binding exonic splicing enhancers, we examined the positional distribution of the in vitro SELEX-derived position weight matrix for three SR proteins: ASF/SF2, SC35, and 9G8 (SI Text) (28, 29). Three hnRNP proteins were also analyzed in this study: hnRNP A1, hnRNP L, and hnRNP C (SI Text) (30–32). This analysis largely supported the role of SR proteins as activators that bind ESEs whereas hnRNP binding sites are located at predominantly intronic locations. Binding motifs for hnRNP C were concentrated around the 3′ss consistent with early reports of the location of hnRNP C dependant functional elements (17). Both hnRNP L and hnRNP A1 also bound intronic elements albeit further away from the splice sites. The analysis of the binding sites of known splicing factors revealed a nonuniform positional distribution that was indicative of their function.
If the position of a splicing motif relative to a splice site is a signature of that motif’s function in splicing, then motifs with similar positional distributions should play similar roles in splicing and motifs with different positional distributions should play different roles in splicing. Therefore, by clustering the motifs according to their positional distribution around splice sites, we expected to organize elements into distinct functional classes.
Clustering Words by Positional Distribution Recovers Splicing Elements.
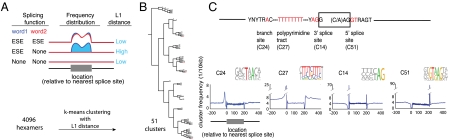
We developed an algorithm to cluster sequence motifs according to their positional distribution around splice sites. We first tabulated the frequency of every possible sequence motif around all the annotated splice sites in the human genome. This was accomplished by mapping 4,096 hexamers to all three hundred nucleotide windows around annotated 3′ss. This mapping associated each hexamer with a vector that contained the genomic occurrence of that hexamer at each position around all the 3′ss. This 300 unit long vector had a first position of -200 and a last position of +99 relative to the 3′ss. Counts were normalized to enable comparisons between hexamer positional distributions based on shape and not frequency. Repeating this procedure for the regions around the 5′ splice sites (5′ss) created a second vector that together with the 3′ss vector were used to summarize the positional distribution of hexamers around exon junctions in the human genome.
The overall goal of this method was to cluster hexamers into subsets that shared a similar positional distribution. This clustering required a method for pairwise comparison of two shapes. The difference in positional distribution shapes between two hexamers was calculated by determining the L1 distances between all possible pairwise combinations of these 4,096 vectors (Fig. 2A and Eq. 1). In a graph of normalized hexamer counts, L1 distance is simply the area between two positional distributions (shaded blue in Fig. 2A). These L1 distances were used to cluster (k-means) the hexamers into 51 distinct groups. The optimal value of k was determined by the CH index (33). The hexamers within each cluster were aligned without gaps and displayed as pictogram motifs (Fig. 2C). The resulting motifs returned by this analysis had distinct positional distributions around the 3′ and 5′ss (Fig. 2C).
Fig. 2.
Clustering motifs according to their positional distribution around splice sites. The positional distributions of all 4,096 possible hexamers were plotted around a database of human splice sites. (A) Several comparisons of two hypothetical hexamers (word 1 and word 2) are drawn to illustrate three different scenarios. L1 distance (shaded blue area) is used to compare normalized frequency distributions. Low L1 distance indicates there are small differences between two positional distributions and the two hexamers have the same or no difference in splicing function. High L1 distance denotes the two positional distributions are vastly different and likely differ in their role in splicing. (B) L1 distance was used to cluster the hexamers into 51 distinct groups based on the shape of their positional distributions around splice sites. Motifs and positional distributions of all 51 clusters can be found in the supplement. The clusters that correspond to the canonical splicing elements are indicated in red. (C) The arrangement of these elements on a prototypical pre-mRNA is annotated on the exon diagram. Hexamers within these clusters were aligned into motifs. Average occurrence frequencies of all the cluster’s hexamer were calculated at each position around the splice site database.
An immediately obvious feature of all 51 clusters was the sequence similarity between the hexamers that clustered together. In other words, hexamers that were highly similar in positional distribution were also highly similar in sequence. Hamming distance (i.e., the number of shifts or mismatches in the optimal ungapped alignment of two hexamers) was used to compare the sequence similarity of hexamers within a cluster. Intracluster similarity of hexamer sequence was much higher than expected by chance (all p values < 0.01; 1,000 trials per cluster, 51 clusters). As there is no a priori reason for similar sequences to share similar positional distributions relative to splice sites, we interpreted the strong sequence motifs found in the clusters as binding motifs of splicing factors that function at an optimal distance from a splice site. Consistent with this observation, we found motifs that match the known canonical splicing elements (i.e., branch point, polypyrimidine tract, 3′ss, and 5′ss) at the correct location relative to exon/intron boundaries (Fig. 2C). Cluster 24 peaks at position -26 nt and represents the branchpoint sequence with a core TRAY motif flanked by extended complimentarity to U2snRNA (i.e., 4 nucleotides upstream and 3 nucleotides downstream of the bulged A). It is important to note that the motif returned by this algorithm is a far better fit to the known mechanism of U2 snRNA mediated branch point recognition than motifs built from alignments of experimentally defined branchpoints. Similarly, the 5′ss motif (cluster 51, Fig. 2C) contains GTAAGT—a perfect stretch of complementarity to the mammalian U1 snRNA. Interestingly, this motif is avoided in the downstream exon proximal to the bona fide 5′ss. The polypyrimidine tracts are U-rich and covered by several clusters. A motif identical to the U2AF65 SELEX result (Fig. 1A) was found. The 3′ss AG and the polypyrimidine tract cluster separately presumably because of the variable spacing often found between these elements in natural splicing substrates and because they are recognized by separate factors.
Point Mutations that Create Mutant Hexamers with Large L1 Distances from Wild-Type Hexamers Alter Splicing in Vivo.
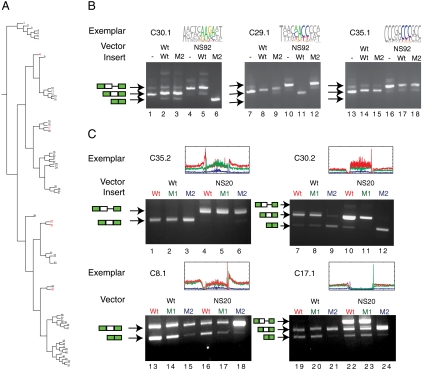
To validate elements from different clusters in vivo we assayed their effect on exon inclusion in a variety of splicing reporter minigenes. Test cases (exemplars) chosen to represent a cluster were cloned into reporter constructs, transfected into 293 cells and assayed by RT-PCR. To determine if the positional distribution distance measurements used in the clustering were predictive in identifying substitutions that disrupt a splicing element, we selected point mutations based on the degree to which they shifted the intraallelic L1 distance of the insert. There are eighteen different point mutations that can be introduced into a hexamer. Each of these mutations creates a new hexamer with a different positional distribution around splice sites. Substitutions with a large L1 distance were predicted to be most likely to disrupt splicing. Ranking all possible point mutations by L1 distance we found the top 25% to have twice as many ESE or exonic splicing silencer (ESS) changing mutations than the bottom 25% of this ranked list (34) (SI Text). We used L1 distance to design predicted splicing mutants for functional analysis in splicing reporter constructs (Fig. 2C). This analysis was performed for exemplars drawn from three clusters that represented unique splicing elements. For all three exemplars, the inserts and mutants spliced normally when ligated into the vector that contained wild-type splice sites (Fig. 3B, lanes 2, 3, 8, 9, 14, and 15). However when introduced into the context of mutation NS92 where the test exon was weakened by a mutation in the 5′ss, two of the three wild-type/mutant pairs displayed divergent splicing phenotypes (i.e., the wild-type sequence spliced differently than the predicted point mutant for cluster 30 and cluster 29—Fig. 3B, lanes 5, 6, 11, and 12). Neither the wild type nor the mutant of cluster 35 affected splicing (C35.1 in Fig. 3B). To see if the results observed in the mutant context of NS92 were general, we repeated the assay with different cluster exemplars (C35.2 and C30.2 in Fig. 3C) and different mutant context (NS20—weakened polypyrimidine tract) with identical results. This consistency between exemplars across different conditions suggested that the clusters are effectively characterizing the splicing activity of sequence elements. It is, however, possible that any variation in the sequence would disrupt this splicing activity. To establish the specificity of this prediction we tested variations that would be predicted to be neutral (i.e., variations in the same hexamer that results in low L1 distances). In all cases examined, these negative control (M1) mutants were spliced similarly to wild-type inserts in the splicing assay. The wild-type splicing pattern was similar to the predicted neutral mutant (Fig. 3C, lanes 7 and 8 and lanes 10 and 11). The mutation with high L1 distance was spliced differently than both the wild type and predicted neutral mutations (Fig. 3C, lane 9 versus lanes 7 and 8).
Fig. 3.
Minigene assay of element function confirms splicing differences between wild-type cluster exemplars and predicted mutants. (A) The clusters selected for functional analysis are indicated in red. (B) Exemplars drawn from each cluster are tested with their variants and no insert controls in several splicing reporter constructs. Total RNA from transfection into 293 cells was analyzed by RT-PCR. Arrows indicate the nature of the splicing product. M2 denotes the point mutant with the highest intraallelic L1 distance predicted to be most deleterious to the splicing function of the wild-type insert. (C) Additional exemplars for clusters 30 and 35, along with exemplars for clusters 8 and 17 were used to contrast the effect of predicted neutral mutations (M1) or the effect of predicted change-of-function mutations (M2) with wild-type splicing. As before, the M2 mutation is the variation with the highest intraallelic L1 distance, and the negative control, the M1 mutation, has the lowest intraallelic L1 distance.
Exemplars were also selected from two additional clusters that represent a variety of intronic splicing enhancers (i.e., positional distributions are enriched in the intronic regions). The predicted neutral mutants (M1) were spliced similarly to wild type (Fig. 3C comparing lanes 13 and 14, 16 and 17, 19 and 20, and 22 and 23), whereas the change-of-function mutants (M2) were spliced differently (Fig. 3C comparing lanes 13 and 15, 16 and 18, 19 and 21, and 22 and 24). In both cases, mutating an intronic element in the exon exhibited positive splicing phenotypes.
High Intraallelic Distance Is Predictive of Splicing Mutations.
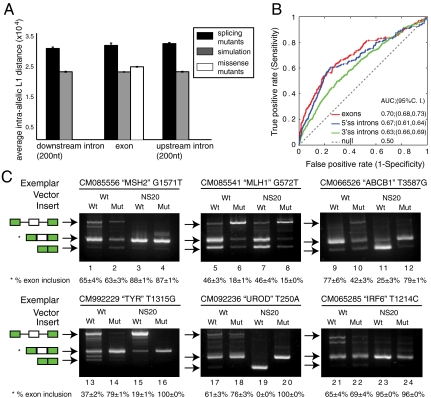
To test the predictive power of using intraallelic L1 distance to discover splicing mutations, we computed the intraallelic L1 distances of splicing mutations that were downloaded from the Human Gene Mutation Database (HGMD). Disease-causing alleles specifically associated with splicing exhibited significantly higher L1 distances than simulated mutations (p-value < 0.001 for the upstream intron, exon, and downstream intron) (Fig. 4A). The simulation incorporated mutational bias toward transitions (see Materials and Methods). Interestingly missense disease alleles downloaded from HGMD also displayed a significantly higher intraallelic L1 distance than expected (p-value < 0.001). This data suggests that even human disease alleles located outside of the canonical splice sites are more likely to cause aberrant splicing than natural variations that do not cause disease. We roughly estimated the fraction of splicing mutants by modeling the missense category of HGMD mutations as a mixture of exonic HGMD mutations that are known to cause splicing defects and simulated mutations (which are presumed not to cause splicing defects). In other words a hypothetical set comprised of 78% simulated mutations and 22% splicing mutants had the same average intraallelic L1 distance as the HGMD missense mutants. Accounting for these mutants along with HGMD entries that were formally classified as splicing mutants suggested that about a third of all disease-causing mutations display some sort of aberrant splicing phenotype. To explore the usefulness of L1 distance in predicting splicing mutations, we performed receiver operating characteristic (ROC) curve analysis, comparing the true to false positive rates at different thresholds of L1 (Fig. 4B). The ROC curve analysis suggests that an L1 prediction threshold that can identify 50% of the exonic splicing mutations in a sample (i.e., y ≈ 0.50 in Fig. 4B), would also return 20% false positives (i.e., x ≈ 0.2). This analysis demonstrated that the model was significantly predictive of splicing mutants—especially 5′ss and exonic mutants (Fig. 4B). As the later category of exonic mutants falls outside of the well-defined canonical splice sites, there are few other options to evaluate the effect of mutations. This method could be applied to finding splicing mutations in exons. To investigate this idea that missense mutations disrupt splicing, we tested six missense mutations with high L1 distances in the minigene splicing assay (Fig. 4). RT-PCR analysis of these exemplars uncovered an obvious difference in splicing between wild-type and mutant inserts in four of the six exemplars tested (Fig. 4C). This data confirmed the presence of processing mutations in exonic mutations. A web interface has been written to facilitate the analysis of variations in human pre-mRNA (http://fairbrother.biomed.brown.edu/data/mutations).
Fig. 4.
Human disease alleles are predicted to disrupt splicing. (A) Average intraallelic L1 distances for each category of mutation (HGMD splicing and HGMD missense/nonsense) and their corresponding background models of simulated mutations divided by location with respect to the splice sites. Error bars denote 95% confidence intervals. (B) Receiver operating characteristics (ROC) curve analysis using HGMD splicing mutants in regions around the 3′ss and 5′ss as “true positives” and simulated mutations as “true negatives.” ROC curve analysis classifies these mutations at decreasing thresholds of L1 stringency plotting the false against true positive rates. The exonic region is shown in red; upstream and downstream intronic regions are shown in green and blue, respectively. (C) Exemplars were selected from the HGMD missense mutants with the highest intraallelic L1 distance. Total RNA from transfection into 293 cells was analyzed by RT-PCR. The HGMD ID, gene name, and the mutational position are shown for each experiment. Quantifications on exon inclusion products are also shown. Arrows indicate the identity of the splicing product.
Discussion
In the output of the clustering, the canonical splicing elements segregated into discrete clusters. Strong 5′ss motifs (cluster 51) and 3′ss motifs (cluster 14) emerged as independent clusters. The hexamers in cluster 27 represented the polypyrimidine tract with their well-characterized signal located 4–20 nucleotides upstream of the 3′ss (Fig. 2C). Clusters 23 and 24 both appeared to fit the T(A/G)A(C/T) of the eukaryotic branchpoint sequence. ESEs mostly fell within 5 clusters (clusters 29–33, Fig. 2B). Further sorting the ESE hexamers into five prime specific ESEs, 3′ splice site ESEs and shared ESEs revealed that ESEs specific to the 3′ss fell mostly within cluster 30 and the smaller 5′ss specific ESEs segregated into cluster 29. In addition to ESEs, a variety of intronic splicing enhancers (ISEs) could be recognized within the cluster results. A prominent ISE, the G triplet, was found in cluster 8 (35–38). We found G triplets and C triplets to possess distinct nonoverlapping positional distributions around human splice sites (compare cluster 8 to cluster 35). Whereas both C and G triplets have a predominantly intronic positional distribution, C triplets tend to occur closer to the splice sites than G triplets. C triplets could be a recognition element for a protein like hnRNP C. Like many intronic enhancers, both C and G triplets occur at lower frequency on the exonic side of splice sites suggesting that they are not tolerated in the constitutively spliced exons that comprise the majority of the database used in this study. We did not find that mutations in exonic C triplets alter their effect on splicing (Fig. 3). C triplets may require other splicing elements for their activity and cannot function in isolation in a minigene. One candidate for this auxiliary element is the G triplet as these elements cooccur. C triplets are predominantly located upstream of the 3′ss, roughly around 30 nucleotides downstream of the local G triplet peak. Across the database, 22% of introns have G triplets between positions -65 and -50 relative to the 3′ss. If the intron contains a C triplet, the likelihood of a G triplet increases from 22% to 34% (p-value ≈ 0, chi-square test). It is possible that this co-occurrence may reflect a function synergy such as their potential to form structure or a larger ribonucleoprotein (RNP) complex through their transacting factors.
The general observation of intronic motifs that increase in frequency with decreasing distance to the splice site and then decrease in frequency when approaching the splice site from the exonic side is not consistent across all motif classes. Certain motifs (cluster 17) appear to increase in frequency with decreasing distance to the splice sites on both the intronic and exonic side of the junction. This type of distinction would not have been discovered by previous computational approaches. One possible explanation for this outlier might be that this motif is not an RNA element but rather a recognition element for a DNA binding protein. Polymerase pausing and chromatin formation with specific histone modifications are two DNA binding phenomena that have been implicated in enhanced splicing (39). A/T rich elements are often found in recognition sites of DNA bending proteins or could form the weak RNA∶DNA duplexes that promote the polymerase backtracking associated with some types of transcriptional pauses (40).
Although describing the mechanism of each element is beyond the scope of this study, we demonstrate that mutations that are disruptive to positional distribution are disruptive to splicing. We also find evidence that missense mutations that cause human disease are more likely to disrupt splicing than simulated mutations. Because of the difficulty of assaying splicing in patients, very little is known about the prevalence of splicing defects in human disease. About 15% of the mutations in the HGMD are described as splicing mutants (9). Some have been validated directly but many of these mutations colocalize with critical regions of splice sites and so are assumed to disrupt splicing. A more problematic class of identification is the set of mutations that fall outside of well-defined sites. It is possible that many of these disease alleles are associated with subtle defects in splicing that could exacerbate the disease phenotype. Using an approach that models the missense mutations as a mixture of exonic splicing mutants and simulated mutations, we estimate that 22% of missense disease alleles alter splicing. A reanalysis of missense mutations supports the notion that many disease alleles originally classified as missense also disrupt splicing (41). Furthermore, another recent study finds a similar fraction (i.e., 25%) of > coding mutations alters splicing (42).This class of “undiagnosed” splicing mutations along with known splicing mutations predicts that about one third of all mutations alter splicing.
It is important to be able to identify the many human disease alleles that alter splicing and characterize missense mutations for their effect on pre-mRNA processing. In the future, new molecular therapies that correct splicing defects may ameliorate many genetic disorders (43). The ability to correctly identify splicing mutations by their elevated L1 distance and the ability to predict mutations in the minigene system demonstrate that this is a useful tool in predicting causal alleles.
Materials and Methods
A more detailed description of these methods can be found in SI Text.
Binding and Splicing Assays.
RNA probes were T7 transcribed from DNA oligos (all sequences listed in SI Text) with incorporating 32P label and incubated in HeLa nuclear extract pre or mock treated with MC2 antibody. Label transfer was visualized by phosphoimager following PAGE. RNA elements were also tested for function in variations of the pZW4 splicing reporter minigene (i.e., Fig. 1 and “wt” vector in Fig. 4) (44). Additional constructs with variations characterized as splicing mutations in prior reports (45) were designed as sensitized reporters. Inserts were selected on the basis of their match to the cluster motif. The most extreme difference between wild type and mutant hexamers represents M2, the point mutation that the method would predict most likely to disrupt a motif. Conversely, the most similar positional distributions, M1, would be predicted to function similarly to the wild-type sequence. These variations were introduced into the reporter, transfected into 293 cells and assayed by RT-PCR. Both alleles of missense mutations were tested with 15nt of flank as a 31-mer ligated into the minigene.
Clustering Algorithm and Computational Prediction.
The positional distributions of all 4,096 hexamers were plotted around a dataset of human splice sites. Normalized counts were compared via the L1 distance metric for all pairwise combinations of hexamers. The data was clustered using the CH index to determine an optimal value k = 51 for K-means clustering (33, 46). In Figs. 3 and 4, for a given point mutation the representative L1 distance was taken to be the largest intraallelic distance of the 6 distances calculated by comparing each tiled wild-type hexamer with its mutant counterpart. L1 distances were calculated in this way for the 8,027 disease-causing splicing mutations and 42,532 missense/nonsense mutations downloaded from the HGMD. Simulated mutations (preserving a twofold higher bias toward transitions) were used to generate background mutations. ROC curves were generated in MatLab on a mutation set that contained equal quantities of background mutations and true positives (HGMD splicing mutants).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101135108/-/DCSupplemental.
References
- 1.Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jurica MS, Sousa D, Moore MJ, Grigorieff N. Three-dimensional structure of C complex spliceosomes by electron microscopy. Nat Struct Mol Biol. 2004;11:265–269. doi: 10.1038/nsmb728. [DOI] [PubMed] [Google Scholar]
- 3.Nilsen TW. The spliceosome: No assembly required? Mol Cell. 2002;9:8–9. doi: 10.1016/s1097-2765(02)00430-6. [DOI] [PubMed] [Google Scholar]
- 4.Chen YI, et al. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 2007;35:3928–3944. doi: 10.1093/nar/gkm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 7.Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci USA. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun H, Chasin LA. Multiple splicing defects in an intronic false exon. Mol Cell Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stenson PD, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Bigas N, Audit B, Ouzounis C, Parra G, Guigo R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005;579:1900–1903. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 11.Kwan T, et al. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 12.Kwan T, et al. Heritability of alternative splicing in the human genome. Genome Res. 2007;17:1210–1218. doi: 10.1101/gr.6281007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 14.Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol Cell Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Contreras R, et al. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swanson MS, Dreyfuss G. RNA binding specificity of hnRNP proteins: A subset bind to the 3′ end of introns. EMBO J. 1988;7:3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook CR, McNally MT. SR protein and snRNP requirements for assembly of the Rous sarcoma virus negative regulator of splicing complex in vitro. Virology. 1998;242:211–220. doi: 10.1006/viro.1997.8983. [DOI] [PubMed] [Google Scholar]
- 19.McNally LM, McNally MT. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J Virol. 1996;70:1163–1172. doi: 10.1128/jvi.70.2.1163-1172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Xiao SH, Manley JL. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 1998;12:2222–2233. doi: 10.1101/gad.12.14.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanopka A, Muhlemann O, Akusjarvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 22.Ibrahim EC, Schaal TD, Hertel KJ, Reed R, Maniatis T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc Natl Acad Sci USA. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goren A, et al. Comparative analysis identifies exonic splicing regulatory sequences—The complex definition of enhancers and silencers. Mol Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 25.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA–protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 28.Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: Significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorlach M, Burd CG, Dreyfuss G. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J Biol Chem. 1994;269:23074–23078. [PubMed] [Google Scholar]
- 32.Hui J, et al. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calinski T, Harabasz J. A dendrite method for cluster analysis. Commun Stat. 1974:1–27. [Google Scholar]
- 34.Stadler MB, et al. Inference of splicing regulatory activities by sequence neighborhood analysis. PLoS Genet. 2006;2(11):e191. doi: 10.1371/journal.pgen.0020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caputi M, Zahler AM. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H'/F/2H9 family. J Biol Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 36.McCullough AJ, Berget SM. An intronic splicing enhancer binds U1 snRNPs to enhance splicing and select 5′ splice sites. Mol Cell Biol. 2000;20:9225–9235. doi: 10.1128/mcb.20.24.9225-9235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid DC, et al. Next-generation SELEX identifies sequence and structural determinants of splicing factor binding in human pre-mRNA sequence. RNA. 2009;15:2385–2397. doi: 10.1261/rna.1821809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeo G, Hoon S, Venkatesh B, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc Natl Acad Sci USA. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadener S, Fededa JP, Rosbash M, Kornblihtt AR. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc Natl Acad Sci USA. 2002;99:8185–8190. doi: 10.1073/pnas.122246099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulish D, Struhl K. TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Mol Cell Biol. 2001;21:4162–4168. doi: 10.1128/MCB.21.13.4162-4168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ars E, et al. Mutations affecting mRNA splicing are the most common molecular defects in patients with nuerofibromatosis type 1. Hum Mol Genet. 2000;9:237–247. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- 42.Sterne-Weiler T, Howard J, Mort M, Cooper DN, Sanford JR. Loss of exon identify is a common mechanism of human inherited disease. Genome Res. 2011 doi: 10.1101/gr.118638.110. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hua Y, et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, et al. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 45.Chen IT, Chasin LA. Direct selection for mutations affecting specific splice sites in a hamster dihydrofolate reductase minigene. Mol Cell Biol. 1993;13:289–300. doi: 10.1128/mcb.13.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacQueen J. Some methods for classification and analysis of multivariate observations; Proceedings of the Fifth Berkeley Symposium on Mathematics, Statististics, and Probability; 1967. pp. 281–297. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.