Abstract
Neither the number of HIV-1 proviruses within individual infected cells in HIV-1–infected patients nor their genetic relatedness within individual infected cells and between cells and plasma virus are well defined. To address these issues we developed a technique to quantify and genetically characterize HIV-1 DNA from single infected cells in vivo. Analysis of peripheral blood CD4+ T cells from nine patients revealed that the majority of infected cells contain only one copy of HIV-1 DNA, implying a limited potential for recombination in virus produced by these cells. The genetic similarity between HIV populations in CD4+ T cells and plasma implies ongoing exchange between these compartments both early and late after infection.
The genetic diversity of HIV (HIV-1) allows the virus to escape immune pressure and rapidly develop drug resistance and has hindered the development of a functional vaccine. Three mechanisms contribute to the genetic diversity of HIV: rapid, high-level virus turnover (ca. 108–109 cells are infected every day), nucleotide misincorporation during replication of the HIV-1 genome by the error-prone reverse transcriptase, and recombination (1–3).
As with other retroviruses, HIV-1 recombination occurs during reverse transcription when reverse transcriptase switches between the two RNA genome templates in the infecting virion and uses information from both of them to generate a hybrid viral DNA. Although recombination can occur in all infection events, only virions that contain two genetically distinct RNAs can generate a recombinant that is genotypically different from either of the two parental strains (4). The production of a genotypically different recombinant is therefore a multistep process. The virus producer cell needs to be infected by two or more genetically distinct viruses, RNAs transcribed from the different proviruses have to be copackaged into a heterodimeric virion, and template switching during reverse transcription must take place to generate recombinant viral DNA (5). It has been estimated that as many as 30 template switches may take place during a single infection event (reviewed in ref. 6). The potential for productive recombination in HIV-1–infected individuals is therefore strongly dependent on both the frequency of multiply infected cells and the genetic relationship of the proviruses they contain.
Isolation of recombinants from infected individuals provides evidence of multiple infected cells (7–10). Furthermore, in vitro studies have shown the occurrence of doubly infected cells (11, 12) and the generation of heterodimeric virions with two different viral RNAs (13). Evidence for multiply HIV-1–infected cells in vivo was first demonstrated in spleen by Gratton et al. (14) and further confirmed in a study by Jung et al. (15). The latter study concluded that CD4+ cells isolated from the spleen harbored between one and eight (with a mean of 3.2) proviruses per cell and that the proviruses within single cells were genetically diverse (15). Although both the in vitro and in vivo studies point to the possibility of extensive multiple infection, recent modeling studies by Neher et al. (16) and Batorsky et al. (17) concluded that, on the basis of the amount of viral recombination observed during chronic HIV-1 infection, only 10% or less of HIV-1–infected cells are multiply infected with genetically distinct virus.
Although the modeling studies indicate low effective recombination rates during disease progression, it is unclear how often infected host cells contain multiple HIV-1 proviruses. Furthermore, the genetic relatedness of proviruses within an infected cell to one another and to the extracellular virus population is unknown. To address these issues we developed the single-cell sequencing assay (SCS), which allows a direct analysis of the number of HIV-1 DNA molecules in single HIV-1–infected cells and reveals their relatedness to one another, to DNA in other cells, and to genome sequences derived from contemporaneous plasma virus RNA.
In the present study, analysis of cells from five recently (<6 mo) and four chronically (2–15 y) infected patients revealed that the majority (>85%) of infected CD4+ T cells in blood contain only one copy of HIV-1 DNA, implying a limited potential for recombination in virus produced by these cells. Sequence analysis revealed that intracellular viral DNA from CD4+ T cells in each of the nine patients was phylogenetically similar to contemporaneous plasma RNA, indicating ongoing exchange between these compartments during early and chronic HIV-1 infection.
Results
The Majority of Infected CD4+ T Cells Contain One DNA Molecule.
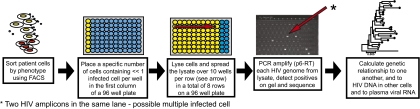
The rate of HIV-1 recombination is dependent on multiply HIV-1–infected cells, the number of which in peripheral blood is unknown. Therefore, we developed the SCS to quantify and genetically characterize HIV-1 DNA molecules from individual infected cells [Fig. 1 and SI Materials and Methods, Detailed Description of Single-Cell Sequencing Assay (SCS)]. The ability of the assay to detect all proviruses in singly and multiply infected cells was validated using 293T cells infected with defined numbers of HIV-based vectors (SI Materials and Methods, Validation of SCS, and Table S1). To evaluate the number of multiply infected CD4+ T cells from peripheral blood a 1.3-kb gag-pol fragment was amplified from samples collected from five recently infected (<6 mo of infection) and four chronically infected patients (2–15 y of infection) using SCS (Table 1). Two time points, ≈6 mo apart, were analyzed for three of the chronically infected patients. The analysis revealed that the majority (>85%) of infected CD4+ T cells contained a single viral DNA molecule (Table 2), but at least one row containing the cell lysate with << one infected cell spread over 10 wells had two or more HIV-1 DNA molecules in eight of the nine patients. There were three possible reasons for the detection of more than one HIV-1 DNA molecule in a row of 10 wells: (i) one cell contained two or more different HIV-1 DNA molecules; (ii) one cell contained a single provirus but was in the process of DNA replication; or (iii) two or more HIV-1 positive cells were lysed and analyzed in the same row. In each case, the number of rows with more than one copy of viral DNA corresponded closely to that predicted by the Poisson distribution under the assumption of no multiple infection [predicted values are shown in parentheses in columns 2–6 (from left) in Table 2 (“Individual patients”)]. For each individual patient, correspondingly, there was insufficient statistical support to reject the hypothesis that no multiple infection was present (P ≥ 0.10 for all patients) [Table 2 (“Individual patients”), column 7]. Additionally, patients sampled early in infection collectively did not demonstrate significant evidence of multiple infection (P = 0.78) [Table 2 (“Combining all early and all chronic patients”), column 7]. However, when results from all patients with chronic infection were combined, the number of rows with more than one DNA molecule significantly exceeded that predicted from a random distribution, implying that multiple infection was present (P = 0.03). These data indicate that, most of the time, the multiple DNA molecules are more likely to have been derived from two or more HIV-positive cells lysed and analyzed in a row of 10 wells than from one cell containing multiple HIV-1 molecules.
Fig. 1.
SCS. CD4+ T cells and monocytes from HIV-1–infected patients were sorted by FACS into PCR plates at dilutions selected so that each well would contain an average of much less than one infected cell. The cells in each well were lysed and their DNA distributed over 10 wells. PCR using primers spanning a 1.3-kb fragment of HIV-1 DNA encompassing part of the gag-pol region was performed, and the products were detected by gel electrophoresis and sequenced. The number of viral DNA molecules per infected cell was estimated from the number of positive wells. Viral DNA amplicons were sequenced and aligned to pNL43. The relatedness of viral DNA sequences to one another and to single-genome sequences derived from contemporaneous plasma virus RNA was determined by phylogenetic analysis.
Table 1.
Patient information
| Patient no. | No. of samples | CD4+ T-cell count (cells/μL) | Viral load(RNA copies/mL) | Length of infection (y) |
| Early Infection | ||||
| 1 | 1 | 512 | 29,700 | 0.5 |
| 2 | 1 | 407 | 3,800 | <0.5 |
| 3 | 1 | 906 | 510 | <0.5 |
| 4 | 1 | 1057 | 1,400,000 | <0.5 |
| 5 | 1 | 482 | 79,800 | 0.5 |
| Chronic Infection | ||||
| 6 | 2 | 704 | 47,787 | 2 |
| 664 | 86,212 | 2.5 | ||
| 7 | 2 | 490 | 12,482 | 3–5.0 |
| 339 | 16,339 | 3–5.5 | ||
| 8 | 2 | 342 | 15,362 | 8.5 |
| 343 | 8,753 | 9 | ||
| 9 | 1 | 101 | 1,801,380 | >15 |
Table 2.
Frequency of infection
| Frequency of infected CD4+ T cells | ||||||||||||
| Observed no. of HIV DNA molecules per row |
Frequency of multiple infection |
Method 1* |
Method 2† |
|||||||||
| Patient | 0 | 1 | 2 | 3 | 4 | P value for null hypothesis of no multiple infection | Upper 95% confidence bound (%) | Most consistent with observed no. of multiple HIV DNA molecules (%) | Cells/HIV DNA | 95% CI | Cells/HIV DNA | 95% CI |
| Individual patients | ||||||||||||
| Early Infection | ||||||||||||
| 1 (1)‡ | 94 (92.1)§ | 20 (22.4) | 4 (4.4) | 1 (0.9) | 1 (0.2) | 0.5 | 40.5 | 2.0 | 869 | 625–1,247 | 1,169 | 798–1,790 |
| 2 (1) | 77 (79.3) | 19 (14.5) | 0 (1.9) | 0 (0.2) | 0 (0) | 1.0 | 14.6 | 0 | 813 | 521–1,349 | 813 | 521–1,349 |
| 3 (1) | 142 (140.5) | 14 (17.2) | 4 (2) | 0 (0.2) | 0 (0) | 0.2 | 43.9 | 10 | 2,847 | 1,881–4,543 | 3,480 | 2,202–5,872 |
| 4 (1) | 58 (61.4) | 21 (15.1) | 1 (2.9) | 0 (0.6) | 0 (0.1) | 0.98 | 19.8 | 0 | 1,078 | 719–1701 | 1,127 | 745–1,798 |
| 5 (1) | 119 (118) | 15 (16.1) | 1 (1.7) | 1 (0.1) | 0 (0) | 0.56 | 32.6 | 0.9 | 3,532 | 2,287–5,782 | 4,155 | 2,595–7,133 |
| Chronic Infection | ||||||||||||
| 6 (2) | 65 (65.6) | 20 (19.1) | 3 (2.9) | 0 (0.3) | 0 (0) | 0.25 | 30.4 | 4.9 | 954 | 651–1,460 | 1,078 | 719–1,701 |
| 142 (139) | 20 (25.4) | 5 (3.2) | 1 (0.3) | 0 (0) | 40.5 | 783 | 558–1,137 | 994 | 678–1,521 | |||
| 7 (2) | 128 (122) | 15 (24.9) | 7 (4.2) | 1 (0.7) | 1 (0.1) | 0.1 | 56.3 | 9.7 | 1,296 | 936–1,850 | 1,943 | 1,306–3,033 |
| 88 (88.8) | 20 (20.1) | 4 (3.2) | 0 (0.4) | 0 (0) | 34.2 | 666 | 461–1,002 | 777 | 522–1,212 | |||
| 8 (2) | 99 (97.3) | 10 (13.4) | 3 (1.2) | 0 (0.1) | 0 (0) | 0.14 | 49.5 | 10.4 | 1,465 | 902–2,563 | 1,803 | 1,055–3,368 |
| 41 (37.4) | 10 (14.8) | 2 (3.2) | 3 (0.5) | 0 (0.1) | 57.7 | 2,191 | 1,461–3,457 | 3,360 | 2,037–6,003 | |||
| 9 (1) | 47 (47.6) | 15 (14.1) | 2 (2.1) | 0 (0.2) | 0 (0) | 0.69 | 32.6 | 0 | 101 | 65–168 | 113 | 71–194 |
| Combining all early and all chronic patients | ||||||||||||
| Early Infection | ||||||||||||
| 490 (491.3) | 89 (85.3) | 10 (12.9) | 2 (2.0) | 1 (0.3) | 0.78 | 19.2 | 2.58 | 1,503¶ | 1,730 | |||
| Chronic Infection | ||||||||||||
| 610 (597.7) | 110 (131.8) | 26 (20.0) | 5 (2.5) | 1 (0.2) | 0.03 | 29.1 | 7.0 | 617 | 771 | |||
*Method 1: Assuming no multiple infection.
†Method 2: Assuming maximal multiple infection.
‡Number of samples per patient in parentheses.
§Poisson predicted values in parentheses.
¶Geometric mean of early and chronic infection values.
Although we cannot definitively determine the frequency of multiply infected cells from the data in Table 2, we can use these results to put bounds on their frequency. If we conservatively assume that all rows containing two or more HIV-1 DNA molecules are the result of multiply infected cells, we can conclude with a confidence level of 95% that fewer than 15–44% of cells during early infection and fewer than 30–58% of cells during chronic infection are multiply infected [Table 2 (“Individual patients”), column 8]. When we combined all of the data from all of the early infected patients we found with 95% confidence that multiple infection occurs in less than 20% of cells [Table 2 (“Combining all early and all chronic patients”), column 8]. For cells from chronically infected patients, multiple infections occur at less than 30%. However, when simulations were conducted to estimate the rate of multiple infection most consistent with the observed distribution of HIV-1 DNA molecules, we found that the most likely number of multiply infected cells was ≤10% for all patients [Table 2 (“Individual patients”), column 9]. Overall, these data suggest that during untreated HIV-1 infection multiple infection of CD4+ T cells in peripheral blood is an uncommon event; however, it seems that multiply infected cells may be more common during chronic infection.
Frequency of Infection of Peripheral CD4+ T Cells Is Similar During Early and Chronic Infection.
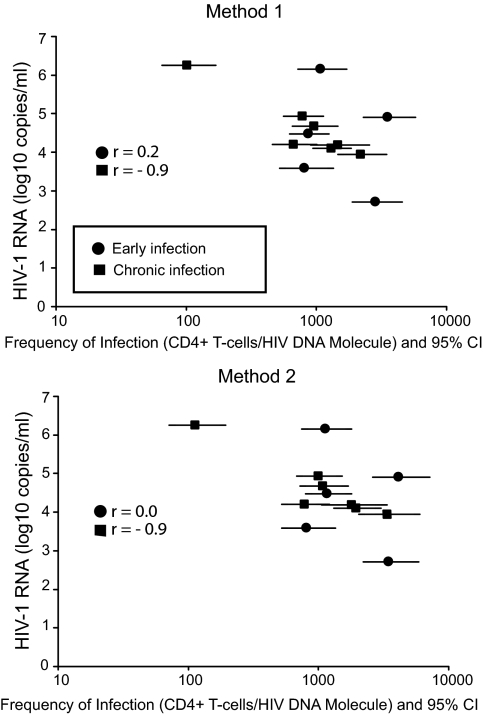
To determine whether viral RNA levels, CD4+ T-cell count, or length of infection influenced the frequency of HIV-1–infected cells, we calculated the frequency of infected CD4+ T cells in all samples from the nine patients. The frequency of infection was determined by dividing the total number of cells analyzed in each sample by the number of infected cells calculated from the number of DNA molecules detected in each row of 10 wells. Because multiple DNA molecules in one row could have been derived from singly or multiply infected cells, we used two methods to account for the range of possible multiple infection rates. The first method, in which all multiple DNA molecules were assumed to derive from singly infected cells (no occurrence of multiple infection), yielding a higher frequency of infection (fewer cells per HIV DNA molecule). The second method, in which all multiple DNA molecules were assumed to derive from multiply infected cells (maximal occurrence of multiple infection), yielding a lower frequency of infection (more cells per HIV DNA molecule) [Table 2 (“Individual patients”), columns 10–13]. These calculations gave a geometric mean of 1,503–1,730 cells per DNA molecule during early infection (patients 1–5) and 617–771 cells per DNA molecule during chronic infection (patients 6–9). When we compared the infection frequency in early infection with chronic infection (geometric mean of the two time points for patients 6–8) we could not show any difference between the two groups for methods 1 or 2 (P > 0.4, Mann–Whitney test). Similarly, when we accounted for the range of infection frequency we found that the ranges during early and chronic infection overlapped, suggesting no difference of infection frequency during early and chronic infection (Fig. 2). However, the ranges of estimated infection frequency demonstrated a large variability of infection frequency across patients (521–7,133 cells per DNA molecule during early infection for patients 1–5 and 65–6,003 cells per DNA molecule during chronic infection for patients 6–9). For example, patient 9, who had been infected for at least 15 y, had a much higher frequency of infected CD4+ T cells compared with the other patients. Further statistical analysis showed no correlation between frequency of infected cells and plasma viral load [Spearman r = 0.20 (method 1) and 0.00 (method 2)] in samples collected from patients during early infection; however, a correlation was found in samples collected from patients during chronic infection [Spearman r = −0.86 (method 1) and −0.89 (method 2)] (Fig. 2), but when patient 9, infected for > 15 y, was taken out from the data set this correlation was not as strong [Spearman r = −0.77 (method 1) and −0.82 (method 2)]. No correlation was found between frequency of infected CD4+ T cells and CD4+ T-cell count in either subset of patients [Spearman r = 0.30 (method 1) and 0.10 (method 2) and 0.13 (method 1) and 0.18 (method 2) in early and chronic infection, respectively]. In addition to analyzing CD4+ T cells, we also analyzed monocytes from peripheral blood. Monocytes were isolated by sorting either for CD14+ or CD14loCD16hi positive cells. This analysis (CD14+ in five of the nine patients and CD16hiCD14lo in four of the nine patients) did not identify any HIV-1–infected monocytes corresponding to less than 1 HIV-1 DNA molecule per 2,300–200,000 cells (SI Discussion and Table S2).
Fig. 2.
Frequency of infection of CD4+ T cells from peripheral blood in early (n = 5) and chronic infection (n = 4) vs. plasma viral RNA level. Frequency of infection for each patient and time point was calculated using two methods. The first method assumed that each observed HIV-1 amplicon represented a unique infected cell, corresponding to an assumption that no multiple infection was present. The second method assumed that any row with multiple proviruses represented only one infected cell, corresponding to an assumption that the maximum possible rate of multiple infection was present. Frequency of infection is shown as total CD4+ T cells per HIV DNA molecule. The frequency of infection with 95% confidence interval in early and chronic infection was compared, as well as the correlation between frequency of infection and plasma viral RNA levels within the two groups. The correlation analysis is represented as Spearman r values in the figure.
Sequences from Cellular DNA and Plasma RNA Are Phylogenetically Similar in Both Early and Chronic Infection.
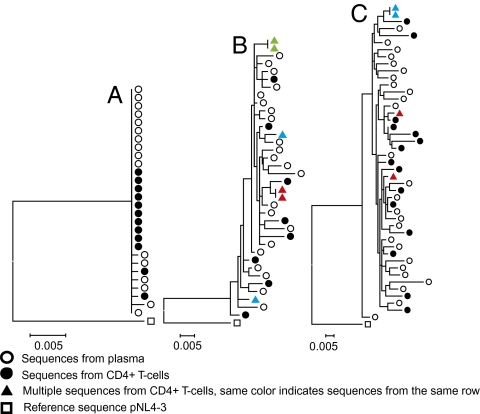
To evaluate the relatedness of viral DNA sequences from CD4+ T cells to one another and to single-genome sequences derived from contemporaneous plasma virus RNA, we conducted phylogenetic analyses (Fig. 3). Sequences from all nine patient samples as well as standard laboratory viruses formed independent populations that were at least 5% different from one another on a phylogenetic tree, with no intermingling, demonstrating that the viruses found in these patients were genetically distinct. As expected, intrapatient comparisons showed that all five early infection patients had nearly monomorphic viral populations derived from both cells and plasma (an example is shown in Fig. 3A). The homogeneity of viral sequences in patients with early infection contrasted with the heterogeneity of sequences found in patients with chronic infection (examples shown in Fig. 3 B and C). In samples from seven of the nine patients we found sequences that were identical for one or more doublet DNA molecules amplified from the same row of 10 wells (examples shown in Fig. 3 B and C), for a total of 23 identical doublets out of 45 multiples. Such doublets could result either from a cell in the process of S-phase DNA synthesis or from one cell infected by two genetically identical virions. For all patients analyzed, the phylogenetic distribution of intracellular DNA sequences was similar to single-genome sequences derived from plasma virus RNA taken at the same time. The genetic similarity between HIV-1 populations in CD4+ T cells and plasma implies ongoing exchange between these compartments.
Fig. 3.
Phylogenetic analysis of viral sequences from cells and plasma. Maximum likelihood trees of plasma (open circles) and cell-derived sequences (filled circles) from early HIV infection (patient 2) (A), chronic HIV infection (patient 7, time point 1) (B), and patient 9, the longest-infected patient (C). All trees are rooted to pNL43 (open square). Colored triangles denote sequences from different wells in the same row.
HIV-1 Genetic Diversity Is Similar in Paired Cell and Plasma Samples.
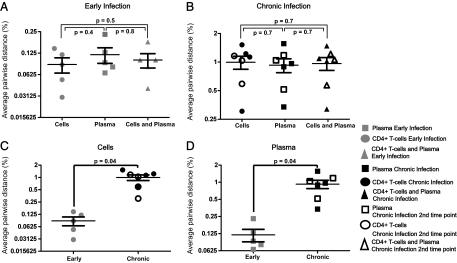
To further assess the genetic relationship between HIV-1 populations found in cells and plasma, the diversity of sequences from all patient samples was calculated using average pairwise distance (APD). The APD of the HIV-1 DNA from CD4+ T cells and plasma RNA was 0.09% and 0.12% in recently (Fig. 4A) and 1.0% and 0.9% in chronically infected patients (both time points were combined for patients 6–8), respectively (Fig. 4B). For each patient, the APD of intracellular and extracellular HIV-1 populations was similar (P ≥ 0.4, Mann–Whitney test) and, consistent with the tree analysis, the APD of the combined cell- and plasma-derived sequences was not different from the APD of either the intracellular or extracellular HIV-1 populations (both P ≥ 0.5, Mann–Whitney test) (Fig. 4 A and B). As expected, the APD of HIV-1 populations from both cells and plasma in early infection was significantly lower than that found in these two compartments during chronic infection (P < 0.04) for both compartments and for both time points compared separately for the chronic infection patients) (Fig. 4 C and D).
Fig. 4.
Genetic diversity of HIV-1 in CD4+ T cells and plasma during early (n = 5) and chronic (n = 4) stages of infection. Diversity was measured as APD and shown as percent differences. Symbols in gray represent values from early infection, and symbols in black represent values from chronic infection. Intrapatient genetic diversity of HIV-1 DNA from CD4+ T cells vs. RNA from plasma and the combined interpopulation APD of cells and plasma during early (A) and chronic infection (B). Comparison of HIV-1 diversity in CD4+ T cells (C) and plasma (D) collected from patients during early vs. the two different time points for chronic infection. Statistical analysis was conducted using the Mann–Whitney test.
Discussion
The production of recombinant HIV-1 variants is strongly dependent on both the frequency of multiply infected cells and the genetic relationship of the proviruses they contain (4). Infection of cells with two different virions has been shown to occur in vitro (11, 12, 18); however, to date in vivo infected cells containing multiple HIV-1 variants have only been identified in spleen tissue from chronically infected patients (14, 15). Despite the lack of direct evidence since the publication of these studies, it has been generally accepted that most cells are multiply infected during HIV-1 disease.
To investigate the numbers of multiply infected cells in the peripheral blood from HIV-1–infected individuals we developed the SCS. We found that the majority (>85%) of CD4+ T cells from the peripheral blood in nine treatment-naïve patients (five in early and four in chronic infection) contain only a single viral DNA molecule. In samples from eight of the nine patients we observed DNA molecules that could arise from multiply infected cells. If we assume that all of the multiple HIV-1 DNA molecules that we observed were derived from multiply infected cells the upper 95% confidence bound for the proportion of multiply HIV-1–infected CD4+ T cells in peripheral blood is 19% and 29% during early and chronic infection, respectively, far below the previous estimate (14, 15). Furthermore, when we compared the actual number of multiple HIV-1 DNA molecules with the values predicted by Poisson distribution, we estimated the most likely frequency of cells containing multiple HIV-1 DNA molecules during early and chronic infection to be 2.6% and 7.0%, respectively, and not significantly different from 0 for any individual patient.
These findings agree with the recent analyses of Neher et al. (16) and Batorsky et al. (17), who used modeling to estimate a rate of recombination consistent with an effective coinfection rate of ≤10%, and Simmonds et al. (19), who found evidence for one provirus per infected peripheral blood mononuclear cell. Several factors having to do with the cell source may explain the discrepancy with the Gratton and Jung results (14, 15). These authors analyzed spleen cells, as opposed to cells from the peripheral blood, and some studies have detected a higher concentration of HIV-1 in lymphoid tissue (20, 21); therefore, spleen cells may encounter HIV-1 virions more frequently than cells in the peripheral circulation. In addition, in vitro studies have shown that cell-to-cell transmission (which is a common feature in lymphoid tissue) more efficiently transfers virions to the acceptor cell, increasing the possibility of multiple infection (11, 12, 22). Furthermore, our analysis was limited to CD4+ T cells from the peripheral blood, and other cells and/or cellular reservoirs such as the spleen may have higher numbers of multiply HIV-1–infected cells. Despite these explanations, our results demonstrate that during productive HIV-1 infection most of the CD4+ T cells in the peripheral blood contain a single HIV-1 DNA molecule with the same population of sequences as found in plasma virus.
According to previous studies, CD4+ T-cell populations harbor the highest, but still relatively low, numbers of HIV-1 proviruses in the peripheral blood (infection rate of one per 100–107 cells) (19, 23, 24). In agreement with these studies, we found low frequencies of infection in CD4+ T cells from peripheral blood. However, in using SCS we achieved more accurate estimates of the HIV-1 infection rate of CD4+ T cells, averaging ≈1,600 cells per HIV DNA molecule (0.06%) during early infection and ≈700 cells per HIV DNA molecule (0.14%) during chronic infection, values not significantly different from each other (P > 0.4). Taken together, these findings indicate that the duration of infection does not seem to affect the frequency of infection of CD4+ T cells as long as the viral RNA levels are relatively low and the CD4+ T-cell count is higher than 300 cells/μL. An earlier study revealed that a high frequency of infection correlates with low CD4+ T-cell counts and high viral loads (25). In agreement with this study, we found that patient 9, infected for 15 y with a low CD4+ T-cell count (101 cells/μL) and a high viral load (1.8 × 106 copies/mL), had a much higher CD4+ T-cell infection rate (average ≈107 cells per HIV DNA molecule, 0.93%) than all of the other patients analyzed. When comparing the CD4+ T-cell frequency of infection with viral load we found a correlation between frequency of infection and viral RNA levels in patients during chronic infection. The lack of correlation between the infection frequency of CD4+ T cells and viral RNA levels in early infection could be explained by the fact that a viral set point has not been established in these patients. Interestingly, we were unable to detect infection of monocytes in this study (SI Discussion and Table S2), despite previous reports to the contrary (26–28).
The genetic makeup of HIV-1 DNA molecules in multiply infected cells is crucial for the production of heterodimeric virions (4). In samples from eight of the nine patients, we detected more than one HIV-1 DNA molecule in the infected cell lysate (<20% of the time), and these molecules could be the result of a multiply infected cell. Analyzing the genetic relationship between these HIV-1 DNA molecules revealed both genetically homogenous and distinct sequences. Because identical viral sequences were rarely found among the sequences from plasma (7 of 203 sequences) in this study or in other studies of untreated chronic infection, we believe that the homogenous sequences found in the cell samples from the chronic patients come from cells with multiple proviruses, either singly infected cells in the process of replication or doubly infected by cell-to-cell transfer of virus from a singly infected cell. Of the 45 potentially multiply infected cells we observed, 23 contained pairs of identical sequences (8 during acute infection and 15 during chronic infection, time points 1 and 2 combined for patients 6–8). However, the rest did contain genetically distinct DNA sequences, which, if they arose from double infection and not multiple singly infected cells, could give rise to heterodimeric virions and new genetic recombinants.
We also analyzed the genetic relationship between intracellular HIV-1 DNA populations and HIV-1 RNA isolated from plasma. The half-life of HIV-1 in blood is very short (on the order of 1 h) (29), indicating that the virions found in plasma have recently been produced and reflect ongoing viral production from cells in the peripheral blood. In agreement with a recent report by Edo-Matas et al. (30), we found that intracellular viral DNA sequences in these infected patients were phylogenetically similar to sequences derived from contemporaneous plasma RNA. We also found that the genetic diversity of the HIV-1 populations in these two compartments was similar. This comparison suggests that the virus found in these patients’ plasma is produced by their peripheral CD4+ T cells. Recently, we and others demonstrated that genetic diversity in virus isolated from plasma in the majority of patients is low in early infection and increases with time of infection (31, 32). In this study we were able to demonstrate the same pattern in sequences isolated from peripheral CD4+ T cells.
In conclusion, our results indicate that most CD4+ T cells in the peripheral blood contain only one copy of HIV-1 DNA, implying limited potential for recombination in viruses produced by this cellular population. The genetic similarity between HIV-1 populations in CD4+ T cells isolated from peripheral blood and plasma implies ongoing exchange between plasma RNA and DNA from peripheral CD4+ T cells. The failure to detect HIV-1–infected monocytes in this study implies that their frequency of infection is very low compared with CD4+ T cells.
Materials and Methods
Clinical Specimens.
Cells and plasma samples from five recently and four chronically infected HIV-1 subtype B-infected individuals were analyzed. The samples were obtained from patients attending the National Institute of Allergy and Infectious Disease Critical Care Medical Department of the National Institutes of Health (NIH), Bethesda, MD (n = 4, patients 6–9) and Venhälsan at Södersjukhuset, Stockholm, Sweden (n = 5, patients 1–5). Informed consent was obtained from all of the patients. The study was approved by the institutional review boards at the NIH and the Karolinska Institutet. All patients were treatment naïve throughout the study (Table 1).
Single-Cell Sequencing.
The single-cell sequencing technique allows for the quantitative and genetic analysis of intracellular HIV-1 viral populations. In brief, pools of cells, each containing << one infected cell, are lysed and distributed across 10 wells per row in a total of eight rows on a 96-well PCR plate. PCR amplification and sequencing of the DNA in each well allows enumeration and analysis of the genetic relationship of viral DNA molecules in each infected cell (Fig. 1, SI Materials and Methods, and Table S1).
Single-Genome Sequencing.
To compare the intracellular populations identified using the SCS with HIV-1 RNA populations found in plasma, we performed single-genome sequencing on the plasma samples from each of the nine patients, as described earlier (31, 33, 34).
Supplementary Material
Acknowledgments
We acknowledge with gratitude the patients participating in this study, G. Bratt and A. Thalme for clinical assistance with patients in Sweden, Diane Rock for assistance with recruitment of the patients attending the clinic at the National Institutes of Health (NIH), and M. Mild for assistance in phylogenetic analysis. This study was supported by an NIH Bench to Bedside Award (to F.M.) in the HIV Drug Resistance Program, the Intramural Research Program of the National Cancer Institute (Center for Cancer Research, NIH) the Swedish Research Council, and American Foundation for AIDS Research Grant 107170-44-RGRL. J.M.C. was a research professor of the American Cancer Society with support from the F. M. Kirby Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107729108/-/DCSupplemental.
References
- 1.Coffin JM. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 2.Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouzine IM, Coffin JM. Evolution of human immunodeficiency virus under selection and weak recombination. Genetics. 2005;170:7–18. doi: 10.1534/genetics.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu WS, Temin HM. Genetic consequences of packaging two RNA genomes in one retroviral particle: Pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu WS, Rhodes T, Dang Q, Pathak V. Retroviral recombination: Review of genetic analyses. Front Biosci. 2003;8:d143–d155. doi: 10.2741/940. [DOI] [PubMed] [Google Scholar]
- 6.Basu VP, et al. Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008;134:19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Charpentier C, Nora T, Tenaillon O, Clavel F, Hance AJ. Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J Virol. 2006;80:2472–2482. doi: 10.1128/JVI.80.5.2472-2482.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, et al. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 2010;6:e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nora T, et al. Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J Virol. 2007;81:7620–7628. doi: 10.1128/JVI.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shriner D, Rodrigo AG, Nickle DC, Mullins JI. Pervasive genomic recombination of HIV-1 in vivo. Genetics. 2004;167:1573–1583. doi: 10.1534/genetics.103.023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, et al. Mechanisms of nonrandom human immunodeficiency virus type 1 infection and double infection: Preference in virus entry is important but is not the sole factor. J Virol. 2005;79:4140–4149. doi: 10.1128/JVI.79.7.4140-4149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang Q, et al. Nonrandom HIV-1 infection and double infection via direct and cell-mediated pathways. Proc Natl Acad Sci USA. 2004;101:632–637. doi: 10.1073/pnas.0307636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, et al. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci USA. 2009;106:13535–13540. doi: 10.1073/pnas.0906822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gratton S, Cheynier R, Dumaurier MJ, Oksenhendler E, Wain-Hobson S. Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers. Proc Natl Acad Sci USA. 2000;97:14566–14571. doi: 10.1073/pnas.97.26.14566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung A, et al. Recombination: Multiply infected spleen cells in HIV patients. Nature. 2002;418:144. doi: 10.1038/418144a. [DOI] [PubMed] [Google Scholar]
- 16.Neher RA, Leitner T. Recombination rate and selection strength in HIV intra-patient evolution. PLOS Comput Biol. 2010;6:e1000660. doi: 10.1371/journal.pcbi.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batorsky R, et al. Estimate of effective recombination rate and average selection coefficient for HIV in chronic infection. Proc Natl Acad Sci USA. 2011;108:5661–5666. doi: 10.1073/pnas.1102036108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quiñones-Mateu ME, et al. In vitro intersubtype recombinants of human immunodeficiency virus type 1: Comparison to recent and circulating in vivo recombinant forms. J Virol. 2002;76:9600–9613. doi: 10.1128/JVI.76.19.9600-9613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simmonds P, et al. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J Virol. 1990;64:864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantaleo G, et al. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature. 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 21.Nuovo GJ, et al. In situ detection of PCR-amplified HIV-1 nucleic acids in lymph nodes and peripheral blood in patients with asymptomatic HIV-1 infection and advanced-stage AIDS. J Acquir Immune Defic Syndr. 1994;7:916–923. [PubMed] [Google Scholar]
- 22.Chen P, Hübner W, Spinelli MA, Chen BK. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol. 2007;81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Psallidopoulos MC, et al. Integrated proviral human immunodeficiency virus type 1 is present in CD4+ peripheral blood lymphocytes in healthy seropositive individuals. J Virol. 1989;63:4626–4631. doi: 10.1128/jvi.63.11.4626-4631.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brinchmann JE, Albert J, Vartdal F. Few infected CD4+ T cells but a high proportion of replication-competent provirus copies in asymptomatic human immunodeficiency virus type 1 infection. J Virol. 1991;65:2019–2023. doi: 10.1128/jvi.65.4.2019-2023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnittman SM, et al. Increasing viral burden in CD4+ T cells from patients with human immunodeficiency virus (HIV) infection reflects rapidly progressive immunosuppression and clinical disease. Ann Intern Med. 1990;113:438–443. doi: 10.7326/0003-4819-113-6-438. [DOI] [PubMed] [Google Scholar]
- 26.Ellery PJ, et al. The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. J Immunol. 2007;178:6581–6589. doi: 10.4049/jimmunol.178.10.6581. [DOI] [PubMed] [Google Scholar]
- 27.Gartner S, et al. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 28.Fulcher JA, et al. Compartmentalization of human immunodeficiency virus type 1 between blood monocytes and CD4+ T cells during infection. J Virol. 2004;78:7883–7893. doi: 10.1128/JVI.78.15.7883-7893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 30.Edo-Matas D, et al. Genetic composition of replication competent clonal HIV-1 variants isolated from peripheral blood mononuclear cells (PBMC), HIV-1 proviral DNA from PBMC and HIV-1 RNA in serum in the course of HIV-1 infection. Virology. 2010;405:492–504. doi: 10.1016/j.virol.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 31.Kearney M, et al. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J Virol. 2009;83:2715–2727. doi: 10.1128/JVI.01960-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer S, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kearney M, et al. Frequent polymorphism at drug resistance sites in HIV-1 protease and reverse transcriptase. AIDS. 2008;22:497–501. doi: 10.1097/QAD.0b013e3282f29478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.