RNA constitutes the central conduit for information storage, conveyance, and manipulation in biological systems (1, 2). RNAs encode their critical information at two levels. First, the primary sequence directs protein synthesis and contains simple cis-acting elements that bind regulatory factors and other RNAs. Second, most RNAs fold to create complex base-paired and higher order structures with intrinsic regulatory functions. Until recently, it has been difficult or impossible to interrogate the structures of most RNAs, especially in complex biological environments. Ongoing advances in nucleotide-resolution RNA structure probing have made possible increasingly rigorous and quantitative analyses (3), and recent large-scale and whole-genome studies have revealed or better defined rules for how RNA structure regulates translation initiation, protein folding, splicing, and access to protein binding sites (4–6). The clear power of next-generation sequencing (NGS) to transform nucleic acid-based analyses (7, 8) has motivated significant efforts (a scientific dash) to meld chemical and enzymatic probing experiments with NGS readouts (termed seq experiments). The melding of RNA structure probing experiments with NGS readout is a potential marriage made in transcriptome heaven. Two papers in PNAS (9, 10) illustrate important progress toward this highly sought goal.
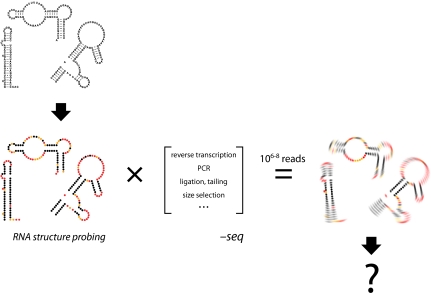
In an RNA structure probing experiment, RNAs are initially incubated with a “reagent” that reacts sparsely and leaves an imprint on the ensemble of RNA molecules (Fig. 1, Left). Features unique to each probe govern the ultimate quality and usefulness of the structural information gleaned. Both RNA-cleaving proteins (RNases) and small chemical probes are widely used. On reaction completion, the RNAs present during probing contain a full and exact imprint of the probing event (Fig. 1, Lower Left). The challenge is to extract this information as accurately as possible.
Fig. 1.
Overview of an RNA structure probing dash seq experiment. An RNA pool (Upper Left) is subjected to RNA structure probing to yield a chemical imprint of the initial structures (Left, colored structures). Red, yellow, and black spheres indicate nucleotides with high, medium, and low reactivities, respectively, toward structure probing. (Center) RNAs are then converted to double-stranded DNA molecules for NGS; not all processing steps shown are used in every NGS experiment. (Right) Sequencing yields a count of the original RNA modifications times the effects of all processing steps, which must be deconvoluted to recapitulate fully the results of the original probing step.
NGS approaches represent a transformative set of technologies that, in principle, make it possible to determine directly all the species present after structural probing. However, NGS requires that the pool of probed RNAs be converted into double-stranded DNAs with known adapter sequences on both ends. In principle, sequencing reads from the ends of these DNAs can be quantified to count the original RNA modification events. The specific steps required to produce a DNA library vary depending on the NGS approach but typically involve using reverse transcriptase to create a cDNA from the probed RNA, one or more ligation steps to create handles for subsequent manipulations, amplification by PCR, and size selection (Fig. 1, Center). Sequences within the resulting DNA library report the original RNA probe imprint but do so imperfectly (11). There are two potential ways to deal with the data blurring (Fig. 1, Right) that results from DNA library construction: (i) design NGS processing to reduce or eliminate these biases and (ii) create bioinformatics tools that fully account for the idiosyncrasies of each manipulation. NGS data are particularly amenable to this latter approach because the data are, in essence, digital and the tens of millions of sequencing reads facilitate statistical deconvolution.
It is in this bioinformatics area that the papers by Aviran et al. (9) and Lucks et al. (10) focus. Reverse transcriptase-mediated primer extension has been used to create cDNAs to read out the results of RNA chemical probing experiments for over 3 decades. Reverse transcription works because the RNA modification or cleavage blocks DNA extension (3). However, reverse transcriptase sometimes stops spontaneously and the enzyme stops at the first modification encountered, even if an RNA has been modified several times. These processes are called drop-off and can be accounted for using an approximate heuristic algorithm (12). Aviran et al. (9) now present an authoritative “maximum likelihood” approach for modeling reverse transcriptase-mediated primer extension and for automatically extracting the probability of RNA modification at each position in a probed RNA. The approach is computationally efficient, provides a measure of overall data quality, and will likely become the new standard for modeling drop-off by polymerase enzymes.
Lucks et al. (10) take on the challenge of melding the SHAPE RNA structure probing technology with an NGS readout to create one approach for SHAPE-seq. SHAPE is an acronym for selective 2′-hydroxyl acylation analyzed by primer extension. SHAPE measures local nucleotide flexibility in RNA, because the 2′-OH group in unconstrained nucleotides reacts preferentially with hydroxyl-selective electrophiles. SHAPE modification of RNA is largely independent of sequence and modification frequencies reflect a true measure of molecular order (13). Previously, sites of modification were detected by generating fluorescently labeled cDNAs that were then analyzed by automated capillary electrophoresis, which remains the gold standard for a SHAPE readout. The results of SHAPE experiments provide a direct measurement of local RNA structure and can be used to calculate pseudo-free energy terms to yield highly accurate RNA secondary structure predictions (13).
In the SHAPE-seq approach used by Lucks et al. (10), three processing steps are used to detect modification sites: (i) reverse transcriptase-mediated primer extension, for which there is now an authoritative approach for extracting the probability of RNA modification (9); (ii) a single-stranded DNA ligation step; and (iii) PCR. The idiosyncrasies of the last two steps are not well understood. In the approach taken by Lucks et al. (10), the RT step begins at a defined site and the complexity of the input RNA is limited to transcripts of ∼400 nt. As proof-of-principle experiments, Lucks et al. (10) analyze the specificity domain of the Bacillus subtilis RNase P enzyme, mutants of this RNA, and other model RNAs. They exploit key advantages of NGS experiments in data analysis: (i) the data are digital and can be related directly to the sequence of the probed RNA; (ii) the dynamic range of the experiment potentially spans several orders of magnitude in RNA concentration; and (iii) experiments are readily multiplexed, such that several hundred RNA are analyzable in a single experiment.
Lucks et al. (10) make an important step in the right direction, but there is more work to be done. These authors carefully note that the precise reactivities measured by SHAPE-seq can differ from those read out by capillary electrophoresis. A correlation plot reveals that there is a tendency for points to cluster near the axes instead of in the center,
The melding of RNA structure probing experiments with NGS readout is a potential marriage made in transcriptome heaven.
which means that nucleotides highly reactive by SHAPE as measured by capillary electrophoresis are scored as unreactive in the current SHAPE-seq experiment, and vice versa. In some cases, the observed changes in SHAPE reactivity resulting from introducing a point mutation into the RNase P RNA fell in opposite directions as measured by SHAPE-seq and capillary electrophoresis. When the SHAPE-seq data are used to constrain RNA secondary structure prediction, the resulting model of the RNase P specificity domain includes only 50% of the accepted base pairs and is roughly the same as if no experimental data were used at all.
Although the NGS library construction steps cause the SHAPE-seq data to be quite different from those obtained by capillary electrophoresis, it might be possible to develop bioinformatics approaches to adjust RNA structure prediction and other analysis algorithms. In general, the SHAPE-seq data seem to have a bimodal distribution, with many highly reactive and unreactive nucleotides and fewer intermediate measurements. Intriguingly, if the SHAPE-seq data are simply scaled up by an order of magnitude, so as to emphasize the intermediate fine structural features, we found that a solid secondary structure prediction results. This, admittedly cursory, analysis suggests that bioinformatics approaches may be able to account for the convolution of SHAPE data by the sample preparation steps required in an NGS experiment.
All available evidence suggests that higher order structure modulates RNA function at every step at which RNA plays a role (1, 2, 4–6). RNA structure probing dash seq experiments are in the early stages of development but are likely to play a transforming role in understanding how biological information is manifested in the higher order structure of RNA. In the past year, two RNase-seq experiments (6, 14) and the SHAPE-seq class experiment in PNAS (10) have been developed. These approaches are likely to prove powerful for understanding RNA structure and dynamics and interactions with RNA, protein, and small molecule ligands in myriad fundamental biological processes. The next big challenges include analysis of full-length RNAs and RNAs in their native cellular or viral environments. Although RNA structure probing dash seq approaches offer enormous promise and opportunities, thus far, the additional enzymatic processing steps required for NGS readouts have had the effect of blurring the lens through which we visualize RNA biology (Fig. 1). Work to date in this field offers a clear and promising glimpse into a future in which experimental and algorithmic innovation may provide “corrective lenses” through which we can view RNA structure at new levels of sophistication.
Acknowledgments
I am indebted to Julius Lucks for sharing SHAPE-seq data prior to publication and to Christine Hajdin for RNA secondary structure calculations. Work on creating high-content molecular microscopes for RNA in my laboratory is supported by the US National Science Foundation and National Institutes of Health.
Footnotes
References
- 1.Nilsen TW. RNA 1997-2007: A remarkable decade of discovery. Mol Cell. 2007;28:715–720. doi: 10.1016/j.molcel.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 2.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Weeks KM. Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol. 2010;20:295–304. doi: 10.1016/j.sbi.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Burge CB. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wold B, Myers RM. Sequence census methods for functional genomics. Nat Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aviran S, et al. Modeling and automation of sequencing-based characterization of RNA structure. Proc Natl Acad Sci USA. 2011;108:11069–11074. doi: 10.1073/pnas.1106541108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucks JB, et al. Multiplexed RNA structure characterization with selective 2'-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc Natl Acad Sci USA. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linsen SE, et al. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods. 2009;6:474–476. doi: 10.1038/nmeth0709-474. [DOI] [PubMed] [Google Scholar]
- 12.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weeks KM, Mauger DM. Exploring RNA structural codes with SHAPE chemistry. Acc Chem Res. May 26, 2011 doi: 10.1021/ar200051h. 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Underwood JG, et al. FragSeq: Transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]