Abstract
Autophagy, a lysosome-mediated catabolic process, contributes to maintenance of intracellular homeostasis and cellular response to metabolic stress. In yeast, genes essential to the execution of autophagy have been defined, including autophagy-related gene 1 (ATG1), a kinase responsible for initiation of autophagy downstream of target of rapamycin. Here we investigate the role of the mammalian Atg1 homologs, uncoordinated family member (unc)-51–like kinase 1 and 2 (ULK1 and ULK2), in autophagy by generating mouse embryo fibroblasts (MEFs) doubly deficient for ULK1 and ULK2. We found that ULK1/2 are required in the autophagy response to amino acid deprivation but not for autophagy induced by deprivation of glucose or inhibition of glucose metabolism. This ULK1/2-independent autophagy was not the simple result of bioenergetic compromise and failed to be induced by AMP-activated protein kinase activators such as 5-aminoimidazole-4-carboxamide riboside and phenformin. Instead we found that autophagy induction upon glucose deprivation correlated with a rise in cellular ammonia levels caused by elevated amino acid catabolism. Even in complete medium, ammonia induced autophagy in WT and Ulk1/2−/− MEFs but not in Atg5-deficient MEFs. The autophagy response to ammonia is abrogated by a cell-permeable form of pyruvate resulting from the scavenging of excess ammonia through pyruvate conversion to alanine. Thus, although ULK1 and/or ULK2 are required for the autophagy response following deprivation of nitrogenous amino acids, the autophagy response to the enhanced amino acid catabolism induced by deprivation of glucose or direct exposure to ammonia does not require ULK1 and/or ULK2. Together, these data suggest that autophagy provides cells with a mechanism to adapt not only to nitrogen deprivation but also to nitrogen excess.
Autophagy is an evolutionarily conserved, catabolic process in which cytosolic components and organelles are sequestered in double membrane-bound vesicles called “autophagosomes,” which are delivered to lysosomes for degradation and recycling. This process occurs constitutively to maintain intracellular homeostasis and is considered an adaptive response and survival mechanism to a variety of environmental changes. Increasing evidence suggests that autophagy is implicated in a wide range of pathological conditions. The role of autophagy in diseases is varied; autophagy can either increase susceptibility or protect cells from pathogenic or therapeutic stress (1, 2). We have reported previously that during nutrient deprivation after growth factor withdrawal, autophagy is able to maintain ATP levels and promote cell survival (3). Moreover, a growing body of evidence suggests that autophagy contributes significantly to metabolic regulation at both cellular and organismal levels (4).
In yeast, nutrient starvation activates autophagy-related gene 1 (Atg1), a central autophagy regulator required for bulk autophagy. Distinct from other Atg proteins, Atg1 is a protein kinase that is negatively regulated by the target of rapamycin (TOR) pathway, and it is evolutionarily conserved. In mammals five uncoordinated family member (unc)-51–like kinase proteins (ULKs) have been reported as Atg1 homologs (5). Among these, ULK1 and ULK2 have been reported to be necessary for autophagy, as tested by elimination of the genes using siRNA transfection (6–8). Similar to the function of Atg1, ULK1 and ULK2 play an important role in autophagy, particularly in the recruitment of other Atg proteins such as microtubule-associated protein 1 light chain 3α (LC3) and Atg16 to the autophagosome formation site (6, 9) and in the subcellular relocalization of membrane protein Atg9 during the autophagy process (10). The regulation of ULK1/2 complex interactions with mTOR and the ULK1/2 binding partners Atg13 and FIP200 has been investigated actively under conditions of nutrient deprivation or rapamycin treatment. Based on these results, it is believed the ULK1/2 complex plays an essential role in the initiation of autophagy.
We generated mice with targeted deletion of ulk1 and ulk2, respectively. Surprisingly, ulk1−/− mice are viable and have a relatively mild autophagy phenotype resulting in defective mitochondria clearance during reticulocyte maturation (11). Here we describe the generation of ulk2−/− mice, which also are viable and have no overt autophagy phenotype. To determine whether ULK1 and ULK2 are redundant, we generated mouse embryonic fibroblast (MEFs) from mice deficient for both ulk1 and ulk2 (Ulk1/2 double-knockout, DKO). In this study we investigated the contribution of ULK1, ULK2, and Atg5 in autophagy induced by nutrient deprivation by using Ulk1/2 DKO MEFs or Atg5-deficient MEFs. We report that ULK1/2 are required for the autophagy response to amino acid deprivation but not for the response to glucose deprivation. Furthermore, we show that ammonia is generated at higher levels when cells are deprived of glucose and that ammonia-induced autophagy is dependent on Atg5 but not on ULK1/2.
Results
Autophagy in Response to Amino Acid or Glucose Starvation Is Regulated Differentially by Ulk1/2 and Atg5.
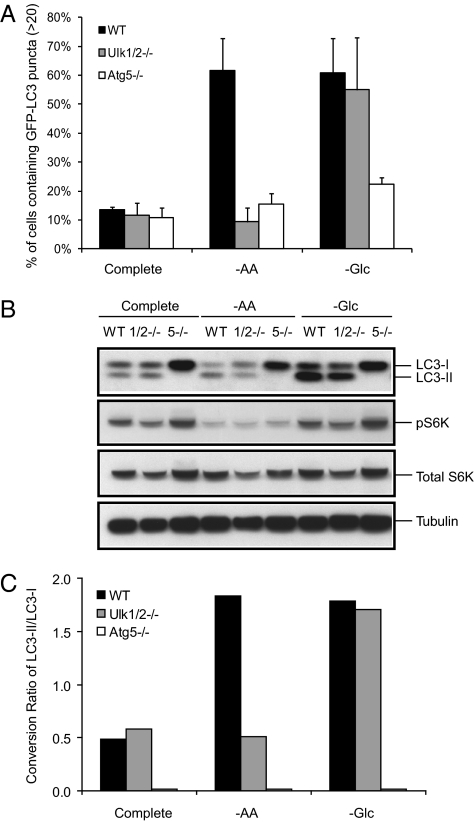
We have shown previously that when nutrient uptake of cells is decreased upon growth factor deprivation, autophagy can provide intracellular carbon sources that can maintain sufficient ATP levels (3). In yeast, the initiation of autophagy following nutrient deprivation is dependent on Atg1, and we set out to determine if mammalian cells exhibit a similar dependency on the mammalian homologs of Atg1. We and others have reported previously the partial autophagic defects of ULK1 deletion in mice (11). Because this partial autophagy defect may result from ULK1 and ULK2 having redundant functions in autophagy induction, we generated ulk2 KO mice as shown in Fig. S1. ulk2 KO mice are viable and are born at expected Mendelian frequency, as also shown previously for ulk1 KO mice (11). Primary Ulk1/2 DKO MEFs were generated from matings of ulk1+/− and ulk2+/− heterozygous mice and were immortalized by transduction with SV40 DNA as previously described (12). ulk1−/−ulk2−/− mice die shortly after birth, and their phenotype will be described elsewhere. To determine the requirement for Ulk1 and Ulk2 in autophagy, a wide range of culture conditions including nutrient starvation and chemical treatments known to activate autophagy were tested. LC3 fused with GFP was expressed ectopically in MEFs to measure autophagy. Autophagy was determined by monitoring the number of GFP-LC3 puncta in treated cells by fluorescence microscopy. The percentage of cells with >20 puncta was scored as a measure of autophagy. The number of GFP-LC3 puncta in WT MEFs was increased under amino acid or glucose deprivation, indicating autophagy activation (Fig. 1A).
Fig. 1.
Ulk1/2 are required for the autophagy response to amino acid deprivation but not for the response to glucose deprivation. GFP-LC3–expressing WT, Ulk1/2 DKO (Ulk1/2−/−) and Atg5 KO (Atg5−/−) MEFs were cultured in complete medium for 24 h and then were left untreated or were subjected to amino acid starvation (−AA) for 6 h or glucose starvation (−Glc) for 24 h as described in Materials and Methods. (A) Autophagy activity was measured by fluorescence microscopy. The percentage of cells containing >20 GFP-LC3 puncta was determined. (B) LC3 conversion from cytosolic LC3 (LC3-I) to lipid-conjugated LC3 (LC3-II) and the presence of phopho-p70 S6 kinase (pS6K), total p70 S6 kinase (S6K), and tubulin were detected by immunoblotting. (C) The conversion ratio of LC3-II/LC3-I of the experiment shown in B was quantified by Image J (National Institutes of Health).
Ulk1/2 DKO MEFs are not able to induce autophagy in response to amino acid starvation for 6 h, but autophagy response after 24 h of glucose deprivation is comparable to that seen in WT MEFs. Autophagy is not detected in Atg5 KO MEFs under either condition (Fig. 1A). In addition to the formation of GFP- LC3 puncta, conversion of LC3 from the cytosolic (LC3-I) to the lipid-bound form (LC3-II) was detected by immunoblotting. The conversion ratio of LC3-I to LC3-II was increased in WT MEFs in response to amino acid or glucose starvation. Ulk1/2 DKO MEFs showed no conversion from LC3-I to LC3-II under conditions of amino acid deprivation over that seen in cells cultured in complete medium, whereas glucose starvation led to LC3 conversion comparable to that seen in WT MEFs deprived of glucose. In contrast, Atg5 KO MEFs showed no LC3 conversion following amino acid or glucose withdrawal (Fig. 1 B and C). The mammalian target of rapamycin (mTOR) complex is considered a major signaling pathway regulating autophagy. Under nutrient starvation, overall phosphorylation of p70 S6 kinase is reduced significantly in WT, Ulk1/2 DKO, and Atg5 KO MEFs, suggesting that mTOR activity is affected by the nutrient status following amino acid or glucose deprivation (Fig. 1B). These data suggest that ULK1/2 mediate autophagy in response to amino acid starvation, and this finding correlates with an observed repression of mTOR activity, as previously reported for Atg1-deficient yeast (13). However, the apparently normal induction of autophagy in response to glucose deprivation in Ulk1/2-deficient cells was unexpected.
Ulk1/2 Are Not Essential Initiators of Autophagy.
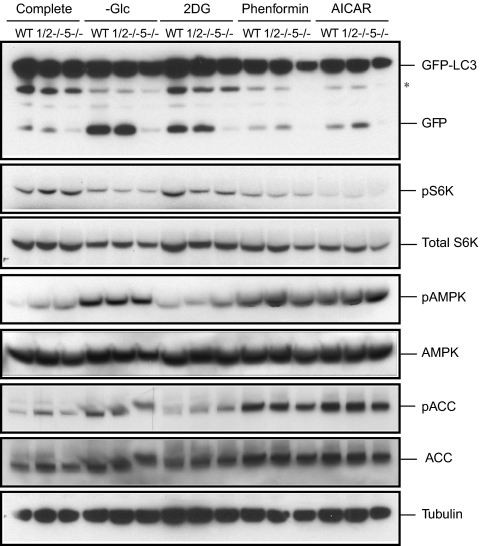
Next, we determined whether ULK1/2 play a role in autophagy induced under conditions of bioenergetic failure. Treatments with 2-deoxyglucose (2DG), phenformin, and 5-aminoimidazole-4-carboxamide riboside (AICAR) were compared with glucose deprivation in WT, Ulk1/2 DKO, and Atg5 KO MEFs. 2DG is a nonmetabolized form of glucose. Phenformin and AICAR were chosen because of their ability to activate AMP-activated protein kinase (AMPK). Autophagy was monitored by GFP-LC3 processing. During autophagy, autophagosomes containing GFP-LC3 fuse with the lysosomes where autophagosome cargo molecules such as GFP-LC3 are degraded. The GFP moiety is removed proteolytically from LC3 in the lysosome. The released GFP moiety remains stable from lysosomal proteolysis. Thus, the appearance of free GFP can be used as a marker to monitor autophagosome delivery to lysosomes and degradation of the cargo (14). Similar to glucose deprivation, treatment with 2DG led to the appearance of cleaved GFP from GFP-LC3 via autophagy in both WT and Ulk1/2 DKO but not in Atg5 KO GFP-LC3–expressing MEFs. However, the addition of 10 mM 2DG to complete medium (25 mM glucose) did not result in either the activation of AMPK phosphorylation or the loss of p70 S6 kinase phosphorylation. Presumably, this impairment of glucose metabolism could be compensated by the catabolism of alternative bioenergetic substrates. In contrast, treatment with phenformin and AICAR failed to enhance GFP processing significantly in any of the genotypes despite a potent reduction in p70 S6 kinase phosphorylation and the induction of phosphorylation of AMPK and acetyl-CoA carboxylase (ACC) (Fig. 2).
Fig. 2.
Ulk1/2 are not required for autophagy in response to bioenergetic stress. WT, Ulk1/2 DKO (1/2−/−), and Atg5 KO (5−/−) MEFs expressing GFP-LC3 were cultured in complete medium and then were subjected to glucose starvation (−Glc) or treatment with 10 mM 2DG, 1 mM phenformin, or 2 mM AICAR for 24 h. GFP-LC3 processing was determined by immunoblotting to monitor autophagy. Immunoblotting for phopho-p70 S6 kinase (pS6K), p70 S6 kinase (S6K), phospho-AMPK (pAMPK), total AMPK, phospho-ACC (pACC), total ACC, and tubulin was performed also. Asterisk indicates nonspecific bands.
Ammonia Stimulates Autophagy.
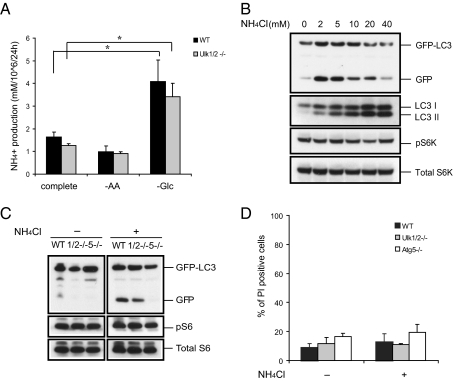
The results described above suggest the ULK1/2-independent autophagy observed in response to glucose withdrawal or 2DG treatment does not simply reflect a bioenergetic compromise that can be mimicked by AMPK activation. Cells deprived of glucose often turn to catabolism of other substrates to maintain their viability. Previously, we have found that glucose-deprived cells depend on autophagy to survive acute glucose withdrawal and require AMPK activation to initiate β-oxidation, a process that takes 24–48 h in proliferating cells (15, 16). During that time, amino acids are the most likely substrate for cells to catabolize. To determine if amino acid catabolism occurred, we measured the production of ammonia, a by-product of amino acid catabolism. The levels of ammonia rose following glucose deprivation in cultures from WT and Ulk1/2 DKO MEFs (Fig. 3A) compared with ammonia generated in cultures of cells incubated in complete medium or medium lacking amino acids. Although some studies (17, 18) have shown that ammonia inhibits autophagy by interfering with lysosomal function, it has been reported recently (19) that ammonia generated from glutaminolysis can contribute to the induction of autophagy. To test dose-dependent effects of ammonia in complete medium, we treated GFP-LC3–expressing WT MEFs with various concentrations (2–40 mM) of NH4Cl and observed autophagy activity by GFP-LC3 processing and LC3 conversion. Autophagy was induced by NH4Cl in a dose-dependent fashion peaking at doses between 2–5 mM in our experiments, but treatment with concentrations above 10 mM NH4Cl significantly reduced the free GFP level (Fig. 3B), as previously reported (19).
Fig. 3.
Ammonia stimulates mTOR-independent autophagy. (A) Glucose deprivation, but not amino acid starvation, increases ammonia production. Ammonia levels in media of cells cultured for 24 h were determined with the Nova Biomedical Flex Analyzer. *P < 0.05. (B) NH4Cl treatment increases autophagy without affecting mTOR activity. WT MEFs expressing GFP-LC3 were treated with varying concentrations of NH4Cl in complete medium for 24 h. Autophagy was determined by GFP-LC3 processing and LC3 conversion. mTOR activity also was assessed by immunoblotting for phospho-p70 S6 kinase (pS6K). (C) Atg5, but not Ulk1/2, is required for ammonia-induced autophagy. The GFP-LC3–expressing WT, Ulk1/2 DKO (1/2−/−), and Atg5 KO (5−/−) MEFs were treated with 2 mM NH4Cl for 24 h. Autophagy was measured by immunoblotting for GFP-LC3 processing. (D) The dose of NH4Cl that induces autophagy has no effect on cell viability. After 48 h incubation at the conditions indicated in C, cell viability was determined by propidium iodide (PI) exclusion.
Next we determined whether ammonia-induced autophagy is dependent on autophagy genes by using GFP-LC3–expressing Ulk1/2 DKO and Atg5 KO MEFs. Treatment with 2 mM NH4Cl reproducibly increased free GFP levels in WT and Ulk1/2 DKO MEFs but not in Atg5 KO cells (Fig. 3C). We also examined if the known upstream regulator of autophagy, mTOR, was affected by ammonia treatment by monitoring phosphorylation of p70 S6 kinase. Phospho-p70 S6 kinase was not reduced significantly by NH4Cl treatment despite the observed induction of autophagy activity in WT and ULK1/2-deficient cells (Fig. 3B and C ). These data suggest that ammonia-induced autophagy is not dependent on mTOR or ULK1/2. The concentration of NH4Cl able to induce autophagy did not induce cell death in WT, Ulk1/2 DKO, or Atg5 KO MEFs over the time course of the experiments (Fig. 3D). Taken together, these results suggest that autophagy can be regulated by ammonia in a dose-dependent manner that does not depend on ULK1/2 or suppression of mTOR activity.
Ammonia Generated from Glucosamine Treatment Induces Autophagy.
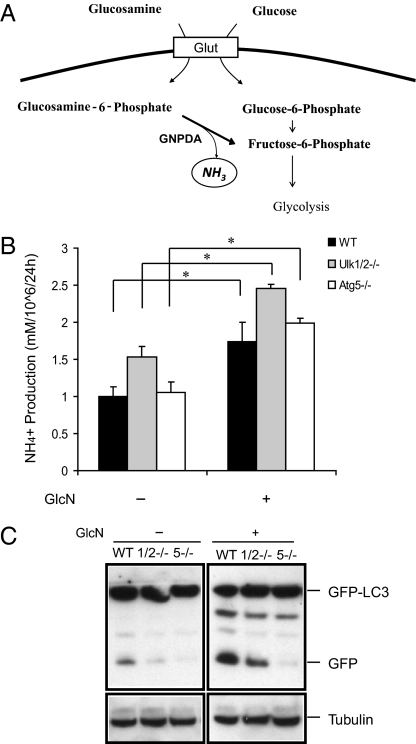
To explore the effect of endogenously produced ammonia on autophagy, we added the amino sugar glucosamine to the culture medium. As outlined in Fig. 4A, glucosamine can be taken up by glucose transporters and phosphorylated by hexokinase (20). Although a portion of glucosamine enters the hexosamine biosynthetic pathway, glucosamine also enters glycolysis after glucosamine-6-phosphate (GlcN-6-P) is deaminated by GlcN-6-P deaminase (GNPDA), resulting in the generation of ammonia and fructose-6-phosphate (21).
Fig. 4.
Ammonia generated following glucosamine treatment induces autophagy. (A) Diagram showing ammonia production during glucosamine metabolism. (B) WT, Ulk1/2 DKO (1/2−/−), and Atg5 KO (5−/−) MEFs expressing GFP-LC3 were cultured in complete medium with 10 mM glucosamine (GlcN) for 24 h. Ammonia levels were measured in supernatants of cell cultures with the Nova Biomedical Flex Analyzer. *P < 0.05. (C) Glucosamine treatment increases autophagy. Autophagy was determined by GFP-LC3 processing at 24 h as indicated in B.
To confirm that glucosamine was being catabolized, we measured ammonia levels in the media of cells cultured with or without glucosamine for 24 h. The concentration of ammonia in the culture medium was increased significantly after 24 h of glucosamine treatment of cultures from WT, Ulk1/2 DKO, and Atg5 KO MEFs (Fig. 4B). To examine whether glucosamine treatment increased autophagy, GFP-LC3 processing was examined using cell lysates from glucosamine-treated WT, Ulk1/2 DKO, and Atg5 KO MEFs. Glucosamine treatment induced an enhanced accumulation of cleaved GFP from GFP-LC3 in WT and ULK1/2-deficient MEFs. However, Atg5 KO MEFs failed to exhibit increased GFP release in response to glucosamine.
Methyl Pyruvate Abrogates Autophagy Induced by Ammonia.
It has been reported that glucose-derived pyruvate plays a role in removing excess ammonia produced by amino acid catabolism (22). Pyruvate can be transaminated to alanine and secreted from cells, thus acting as a sink to reduce the cellular level of reduced nitrogen. Accordingly, we hypothesized that a cell-permeable form of pyruvate, methyl pyruvate (MP), might suppress ammonia-induced autophagy. To test this hypothesis, we investigated whether treatment with exogenous MP suppressed the autophagy response to ammonia derived either from glucosamine treatment or from direct treatment with NH4Cl. MP (20 mM) in combination with 10 mM glucosamine or 2 mM NH4Cl was added to the GFP-LC3–expressing WT, Ulk1/2 DKO, and Atg5 KO MEFs. The free GFP accumulation induced by glucosamine treatment was reduced significantly by treatment with 20 mM MP in WT and Ulk1/2 DKO MEFs (Fig. 5A). Exogenous ammonia-induced GFP cleavage also was suppressed by treatment with MP (Fig. 5B).
Fig. 5.
MP abrogates autophagy induced by ammonia. Autophagy induction by (A) glucosamine (GlcN) or (B) NH4Cl is suppressed by MP treatment. WT, Ulk1/2 DKO (1/2−/−), and Atg5 KO (5−/−) MEFs expressing GFP-LC3 were cultured in complete medium for 24 h before drug treatments. MP (20 mM) was added for an additional 24 h with either 10 mM glucosamine (A) or 2 mM NH4Cl (B). GFP-LC3 processing was detected by immunoblotting to assess autophagy. (C) Relative alanine levels in the culture media of the cells following 24 h incubation as indicated in A were measured by GC-MS analysis in WT MEFs. Com, complete medium. *P < 0.05.
To confirm that MP can be used as a substrate for ammonia detoxification, the alanine levels in the culture media of cells cultured in the presence or absence of 10 mM glucosamine and/or 20 mM MP were measured by GC-MS analysis (Fig. 5C). Alanine levels were significantly higher after treatment with both MP and glucosamine than after treatment with glucosamine alone (Fig. 5C), correlating with the ability of MP to suppress glucosamine-induced autophagy (Fig. 5A). In addition, ammonia levels were decreased following treatment with glucosamine plus MP compared with treatment with glucosamine alone (Fig. S2).
Discussion
Autophagy is considered an essential process for cell growth and survival in response to nutrient deprivation and environmental stress. In this study we compared the autophagy response to various known autophagy-inducing treatments in MEFs generated from mice deficient for Ulk1 and Ulk2 with the previously described response in Atg5-deficient MEFs. We initially found that although Ulk1/2 DKO are selectively impaired in the autophagy response to amino acid deprivation, the autophagic response to glucose starvation is comparable to that of WT MEFs. It has been reported that amino acid starvation or mTOR inhibition can lead to suppression of general anabolism (23). These results imply that Ulk1/2 are necessary for autophagy signaling, which might block cellular anabolism during amino acid starvation. Recent work by others supports this conclusion (6–8). On the other hand, the present work demonstrates the existence of an independent pathway to induce autophagy initiated by the cellular catabolism of nitrogenous compounds and the accumulation of ammonia. Unlike autophagy initiated by amino acid deprivation, autophagy initiated by amino acid catabolism does not depend on the suppression of mTOR activity or on the function of ULK1 or ULK2. However, both forms of autophagy involve the common autophagic program that includes Atg8 (LC3) processing and Atg5 function.
Prior work may have failed to identify this alternative pathway of autophagy initiation because during glucose deprivation transformed cell lines undergo a bioenergetic crisis that activates autophagy through AMPK inhibition of mTOR (24, 25). In contrast, nontransformed cells such as the MEFs studied here can use alternative energy sources to maintain cell integrity following glucose withdrawal. Kim et al. (25) reported a defective autophagy response to short-term (5 h) glucose deprivation in Ulk1-deficient MEFs. Although, acute glucose withdrawal induces an AMPK-dependent inactivation of mTOR that can initiate autophagy in an ULK1-dependent fashion (25), it has been demonstrated that by 24 h nontransformed cells can reactivate mTOR-dependent growth by turning to alternative bioenergetic substrates (26). The present data demonstrate that amino acid catabolism provides the bioenergetic substrate for recovery and that the ongoing autophagy observed at 24 h is initiated by the ammonia released through the catabolism of amino acids. This ammonia-dependent autophagy is ULK1/2 independent. Consistent with this finding, we found that ammonia levels in MEFs increased significantly during glucose deprivation, whereas ammonia levels fell during amino acid deprivation. These data suggest that cells increase their amino acid catabolism to supply bioenergetic fuel when the major energy source—glucose—is absent. The resulting increase in ammonia is in the range that optimally stimulates autophagy (Fig. 3) to promote adaptation and survival. However, prolonged glucose deprivation in transformed cells leads to activation of β-oxidation in an AMPK-dependent fashion and ultimately to the suppression of mTOR activation (16).
In 2010, Eng et al. (19) reported that ammonia derived from glutaminolysis can contribute to the activation of autophagy. The U2OS cells they studied were transformed cells that exhibit glutaminolysis, which both rapidly depletes the medium of glutamine and generates ammonia as a byproduct of glutamine catabolism. These cells exhibit a progressive increase in autophagy over time in culture, and part of this increase is suppressed by RNAi-mediated suppression of ULK1. The data presented here suggest that the residual autophagy exhibited by U2OS cells after 96 h in culture is the result of the accumulating ammonia, whereas the ULK1-dependent autophagy may have resulted from the concomitant depletion of glutamine. In proliferating cells, glutamine has been identified as an alternative carbon source that can be used as a tricarboxylic acid cycle intermediate (27, 28), leading to the major source of ammonia production. In the presence of glucose, the resulting ammonia is buffered by the ability of the cell to convert glucose-derived pyruvate into alanine. However, when glucose supplies are depleted, the resulting ammonia triggers autophagy. When cells are treated with exogenous ammonia, they react by initiating autophagy, a process that can be mitigated by supplying the cells with a cell-permeable form of pyruvate (Fig. 5). Comparable observations have been reported in yeast, where treatment with ammonia results in a cellular growth defect and massive secretion of amino acids including alanine (29).
It has been reported recently that expression of Myc in cancer cells leads to increased glutamine metabolism by increasing the level of glutamine transporters and the enzymes involved in glutaminolysis in certain cancer cells (27, 28). Because glutamine is a major nitrogen donor, Myc expression increases the ammonia levels derived from activation of glutaminolysis, thus possibly leading to an increase of basal autophagy and the prominent level of autophagy vesicles seen in Myc-derived tumors such as Burkitt lymphoma (30). Although the exact mechanism of how ammonia stimulates autophagy is largely unknown, if ammonia-induced autophagy has a cellular-protective role in cancer, inhibition of autophagy may be beneficial for the treatment of Myc-induced malignancies.
In summary, creating MEFs deficient for both Ulk1 and Ulk2 through targeted deletions has allowed us to demonstrate the existence of multiple pathways of autophagy. Amino acid deprivation leads to autophagy dependent on both ULK1/2 and Atg5. However, glucose deprivation results in the accumulation of ammonia derived from the catabolism of amino acids and autophagy which requires Atg5 but not ULK1/2 or mTOR. In addition to its role as bioenergetic fuel, glucose plays a critical role in maintaining homeostasis of intracellular ammonia level, further contributing to the regulation of ammonia-induced autophagy.
Materials and Methods
Generation of Floxed Mice and MEFs.
The ulk1−/− mice were generated as previously described (11). To generate ulk2−/− mice, a targeting construct including a neomycin resistance cassette and a 4.1-kb NotI-EcoRI fragment containing exons I, II, and III, each flanked by loxP sites, was generated a shown in Fig. S1. This construct was electroporated into R1 embryonic stem cells (31). Electroporation, selection, maintenance, and identification of positive ES cell clones by Southern blotting were performed as described (32). Heterozygous ulk2+/fl mice were mated with EIIa-cre transgenic mice (Jackson Laboratory) to generate ulk2+/− mice. The ulk2−/− mice were generated at the expected Mendelian frequency from heterozygous parents, were viable, and had no overt phenotype. Ulk1/2 DKO MEFs were generated from matings of ulk1+/− and ulk2+/− heterozygous mice. MEFs were established from individual embryos of specified genotypes and immortalized as previously described (12) and were maintained in DMEM with 10% FBS (Gemini), 100 U/mL penicillin, and 100 μg/mL streptomycin. Primary Atg5 KO MEFs were generated from matings of Atg5 heterozygous mice generously provided by Noboru Mizushima (Tokyo Medical and Dental University, Bunkyo-Ku, Japan) and then were immortalized by transfection with SV40 DNA.
Cell Culture.
MEFs were cultured in DMEM (Invitrogen) with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. For glucose starvation, DMEM without glucose (Invitrogen) was supplemented with 10% dialyzed FBS. For the amino acid starvation medium, Earle's balanced saline solution was supplemented with 10% dialyzed FBS, glucose, vitamins, and minerals at the same concentration as in DMEM.
Reagents.
2DG (used at 10 mM), phenformin (used at 1 mM), glucosamine (used at 10 mM), and MP (used at 20 mM) were purchased from Sigma. AICAR (used at 2 mM) was obtained from Toronto Research Chemicals, Inc.
Plasmid Constructs.
GFP-LC3 was described previously (30). WT GFP-LC3, Ulk1/2 DKO GFP-LC3, and Atg5 KO GFP-LC3 MEF were generated by retroviral transduction and sorted for GFP+ clones by MoFlow as previously described (30). Individual clones were used in experiments using transduced cells.
Immunoblotting.
MEFs or MEFs stably expressing GFP-LC3 were plated in nutrient-replete medium 1 d before treatment. After indicated treatments, cells were lysed in RIPA buffer (50 mM TRIS-Cl pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, 1 mM EDTA) with protease inhibitor cocktail (Roche Aplied Bioscience) and phosphatase inhibitor (Sigma-Aldrich). Lysates were resolved by SDS/PAGE, and blots were probed using the following primary antibodies: anti-GFP monoclonal antibody (Roche Applied Science); anti-LC3 (generated by Quality Control Biochemicals); anti-ULK1 (N17; Santa Cruz Biotechnology or generated by YenZym custom service); anti-ULK2 (generated by YenZym custom service); anti-p70 S6 kinase; anti–phospho-p70 S6 kinase (Thr389); anti-AMPK; anti–phospho-AMPK T172; anti-ACC; anti-tubulin (Sigma-Aldrich) and anti–phospho-ACC (Cell Signaling Technology). Bands were detected using HRP-labeled secondary antibodies and the enhanced chemiluminescence detection kit (Amersham). The band intensities of LC3-I and LC3-II were quantified by Image J (National Institutes of Health).
Cell Viability.
Cell viability was determined by propidium iodide exclusion using flow cytometry. MEFs were trypsinized and resuspended in propidium iodide solution at 1 μg/mL (Invitrogen) before analysis.
Fluorescence Microscopy.
For fluorescence microscopy, GFP-LC3–expressing MEFs were imaged live on an inverted Nikon Eclipse (TE300) fluorescent microscope and analyzed by Metamorph software (Universal Imaging).
Measurements of Metabolites.
Glucose uptake, lactate production, glutamine consumption, and ammonia production were measured using the Nova Biomedical Flex Analyzer. Alternatively, ammonia concentrations were measured using the Sigma Ammonia Assay Kit. Secreted alanine levels in the culture medium were determined by GC-MS analysis as previously described (33). Identification of the alanine peak was confirmed by using an alanine standard peak. Alanine levels were quantified by integration of peak areas and were compared with those in the control medium.
Statistics.
An unpaired, two-tailed Student's t test was used.
Supplementary Material
Acknowledgments
We thank the members of the C.B.T. laboratory for technical assistance and comments on the manuscript. This work was supported by grants from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107969108/-/DCSupplemental.
References
- 1.Yang Z, Klionsky DJ. Eaten alive: A history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lum JJ, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 4.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young AR, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 11.Kundu M, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei MC, et al. Proapoptotic BAX and BAK: A requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamada Y, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheong H, et al. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buzzai M, et al. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67:6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- 16.Buzzai M, et al. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation. Oncogene. 2005;24:4165–4173. doi: 10.1038/sj.onc.1208622. [DOI] [PubMed] [Google Scholar]
- 17.Seglen PO, Reith A. Ammonia inhibition of protein degradation in isolated rat hepatocytes. Quantitative ultrastructural alterations in the lysosomal system. Exp Cell Res. 1976;100:276–280. doi: 10.1016/0014-4827(76)90148-8. [DOI] [PubMed] [Google Scholar]
- 18.Bates PJ, Coetzee GA, Van der Westhuyzen DR. The degradation of endogenous and exogenous proteins in cultured smooth muscle cells. Biochim Biophys Acta. 1982;719:377–387. doi: 10.1016/0304-4165(82)90113-1. [DOI] [PubMed] [Google Scholar]
- 19.Eng CH, Yu K, Lucas J, White E, Abraham RT. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal 3:ra31. 2010 doi: 10.1126/scisignal.2000911. [DOI] [PubMed] [Google Scholar]
- 20.Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- 21.Wolosker H, et al. Molecularly cloned mammalian glucosamine-6-phosphate deaminase localizes to transporting epithelium and lacks oscillin activity. FASEB J. 1998;12:91–99. doi: 10.1096/fasebj.12.1.91. [DOI] [PubMed] [Google Scholar]
- 22.DeBerardinis RJ, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009;19:R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones RG, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 27.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hess DC, Lu W, Rabinowitz JD, Botstein D. Ammonium toxicity and potassium limitation in yeast. PLoS Biol. 2006;4:e351. doi: 10.1371/journal.pbio.0040351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amaravadi RK, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiels H, et al. TRAF4 deficiency leads to tracheal malformation with resulting alterations in air flow to the lungs. Am J Pathol. 2000;157:679–688. doi: 10.1016/S0002-9440(10)64578-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.