Abstract
MspJI is a novel modification-dependent restriction endonuclease that cleaves at a fixed distance away from the modification site. Here, we present the biochemical characterization of several MspJI homologs, including FspEI, LpnPI, AspBHI, RlaI, and SgrTI. All of the enzymes specifically recognize cytosine C5 modification (methylation or hydroxymethylation) in DNA and cleave at a constant distance (N12/N16) away from the modified cytosine. Each displays its own sequence context preference, favoring different nucleotides flanking the modified cytosine. By cleaving on both sides of fully modified CpG sites, they allow the extraction of 32-base long fragments around the modified sites from the genomic DNA. These enzymes provide powerful tools for direct interrogation of the epigenome. For example, we show that RlaI, an enzyme that prefers mCWG but not mCpG sites, generates digestion patterns that differ between plant and mammalian genomic DNA, highlighting the difference between their epigenomic patterns. In addition, we demonstrate that deep sequencing of the digested DNA fragments generated from these enzymes provides a feasible method to map the modified sites in the genome. Altogether, the MspJI family of enzymes represent appealing tools of choice for method development in DNA epigenetic studies.
Keywords: 5-methylcytosine, methylome
Modified DNA bases appear in genomic DNAs in all domains of life, spanning the evolutionary distance from viruses to eukaryotic species. DNA base modifications vary in form and genomic location enriching the information content encoded by genomes. The biological role of base modifications varies, ranging from protection against restriction endonucleases in bacteria and bacteriophages to transcriptional regulation in mammals. In prokaryotes, DNA methyltransferases in restriction-modification systems modify the host genomic DNA, so that restriction endonucleases can target foreign DNA and protect the host cell from invaders (1). However, a few bacteriophages respond by incorporating modified bases into their genomes as a way to block restriction endonuclease cleavage (2). For example, in Xanthomonas oryzae phage XP12, all cytosines exist in the form of 5-methylcytosine (5mC) (3). Another example is the well-studied T4 phage, in which 5-hydroxymethylcytosine (5hmC) is incorporated into the DNA during replication and additional glucosyltransferases further modify all 5hmC to glucosylated-hydroxymethylcytosine (5ghmC). T4 genomic DNA containing 5ghmC is resistant to cleavage by most restriction endonucleases, with the exception of Type IV modification-dependent endonucleases (4, 5).
Several different types of modification-dependent endonucleases are found in prokaryotes. For example, N6-adenosine methylation is recognized by a few known enzymes, e.g., DpnI (GmATC). A group of sequence-specific cytosine methylation-dependent restriction endonucleases including GlaI (GmCGmC), BisI (GmCNGC), etc., have been reported recently, which cleave within the recognition site in a Type IIP-like manner (6). McrA has been shown to restrict CmCGG-containing DNA in vivo (7, 8) and to bind to (Y > R)mCGR in vitro (9). McrBC recognizes pairs of (A/G)mC separated by 30–3,000 base pairs and cleaves 30–35 base pairs from one recognition element (10–12). Mrr in Escherichia coli is known to restrict both cytosine- and adenine-methylated DNA, although its consensus recognition sequence remains elusive (13). Homologs of these modification-dependent endonucleases can be found in numerous bacterial species, yet few have been studied.
Recently, our group reported the discovery of a unique group of Mrr-like modification-dependent restriction endonucleases, represented by MspJI (14). MspJI recognizes 5mC in the context of mCNNR (R = G or A) and introduces double-stranded breaks at fixed distances (N12/N16 from mC) on the 3′ side of the mC, leaving a four-base 5′ overhang. A unique feature of these enzymes is that with symmetrically methylated sequences [e.g., mCpG or mCHG sites, (H = C, T, or A)], cleavages elicited by two methylated half-sites result in DNA fragments about 32 bp in size being extracted from the genomic DNA, with the methylated site in the middle. This property allows the development of sequencing-based applications for investigating the epigenomes of higher organisms.
In many eukaryotic species, 5-methylcytosine is one of the epigenetic marks crucial for transcriptional programming in development as well as disease pathology. Epigenetic DNA modifications are thought to affect DNA-protein interactions and are found in promoter regions as well as gene bodies, in both CpG and non-CpG contexts (15). Theoretically, MspJI allows interrogation of up to half of all the methylated CpG sites, and up to a quarter of all the fully methylated CpG sites can be extracted in the form of the 32-bp fragments (including the overhangs). However, its coverage on the entire methylome is still limited. Thus, it would be advantageous to have multiple MspJI-like enzymes that can recognize a wider set of methylated sites to reach higher coverage of the entire epigenome.
Based on our bioinformatic analysis, we have identified a number of MspJI homologs in GenBank (14). In this paper, we present the detailed characterization of additional MspJI homologs. Our results suggest that although many of the biochemical properties of the MspJI family members are similar, they display a diversity of recognition specificities in the flanking nucleotides of the modified cytosine. We demonstrate the ability to differentiate methylation levels in genomic DNA using this family of enzymes. We also illustrate a unique application for recognition of the predominant methylation type in a specific epigenome, such as distinguishing the common CpG methylation in mammals from CHG methylation in plants. Lastly, as proof of principle, we show that direct sequencing of the extracted 32-mer pool using high-throughput technology provides a quick and reliable approach to epigenomic mapping.
With a growing sequence collection in databases and continuing biochemical characterization efforts, we expect that more MspJI-like DNA modification-dependent enzymes, possibly with diversified properties, will emerge. As a result, these enzymes should provide a set of useful tools in developing enzyme-based methods for mapping dynamic epigenomes.
Results
Modification-Dependent Endonuclease Activity.
MspJI and its homologs, including FspEI, LpnPI, AspBHI, RlaI, and SgrTI, were synthesized using the overlapping oligonucleotide assembly method as previously described (14). Recombinant enzymes, with or without N-terminal His-tags, were then expressed in the dcm- E. coli strain T7 Express and purified to apparent homogeneity (SI Appendix, Fig. S1) (Materials and Methods).
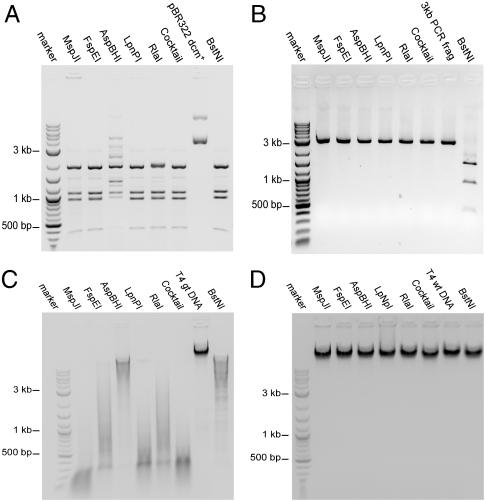
We have previously characterized MspJI as a modification-dependent endonuclease that recognizes 5-methylcytosine (5mC) or 5-hydroxymethylcytosine (5hmC) and cleaves both strands at specific positions (N12/N16) on the 3′ side of the modified base (14). The activity of the homologs was assessed using a set of modified plasmids or genomic DNA. Fig. 1A shows the cleavage activity on the Dcm-methylated (CmCWGG) pBR322 plasmid DNA. By comparing the resulting digest with the digestion pattern of the known methylation-insensitive restriction endonuclease, BstNI (CC↓WGG), we inferred that all of the enzymes cleave the methylated pBR322 DNA near the dcm sites. The cleavage is methylation-dependent, because no cleavage is observed on an unmethylated 3 kb PCR fragment (Fig. 1B).
Fig. 1.
Modification-dependent endonuclease activity of MspJI and homologs. DNA was incubated at 37 °C with 1 unit of each of the indicated homolog, a cocktail of the homologs (0.2 unit each), or BstNI (CC↓WGG). Control DNA was incubated in parallel without enzyme addition. (A) pBR322 dcm+ (CmCWGG methylated) plasmid DNA. (B) A 3-kb PCR amplified DNA fragment (unmodified) DNA. (C). T4gt (hydroxymethylated) genomic DNA. (D) T4wt (glucosylated) genomic DNA.
All enzymes extensively cleave T4gt genomic DNA, which exclusively contains 5hmC (Fig. 1C). However, none of the enzymes appear to cleave the wild-type T4 genomic DNA, which contains glucosylated 5hmC (Fig. 1D). This observation suggests a potential basis for mapping 5hmC locations in genomic DNA (see Discussion). The extent of digestion on T4gt DNA varies for each enzyme and is consistent with their individual recognition specificity (see Table 1). In addition to C5 cytosine modifications, we investigated the activity on DNA containing N4 methylated cytosine, but no cleavage was observed.
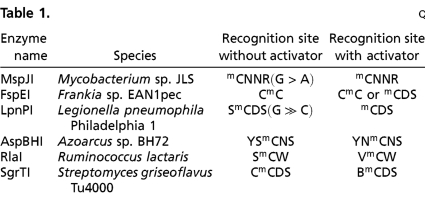
Table 1.
Consensus recognition sequences
N = A or C or G or T; D = A or G or T; B = C or G or T; V = A or C or G; R = A or G; S = C or G; W = A or T; Y = C or T.
In our previous work, we observed that introducing a short double-stranded oligonucleotide containing a methylated site stimulates MspJI digestion. The activator is designed to form a stem-loop structure, and the sequence flanking the methylated sites is shorter than 12/16 bp and thus is too short to be cleaved (SI Appendix, Fig. S2). The same stimulatory effect was observed for all the homologous enzymes, further suggesting a common reaction mechanism.
Determination of the Recognition Sequence and Cleavage Site.
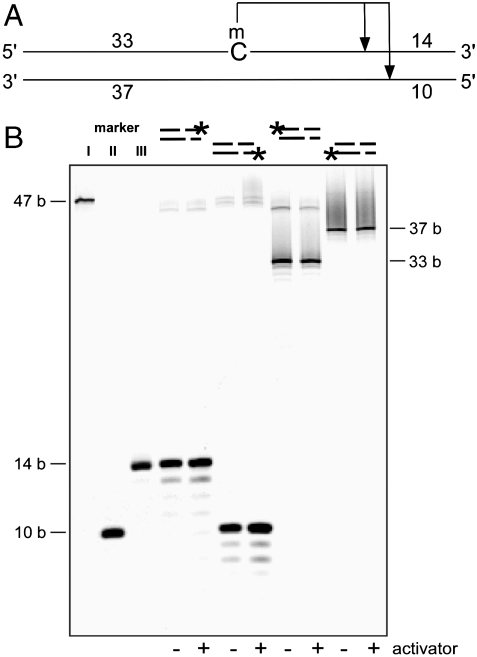
To determine the cleavage sites of each of the homologs, a set of hemimethylated oligonucleotides were labeled at either the 5′ or 3′ end of the top or bottom strand to allow detection of each of the cleavage products (Fig. 2A). The digested products were resolved by denaturing PAGE. Fig. 2B represents the typical digestion pattern, showing the result of LpnPI digestion. By comparing the cleavage pattern with markers and with that produced by MspJI, we concluded that all homologous enzymes cleave at the same position (i.e., N12/N16 on the 3′ side of the modified cytosine). In addition to the predominant cleavage position at N12/N16 from mC, a low percentage of cleavage events appear at additional positions, mostly one nucleotide from the major cleavage site. Although this cleavage-distance wobble may affect the interpretation of relative position of the modification within the resolved fragment, only a very small percentage of the products appear to be affected (see Mapping the Human Epigenome Using MspJI section).
Fig. 2.
The cleavage position is N12/N16 on the 3′ side of the modified cytosine. (A) Schematic diagram of the hemimethylated oligonucleotide structure used for determination of the cleavage position. The full-length oligonucleotide is 47 bp. A cut in the illustrated positions will result in four fragments: 14, 10, 33, and 37 bases. (B). LpnPI digestion of the hemimethylated oligonucleotide. Each oligonucleotide was fluorescein amidite (FAM) labeled at one end: 3′ top, 5′ bottom, 5′ top, or 3′ bottom. Markers I, II, and III are FAM labeled 47, 10, and 14 bases, respectively. The digestion reaction was carried out at 37 °C with or without activator present, and resolved on a denaturing gel. The cleavage position is predominantly N12/N16 on the 3′ side of the modified cytosine. The additional bands observed are due to wobbling, resulting in a cut further away to the 3′ side.
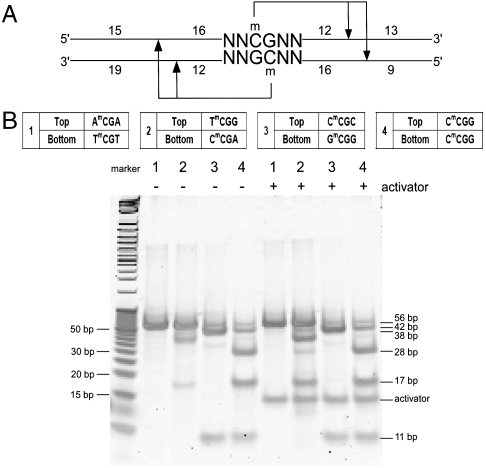
To determine the sequence recognition preference of these enzymes, we utilized a few sets of synthetic oligonucleotides containing methylated cytosines flanked by varying sequences. As we envision one of the main applications for these enzymes in mapping eukaryotic epigenomes, we designed the oligonucleotides based on the common methylated sites observed in genomes of higher organisms, including NmCGN, NCmCGGN, NmCNGN, NCmCWGGN, NGmCN, etc. (where N represents any nucleotide among A, T, C, or G, not a randomized nucleotide mix). Fig. 3A shows a schematic structure of the oligonucleotides used. The oligonucleotides length is 56 bp, with one methylated cytosine on the top strand and the other on the opposite strand (the full sequences used are listed in SI Appendix, Table S1). Even though these enzymes recognize hemimethylated sites, the oligonucleotides were designed to have methylation on both strands, providing the advantage of testing two different methylated sites simultaneously. If the enzyme recognizes the top strand methylated site, cleavage will result in two fragments of 42 and 11 base pairs; if the enzyme recognizes the bottom strand methylated site, cleavage will result in two fragments of 38 and 17 base pairs; if the enzyme recognizes both top and bottom strand methylated site, cleavages on both sides will result in fragments of average length of 28, 17, and 11 base pairs (Fig. 3B). We used an end-point digestion assay either with or without the oligonucleotide activator, and we resolved the enzymatic cleavage products using 20% PAGE (Materials and Methods). Fig. 3B shows a representative gel of FspEI digestion of the NmCGN set. Based on the cleavage pattern, we compiled a list of cleavable and noncleavable sites (SI Appendix, Tables S2–S7), from which we infer the consensus recognition sequence for each enzyme. Table 1 lists the consensus recognition sequence of each homolog either with or without the activator. Overall, the recognition sequences for each enzyme are different, with some enzymes requiring particular nucleotides on the 5′ side of the mC (e.g., FspEI), others on the 3′ side of the mC (e.g., MspJI), and some on both sides (e.g., RlaI). For some enzymes, such as FspEI, LpnPI, and AspBHI, it appears that the promiscuity in recognition sites increases in the presence of the oligo activator. For others, such as MspJI, even though there is no obvious change in specificity due to activator presence, we observed that cleavage of some sites appears more efficient in the presence of the activator.
Fig. 3.
Sequence recognition determination. (A) Schematic diagram of the fully methylated synthetic oligonucleotide structure used for sequence specificity determination. A 56-bp oligonucleotide, methylated in position 31 (C) on the top strand and on the opposite strand across from position 32 (G). Recognition of the top strand site will result in two fragments at 42 and 11. Recognition of the bottom strand site will result in 38- and 17-bp fragments, and recognition of both sites will yield fragments of average length of 28, 17, and 11 bp. The N in the sequence represents each of the four bases A, T, C, or G in separate tests, and not a randomized position. B. Representative digestion by FspEI of some oligonucleotides from the NmCGN set, with and without activator. An undigested oligonucleotide is presented in lane 1. The representative oligonucleotide variable sequence region is listed in the table (the full oligonucleotide sequence is listed in SI Appendix, Table S1). The digested products were resolved on a TBE native polyacrylamide gel.
Differential Digestion of Genomic DNA.
As the availability of regular restriction endonucleases in the 1970s helped to expedite the mapping of genomic structure, the availability of the modification-dependent restriction enzyme, MspJI, along with its characterized homologs may provide a powerful tool set for studying the epigenomes of higher organisms. Because of the diversity in their specificity, they may be used either individually or combined in a “cocktail” form to probe the epigenetic status either genome-wide or at specific loci.
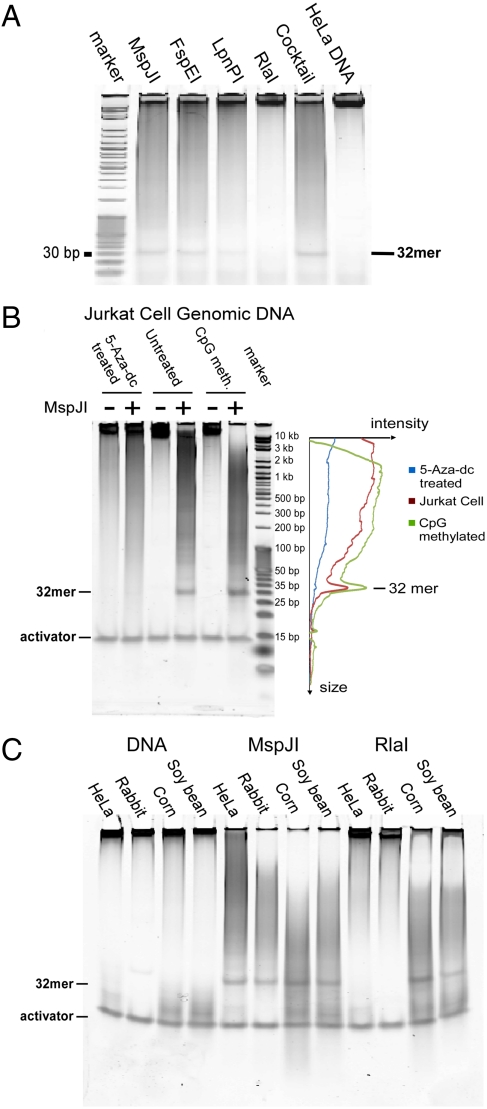
Fig. 4A demonstrates the activities of MspJI and its homologs on mammalian genomic DNA. Notice the prominent 32-mer band in the polyacrylamide gel. This band can be isolated and sequenced, with most of the fragments containing a fully modified site (see Mapping the Human Epigenome Using MspJI section). Because of the different recognition specificity for each enzyme, the 32-mer isolated from each digestion will cover a different subset of modified sites in the genome. In addition, global genomic methylation density can be revealed directly from the gel. A comparison of MspJI digestion of fully modified and mC-depleted genomic DNA illustrates this. An mC-depleted sample [Jurkat cell genomic DNA isolated following 5-aza-2-deoxycytidine (5-Aza-dC) treatment of the cell culture] was compared with a control sample from the untreated culture and with a fully modified sample obtained by treating DNA isolated from an untreated culture with M.SssI (thus enzymatically methylating all of the CpG sites). These samples were digested with MspJI, and the results are presented in Fig. 4B. As 5-Aza-dC is a potent inhibitor to the methyltransferases, the methylation level dramatically decreases in the 5-Aza-dC treated cells compared with the other DNAs (16). The difference in methylation density among the three is also discernable in both the intensity of the 32-mer band, and in the different extent of disappearance of the input DNA band. In addition, as shown in the densitometry profile of each lane, the average size of the digested fragment is smaller for the CpG-methylated Jurkat genomic DNA than for the native Jurkat genomic DNA, consistent with a higher methylation level in the former sample. Note that the lanes of undigested input DNA and the presence of equal amounts of activator in each sample provide a normalization standard.
Fig. 4.
Methylation analysis in genomic DNA using MspJI and homologs. (A) Formation of a 32-mer. HeLa genomic DNA was incubated with 1 unit of MspJI, FspEI, LpnPI, RlaI, or a cocktail (0.2 unit each) as indicated, overnight at 37 °C without activator. The reaction was resolved on a 20% TBE native gel. The apparent band is the 32-mer formed as a result of digestion of fully methylated CpG sites. (B) Methylation level comparison. Jurkat cell DNA, after 5-Aza-dC treatment, untreated or enzymatically CpG methylated, all in the presence of activator, incubated with buffer only (undigested) or with MspJI (as indicated) were resolved on a native gel. Lane profiling was generated by densitometry. The activator in each lane can be used as normalization control, because it is added in known amount and is not digested by MspJI or its homologs. Three indicators of the methylation level can be observed: The enzymatically methylated CpG sample is more extensively digested as judged by the disappearance of the substrate, yields a shorter average fragment, and displays a higher density of the 32-mer band. (C) Comparison of methylation in plants vs. mammals. DNA from HeLa cells, rabbit liver, corn, or soy bean were incubated with buffer only (undigested), MspJI, or RlaI (in the presence of activator). Digestion with MspJI, which recognizes a subset of CN methylated sites, produces a 32-mer band from both mammalian DNA (which contains predominantly methylated CpG sites), and plant genomic DNA (which contains both methylated CpG sites and methylated CHG sites). RlaI, which recognizes a subset of CWG methylated sites, can produce the 32-mer band only from plant genomic DNA (because it can digest some methylated CHG sites but not CpG sites).
One of the MspJI homologs, RlaI, has a unique recognition sequence (VmCW, Table 1), so that it does not act on mCpG sites, but only on the relatively infrequent mCWG sites. This property may be used to differentiate methylation patterns in genomes. Fig. 4C compares MspJI and RlaI digestion of four genomes: two mammalian and two plant genomes. For both HeLa and rabbit liver DNA, which have predominantly CpG methylation, the 32-mer band is clearly visible after MspJI digestion, but is not discernable after RlaI digestion. However, in both plant DNAs (corn and soy bean), which are known to contain CHG methylation (17), the 32-mer is prominent after both MspJI and RlaI digestion. Thus, the ability of RlaI to distinguish the mCHG sites from mCG sites may find applications in plant epigenomics as well as stem cell epigenomics, where CHG methylation may also be significant (15).
Mapping the Human Epigenome Using MspJI.
To test the feasibility of using MspJI-like enzymes in mapping epigenomes, we sequenced the gel-extracted 32-mer pool from MspJI digested IMR90 human lung fibroblast genomic DNA (Materials and Methods). We chose IMR90 cell line as a benchmark because its entire methylome was decoded using the bisulfite-sequencing method (referred as the “Salk reference” hereafter) (15). From one sequencing run on the SOLiD platform using only a quarter of a slide, we generated 71.8 M reads of 35 bases, among which 23.5 M (32.7%) reads were aligned unambiguously to about 10.4 M distinct genomic locations on the reference genome (hg18) (details in Materials and Methods).
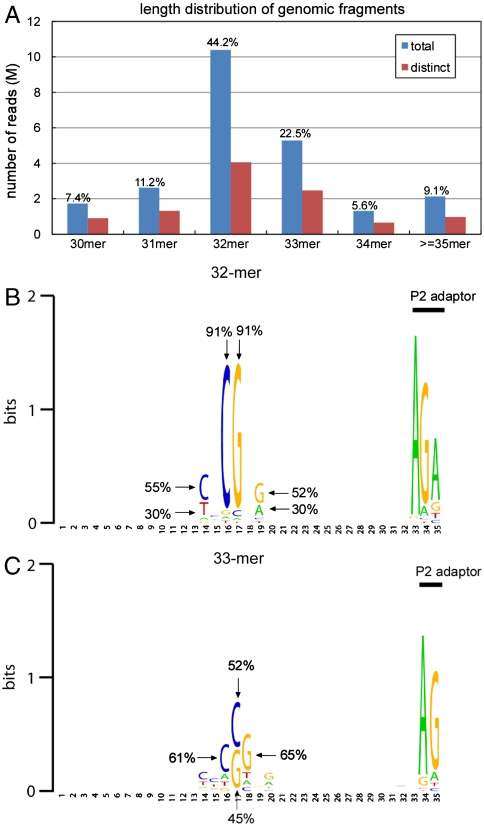
Fig. 5A shows the length distribution of the sequenced genomic fragments for the total 23.5 M mapped reads and the 10.4 M distinct reads. The majority (> 60%) of the sequenced genomic fragments is 32–33 nucleotides long. We then investigated the base composition within each size group. Fig. 5B shows the sequence logo representation constructed from the 4.1 M distinct sequencing reads with 32-mer genomic fragments. Over 90% of the reads contain CG in the center of the 32-mers (Fig. 5B). The overrepresented sequence motif, YNCGNR (Y = C/T, R = G/A), reflects the expected fully methylated CpG sites, from which MspJI extracts the 32-bp fragments. In addition, consistent with the cleavage preference observed on synthetic oligonucleotides, mCNNG frequency is almost twice as high as that of mCNNA, despite the similar frequency of mCNNG to that of mCNNA in the genome. These results support the conclusion that most of the fragments are generated by MspJI digestion on a single fully methylated CG site. The fold coverage for this quad-slide sequencing run resulted in an approximation of 5.0 X coverage for the extracted 32-mers. SI Appendix, Table S8 lists the top ten overrepresented 32-mers in our dataset. All of the overrepresented 32-mers are from repetitive elements, which are known to be heavily methylated (15).
Fig. 5.
Sequencing human IMR90 methylome using MspJI. (A) Length distribution of the genomic fragments in all the uniquely mapped reads (blue) and after grouping by distinct genomic locations (red). (B) Sequence logo of the sequenced 32-mers. The logo was generated by WebLogo (23). (C) Sequence logo of the sequenced 33-mers.
Fig. 5C shows the sequence logo representation constructed from the 2.5 M distinct reads with 33-bp genomic fragments. Previously, we have observed that the cleavage position of MspJI can wobble at low frequency, mostly by 1 base (14). The motif in Fig. 5C is consistent with this finding, with the fully methylated CG site either in position 16/17 or in position 17/18. For both types of reads, MspJI cleaves at 16 base pairs on one side and 17 base pairs on the other side. Altogether, we conclude that the 32-bp and 33-bp reads reliably report the full methylation status for the central CpG sites.
Besides 32-mers and 33 mers, we also analyzed the reads that contain genomic fragments of other sizes (30, 31, 34, ≥35 base pairs). Sequence logo representations are shown in SI Appendix, Fig. S5. For these groups, the overrepresented motifs are generally much weaker than those in the 32- and 33-bp groups, suggesting that most of them are likely generated from MspJI cleavages on two nearby methylated CG sites rather than on the same methylated CG site. Thus, interpretation of these reads would require aligning them to the reference genome and searching for the potential MspJI sites in the right distance from the ends. Nonetheless, the motifs are reminiscent of the MspJI recognition sites, especially on the P1 sequencing adaptor end (SI Appendix, Fig. S5).
We next compared the combined fully methylated CpG sites extracted from the pool of 32- and 33-bp reads with the Salk reference methylome. Because MspJI can excise 32- to 33-bp fragments only on the YNmCGNR (Y = C/T, R = G/A) sites in the genome, we selected a subset of the fully methylated CG sites in the Salk reference that has the Y and R in the flanking positions (i.e., YNmCGNR). In addition, we require that the 32-bp fragments encompassing the selected YNmCGNR sites are unique in the genome. The Salk reference subset contains about 4.2 M fully methylated CG sites. Approximately 87% of the MspJI generated 32/33 bp sequences were identified in the Salk reference subset, and approximately 55% of the 4.2 M Salk sequences were present in the 32/33 bp pool (SI Appendix, Fig. S6A). The high specificity confirms the effectiveness of our approach and the accuracy of the sequencing data. The sensitivity may be further improved by increasing the sequencing depth. SI Appendix, Fig. S6B shows that the average fold coverage of the sequenced MspJI recognition sites (CNmCGNG in SI Appendix, Fig. S6B) correlates with the methylation levels reported in the Salk data.
Discussion
Base modification of genomic DNA is widely adopted by many living species, from bacteriophages to eukaryotic organisms, with a broad range of biological functions that can be distinctly different. In the genomes of higher organisms, it is generally accepted that changes in the dynamic epigenomic marks play crucial roles in differential transcriptional regulation during development and pathological processes (15, 18, 19). One of the challenges in mapping genomic DNA modification is to distinguish the modified nucleotides from the majority of the nonmodified DNA. A widely used principle is bisulfite conversion, in which unmodified cytosines are selectively converted to uracils whereas methylated cytosines remain unchanged. Sequencing of the converted DNA would then reveal the modified cytosines to the nucleotide-level resolution. However, genome-wide application of this method remains technically challenging (20) and costly. Other methods include those based on the selective enrichment of the 5mC-containing DNA by using antibodies or 5mC-binding agents, but these methods are heavily dependent on the specificity of the affinity reagents. Compared with these methods, enzyme-based methods have their unique advantages due to their high specificities, mild reaction conditions, and convenient usage.
Following our discovery of MspJI, a unique modification-dependent restriction endonuclease (14), we present here the enzymatic properties of some of its homologs from other prokaryotic species. In brief, these enzymes recognize hemimodified sites and cleave on the 3′ side of the modified cytosine, at a distance N12/N16 away from it. In symmetrically modified sites (such as fully methylated CpG sites), each strand directs cleavage independently, forming a 32-nucleotide long fragment (Fig. 4). This property of long-reaching cleavage at fixed positions allows isolation and direct sequencing of the 32-mer product, or end-sequencing of the longer fragments to reveal the position of the modified cytosine in the genome. The availability of multiple MspJI-like enzymes with different recognition preferences increases the coverage of the modified sites in the epigenome.
As shown in Fig. 4, these enzymes should be useful in detecting the presence of modified cytosines and examining the global structure of the epigenome. Roughly, the relative level of methylation in the input DNA correlates well with disappearance of substrate, the average size of the digested fragments, and the amount of the accumulated 32-mer (Fig. 4B). By using a panel of MspJI-like enzymes, specific methylation patterns can be inferred directly from the digestion pattern. For example, RlaI is able to specifically detect methylated CWG sites, which are abundantly present in plants but not in mammalian genomes (Fig. 4C).
We further demonstrated the utility of these enzymes in mapping the reference methylome by directly sequencing the pool of gel-extracted 32-mers. With a single sequencing run, we achieve approximately 55% sensitivity and approximately 87% specificity compared with a subset of the Salk reference. A few factors should be considered here in the interpretation of sensitivity: First, the enzyme specificity may introduce bias toward those methylated CG sites with favorable sequence context; second, the resolution of the 32-mer in human genome may still be limited so that in some cases it may be difficult to uniquely align them to the genome, especially when the repetitive elements in the genome tend to be highly methylated. In the Salk reference methylome, we have determined about 15% of the theoretical 32-mer generated by MspJI have at least another duplicated copy elsewhere in the genome. In addition, when the distance between the two methylated CG sites is short, it may interfere with the MspJI digestion so that fragments shorter than 32 base pairs can be formed. SI Appendix, Fig. S3 shows different digestion scenarios depending on the distance between the neighboring methylated CG. Nevertheless, each of these scenarios may produce several possibilities that may be resolved by performing the digestion using different combinations of MspJI homologs that detect different sets of methylated sites.
Recently, another form of modified cytosine, 5-hydroxymethylcytosine (5hmC), was found to be present in human and mouse genomes (18, 21), although their genomic locations remain largely unknown. Although bisulfite sequencing yields single nucleotide resolution information, it does not allow differentiation between 5mC and 5hmC (22). We have found that the MspJI-like enzymes are able to recognize and cleave 5hmC, but not glucosylated 5hmC (Fig. 1). This may provide a simple basis for different strategies to map the genomic locations of 5hmC. In addition to genome-wide methylation analysis, MspJI-like enzymes may also be used as a quick assay to probe the modification status in specific genomic loci. In this regard, common analytical methods, such as PCR or ligation-mediated methods, can be used to detect the enzymatic DNA cleavage before and after glucosylation reaction. An added advantage in some cases is the ability to distinguish the strand-specific modification status.
At the sequence level, families of modification-dependent restriction endonucleases (e.g., MspJI, Mrr, McrBC, McrA, etc.) appear to be rather conserved, and homologs can be readily identified in many prokaryotic species (SI Appendix, Fig. S4). In contrast, each specificity of the more-studied type IIP restriction enzymes is one of nature’s unique inventions—they typically do not have widespread homologs in other species. Despite sequence similarity, members of the MspJI family have differences in their dependence on the flanking nucleotide of the recognized modified base. Although currently it appears that modification-dependent restriction endonucleases are not as diverse as their type IIP counterparts, given the abundance of various DNA modifications in bacteriophages, future studies may show that bacterial hosts have many specialized families of enzymes that recognize these alien modifications.
Materials and Methods
All enzymes, plasmids and bacterial strains, if not otherwise specified, were obtained from New England Biolabs Inc. (NEB).
Cloning, protein expression and purification, unit definition, and endonuclease assays are detailed in the SI Appendix.
All genomic DNA digestion reactions were carried out in standard NEB buffer 4. Genomic DNA samples of HeLa and Jurkat cell DNA as well as 5-Aza-dC-treated (16) and enzymatically mCpG-methylated Jurkat cell DNA were obtained from NEB, and other genomic DNAs were purchased from BioChain [rabbit liver DNA (#D1834149), corn DNA (#D1634330), soy bean DNA (#D1634370)]. In the digestion series presented in Fig. 4, approximately 1 μg of each genomic DNA sample was digested by 1 unit of each enzyme in the presence of 0.5 μM double-stranded DNA activator in a 30-μL volume. All reactions were incubated at 37 °C for 16 h. The reactions were then subjected to a 20% TBE (89 mM Tris/89 mM boric acid/2 mM EDTA, pH 8) native polyacrylamide gel electrophoresis (PAGE), and visualized by SYBR GOLD staining.
Sequencing and bioinformatic analysis are detailed in the SI Appendix.
Supplementary Material
Acknowledgments.
We thank our colleagues Drs. Mala Samaranayake, Harriet Strimpel, Katherine Marks, Romualdas Vaisvila, Janos Posfai, and members in the Restriction Enzyme Division at NEB for helpful discussions throughout this project and sharing of reagents. We thank Drs. Elisabeth Raleigh and William Jack for critical reading of this manuscript. This work is supported by NEB and by National Institute of General Medical Sciences Small Business Innovation Research Grant 1R44GM095209-01 (Y.Z.).
Footnotes
Conflict of interest statement: Subjects of this paper are potential products of New England Biolabs.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018448108/-/DCSupplemental.
References
- 1.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—A database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warren RA. Modified bases in bacteriophage DNAs. Annu Rev Microbiol. 1980;34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- 3.Ehrlich M, Ehrlich K, Mayo JA. Unusual properties of the DNA from Xanthomonas phage XP-12 in which 5-methylcytosine completely replaces cytosine. Biochim Biophys Acta. 1975;395:109–119. doi: 10.1016/0005-2787(75)90149-5. [DOI] [PubMed] [Google Scholar]
- 4.Revel HR. Restriction of nonglucosylated T-even bacteriophage: Properties of permissive mutants of Escherichia coli B and K12. Virology. 1967;31:688–701. doi: 10.1016/0042-6822(67)90197-3. [DOI] [PubMed] [Google Scholar]
- 5.Bair CL, Black LW. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J Mol Biol. 2007;366:768–778. doi: 10.1016/j.jmb.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarasova GV, Nayakshina TN, Degtyarev SK. Substrate specificity of new methyl-directed DNA endonuclease GlaI. BMC Mol Biol. 2008;9:7. doi: 10.1186/1471-2199-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raleigh EA, Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci USA. 1986;83:9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raleigh EA, Trimarchi R, Revel H. Genetic and physical mapping of the mcrA (rglA) and mcrB (rglB) loci of Escherichia coli K-12. Genetics. 1989;122:279–296. doi: 10.1093/genetics/122.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulligan EA, Hatchwell E, McCorkle SR, Dunn JJ. Differential binding of Escherichia coli McrA protein to DNA sequences that contain the dinucleotide m5CpG. Nucleic Acids Res. 2010;38:1997–2005. doi: 10.1093/nar/gkp1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panne D, Raleigh EA, Bickle TA. The McrBC endonuclease translocates DNA in a reaction dependent on GTP hydrolysis. J Mol Biol. 1999;290:49–60. doi: 10.1006/jmbi.1999.2894. [DOI] [PubMed] [Google Scholar]
- 11.Sutherland E, Coe L, Raleigh EA. McrBC: A multisubunit GTP-dependent restriction endonuclease. J Mol Biol. 1992;225:327–348. doi: 10.1016/0022-2836(92)90925-a. [DOI] [PubMed] [Google Scholar]
- 12.Raleigh EA. Organization and function of the mcrBC genes of Escherichia coli K-12. Mol Microbiol. 1992;6:1079–1086. doi: 10.1111/j.1365-2958.1992.tb01546.x. [DOI] [PubMed] [Google Scholar]
- 13.Waite-Rees PA, et al. Characterization and expression of the Escherichia coli Mrr restriction system. J Bacteriol. 1991;173:5207–5219. doi: 10.1128/jb.173.16.5207-5219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y, et al. A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res. 2010;38:5527–5534. doi: 10.1093/nar/gkq327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 17.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 18.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: Systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.