Abstract
The parallel β-helix is a geometrically regular fold commonly found in the proteomes of bacteria, viruses, fungi, archaea, and some vertebrates. β-helix structure has been observed in monomeric units of some aggregated amyloid fibers. In contrast, soluble β-helices, both right- and left-handed, are usually “capped” on each end by one or more secondary structures. Here, an in-depth classification of the diverse range of β-helix cap structures reveals subtle commonalities in structural components and in interactions with the β-helix core. Based on these uncovered commonalities, a toolkit of automated predictors was developed for the two distinct types of cap structures. In vitro deletion of the toolkit-predicted C-terminal cap from the pertactin β-helix resulted in increased aggregation and the formation of soluble oligomeric species. These results suggest that β-helix cap motifs can prevent specific, β-sheet-mediated oligomeric interactions, similar to those observed in amyloid formation.
Keywords: beta-helix, hidden Markov model, threading, aggregation prediction, beta-sheet oligomerization
Parallel β-helices (1–3) are defined by the regular nature of their rungs, each of which consists of two or three β-strands arranged in sequential repeats separated by loops of various lengths (Fig. 1). Helical stacking of these rungs produces two or three parallel β-sheets surrounding a central core filled with inward-facing amino acid side chains. β-helices form structurally and geometrically regular domains, despite the presence of loops of various lengths, which can themselves include regular structure. The regularity of the β-helix structure persists despite great disparity in primary sequence (4–6). While right-handed parallel β-helices were the first to be described (3), left-handed (4, 7) and two-stranded (8) β-helices have now also been identified. Notably, β-helices are overrepresented in bacterial Protein Data Bank (PDB) (9) entries but rare in eukaryotic entries (5).
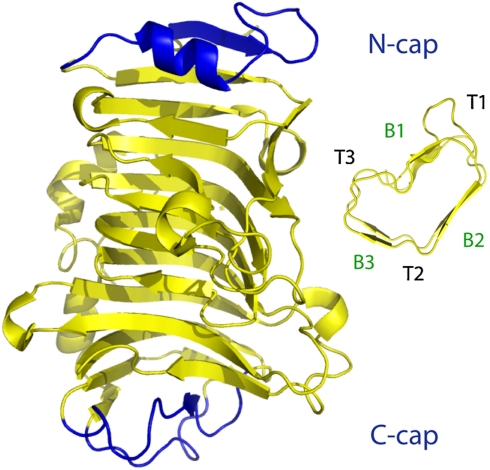
Fig. 1.
Pectin methylesterase, 1GQ8; a typical β-helix. 1GQ8 is a right-handed β-helix, three-sided, with a single α-helix cap at its N terminus and a previous-strand visor cap at its C terminus. Inset, the assignment of β-strand and turn names in a β-helix rung as seen in residues 167-225 of pectate lyase C (2PEC), a similar β-helix.
Richardson and Richardson (10) noted that β-helices often begin and end with a loop at either end, termed a β-helix cap. This cap is amphipathic: one side, sometimes incorporating charged residues, is exposed to solvent, while the other side caps the hydrophobic core of the β-helix. Caps thus protect β-helix cores from solvent exposure. Richardson and Richardson, among others (11), speculated that caps could also prevent aggregation of β-helices. Many agglutinative proteins, including prion and amyloid proteins, are suspected to consist of repeating and indefinitely extendable β-sheets assembled from monomers. Without a mechanism to interrupt formation of potential intermolecular hydrogen bonds at the ends of β-helices, β-helix-forming peptides could associate to form multimeric fibers similar to amyloid. Thus, disruption of β-helix caps could sequester β-helices into aggregate fibrils. However, despite the possible importance of β-helix caps as preventers of aggregation, and despite interest in β-helices as potential models of prion and aggregative protein assembly (12–15), no survey of the presumably analogous assembly interfaces—the known caps and the adjacent structures—has been made.
In this paper we present an in-depth study of β-helix cap structures, describe beta-helix β-helix cap detectors based on our structural results, and report experimental evidence demonstrating a role for cap structures in preventing β-helix aggregation. The available structures in the PDB are classified by β-helix cap fold into α-helix and visor cap motifs that cross-correlate with established β-helix families. Despite wide variety in both sequence and structure, these motifs display subtle but consistent themes that were revealed by focused modeling using hidden Markov models (HMMs) and threading approaches. These models were compiled into a prediction toolkit that accurately identifies β-helix caps from protein sequences with high specificity, even across superfamilies. In vitro deletion of a toolkit-predicted result, the C-terminal cap of the pertactin β-helix, is shown to promote intermolecular interactions and aggregation.
Results
Structural Characterization of β-Helix Caps.
In order to understand β-helix cap structures in sufficient detail to make β-helix cap classification possible, clear definitions and characterization of β-helix caps were required. Therefore, a survey of available β-helix structures from the PDB was conducted (see SI Text). For purposes of analysis, the extent of each cap was defined as the minimum continuous set of residues necessary to fulfill three requirements: (i) at least one continuous subset of cap residues maintains van der Waals contact with the hydrophobic core of the β-helix; (ii) at least one continuous subset of cap residues maintains van der Waals contact with at least one strand of the terminal rung of the β-helix such that the cap backbone intersects the plane of its β-sheet, and (iii) caps do not begin or end within an element of regular secondary structure. The first two requirements reflect the functions of β-helix caps. Contact with the hydrophobic core adds stability and solubility to the β-helix, while intersecting at least one β-sheet plane provides steric hindrance against H-bonding of the terminal rung with other proteins, especially other monomers or oligomers of the β-helix. The third requirement forced the inclusion of the entirety of each element of secondary structure, ensuring sufficient data for accurate sequence/secondary structure comparison.
This survey revealed the vast majority of β-helix caps to follow one of two loose structural patterns. The definitions of these patterns, the α-helix and visor caps, are derived from the general definition above and described in more detail in SI Text. The α-helix caps (Fig. 2 A–C) are characterized by two secondary structures, at least one of which is an α-helix, lying approximately parallel to each other and to β-strand(s) of the adjacent β-helix rung. In contrast, visor caps (Fig. 2 D–I) have in common a more acute angle between structural elements than the angles connecting β-strands in the adjacent β-helix, and a near-perpendicular, as opposed to aligned, intersection with the plane of at least one β-sheet. Aside from the common patterns of turns and contact points, and the presence of at least one α-helix in α-helix caps, β-helix caps display a wide variety of sequence and structure diversity, incorporating loops, additional α-helices, and short β-strands.
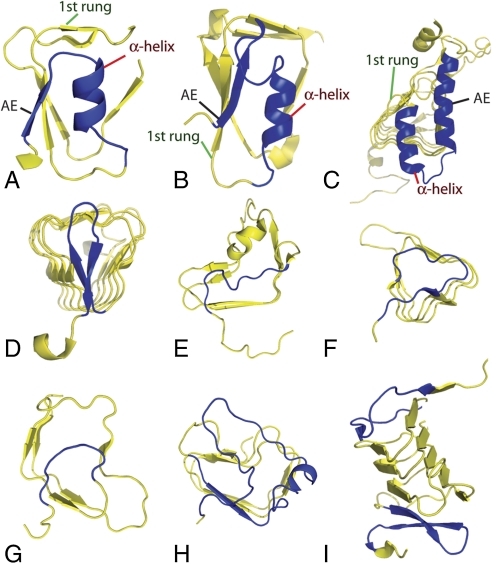
Fig. 2.
α-helix and visor cap structures. All α-helix caps displayed are N-caps and all visor caps displayed are C-caps, except for (I), where both N- and C-caps are visors. For visibility, other domains and distant portions of the β-helices have been removed. All images were produced using PyMol (www.pymol.org). (A–C) Representative α-helix N-caps. Structural components used in HELIXCAP-HMM prediction are labeled: black labels, AE; red labels, α-helix; and green labels, first rung. (A) 1BN8 (residues 33–46 shown); (B) 1GQ8 (residues 8–32 shown); (C) 1KQA (residues 22–54 shown). (D–I) Representative visor family caps. (D) 1KQA (residues 53–190 shown), a previous-strand visor on a left-handed β-helix; (E) 1JTA (residues 260–340 shown), a previous-strand visor on a right-handed β-helix; (F) 1G95 (residues 376–441 shown), a cross-helix visor on a left-handed β-helix; (G) 1DBG (residues 379–433 shown), a cross-helix visor on a right-handed β-helix; (H) 2PEC (residues 217–316 shown), a cross-helix visor containing an α-helix on a right-handed β-helix; (I) 1HF2 (residues 90–206 shown), a structure with visor caps at both the N- and C-terminal ends. Detailed discussion of these cap types may be found in SI Text.
Despite this diversity, models were effectively developed to describe the commonalities of each type of cap. The α-helix caps are composed of sequentially arranged secondary structures, making them compatible with the linear arrangement of states in a Markov model. Hence the α-helix caps were analyzed using global structural alignment (DALI), which could be described by a HMM. The visor caps, in contrast, have more diverse secondary and tertiary structure arrangements. To better capture the diversity of possible structural elements and arrangements of supersecondary structure that characterize visor caps, we used a library of visor cap templates with the RAPTOR threader (16). In addition to identifying the structural commonalities of these cap motifs, each of the predictive models (designated HELIXCAP-HMM and HELIXCAP-visor, respectively) are shown to function as a detector of their respective motifs.
HELIXCAP-HMM: HMM-Based Predictive Model of α-Helix Caps.
Our initial dataset of caps (Table S1) contained 44 β-helix proteins, representing 32 families from the Structural Classification of Proteins (SCOP) (1) of β-helix structures as represented in the PDB. This set was manually divided into classes by N-terminal cap structure, using the terms defined in SI Text, as follows: single α-helix, 24 structures (17 from pectate lyase superfamily, 7 from left-handed superfamily); double α-helix, 2 structures; previous-strand visor, 1 structure; cross-strand visor, 6 structures; interleaved oligomers, 1 structure; and cap not found, 7 structures (see Table S1, N-cap). Of these detected cap types, the single and double α-helix caps had sufficiently similar structures to allow initial structure and sequence alignment (Fig. 3A); the composite structure and sequence alignments of the α-helix caps comprised representatives of 17 SCOP families. Within the α-helix cap, the conserved α-helix and the turns at either end of it—one to an “additional element” (AE) of any secondary structure, the other to the adjacent rung of β-helix structure—anchor this alignment. The AE itself may be any secondary structure that provides the contacts with the β-helix rung and core defined in Structural Characterization of β-Helix Caps, above.
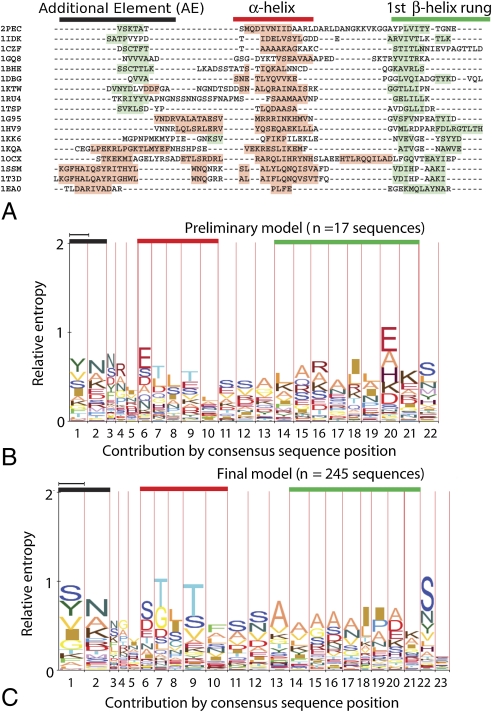
Fig. 3.
Alignment and prediction of α-helix caps. Black bars denote the AE, red the α-helix, and green the first rung of the β-helix. (A) Structurally based alignment. At top, single α-helix caps from the right-handed β-helix pectate lyase superfamily; at bottom, single and double α-helix caps from the left-handed β-helix superfamily. Shading denotes secondary structure by PDB annotation: pink, α-helix; light green, β-strand. (B, C) HMM-Logo representations (18) of the α-helix-cap predictive model. Narrow-column positions are more likely to align with gaps than wide columns. (B) The initial model constructed from 26 aligned crystal structures in 17 families. (C) The augmented model constructed from 1,084 sequences aligned to the initial model.
The sequence and structure patterns of these α-helix caps were used to create an initial descriptive HMM (17). A logo of the HMM, as generated by Logomat-M (18), is shown in Fig. 3B. The most prominent features of the model are the high incidence of residues with side-chain hydroxyl groups (Ser, Thr) within the conserved α-helix and subsequent turn (residues 7, 9, 11, and 12) and the tendency towards residues with hydrophobic side chains (Ala, Leu, Ile) at the N terminus of the first rung (residues 14–21).
In order to produce an α-helix cap HMM with greater statistical support, BLAST (19) was used to search the GenBank (20) protein database for sequence homologs of the 26 single and double α-helix cap structures from the 17 families, resulting in an expanded database of 1,084 sequences. These sequences were aligned by hmmalign to the HMM described above and used to generate a second HMM, depicted in logo format in Fig. 3C. This second model displays moderate but significant signal at all positions, except for low contributions at the turn positions (residues 3–5, 21, and 23). Compared to the first model, there are stronger signals in positions 7 and 9 for serine and threonine residues. In addition, the first rung (residues 13–17) displays a slight but continuous preference for alanine. In positions 18 and 19, where the first rung is crossed by the α-helix above, the preference switches to bulky hydrophobics (isoleucine, leucine, and proline).
To validate our HMM-based model of α-helix caps, several target sets were analyzed. First, as a negative control, the sequences of nonredundant structures in the PDB (9) with all β-helices removed (the “PDB-minus” dataset) was analyzed. None of the sequences in this set (n = 18,659) resulted in an hmmsearch score above threshold (E < 0.5; α = 0). As a positive test and a demonstration of model robustness, leave-one-out cross-validation was performed across each sequence-similarity cluster of the 1,084 source sequences. To ensure that performance was not due to clustering parameters, validation was performed on clusters generated at the lower and upper end of the range of uncertain structural similarity. Below 25% sequence similarity, few structures are similar; conversely, above 75% sequence similarity, few structures fail to exhibit clear commonalities. Therefore, these values were chosen so as to bracket the possible range of clustering parameters. At 25% cluster similarity, the model detected 757 caps (70%), while at 75% similarity, 943 caps (87%) were detected. Finally, to guard against the possibility of overtraining, a model generated without the 26 initial sequences was tested on them; 22 of the 26 sequences were detected (85%).
The predictive performance of the HMM was evaluated on the full set of GenPept bacterial coding open reading frames (ORFs) as of July 27, 2010 (release 177, downloaded from ftp://ftp.ncbi.nih.gov/ncbi-asn1/protein_fasta). After assembly, this set was analyzed with hmmsearch as detailed in Methods. From the GenPept dataset, the model detected 371 potential caps above threshold at 25% cluster similarity and 518 potential caps above threshold at 75% cluster similarity.
HELIXCAP-Visor: Prediction of “Visor” Caps.
Functional similarities of visor caps were investigated as follows. The set of available visor cap structures was determined as shown in Table S1 for both N-terminal and C-terminal structures. The evaluated N-terminal structures are listed under HELIXCAP-HMM: HMM-Based Predictive Model of α-Helix Caps above. The C-terminal structures were divided as follows: single α-helix, 1; previous-strand visor, 20; cross-strand visor, 14; interleaved β-strands, 1; and structure not found, 5. The β-helix domains were extracted from all proteins containing more than one domain, and the set of domains obtained were analyzed using RAPTOR (16) for global β-helix domain Z-scores and for focused cap-to-cap alignments.
The resulting data, presented in full in SI Text, are summarized in graphic form in Fig. 4A. RAPTOR threading results with Z-scores above threshold and with over 50% visor cap-on-cap alignments are depicted with lighter shading indicating lower rmsd for that alignment (see Methods). Because each structure is a representative of a family of β-helices, each hit demonstrates cross-family alignment of cap structures. In addition, beyond the family-to-family alignments (those groups of hits close to the diagonal), a significant number of alignments across superfamilies are observed.
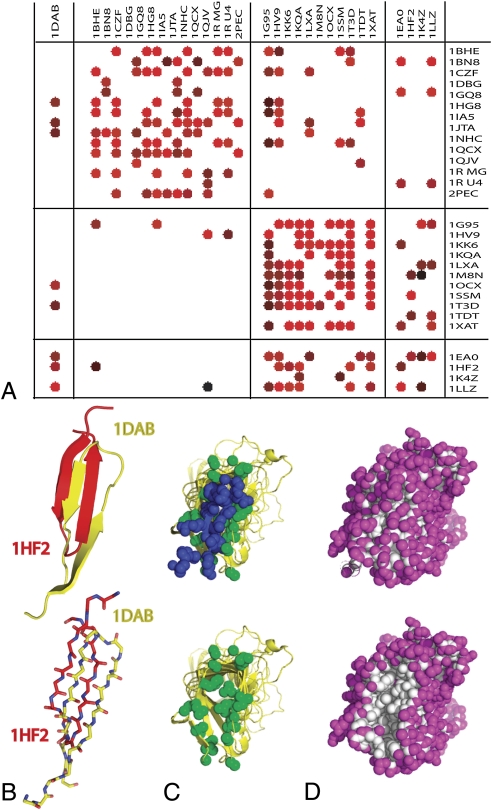
Fig. 4.
Prediction of visor caps. (A) Detection of cap alignment by RAPTOR. Template structures are depicted in rows; query sequences are depicted in columns, with rmsd values from 0.3 to 14.2 angstroms indicated by colors ranging from light red (small rmsd) through dark red to black (large deviations). White spaces indicate no cap-on-cap alignment was found. The sequences and structures shown were used for training, except 1DAB, which was excluded to be a test case. (B) The visor C-cap of 1HF2 (red) superimposed on the visor C-cap of 1DAB (yellow), as aligned by MATT (21), demonstrating the similar but oppositely oriented, hairpin turns of the two visor caps. Top: Ribbon diagrams of C-caps and terminal rungs of the β-helices. Bottom: wire frames of cap backbones, showing alignment of hydrogen bonds. (C) Pertactin (1DAB) shown with (top) and without (bottom) the C-terminal cap (in blue), which protects the hydrophobic core of the β-helix. Hydrophobic residues in the C-terminal rung of the β-helix are shown in green. (D) The C-terminal cap of pertactin protects the core of the β-helix from solvent exposure. Surface exposed residues (shown in magenta), were determined using the PyMol “FindSurfaceResidues” script with a > 2.5 Å2 cutoff. Removal of the C-terminal cap (bottom) reveals a patch of buried residues (shown in white).
Inspection of the alignments revealed previously unapparent structural similarities in that visor-type caps are aligned by the RAPTOR algorithm according to contacts between the cap and the adjacent β-helix. For instance, the hairpins of the pertactin (1DAB) C-terminal visor cap and the 1HF2 visor cap (Fig. 4B) align with an rmsd of less than 5 Å in the RAPTOR threading. A global structural alignment of the C-terminal portions of these proteins, as conducted by MATT (21) reveals these hairpins are oriented in opposite directions. Despite these different orientations relative to the adjacent rung and different loop lengths connecting them to that rung, the locations of alpha carbon atoms for residues within the hairpins, and the hydrogen-bonding patterns, are closely aligned due to a one-residue offset in the positions of the β-strands in the hairpins. This offset reverses the opposite orientation of backbone geometry caused by the reversal of the hairpin. However, further analysis of the pertactin visor cap using the most recent RAPTOR version, RAPTORX (22), shows that the hairpins are similarly oriented in the two next best low-homology matches to 1DAB (see SI Text, Figs. S1 and S2).
The only available example in the PDB of a naturally occurring β-helix aggregate is the Het-s prion from Podospora anserina. Because Het-s is known to aggregate, by our reasoning nothing that approximates a visor cap should be present in its structure. To verify that no visor cap appears in this structure, RAPTOR was used to thread the Het-s prion domain onto the known visor caps. The resulting alignments had poor Z-scores and contained large gaps between each β-strand. An attempt was made to force alignment of Het-s onto only the terminal rungs of the visor-containing templates using MUSCLE (23). Alignment either failed or (in three cases) formed very poor alignments (rmsd > 9 angstroms) with no correspondence to the visor cap turns. Therefore, in the one verifiable case of the Het-s aggregate, no visor cap is detected.
Deletion of the C-Terminal Cap of Pertactin Leads to Oligomerization and Aggregation.
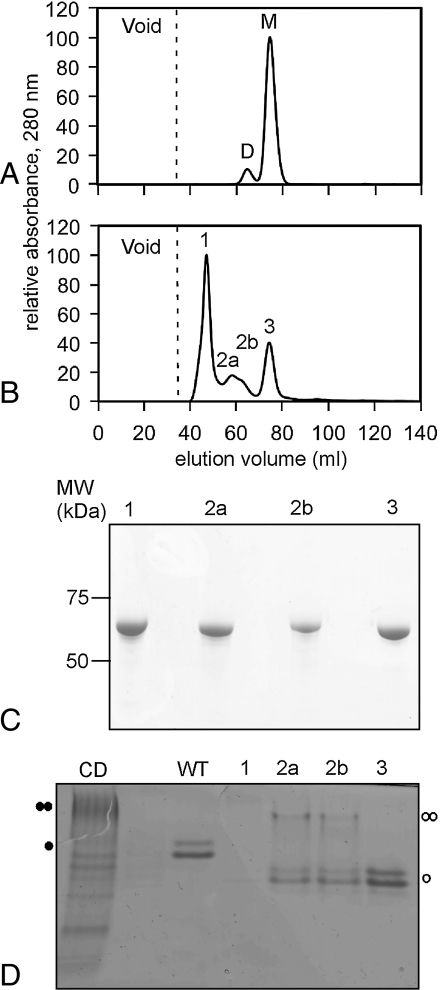
To experimentally investigate the contribution of a β-helix cap to the folding and aggregation properties of a β-helix structure, we deleted the C-terminal visor cap (as observed in its crystal structure, and detected by the HELIXCAP-visor algorithm detailed above) of pertactin, a 16-rung right-handed β-helix structure with well characterized folding properties and very low aggregation propensity (2, 24). Fig. 4 C and D show that removal of the C-cap from the native pertactin structure would lead to the exposure of hydrophobic residues and β-helix surfaces. Removal of the C-terminal cap led to significantly increased aggregation during protein purification, relative to wild type pertactin (see Methods). Moreover, the small fraction of the ΔC-terminal cap construct that did fold into a compact, soluble structure contained more than 50% oligomeric species of various sizes, as judged by size exclusion chromatography (Fig. 5B). These oligomers ranged in size from a presumably dimeric species that eluted at a position identical to the elution position of a covalent disulfide-bonded pertactin T490C dimer (Fig. 5A), to larger soluble oligomers that eluted near the void volume of the size exclusion column (Fig. 5B, peak 1). SDS-PAGE and Western blotting with an antipertactin polyclonal antibody confirmed that all elution peaks contained pertactin (Fig. 5C). Native gel electrophoresis confirmed that the shape/charge properties of a portion of the pertactin ΔC-terminal cap species detected in peaks 2a and 2b are similar to the covalent dimer; the slightly faster gel migration likely reflects the decrease in molecular weight from the deletion of the C-terminal cap. In contrast, the majority of the material in peak 1 is in an aggregated state that is too large to enter the separating gel (Fig. 5D).
Fig. 5.
Removal of the pertactin C-terminal cap leads to formation of soluble oligomeric species as determined by size exclusion chromatography. (A) Elution profile of the single cysteine pertactin mutant T490C: M, monomers; D, disulfide-bonded dimers. (B) Elution profile of the pertactin ΔC-terminal cap construct. Peak 1 elutes near the theoretical void volume of the column (dotted line), suggesting large oligomeric species. Peak 2 elutes at a position similar to the disulfide dimer shown in (A), while peak 3 corresponds to monomeric pertactin. (C) Chromatography results were corroborated with Coomassie-stained SDS-PAGE. Only ΔC-terminal cap was detected in each chromatography peak. Molecular mass (kDa) is indicated on the left. (D) Formation of oligomeric species was confirmed with Coomassie-stained polyacrylamide native gel electrophoresis. Lanes corresponding to the single-cysteine pertactin (CD), wild-type pertactin (WT), and samples from each chromatography peak are indicated. The migration positions of wild type monomeric pertactin (one closed circle) and the covalent dimer (two closed circles) are indicated. Bands corresponding to putative monomeric and dimeric species for the ΔC-terminal cap construct are indicated with open circles.
Discussion
Understanding cap motifs as a key component of the β-helix fold advances our knowledge of the mechanisms that shield this fold from aggregation. As previously noted by other authors (13), the rungs of β-helices are quite similar to the resolved and theorized structures of amyloid protofibrils. To form soluble secretion products, β-helices must avoid forming similar aggregates. Two major forces act to bring together the monomer β-strands of amyloids: the hydrophobic effect and the hydrogen-bonding patterns of β-sheets (25). Secondary effects observed to stabilize amyloid structures, such as tight side-chain packing, side-chain to side-chain hydrogen bonding and packing interactions, and side chain to backbone hydrogen bonding, each depend on the alignment of β-strands. The unifying structural functions of the β-helix caps appear to be to preclude extension of one or more β-sheets via a stably folded physical obstruction.
Because of the broad nature of this structural function, evolution appears to have found a range of solutions to the β-helix-capping problem. For example, the diversity of visor cap shapes illustrate that no one supersecondary structure or motif is required at the ends of a β-helix domain.
It was therefore something of a surprise that a significant number of β-helices do indeed have common, loosely conserved, low-homology motifs. The range of α-helix motifs found in both right- and left-handed β-helices suggest the possibility of either the loose conservation of an ancient motif or convergent evolution in β-helix caps. Likewise, the large number of visor caps that thread atop each other despite their sequence and structure diversity argues for an evolutionary convergence of structure to serve the function of β-helix capping. Close analysis of specific HELIXCAP-visor results, such as the MINC/pertactin match (Fig. 4B), demonstrates that RAPTOR’s loose threading-based detection approach can adapt to variations in orientation and arrangement of structural elements within visor caps.
Oligomerization, including specific dimerization, upon removal of the C-terminal cap of pertactin demonstrates the importance of capping to the prevention of β-helix self-assembly. While the structures of the pertactin oligomeric species are unknown, the specific dimer peak implies a preference for a specific interaction, most likely at or near the deletion site.
The HELIXCAP-HMM and HELIXCAP-visor detectors, which identify α-helix and visor β-helix caps respectively, have been presented here in hopes of aiding future studies of β-helices and amyloidogenic sequences. Beyond their immediate role to detect cap motifs similar to those noted here, we suggest HELIXCAP can be used to scan genomic data. Hitherto unidentified β-helices may be identified by this method, and insight may be gained into β-helices and similar structures, such as leucine-rich repeats, several of which also have caps and cap-like motifs (see SI Text). Further research may reveal other categories of cap-like mechanisms. Because of the low sequence homology of β-helices, their detection from sequence data has often depended on the detection of motifs specific to a particular SCOP family (5). The looser, yet still specific, detectors developed here may have a broader capacity to detect as-yet unrecognized sequence and structure patterns that fit within the definitions of these motifs.
A further future application for studies using the HELIXCAP detectors is to investigate ways to duplicate the function of natural caps. As the function of the β-helix cap depends on its interface to the terminal rung of the β-helix domain, any disruption thereof may destabilize the domain or open it to potential aggregation. The identification of these critical structural features in a wide variety of β-helical proteins may be useful in future efforts to design small molecule or peptide-based caps specifically targeted to block intermolecular interactions, and thereby inhibit amyloid fiber and/or aggregate growth.
Methods
Structural and Sequence Alignments.
β-helix families and superfamilies were determined according to SCOP. A redundant set of structures, containing one PDB structure per protein member of each SCOP family of β-helix structures (44 total proteins from 32 families) was downloaded from the March 16, 2009 PDB release and used for analysis; the latest structure with the fewest ligands, heavy atoms, or other molecules incorporated into the crystal was selected as the representative structure. The structures were grouped by SCOP superfamily. The full list of structures used is found in Table S1.
Structural alignments were generated using DALI (26). All generated alignments were of secondary structures in the β-helix cap and the adjacent rung of the β-helix, along with turns connecting these secondary structures. Alignments were first made in a pair-wise fashion to a template structure: 1DBG (short single helix), 1GQ8 (single helix, pectate lyase superfamily), 1G95 (single helix, left-handed superfamily), 1JTA (previous-strand visor), and 1QCX (cross-strand visor). The rotation and translation matrices derived from the DALI alignment were applied to the original PDB files. The rotated PDB structures were combined to produce a general alignment. The orientations of the secondary structure elements, as well as the long axes of the β-helices, were examined to confirm the accuracy of the alignments. Images of the caps were generated using PyMol (www.pymol.org). Sequence alignments for each superfamily were derived from the DALI alignments. The general sequence alignment of helix caps was arranged to optimize correspondence of secondary structure elements as determined by PSIPRED (27) and turns as determined by inspection of DALI alignments.
HELIXCAP-HMM Hidden Markov Model Generation and Testing.
HMMer (17) was used to compile and calibrate HMMs. An initial seed model was generated from the sequence alignment of α-helix caps using hmmbuild. This model was deemed to have insufficient sequence n for statistical validation. Therefore, a larger model was constructed as follows. A database of β-helix sequences was generated by using BLAST (19) to search the National Institutes of Health (NIH) Entrez nonredundant database for matches to sequences in the alignment of Fig. 3A, with a minimum E-value of 1.0 × 10-60. A total of 1,084 sequences were included in the database. hmmalign was used to align the database to the initial seed model, and hmmbuild was run on the alignment to build the large model. To use the large model for detection of an α-helix cap, hmmsearch was executed, using the forward (as opposed to the default Viterbi) algorithm to calculate the score. E-values of 0.5 and above were considered as positive hits for single sequences; for database searches, the Z parameter was set to 1 (enabling cross-comparison between databases of different sizes), and an E-value of 0.0005 was used as an upper threshold. The forward algorithm was chosen because of the high specificity and low sensitivity of the detector; Eddy (1998) (17) noted that the forward algorithm is more sensitive to subtle patterns.
Statistical Validation and Analysis of α-Helix Cap Predictor.
An updated version of the PDB- dataset from BETAWRAP (5) was used as a negative control dataset. The PDB- database used for HELIXCAP-HMM is the nonredundant database of all sequences in the October 12, 2007 release of the Protein Data Bank (9) with corresponding elements in SCOP (1), excluding all sequences identified as β-helices. PDB--HELIXCAP contains n = 18,659 sequences. For a positive control, cross-validation was conducted as follows. The database of β-helix sequences was clustered using BLASTCLUST (19), creating 69 clusters with greater than 25% sequence identity and 498 clusters with greater than 75% sequence identity. For each cluster, a corresponding cross-validation model was constructed. Cross-validation models were constructed in the same manner as the full model, except that the sequences in the cluster were removed from the database before alignment. Each cluster was then analyzed using its corresponding cross-validation model. Results were pooled for statistical validation.
HELIXCAP-Visor: Visor Cap Predictor.
Because a number of PDB structures with visor caps contain multiple domains (1SSM, 1T3D, 1TDT, 1EA0, 1LLZ, 1K28, 1WMR, 2ARA), portions of these structures more than 20 residues beyond the end of the cap were not analyzed further. The set of isolated β-helix domain structures so created was then analyzed using RAPTOR (16) to globally align each protein’s sequence (the query) onto every other structure in the set (the templates). For these runs, RAPTOR was executed using standard settings. Z-scores were recorded for each alignment. The alignments thus obtained were then tested for alignment of the query sequence’s visor cap(s) onto the corresponding template cap(s). If at least 50% of the query cap was aligned to a region extending five residues beyond either end of the terminus cap, the cap-on-cap alignment was evaluated by calculating the Cα rmsd calculated for that region of the terminus cap. A plot of global Z-score values and Z-score vs. cap-on-cap rmsd for all alignments with cap-on-cap threading, and a histogram of Z-scores for Genbank nonredundant sequences with BETAWRAP scores above -18 may be found in Figs. S3, S4, and S5, respectively.
Web Interface.
The detectors used in this paper are accessible at http://helixcap.csail.mit.edu.
Pertactin Mutant Design.
The pertactin ΔC-terminal cap construct was created by introducing a stop codon in plasmid pPERPLC02 (24) at the codon encoding amino acid residue 520 of wild type pertactin.
Pertactin Mutant Expression and Purification.
T490C and ΔC-terminal cap pertactin were expressed and purified as described previously (24), with the following modifications. As the ΔC-terminal cap construct was more prone to aggregation than wild type pertactin, more stringent refolding conditions were required to obtain soluble protein. After overexpression and cell lysis, the cell lysate pellet was collected by centrifugation at 12,000 × g for 20 min and solubilized overnight in 6 M GdnHCl. ΔC-terminal cap pertactin was refolded at 4 °C by drop-wise addition of 5 mL of the solubilized pellet into 4 L of 50 mM Tris pH 8.8 with gentle stirring. The refolding solution was centrifuged at 12,000 × g for 20 min and filtered through a 0.22 μM membrane to remove aggregates. The remaining soluble protein was then concentrated using a centrifuge concentrator (Millipore).
Size Exclusion Chromatography.
Approximately 6.5 mg of protein was loaded onto a HiLoad 16/60 Superdex 200 size exclusion column (GE Healthcare), preequilibrated with 50 mM Tris pH 8.8. Fractions (1.5 mL) were collected with a flow rate of 0.8 mL/ min. Proteins in chromatography fractions were separated by SDS-PAGE and either stained with Coomassie Brilliant Blue or immunoblotted using an antipertactin polyclonal antibody. Putative oligomeric species were resolved on 7.5% native (no SDS) polyacrylamide gels. Native gels were stained with Coomassie Brilliant Blue.
Supplementary Material
Acknowledgments.
The authors acknowledge helpful discussions with Andrew McDonnell, Charlie O’Donnell, and members of the Berger and Clark labs, Matt Menke for advice on structural alignment and help generating the new PDB- database, Jinbo Xu and Jian Peng for assistance with RAPTOR and RAPTORX, and Ryan Simkovsky for discussions regarding P22 tailspike protein. This research was supported by NIH grants U54-LM008748, R01-GM25874, and R01GM081871.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017504108/-/DCSupplemental.
References
- 1.Andreeva A, et al. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Junker M, et al. Pertactin beta-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc Natl Acad Sci USA. 2006;103:4918–4923. doi: 10.1073/pnas.0507923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoder M, Lietzke S, Jurnak F. Unusual structural features in the parallel beta-helix in pectate lyases. Structure. 1993;1:241–251. doi: 10.1016/0969-2126(93)90013-7. [DOI] [PubMed] [Google Scholar]
- 4.Liou YC, Tocilj A, Davies PL, Jia Z. Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein. Nature. 2000;406:322–324. doi: 10.1038/35018604. [DOI] [PubMed] [Google Scholar]
- 5.Bradley P, Cowen L, Menke M, King J, Berger B. BETAWRAP: successful prediction of parallel beta -helices from primary sequence reveals an association with many microbial pathogens. Proc Nat'l Acad Sci USA. 2001;98:14819–14824. doi: 10.1073/pnas.251267298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaman T, Sugantino M, Roderick S. Structure of the hexapeptide xenobiotic acetyltransferase from Pseudomonas aeruginosa. Biochemistry. 1998;37:6689–6696. doi: 10.1021/bi980106v. [DOI] [PubMed] [Google Scholar]
- 7.Raetz CR, Roderick SL. A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 8.Baumann U, Wu S, Flaherty KM, McKay DB. Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif. EMBO J. 1993;12:3357–3364. doi: 10.1002/j.1460-2075.1993.tb06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Nat'l Acad Sci USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Hecht MH. Rationally designed mutations convert de novo amyloid-like fibrils into monomeric beta-sheet proteins. Proc Nat'l Acad Sci USA. 2002;99:2760–2765. doi: 10.1073/pnas.052706199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perutz MF, Finch JT, Berriman J, Lesk A. Amyloid fibers are water-filled nanotubes. Proc Nat'l Acad Sci USA. 2002;99:5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wasmer C, et al. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 15.Williams AD, et al. Mapping abeta amyloid fibril secondary structure using scanning proline mutagenesis. J Mol Biol. 2004;335:833–842. doi: 10.1016/j.jmb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Xu J, Li M, Kim D, Xu Y. RAPTOR: optimal protein threading by linear programming. Journal of Bioinformatics and Computational Biology. 2003;1:95–117. doi: 10.1142/s0219720003000186. [DOI] [PubMed] [Google Scholar]
- 17.Eddy S. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 18.Schuster-Bockler B, Schultz J, Rahmann S. HMM Logos for visualization of protein families. BMC Bioinformatics. 2004;5:7. doi: 10.1186/1471-2105-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menke M, Berger B, Cowen L. Matt: local flexibility aids protein multiple structure alignment. PLoS Comput Biol. 2008;4:e10. doi: 10.1371/journal.pcbi.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng J, Xu J. Low-homology protein threading. Bioinformatics. 2010;26:i294–300. doi: 10.1093/bioinformatics/btq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Junker M, Clark PL. Slow formation of aggregation-resistant beta-sheet folding intermediates. Proteins. 2010;78:812–824. doi: 10.1002/prot.22609. [DOI] [PubMed] [Google Scholar]
- 25.Bryan AW, Jr, Menke M, Cowen LJ, Lindquist SL, Berger B. BETASCAN: probable beta-amyloids identified by pairwise probabilistic analysis. PLoS Comput Biol. 2009;5:e1000333. doi: 10.1371/journal.pcbi.1000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holm L, Sander C. Alignment of three-dimensional protein structures: network server for database searching. Methods Enzymol. 1996;266:653–662. doi: 10.1016/s0076-6879(96)66041-8. [DOI] [PubMed] [Google Scholar]
- 27.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.