Abstract
The biogenesis and maintenance of the endoplasmic reticulum (ER) requires membrane fusion. ER homotypic fusion is driven by the large GTPase atlastin. Domain analysis of atlastin shows that a conserved region of the C-terminal cytoplasmic tail is absolutely required for fusion activity. Atlastin in adjacent membranes must associate to bring the ER membranes into molecular contact. Drosophila atlastin dimerizes in the presence of GTPγS but is monomeric with GDP or without nucleotide. Oligomerization requires the juxtamembrane middle domain three-helix bundle, as does efficient GTPase activity. A soluble version of the N-terminal cytoplasmic domain that contains the GTPase domain and the middle domain three-helix bundle serves as a potent, concentration-dependent inhibitor of membrane fusion both in vitro and in vivo. However, atlastin domains lacking the middle domain are without effect. GTP-dependent dimerization of atlastin generates an enzymatically active protein that drives membrane fusion after nucleotide hydrolysis and conformational reorganization.
Keywords: endoplasmic reticulum biogenesis, organelle fusion, hereditary spastic paraplegia, spastic paraplegia gene 3A
Membrane fusion reactions are vitally important for many aspects of eukaryotic cell biology, including vesicular traffic within the secretory pathway as well as the biogenesis and maintenance of the entire endomembrane system. SNARE proteins are responsible for membrane fusion within the secretory pathway as well as homotypic fusion of endosomes and lysosomes (1, 2). The fusion of other organelles such as mitochondria and the endoplasmic reticulum (ER) are less well characterized. Many observations suggest that mitochondrial fusion is driven by proteins called mitofusins (3–5). Although ER-resident SNARE proteins are required for vesicular transport back to the ER (6, 7), the protein(s) responsible for the generation and maintenance of the ER has only recently been discovered. We have recently shown that Atlastin is a GTP-dependent membrane fusion protein that is responsible for ER homotypic fusion (8). Membrane fusion provided by atlastin helps shape and maintain the dynamic nature of the ER membrane tubule network (9–13).
Atlastin is the product of the human SPG3A locus (14). SPG3A (Atl1) is a member of a larger family of genes that are responsible for a group of inherited neurological disorders called hereditary spastic paraplegia (HSP) (15, 16). This disease is characterized by progressive lower-extremity weakness and spasticity. The neuropathological basis for compromised motor function in HSP is likely length-dependent axonopathy of the corticospinal tract (16). Twenty HSP gene products have been identified, and their molecular analysis has suggested that three general categories of proteins may be responsible for HSP. These gene functions fall into three broad groups, including intracellular trafficking, mitochondrial function, and axonal pathfinding and myelination (17). More than half of all HSP cases are caused by mutation in ER-resident or ER-associated proteins (18–20). Atlastin and the ER tubule-forming protein receptor expression-enhancing protein 1 (REEP1), as well as other reticulons, are all ER-resident proteins (20, 21). Spastin, a microtubule-severing protein, has been shown to associate with REEP1 and atlastin in some contexts (20, 22, 23), and recent work has also demonstrated that atlastin interacts with all of the ER tubule-forming proteins in the Reticulon and REEP/Yop/DP1 family (20). These observations place the ER at the nexus of HSP pathology.
Recently, atlastin function has been examined in model organisms (24), specifically in the fruit fly Drosophila melanogaster. The Drosophila genome produces a single atlastin protein. Drosophila atlastin (atl) is 541 aa long and has a predicted molecular mass of 61 kDa (Fig. 1A). The atl sequence is highly homologous with all three human isoforms, ranging between 44% and 49% identical (61% and 68% similar) over the entire length of the protein. Overall homology is significantly improved (55–59% identity) when the highly variable N-terminal (17–45 residues) and C-terminal (29–50 residues) regions are excluded.
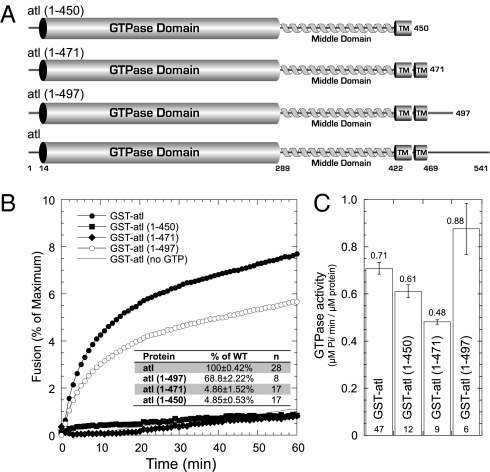
Fig. 1.
The atlastin C-terminal cytoplasmic tail is required for membrane fusion in vitro. (A) Domain structure of atl and C-terminal truncations. The GTPase domain is represented as a cylinder, the middle domain 3HB is shown as a helix, and the transmembrane domains are cylinders labeled “TM.” Relevant domain boundaries are indicated by residue numbers above the diagram. (B) Kinetic fusion graph of unlabeled atl acceptor proteoliposomes fused with equimolar amounts of fluorescently labeled atl donor proteoliposomes. NBD fluorescence was measured at 1-min intervals, and detergent was added at 60 min to determine maximum fluorescence. WT GST-atl-His8, ●; GST-atl(1–450)-His8, ■; GST-atl(1–471)-His8, ◆; GST-atl(1–497)-His8, ○; control reaction in the absence of GTP, gray line. In all cases, the same atl mutant is reconstituted into both liposome populations. (Inset) The average extent of fusion at 60 min is represented in tabular form as percentage of WT fusion for all mutants. WT GST-atl-His8 (n = 28) was set to 100 ± 0.42%. GST-atl(1–450)-His8 (n = 17) was 4.85 ± 0.53%, GST-atl(1–471)-His8 (n = 17) was 4.86 ± 1.52%, and GST-atl(1–497)-His8 (n = 8) was 68.8 ± 2.22%. (C) GTPase activity is represented as a histogram for all mutants. All activity is reported as μM Pi/min per μM enzyme. WT GST-atl-His8 (n = 47) was 0.71 ± 0.03, GST-atl(1–450)-His8 (n = 12) was 0.61 ± 0.03, GST-atl(1–471)-His8 (n = 19) was 0.48 ± 0.01, and GST-atl(1–497)-His8 (n = 6) was 0.88 ± 0.11. (Error bars: SEM. Numbers in histograms = number of replicates.)
We now examine the mechanistic basis for nucleotide-dependent membrane fusion by atl using a structure–function analysis of truncated proteins and a detailed kinetic analysis of nucleotide hydrolysis. Finally, we determine the oligomeric state of the N-terminal cytoplasmic domain and isolated GTPase domain in the presence and absence of nucleotide.
Results
atl is a 61-kDa multidomain protein (Fig. 1A). It has a short N-terminal variable domain, followed by a well-conserved GTPase domain, a middle domain of undefined function, two tandem transmembrane domains, and a C-terminal cytoplasmic domain. We previously documented that atlastin requires GTPase activity for membrane fusion in vitro; however, the function of the other domains remains to be explored. In this article, we extend our previous work by conducting a systematic structure–function analysis of atlastin with the D. melanogaster homolog.
Atlastin C-Terminal Cytoplasmic Tail Is Required for Membrane Fusion.
We began our analysis by determining the region of atlastin required for membrane fusion by producing C-terminal truncations. We have previously shown that GTPase activity is absolutely required for membrane fusion (8), so we reasoned that significant N-terminal deletions would abolish fusion activity by impairing GTPase activity given that atl contains only a 14-residue N-terminal extension beyond the boundary of the GTPase domain. We produced a truncation that removed the entire C-terminal cytoplasmic domain (residues 472–542) and a truncation that eliminated the second transmembrane domain as well as the C-terminal cytoplasmic domain (residues 451–542); both atl(1–471) and atl(1–450) retain an intact GTPase domain and at least one transmembrane anchor. We expressed these proteins in Escherichia coli, reconstituted them into synthetic phosphatidylcholine:phosphatidylserine (PCPS) liposome, and measured membrane fusion by lipid mixing (8). When either of these two proteins was reconstituted into proteoliposomes, no fusion was observed (Fig. 1B). The functionality of these enzymes was confirmed by examining GTPase activity (Fig. 1C). Both atl(1–450) and atl(1–471) were able to cleave GTP at rates that were within ∼30% of WT rates, yet they were incapable of driving membrane fusion. The small reduction in GTPase activity for the C-terminal deletion mutants may reflect a subtle effect of the C-terminal tail on atlastin oligomerization.
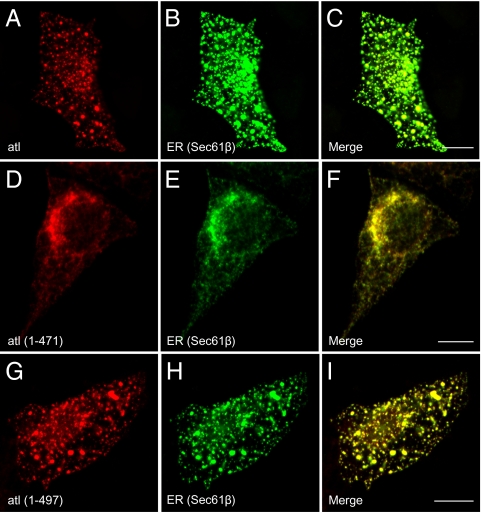
The inability of these proteins to produce a functional atlastin was further explored in a tissue culture model in vivo. When functional atl is overexpressed in mammalian cells, it correctly sorts to the ER, causes the formation of an enlarged ER compartment, and redistributes Golgi-resident proteins to the ER in much the same way as overexpression in intact animals (8). Fig. 2 A–C shows the result of expressing WT atlastin in Cos7 cells. atl colocalizes with the ER marker Sec61β and produces significant changes in ER (Fig. 2B) morphology. When functional atl is expressed, the normally reticular ER (Fig. S1) is lost, and variably sized puncta are now stained with ER markers. However, overexpressing ER-localized atl(1–471) minimally disrupts ER morphology, demonstrating that it is a less or nonfunctional protein in vivo (Fig. 2 D–F).
Fig. 2.
Deletion of the atlastin C-terminal cytoplasmic tail results in a nonfunctional protein in vivo. Cos7 cells were cotransfected with WT atlastin-myc (A–C), atl(1–471)-myc (D–F), or atl(1–497)-myc (G–I) and GFP-Sec61β. WT atlastin and atl(1–497) show an abnormal, punctate ER, indicating a functional atl protein, whereas atl(1–471) localizes to a normally reticular ER, suggesting that it is a nonfunctional protein. (Scale bar: 10 μm.)
These results suggest that the C-terminal cytoplasmic domain of atlastin is critical for membrane fusion. However, this region of atlastin is among the most divergent between atlastin homologs and paralogs. Closer inspection of the sequences that comprise the C-terminal cytoplasmic domain revealed an ∼25-aa stretch of more highly conserved residues membrane-proximal to the second transmembrane domain followed by a very divergent extreme C terminus (Fig. S2). We further subdivided the C-terminal cytoplasmic domain to add back the more conserved juxtamembrane region (residues 471–497). The resulting construct, atl(1–497), was also expressed, reconstituted, and assayed for membrane fusion. Fig. 1 B and C demonstrate that the reintroduction of these 27 residues restores ∼70% of WT fusion activity. Additionally, overexpression of atl(1–497) in Cos7 cells results in a phenotype similar to the overexpression of WT atlastin, confirming that atl(1–497) is functional in vivo (Fig. 2 G–I).
Soluble N-Terminal Cytoplasmic Domain of Atlastin Is a Concentration-Dependent Inhibitor of ER Membrane Fusion.
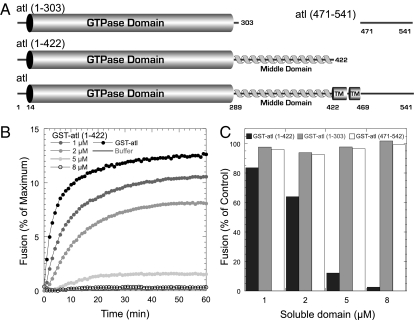
All membrane fusion proteins characterized to date require membrane integration. However, liberated soluble domains that retain the ability to productively interact with membrane integral components are often inhibitors of fusion. This is definitely the case for SNARE proteins, and inclusion of a soluble fragment of either v-SNARE or t-SNARE component will inhibit fusion (25, 26). We tested the functionality of C-terminal truncations that lack a membrane-spanning domain by determining their capacity to inhibit fusion between proteoliposomes that contain WT atlastin. We tested atl(1–422), which encompassed the entire N-terminal cytoplasmic domain; atl(1–303), which contains the isolated GTPase domain; and atl(471–542), the soluble C-terminal fragment (Fig. 3A). When atl(1–422) was titrated into a fusion reaction containing WT atlastin in the membrane, progressive inhibition of both the rate and extent of membrane fusion was seen (Fig. 3B). Inhibition was concentration-dependent, and fusion was completely inhibited with an eightfold molar excess of the atl(1–422) soluble domain (Fig. 3C). Although the full-length N-terminal cytoplasmic domain was a potent inhibitor, neither the isolated GTPase domain [atl(1–303); Fig. 3C, gray column] nor the soluble C-terminal tail [atl(471–542); Fig. 3C, white column] inhibited fusion.
Fig. 3.
The middle domain is required for the in vitro inhibition of fusion by the N-terminal cytoplasmic domain of atl. (A) Domain structure of atl and the soluble atl fragments added acutely to fusion reactions between WT atlastin proteoliposomes. (B) Kinetic fusion graph of unlabeled atl acceptor proteoliposomes fused with equimolar amounts of fluorescently labeled atl donor proteoliposomes (black) with increasing amounts of GST-atl(1–422)-His8 (shades of gray) added to the reaction. (C) The extent of fusion at 60 min is represented in a histogram as the percentage of fusion without any soluble domains added to the reaction.
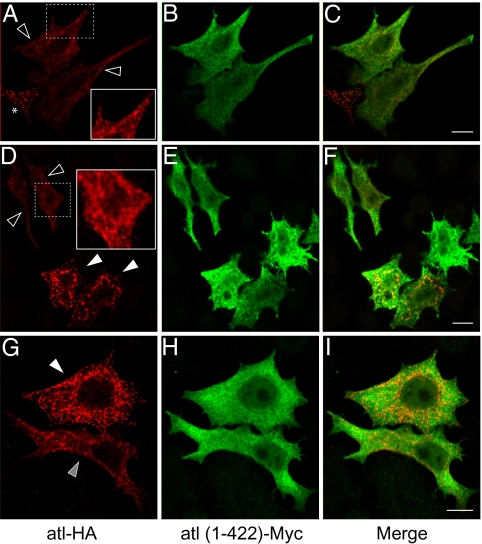
The ability of the isolated N-terminal cytoplasmic domain to inhibit fusion was further explored by co-overexpressing atl(1–422) in Cos7 cells with WT atlastin. As shown in Fig. 2A, overexpressing WT atlastin in Cos7 cells disrupts ER morphology, presumably because of inappropriate and excessive ER fusion. However, simultaneous overexpression of the soluble N-terminal cytoplasmic domain of atlastin, atl(1–422), reduces the severity or largely prevents the aberrant ER structures generated by WT atlastin (Fig. 4). A spectrum of inhibition is seen with atl(1–422), which likely reflects the relative degree of overexpression. When cells were subjectively binned into three groups, defined as normal ER morphology (Fig. 4, empty arrowheads), fused ER (Fig. 4, white arrowheads), and partially fused ER (Fig. 4, gray arrowhead), more than half (56%) of the cells expressing atl(1–422) exhibited a normal ER morphology, whereas 43% displayed a partially fused ER (Fig. S3). We interpret these results to suggest that atl(1–422) productively and preferentially interacts with full-length WT atlastin to inhibit its fusion activity in vivo.
Fig. 4.
Variable overexpression of the atlastin N-terminal cytoplasmic domain dominantly inhibits WT atlastin function in vivo. Cos7 cells overexpressing WT, HA-tagged atl, and myc-tagged atl(1–422) show a reduction in a punctate, fused ER morphology phenotype. (A–C) Cos7 cells expressing both full-length atl and atl(1–422) show a relatively normal ER phenotype (empty arrowheads). Asterisk indicates a cell expressing full-length atl only and shows an abnormal, punctate ER phenotype. (D–F) Cos7 cells expressing full-length atl and varying levels of atl(1–422) show a mix of a normal ER phenotype (empty arrowheads) and a fused ER phenotype (white arrowheads). (G–I) Cos7 cells coexpressing atl and atl(1–422) representing a fused ER phenotype (white arrowhead) and a partially fused ER (gray arrowhead). Insets in A and D give a magnified view and increased color contrast of the outlined region of a cell showing the normal ER phenotype. Empty arrowheads indicate WT ER, where WT atlastin is inhibited by coexpression of atl(1–422). Gray arrowheads show the partially fused ER phenotype, where WT atlastin activity is partially inhibited by atl(1–422). White arrowheads illustrate fully fused ER, where there is minimal or no effect of atl(1–422). (Scale bar: 10 μm.)
Atlastin Dimerization Is Required for Enzymatic Activity.
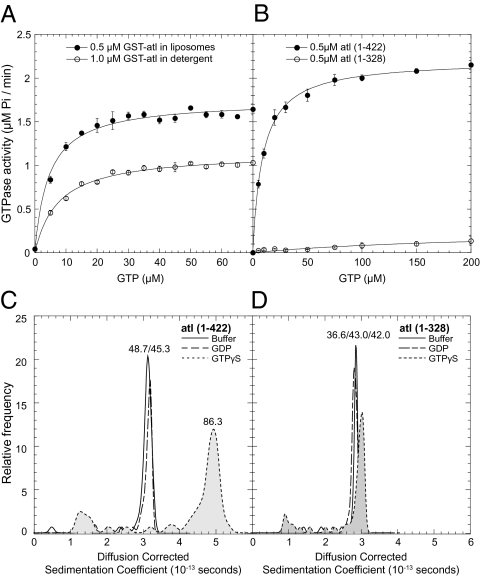
Although we know that atlastin GTPase activity is required for membrane fusion, we know very little regarding the enzymatic properties of atlastin. To address this issue, we measured Michaelis–Menten kinetics for full-length GST-atl in detergent and reconstituted into proteoliposomes by using a coupled enzymatic assay that quantifies inorganic phosphate production. We ensured that quantifying inorganic phosphate production was a reliable measure of GTPase activity by examining all of the guanine nucleotide reaction products by HPLC because guanylate binding protein 1 (GBP1), another large GTPase family member similar in sequence to atlastin, is capable of sequentially cleaving GTP to GMP (Fig. S4). Fig. 5A shows a plot of initial velocity versus GTP concentration. Surprisingly, we observed that reconstitution significantly improved the maximum velocity of this enzyme even though the amount of enzyme assayed in proteoliposomes is half (0.5 μM) of that in detergent (1 μM) (Table 1). These data demonstrate that atlastin is a more active enzyme in a phospholipid bilayer compared with a detergent micelle. The most straightforward interpretation of this activity increase is that atlastin has an improved ability to dimerize in the plane of a membrane after reconstitution (a cis dimer) or that trans dimerization between liposomes stimulates GTPase activity.
Fig. 5.
The atlastin middle domain is required for nucleotide-dependent atlastin dimerization and normal GTP activity. (A) Graph of the initial velocities of GTP hydrolysis versus GTP concentration of WT atlastin solubilized in detergent or reconstituted into synthetic liposomes. The turnover number (Kcat) is increased about threefold when atl is reconstituted into liposomes, whereas the nucleotide affinity is largely unaffected. (B) Initial velocities of GTP hydrolysis by the N-terminal cytoplasmic domain, atl(1–422), and the isolated GTPase domain, atl(1–328). The maximum velocity of atl(1–422) is greater than full-length atlastin in detergent. The GTPase activity is almost completely eliminated in atl(1–328), which lacks the middle domain. The data are fitted with the Michaelis–Menten equation. (Error bars: SEM; n = 3.) (C and D) Distributions of sedimentation coefficients of atl(1–422) and atl(1–328) determined by analytical ultracentrifugation data analyzed by van Holde–Weischet analysis. (C) atl(1–422) in buffer or with GDP is monomeric, whereas it forms a dimer when incubated with GTPγS. (D) atl(1–328) is monomeric under all conditions. Molecular masses were determined by using the genetic algorithm analysis in Ultrascan II (34) (Figs. S6 and S7).
Table 1.
Kinetic parameters for atlastin
| Protein | Km, μM | Vmax, μM Pi/min |
| GST-atl (liposomes) | 4.5 ± 0.5 | 1.7 ± 0.03 |
| GST-atl (TX-100) | 7.5 ± 0.5 | 1.1 ± 0.02 |
| atl(1–422) | 9.3 ± 0.4 | 2.2 ± 0.02 |
| atl(1–328) | 218 ± 145 | 0.3 ± 0.11 |
Next, we examined the activity of the N-terminal cytoplasmic domain, atl(1–422). This protein was produced as a GST-SUMO fusion protein, and the N-terminal tags were removed before analysis. In this case, the liberated N-terminal cytoplasmic domain displayed comparable activity to the reconstituted full-length protein (Fig. 5B and Table 1), suggesting that the regions of atlastin responsible for oligomerization and enzymatic activation reside in this domain. Given that other large GTPases such as dynamin and GBP1 dimerize through contacts within the G domain, we produced an additional construct that expressed the isolated GTPase domain, atl(1–328). However, we found that the GTPase domain alone was not sufficient to produce an active enzyme at the same concentration (0.5 μM) as the other protein examined (Fig. 5B). The atl(1–328) protein is not simply a misfolded protein because it shows a concentration-dependent increase in activity at higher GTP concentrations (up to 1 mM; Fig. S5). These results suggest that the GTPase domain alone is not sufficient to provide a stable interaction platform for GTP hydrolysis.
Nucleotide-Dependent Atlastin Oligomerization Is Driven by the Middle Domain.
GTPase activity measurements and soluble domain inhibitor results suggest that the middle domain of atlastin may be important for oligomerization. To address the question of oligomerization directly, we measured the molecular mass of atl(1–422) and atl(1–328) by analytical ultracentrifugation. Fig. 5 C and D shows the results of velocity sedimentation experiments for these proteins: a van Holde–Weischet analysis (27) produced diffusion-corrected sedimentation coefficients for atl(1–422) in the absence of nucleotide (solid black trace) or in the presence of GDP (large dashed trace), or the nonhydrolyzable GTP analog GTPγS (small dashed trace) revealed a dimer only in the presence of GTPγS. Nucleotide-free atl(1–422) moved as a 3.14 S particle, GDP-bound atl(1–422) migrated similarly to the apo form (3.21 S), and GTPγS sedimented faster at 4.95 S. These sedimentation values correspond to molecular masses of 48.7 (Apo), 45.3 (GDP), and 86.3 (GTPγS) kDa, respectively (Fig. S6). The predicted monomer molecular mass of atl(1–422) is 49,317 Da, strongly suggesting that both the apo and GDP-bound protein is monomeric, whereas the GTPγS-bound protein forms a dimer. A similar analysis with atl(1–328) produced a different result. In this case, all three species (apo, GDP, and GTPγS) migrated at a size most consistent with a monomer molecular mass (36.6, 43.0, and 41.0 kDa, respectively). The predicted monomer molecular mass of atl(1–328) is 37,243 Da. Together, GTPase activity measurements and molecular mass analysis by analytical ultracentrifugation suggests that the N-terminal cytoplasmic domain of atl can dimerize in the presence of GTP and that nucleotide binding promotes a conformation that requires the middle domain for stable oligomerization.
Discussion
Traditional membrane fusion proteins such as viral fusion proteins and SNAREs use energy derived from metastable protein folding intermediates to drive fusion. The use of chemical energy in the form of nucleotide hydrolysis at the point of membrane fusion is unique to atlastin, and perhaps mitofusins, and defines this class of membrane fusion proteins. Detailed analysis of this type of mechanism has only recently begun to be explored. We have determined that a short region of ∼27 aa located within the cytoplasmic C-terminal tail is critically required for atlastin-dependent membrane fusion in vitro (Fig. 1) and in vivo (Fig. 2). Interestingly, three frameshift mutations in human Atlastin-1 (Atl1) [A492fsX522 (28), E502fsX522 (29), and I507fsX522 (30)] known to cause HSP prematurely truncate Atl1 and scramble the C-terminal juxtamembrane region that we have shown is required for membrane fusion.
The N-terminal cytoplasmic domain (residues 1–422), when freed from membrane attachment, functions as a fusion inhibitor both in vitro (Fig. 3) and in vivo (Fig. 4). Quantitative analysis of the enzymatic properties of atlastin revealed that the N-terminal cytoplasmic domain is a more active enzyme than the full-length protein in detergent solution. However, reconstitution of the full-length protein into a phospholipid bilayer improved activity but not to the level of the soluble protein (Fig. 5 A and B). Additionally, GTPase activity required protein sequences outside the GTPase domain, suggesting that this domain alone is not sufficient to a stable dimer. Analytical ultracentrifugation revealed that the N-terminal cytoplasmic domain of atl could dimerize in the presence of GTPγS but not GDP (Fig. 5C and Fig. S6). Furthermore, GTP-dependent dimerization requires the middle domain because the isolated GTPase domain (residues 1–328) was monomer under all nucleotide conditions (Fig. 5D and Fig. S7).
Very recently, two reports detailing the X-ray structure of the N-terminal cytoplasmic domain of human Atl1 were published (31, 32). These data provide important insights into both the biochemical properties and potential fusion mechanisms of atlastin. The large GTPase domain shares significant structural similarity to human GBP1 (33), and the middle domain folds into an antiparallel three-helix bundle (3HB) that would connect the GTPase domain to the tandem transmembrane segments. The GTPase domain is connected to the 3HB by a flexible linker. Both groups identified two crystal forms of Atl1 (1–446) differing by the position of the 3HB relative to the GTPase domain. Crystal form 1 (32) would likely position the transmembrane domains in a “postfusion” conformation (31), whereas crystal form 2 probably represents a “prefusion” structure. All of the current structures contained bound GDP, although attempts were made to bind nonhydrolyzable GTP analogs such as GTPγS or GMPPNP.
Biochemical analysis of the Atl1 (1–446) N-terminal cytoplasmic domain showed that GTPase activity of the WT human protein was comparable to our Drosophila protein [5.33 μM Pi/min per μM enzyme for Atl1 (1–446) vs. 4.4 μM Pi/min per μm enzyme for atl(1–422)] and that GTP promoted dimerization measured by small-angle X-ray scattering (32) or analytical ultracentrifugation (31). Additionally, both groups measured dimerization by gel filtration in the presence of GDP that yielded different outcomes. Bian et al. (31) identified atlastin dimers in the presence of GDP, whereas Byrnes and Sondermann (32) found only monomers with GDP. Small-angle X-ray scattering analysis also supported a monomer in the presence of GDP. This apparent discrepancy may be explained by different experimental conditions such as protein or nucleotide concentration. We detected only monomeric atl(1–422) with GDP (Fig. 5D and Figs. S6 and S8).
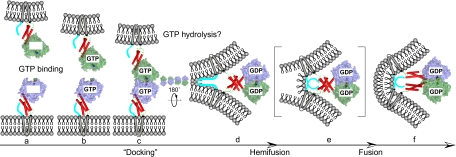
Membrane fusion by atlastin likely involves many steps. The schematic shown in Fig. 6 describes our working model for atlastin function. Atlastin almost entirely faces the cytoplasm with as few as 5 residues in an ER luminal loop. We suggest that the fusion cycle begins with nucleotide-free atlastin monomers (Fig. 6A) in opposing ER membranes. These monomer forms are cartooned as the prefusion (31) or crystal form 2 (32) structures. Then we suggest that nucleotide binding (Fig. 6B) results in a permissive state for association between the GTPase domains (Fig. 6C). The N-terminal cytoplasmic domain of atl, atl(1–422), forms a dimer when bound to nonhydrolyzable GTPγS but not with GDP (Fig. 5C and Fig. S6). We assume that this association also occurs in the context of full-length atlastin. Although this interaction between the GTPase domains is presumably a required intermediate, its stability may be limited. We propose that the interaction between GTPase domains matures to a more stable dimer facilitated primarily by an interaction between the middle domain 3HB segments (Fig. 6D). This conformational change is achieved, perhaps driven by nucleotide hydrolysis, by rotating the GTPase domain dimer 180°, which forces the 3HBs into close proximity. The new association between adjacent 3HBs liberates the C-terminal tail domain to perform its required role. We have localized this required function to a region of 27 aa immediately adjacent to the second transmembrane span (Fig. 1A). The activity of this C-terminal domain may be accomplished by forming a new association with the dimeric 3HB or by direct interaction with lipid (Fig. 6D, shown in cyan). We currently favor a direct interaction with lipid based on the amphipathic nature of this protein sequence (Fig. S2). An interaction between the membrane surface and the amphipathic C-terminal tail could destabilize the bilayer and provide the driving force for outer leaflet mixing, resulting in a hemifusion intermediate (Fig. 6E) that resolves by inner leaflet mixing to full fusion (Fig. 6F). The resulting cis dimer resembles the postfusion (31) or crystal form 1 (32) structure. Finally, we hypothesize that GDP release could then promote dissociation.
Fig. 6.
Model for atlastin-mediated fusion. The GTPase domains are cartooned as surface representations, the middle domains are shown as red cylinders, the transmembrane domains are illustrated as gray cylinders, and the C-terminal tails are shown as thick cyan lines. (A) Bilayer containing nucleotide-free prefusion (form2) monomers. (B) GTP-bound prefusion monomers. (C) Initial, unstable docking intermediate between GTP-bound monomers through surfaces on the GTPase domain. (D) Stabilized dimer formed by domain rotation and 3HB interaction resulting from GTP hydrolysis. (E) Putative hemifusion intermediate. (F) Postfusion bilayer with the form 1 (postfusion) dimer.
Although the precise location and timing of GTP hydrolysis during the cycle is speculative, some biochemical experiments can be used to place constraints on the model. We suggest that the large conformational change postulated to occur after docking (Fig. 6 C and D) uses the energy provided by GTP hydrolysis. However, our oligomerization data suggest that GTPγS, but not GDP, results in the stable dimeric conformation cartooned in Fig. 6D when soluble fragments are examined, indicating that this intermediate may contain bound GTP. Functional analysis revealed that GTP hydrolysis is required to fuse proteoliposomes in vitro because GTPγS or GMPPNP do not support fusion (8). These lipid-mixing results demonstrate that nucleotide hydrolysis is required before hemifusion (Fig. 6E); however, we cannot exclude the possibility that events downstream of GTP hydrolysis (for example, inorganic phosphate release) play an important role (31). Ongoing efforts are directed at experimentally validating the key features of our model.
Materials and Methods
Recombinant Protein Purification.
GST-tagged proteins were produced as previously described (8) with the exception that the truncation mutants lacking transmembrane domains were purified in the absence of detergent. atl(1–422) (pJM781) and atl(1–328) (pJM841) were expressed as GST-SUMO fusion proteins. The N-terminal GST-SUMO–tagged fusion proteins were bound to glutathione resin and purified in Tris buffer with 0.5% Triton X-100 by cleaving off the tag with SENP2 protease. (See SI Materials and Methods for SENP2 production.) The cleaved proteins were further purified by anion-exchange chromatography in Tris buffer [20 mM Tris (pH 7.5), 200 mM KCl, 5% glycerol, and 0.5 mM tris(2-carboxyethyl)phosphine (TCEP)]. Purified proteins were stored as aliquots at −80 °C.
Atlastin Reconstitution and in Vitro Fusion Assays.
WT atlastin, atl(1–450), atl(1–471), and atl(1–497) were reconstituted into preformed 100-nm liposomes as previously described (8). In vitro fusion assays were performed by mixing labeled and unlabeled atl proteoliposomes (0.3 mM lipid each) in 96-well FluoroNunc PolySorp plates (Nunc) and measuring nitrobenzoxadiazole (NBD) fluorescence over time after the addition of 0.5 mM GTP and Mg2+. Inhibition studies with soluble domains of atl were performed by adding the soluble proteins to wells containing atl proteoliposomes before the addition of 1–2 mM GTP and Mg2+.
GTPase Assays.
GTPase activities were measured as previously described (8) by measuring the release of inorganic phosphate from GTP as suggested in the EnzChek Phosphate Assay Kit (Molecular Probes).
Analytical Ultracentrifugation.
atl(1–422) and atl(1–328) were sized by sedimentation velocity experiments carried out in a Beckman Optima XLA analytical ultracentrifuge with 3.5 μM protein and 20 μM nucleotide in cells assembled with 12-mm, two-channel aluminum centerpieces. Sedimentation was carried out at 50,000 or 55,000 rpm in an AN60 rotor at 20 °C. The intensity at 235 nm versus radial position was measured over 4–5 h at 4-min intervals. All data analysis was done with Ultrascan II (34).
Cell Culture, Transfection, and Immunocytochemistry.
Expression plasmids for HA-tagged or Myc-tagged WT atlastin, atl(1–471), atl(1–497), and atl(1–422) were transfected into Cos7 cells and immunostained by using standard protocols. Cells cotransfected with WT atlastin and GFP or WT atlastin and atl(1–422) were scored for ER morphology and binned in three categories: fused ER, partially fused ER, normal ER. Seven independent transfection experiments were performed, and ∼100 cells were scored in each experiment.
The following antibodies were used: mouse anti-Myc (1:1,000; Sigma) and rabbit anti-HA (1:500; Santa Cruz Biotechnology). Secondary antibodies for immunofluorescence (Cy3 conjugates from Jackson Laboratories and Alexa Fluor 488 conjugates from Invitrogen) were used at 1:1,000. Confocal images were acquired with a Nikon C1 confocal microscope.
Detailed methods are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Borries Demeler for advice regarding analytical ultracentrifugation and data analysis with Ultrascan II, as well as Mike Stern and Joseph Faust for comments on the manuscript. Work in J.A.M.’s laboratory is supported by funds from the National Institutes of Health (Grant GM071832) and the G. Harold and Leila Y. Mathers Foundation. Work in A.D.’s laboratory is supported by grants from the Italian Ministry of Health, the Association Française contre les Myopathies, and the Fondazione Telethon.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105056108/-/DCSupplemental.
References
- 1.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McNew JA. Regulation of SNARE-mediated membrane fusion during exocytosis. Chem Rev. 2008;108:1669–1686. doi: 10.1021/cr0782325. [DOI] [PubMed] [Google Scholar]
- 3.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 4.Griffin EE, Detmer SA, Chan DC. Molecular mechanism of mitochondrial membrane fusion. Biochim Biophys Acta. 2006;1763:482–489. doi: 10.1016/j.bbamcr.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 6.Burri L, et al. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc Natl Acad Sci USA. 2003;100:9873–9877. doi: 10.1073/pnas.1734000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dilcher M, et al. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 2003;22:3664–3674. doi: 10.1093/emboj/cdg339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase Atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 9.Farhan H, Hauri HP. Membrane biogenesis: Networking at the ER with atlastin. Curr Biol. 2009;19:R906–R908. doi: 10.1016/j.cub.2009.08.029. [DOI] [PubMed] [Google Scholar]
- 10.Barlowe C. Atlasin GTPases shape up ER networks. Dev Cell. 2009;17:157–158. doi: 10.1016/j.devcel.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 11.English AR, Zurek N, Voeltz GK. Peripheral ER structure and function. Curr Opin Cell Biol. 2009;21:596–602. doi: 10.1016/j.ceb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moss TJ, Daga A, McNew JA. Fusing a lasting relationship between ER tubules. Trends Cell Biol. May 6, 2011 doi: 10.1016/j.tcb.2011.03.009. 10.1016/j.tcb.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pendin D, McNew JA, Daga A. Balancing ER dynamics: Shaping, bending, severing, and mending membranes. Curr Opin Cell Biol. 2011 doi: 10.1016/j.ceb.2011.04.007. 10.1016/j.ceb.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X, et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet. 2001;29:326–331. doi: 10.1038/ng758. [DOI] [PubMed] [Google Scholar]
- 15.Fink JK. Advances in the hereditary spastic paraplegias. Exp Neurol. 2003;184(Suppl 1):S106–S110. doi: 10.1016/j.expneurol.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Salinas S, Proukakis C, Crosby A, Warner TT. Hereditary spastic paraplegia: Clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7:1127–1138. doi: 10.1016/S1474-4422(08)70258-8. [DOI] [PubMed] [Google Scholar]
- 17.Soderblom C, Blackstone C. Traffic accidents: Molecular genetic insights into the pathogenesis of the hereditary spastic paraplegias. Pharmacol Ther. 2006;109:42–56. doi: 10.1016/j.pharmthera.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Hazan J, et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet. 1999;23:296–303. doi: 10.1038/15472. [DOI] [PubMed] [Google Scholar]
- 19.Namekawa M, et al. SPG3A is the most frequent cause of hereditary spastic paraplegia with onset before age 10 years. Neurology. 2006;66:112–114. doi: 10.1212/01.wnl.0000191390.20564.8e. [DOI] [PubMed] [Google Scholar]
- 20.Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120:1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 22.Evans K, et al. Interaction of two hereditary spastic paraplegia gene products, spastin and atlastin, suggests a common pathway for axonal maintenance. Proc Natl Acad Sci USA. 2006;103:10666–10671. doi: 10.1073/pnas.0510863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanderson CM, et al. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum Mol Genet. 2006;15:307–318. doi: 10.1093/hmg/ddi447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fassier C, et al. Zebrafish atlastin controls motility and spinal motor axon architecture via inhibition of the BMP pathway. Nat Neurosci. 2010;13:1380–1387. doi: 10.1038/nn.2662. [DOI] [PubMed] [Google Scholar]
- 25.Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 26.McNew JA, et al. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 27.Demeler B, van Holde KE. Sedimentation velocity analysis of highly heterogeneous systems. Anal Biochem. 2004;335:279–288. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 28.Ivanova N, et al. Hereditary spastic paraplegia 3A associated with axonal neuropathy. Arch Neurol. 2007;64:706–713. doi: 10.1001/archneur.64.5.706. [DOI] [PubMed] [Google Scholar]
- 29.Loureiro JL, et al. Novel SPG3A and SPG4 mutations in dominant spastic paraplegia families. Acta Neurol Scand. 2009;119:113–118. doi: 10.1111/j.1600-0404.2008.01074.x. [DOI] [PubMed] [Google Scholar]
- 30.Tessa A, et al. SPG3A: An additional family carrying a new atlastin mutation. Neurology. 2002;59:2002–2005. doi: 10.1212/01.wnl.0000036902.21438.98. [DOI] [PubMed] [Google Scholar]
- 31.Bian X, et al. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc Natl Acad Sci USA. 2011;108:3976–3981. doi: 10.1073/pnas.1101643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byrnes LJ, Sondermann H. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc Natl Acad Sci USA. 2011;108:2216–2221. doi: 10.1073/pnas.1012792108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature. 2006;440:101–104. doi: 10.1038/nature04510. [DOI] [PubMed] [Google Scholar]
- 34.Demeler B. Ultrascan: A comprehensive data analysis software package for analytical ultracentrifugation experiments. In: Scott DJ, Harding SE, Rowe AJ, editors. Modern Analytical Ultracentrifugation: Techniques and Methods. Cambridge, UK: RSC Publishing; 2005. pp. 210–230. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.