Abstract
Ebolavirus (Ebov), an enveloped virus of the family Filoviridae, causes hemorrhagic fever in humans and nonhuman primates. The viral glycoprotein (GP) is solely responsible for virus–host membrane fusion, but how it does so remains elusive. Fusion occurs after virions reach an endosomal compartment where GP is proteolytically primed by cathepsins. Fusion by primed GP is governed by an internal fusion loop found in GP2, the fusion subunit. This fusion loop contains a stretch of hydrophobic residues, some of which have been shown to be critical for GP-mediated infection. Here we present liposome fusion data and NMR structures for a complete (54-residue) disulfide-bonded internal fusion loop (Ebov FL) in a membrane mimetic. The Ebov FL induced rapid fusion of liposomes of varying compositions at pH values at or below 5.5. Consistently, circular dichroism experiments indicated that the α-helical content of the Ebov FL in the presence of either lipid-mimetic micelles or small liposomes increases in samples exposed to pH ≤5.5. NMR structures in dodecylphosphocholine micelles at pH 7.0 and 5.5 revealed a conformational change from a relatively flat extended loop structure at pH 7.0 to a structure with an ∼90° bend at pH 5.5. Induction of the bend at low pH reorients and compacts the hydrophobic patch at the tip of the FL. We propose that these changes facilitate disruption of lipids at the site of virus–host cell membrane contact and, hence, initiate Ebov fusion.
Keywords: NMR solution structure, virus entry, lipid–protein interaction, membrane protein
Ebolavirus (Ebov), a member of the Filoviridae family, is an enveloped virus capable of causing severe hemorrhagic fever. The virus initially targets dendritic cells and macrophages and then infects a large variety of cell types. This overwhelming invasion results in a fast onset of illness and, in the case of Zaire Ebov, the most fatal species, up to 90% mortality (1, 2). Although there are promising reports of vaccine strategies and liposome-encapsulated siRNA formulations, there are currently no approved vaccines or antiviral drugs to combat Ebov infections (3, 4). A complementary therapeutic approach is to target the viral entry and fusion machinery using either small-molecule or antibody-based strategies.
The glycoprotein (GP) spikes that stud the filamentous Ebov particle are solely responsible for entry and fusion (5, 6). Following binding of the receptor binding subunit (GP1) of GP to an as-of-yet unknown host cell receptor(s), Ebov is endocytosed and delivered to endosomes, where GP is cleaved by cathepsins B and L to a key intermediate form (7–9). A subsequent trigger (8) is then thought to cause conformational changes in GP that expose the internal fusion loop found in the GP2 subunit so that the fusion loop can engage the target bilayer. After the loop penetrates the host cell membrane, GP2 is thought to fold roughly in half, bringing the host and viral membranes in close proximity, resulting in fusion and release of the viral replication machinery into the host cell (10–12). The internal fusion loop of Ebov GP contains a centrally located stretch of hydrophobic residues, some of which, when altered in the context of the full-length GP protein, inhibit the infectivity of GP pseudovirus particles (13).
Crystallographic studies show the structure of the fusion loop in the context of the entire Ebov GP spike in its prefusion trimeric form (14). In this structure, the fusion loop interacts with many residues of the neighboring GP subunit. The crystal structure also confirms the presence of a predicted disulfide bond between the well-conserved cysteine residues C511 and C556, which tether the fusion loop (15–17). Mutation of either Cys abrogated transduction of murine leukemia virus particles pseudotyped with Zaire Ebov GP (16). By analogy with a similar internal fusion loop in the envelope (Env) glycoprotein of the avian sarcoma/leukosis virus (ASLV), this disulfide bond is thought to be critical for fusion; in the absence of either or both of the tethering cysteines (in the context of the full-length protein), ASLV Env-mediated fusion is aborted during the lipid-mixing stage of fusion (18–20). Crystal structures have also been obtained for GP2 in its postfusion conformation, but the fusion loop was not included in the proteins used for crystallization (15, 21). An NMR structure has been presented for a 16-residue linear peptide comprising the central hydrophobic region of the Ebov fusion loop in SDS micelles at pH 7.0 (22). However, this short peptide did not include either of the essential cysteines that tether the fusion loop or the additional 38 residues of the loop.
To investigate how the Ebov disulfide-bonded fusion loop contributes to membrane fusion, we generated a 54-residue construct representing the entire disulfide-bonded internal Ebov fusion loop (Ebov FL; Fig. 1). We show that this Ebov FL construct is capable of inducing liposome fusion in a low pH-dependent manner, and that it undergoes a major conformational change upon insertion into liposomes and phospholipid-mimicking dodecylphosphocholine (DPC) micelles at pH 5.5.
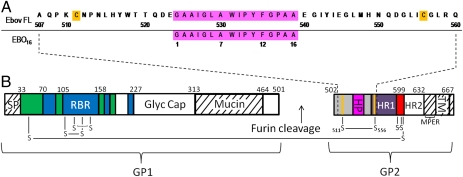
Fig. 1.
Sequence and topology of Ebov glycoprotein containing the fusion loop region. (A) Sequence numbering of (Upper) the Ebov FL used in this study and (Lower) the EBO16 peptide used in a previous NMR study (22). The hydrophobic patch residues are highlighted in magenta and the cysteines forming a disulfide are highlighted in orange. (B) Domain topology of Ebov GP; nomenclature and coloring are as in the published crystal structure (14). White and hatch-marked regions correspond to disordered and construct-deleted regions, respectively. GP1: green, GP1 base; blue, GP1 head; Glyc Cap, glycan cap; Mucin, mucin-like domain; RBR, receptor binding region; SP, signal peptide. GP2: red, chain-reversal region; HP, hydrophobic patch; HR1 and HR2, heptad repeats 1 and 2; MPER, membrane-proximal external region; TM, transmembrane domain. The orange dividing lines represent the cysteine residues of the internal FL (gray). The dashed lines represent the residues making up our internal Ebov FL construct.
Results
Liposome Fusion Induced Under Acidic Conditions.
The ability of the 54-residue disulfide-bonded Ebov FL construct to induce liposome fusion was tested in a FRET-based lipid-mixing assay (23). The Ebov FL was mixed with liposomes composed of POPC:POPG [1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine to 1-palmitoyl-2-oleoyl-sn-glycero-3-(1-phosphoglycerol)] (7:3) at a ratio of 1:20 (protein:lipid). No fusion was observed at pH 7.4. When equilibrated samples were acidified to pH 7.0, 6.5, or 6.0, a slow rate of lipid mixing was observed (Fig. 2). In contrast, when the pH was lowered to 5.5 or less, rapid lipid mixing was observed, albeit reaching a maximum of only ∼5%. More extensive fusion (reaching ∼30%) was observed in reactions in which either the protein:lipid ratio or the percentage of POPG was increased (Figs. S1 and S2). These findings indicate that the Ebov FL can induce liposome fusion, but only at low pH.
Fig. 2.
pH-dependent lipid mixing induced by Ebov FL. Vesicles were composed of POPC:POPG (7:3). Experiments were performed with 50 μM unlabeled:labeled vesicles at a ratio of 9:1 and with 2.5 μM Ebov FL. The labeled vesicles contained 1 mol % 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (NBD-POPE) and 1 mol % 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (Rh-POPE). Percent lipid mixing was derived from FRET of vesicles in the presence or absence (black trace) of Ebov FL upon acidification with 1 M HCl to the indicated pH values.
Secondary Structure Determination by CD Spectroscopy.
The effect of low pH on the secondary structure of the FL was investigated using CD spectroscopy. In the presence of POPC:POPG (4:1) small unilamellar vesicles (SUVs), the FL showed minimal helical content, ∼9% at pH 7.0–6.0, similar to CD spectra recorded in the absence of lipid (Fig. 3A). As the pH was lowered to 5.5–4.5, the amount of helical structure increased to 15–22%. CD spectra were also recorded for the FL in the presence of a large excess of DPC micelles at different pH values (Fig. 3B). DPC caused an increase in helical structure, 18% at pH 7.4–6.0, but the helicity was further enhanced to 25% when samples were acidified to pH 5.5–4.5. The FL was also studied in the absence of SUVs or detergent micelles. At neutral pH the helical content was minimal, but it increased to 22% under acidic solution conditions (Fig. S3).
Fig. 3.
CD spectra of Ebov FL in the presence or absence of (A) SUVs and (B) DPC micelles at different pH values. (A) Ebov FL (42 μM; 0.25 mg/mL) incubated with 1 mM POPC:POPG (4:1) SUVs. The dashed line indicates the CD spectrum of Ebov FL in pH 7.4 buffer without lipid. (B) FL (42 μM) incubated with 5 mM DPC micelles. All spectra were recorded at 22 ± 2 °C.
Structure Determination by Solution NMR.
NMR spectroscopy was used to obtain structures of the disulfide-bonded Ebov FL at pH 7.0 and 5.5 in DPC micelles. Experimental conditions were selected based on the lipid-mixing and CD results. DPC micelles were used for structure determination because SUVs are too large to obtain highly resolved NMR spectra. According to our CD results, the secondary structure of the FL appears to be similar whether bound to DPC micelles or POPC:POPG (4:1) liposomes at pH 5.5. Representative fully assigned heteronuclear single-quantum correlation (HSQC) spectra for the FL in DPC micelles at pH 7.0 and 5.5 are shown in Fig. S4. Standard triple-resonance NMR spectra were acquired and assigned with 15N, 13C-labeled samples in fully deuterated DPC as described in full detail in SI Materials and Methods. The 13Cβ chemical shifts of Cys511 and Cys556 at 41.9 and 41.2 ppm, respectively, confirmed that these residues are in an oxidized state and that the fusion peptide indeed formed a loop (Fig. S5). The reduced form would have shown chemical shifts for these Cys resonances in the 26- to 32-ppm range; no peaks corresponding to Cys residues were detected in this region of the spectra. Moreover, a large fraction of the cross-peaks in the HSQC spectra were significantly changed after reduction of the NMR samples with DTT, further confirming the oxidized disulfide-bonded state of our protein. Structural restraints were obtained from 15N- and 13C-edited NOESY and HNHA experiments as well as from the chemical shift information as described in SI Materials and Methods.
The 20 lowest-energy conformers representing the pH 7.0 “inactive” and 5.5 “active” conformations are depicted in Fig. 4 A and C, respectively. The structural statistics are reported in Table 1. At pH 7.0 there was no standard regular secondary structure detected by MOLMOL secondary structure recognition software (24). Despite this lack of standard secondary structure recognized by NMR, the tight coils and turns that are apparent in the structure may be the reason for the apparent helix detected in the CD experiments for both pH 7.0 and 5.5 conditions in the presence of DPC micelles. Because the Kabsch–Sander routine that is used in MOLMOL defines helix by the presence of intramolecular residue i to i+4 hydrogen bonds and because the molar ellipticity measured in CD responds to φ, ψ backbone torsion angles, it is not surprising that NMR structure representations may underestimate and CD may overestimate actual extents of helix, especially if polypeptides that are localized in interfacial membrane regions can make alternate hydrogen bonds to lipid headgroups and interfacial water molecules while still being helical or near-helical.
Fig. 4.
NMR structures of Ebov FL in DPC micelles at pH 7.0 and 5.5. The 20 lowest-energy conformers representing the Ebov FL structure in DPC micelles at (A) pH 7.0 and (C) pH 5.5. Black dashed lines depict distances between Cα atoms of Ile544 and Trp518 (8.8 ± 0.4 Å at pH 7.0 and 11.3 ± 0.4 Å at pH 5.5) and Ile544 and Gly528 (12.7 ± 0.25 Å at pH 7.0 and 7.1 ± 0.65 Å at pH 5.5). B and D show the lowest-energy conformers at pH 7.0 and pH 5.5, respectively, with side chains of indicated residues shown in blue stick figure representations. Unstructured loops and turn regions are shown in gray, α-helix in green, β-sheet in blue, and the disulfide-linked residues Cys511 and Cys556 in orange. The hydrophobic patch residues defined in Fig. 1 are highlighted in magenta.
Table 1.
NMR and refinement statistics for Ebola fusion loop in DPC micelles
| pH 7.0 | pH 5.5 | |
| NMR distance and dihedral restraints | ||
| Distance restraints | ||
| Total NOE | 665 | 517 |
| Intraresidue | 151 | 155 |
| Interresidue | 514 | 362 |
| Sequential (i − j = 1) | 281 | 216 |
| Medium-range (i − j ≤ 4) | 188 | 114 |
| Long-range (i − j ≥ 5) | 45 | 32 |
| Total dihedral angle restraints | ||
| φ | 8 | 10 |
| ψ | 5 | 10 |
| Structure statistics | ||
| Violations (mean and SD) | ||
| Distance restraints (Å) | 0.029 ± 0.001 | 0.024 ± 0.001 |
| Dihedral angle restraints (°) | 0.28 ± 0.10 | 0.20 ± 0.05 |
| Maximum dihedral angle violation (°) | 0 | 0 |
| Maximum distance restraint violation (Å) | 0 | 0 |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.0037 ± 0.0001 | 0.0029 ± 0.0001 |
| Bond angles (°) | 0.52 ± 0.01 | 0.48 ± 0.11 |
| Impropers (°) | 0.35 ± 0.01 | 0.29 ± 0.02 |
| Average pairwise rmsd* (Å) | ||
| Heavy atoms | 1.45 ± 0.43 | 1.06 ± 0.30 |
| Backbone | 1.95 ± 0.48 | 1.54 ± 0.31 |
*Calculated from the 20 lowest-energy structures out of 200.
The pH 7.0 structures in Fig. 4 A and B depict an elongated loop confined by a disulfide bond. The tip of the loop is composed of a somewhat flexible hydrophobic patch, which is in a planar conformation. The pH 7.0 conformation can be described as a hand, where the wrist represents the disulfide bond and the fingers represent the hydrophobic patch. In the pH 5.5 structure, the FL shows a reorientation of the hydrophobic patch about residues Ile544 and Gly528. The change in structure between pH 7.0 and 5.5 can be thought of as the formation of a fist, where the knuckles represent the hinged region, indicated by a red dashed line in Fig. 4C. The distances between the backbone Cα atoms of Trp518, Gly528, and Ile544 are provided (black dashed lines) to show the major reorientation of the hydrophobic patch relative to the supporting “palm” of the hand formed by the two disulfide-bonded chains. To further illustrate the differences between the structures, Fig. 5A shows a comparison of the lowest-energy conformers at pH 7.0 and 5.5. This comparison clearly shows that the pH 7.0 structure is more elongated and the pH 5.5 structure contains a roughly 90° bend in the hinge region.
Fig. 5.
Comparisons of the Ebov FL NMR structures and a portion of the prefusion crystal structure of the trimeric GP ectodomain [Protein Data Bank (PDB) ID code 3CSY]. (A) Overlay of lowest-energy NMR conformers at pH 7.0 (cyan) and 5.5 (gray) with its helix in green. The structures are aligned by the disulfide bond shown in orange. (B) Overlay of lowest-energy NMR conformer at pH 7.0 (cyan) with Ebov fusion loop residues and HR1 from PDB ID code 3CSY (fusion loop, gray; HR1, rainbow). β-Sheet strand 6 of GP1, shown in black, presumably stabilizes the two strands of β-sheet seen in the FL of the crystal structure (corresponding to residues 515–520 and 543–548 of Fig. 1). Constructs are aligned by residues at the tip of the hydrophobic patch.
The differences between the pH 7.0 and pH 5.5 structures include some elements of standard regular secondary structure as detected by MOLMOL. Most notable is the helix from Gly536 to Ala539. The increased formation of a coil between residues His516 and Gly528 also contributes to the hinge motion or “clenching of the fist.” These conformational changes result in the repositioning of the hydrophobic Ile542-Tyr543-Ile544 region from an inward-facing Trp518 at pH 7.0 (Fig. 4B) over to Ile527-Gly528-Leu529 located in the hydrophobic patch at pH 5.5 (Fig. 4D). Interestingly, this rearrangement moves the aromatic residue Tyr543 toward the hydrophobic front end of the fist. Taken together, these conformational changes result in a redistribution of hydrophobic residues on the surface of the FL, as depicted in Fig. 6. At pH 7.0, the hydrophobic residues are fairly evenly distributed over the surface of the structure (Fig. 6 A and B), but they are more, but not exclusively, focused to the front end of the fist at pH 5.5 (Fig. 6 C and D). This clenching motion compacts the structure and reorients the enlarged hydrophobic patch at the tip, potentially allowing for better membrane accessibility at low than at neutral pH.
Fig. 6.
Hydrophobicity surface map of the lowest-energy conformers at pH 7.0 and pH 5.5. Side view representations at (A) pH 7.0 and (C) pH 5.5. Front view representations with the hydrophobic patch facing forward at (B) pH 7.0 and (D) pH 5.5. Trp, Tyr, Phe, Leu, and Ile are classified as hydrophobic, and their surfaces are shown in red in these representations.
We also aligned the pH 7.0 NMR structure of the Ebov FL with the corresponding region of the pH 8.5 prefusion crystal structure of the trimeric GP ectodomain including heptad repeat 1 (Fig. 5B). Despite many differences between the two structures, the overall elongated shapes of the fusion loops are similar. In the crystal structure, the fusion loop is stabilized by residues in GP1 and GP2 that are absent in the construct used for NMR. In addition, the NMR structure was determined in the presence of DPC micelles, whereas no lipids were present in the crystal. The most striking difference between the two structures is perhaps the β-sheet that is observed at the beginning and end of the fusion loop in the native prefusion crystal structure but is absent in the extended pH 7.0 solution structure in DPC. This small sheet is most likely stabilized by the neighboring β-strand 6 of GP1 in the crystal structure, as shown in Fig. 5B (14).
Discussion
All enveloped viral fusion proteins, be they class I, II, or III, contain a fusion peptide or fusion loop, a relatively hydrophobic stretch of amino acids that engages the target membrane to initiate fusion. The fusion peptides of most class I fusion proteins, for example those of the hemagglutinin (HA) of influenza virus and the Env glycoprotein of HIV, are located at the amino terminus of their transmembrane-anchored fusion subunits. In contrast, two class I viral fusion proteins, the Env glycoprotein of the ASLV and the GP of Ebov, contain an internal fusion loop flanked by conserved Cys residues. Class II and III viral fusion proteins also contain internal fusion loops that appear to be stabilized by disulfide bonds (25, 26). Whereas considerable information is available on the structure and function of N-terminal fusion peptides, less is known about internal fusion loops, especially about their structure as they engage membranes. Although information has been presented on the structure and fusion activity of short (e.g., 15- to 16-residue) linear portions of these loops, no study has yet characterized an entire disulfide-bonded fusion loop in a lipid environment. Here we generated and analyzed a construct, Ebov FL, corresponding to the 54-residue disulfide-bonded fusion loop of Ebov GP. We first demonstrated that the Ebov FL changes conformation (increases its α-helicity) and induces liposome fusion at low, but not neutral, pH. We next determined NMR structures for the loop in DPC micelles at both neutral and acidic pH. Our NMR studies reveal a pronounced low pH-dependent reorientation of the tip of the FL and a concomitant repositioning of the hydrophobic residues at this tip, a region containing amino acids critical for Ebov GP-mediated infection (13). In addition to their relevance to Ebov GP-mediated fusion, our findings may provide more general insight into the mechanism of other viral fusion proteins that contain internal fusion loops.
Following proteolytic priming by endosomal cathepsins, Ebov GP-mediated fusion is thought to follow a canonical fusion cascade used by all characterized viral fusion proteins: The trimeric GP spike is thought to change conformation, insert its fusion loop into the target membrane, and then fold roughly in half, to bring the viral and target membranes together and initiate their merger (10–12). In the native Ebov GP trimer, the fusion loop is extended and relatively flat. Our NMR structure of the disulfide-bonded Ebov FL in DPC micelles at pH 7.0 reveals a similar elongated and relatively flat loop with a hydrophobic patch at its tip. The Ebov FL induces liposome fusion, but only at low pH, an observation that suggests that Ebov fusion requires low pH per se (i.e., that low pH is not just needed for optimal activity of endosomal cathepsins). In its fusion-active (pH 5.5) state, the hydrophobic patch at the tip of the FL is bent ∼90° and endowed with an increased hydrophobic surface area. The overall change can be likened to the clenching of a fist. Although the orientation of the Ebov FL relative to the plane of the membrane is not yet known, the low pH-induced ∼90° bend may drive the hydrophobic tip of the FL deeper into the membrane. This proposal is reminiscent of our model for how the N-terminal fusion peptide of influenza HA engages target membranes (27–30). Our HA fusion peptide construct contains the first 20 residues of the fusion peptide from the X:31 HA (subtype H3) and adopts a “boomerang” shape with a 105° angle separating two helical sections. At low pH, one helical section lies approximately parallel to the membrane while the other penetrates it more steeply and deeply. Analysis of mutant forms of this HA fusion peptide suggests that the 105° angle is required for proper fusion: No fusion is observed with a point mutant that forms a single straight interfacial helix (28), and leaky fusion is observed with a mutant peptide in which the angle of the boomerang increased to ∼150° (30). A 23-residue fusion peptide from a different subtype HA, which has two amino acid differences compared with X:31 HA, was reported to form a tighter bend at a corresponding position (31).
In the hydrophobic patch at its tip, the fusion loop of Ebov GP contains an aromatic-hydrophobic-glycine tripeptide motif (Tyr534-Phe535-Gly536), as has been seen in the fusion peptides/loops of other viral fusion proteins (12, 32, 33), for example those of Marburg GP, HIV Env, influenza HA, Dengue E, Severe Acute Respiratory Syndrome S, and even a fusion protein encoded by a nonenveloped virus (34). This type of motif is well-suited to interact with membrane–water interfaces. In the Dengue E protein, a class II fusion protein, the tripeptide motif (Leu-Phe-Gly) is located at the tip of its internal fusion loop and is thought to embed in the membrane ∼6 Å deep, with the Phe of its aromatic-hydrophobic-glycine tripeptide motif and an upstream Trp serving as “aromatic anchors” at the membrane interface (35). When the upstream Trp was mutated in a linear 15-residue model of part of the Dengue fusion loop, no lipid mixing was observed (36). The bipartite fusion loops of class III viral fusion proteins also contain a combination of aromatic and hydrophobic residues (37–40). Interestingly, one of the fusion loops of HSV glycoprotein B, a class III fusion protein, has been shown to undergo a local change in structure at low pH (41).
Analogous to the Dengue virus fusion loop, the Ebov fusion loop contains a Trp (Trp531 in our numbering system) upstream of the aromatic-hydrophobic-glycine tripeptide motif (Tyr534-Phe536-Gly537) found in the hydrophobic patch at the tip of the Ebov fusion loop. The linear EBO16 construct (Fig. 1) with an Ala substituted for this Trp lacked secondary structure in SDS micelles compared with the wild-type peptide (22, 42). EBO16 was modeled to lie parallel to the membrane surface, with Trp8 and Phe12 (Trp531 and Phe535 in our numbering system) serving as aromatic anchors (Fig. S6) (42). Ebov FL Ile542-Tyr543-Ile544, which moves into the hydrophobic patch at low pH (Fig. 6D), may provide an additional aromatic-hydrophobic anchor. It will be interesting to determine whether Trp531 (with Ile532) and Tyr543 (with Ile542 and Ile544) function in concert with Tyr534 and Phe535 to form aromatic-hydrophobic anchors that help the fusion loop penetrate deeper into the membrane at low pH. Although located in topologically different positions, a dual aromatic-hydrophobic (Phe/Ile plus Trp) anchor mechanism was previously proposed to determine the angle of the boomerang structure of the influenza HA fusion peptide (29).
It was previously speculated that a high turn propensity at the tips of the Ebov GP and ASLV Env fusion loops might be important for fusion function (43, 44). There are two proline residues near the tip of the Ebov fusion loop. These two Pro residues (Pro533 and Pro537), which are conserved among all filovirus GPs, flank the aromatic-hydrophobic-glycine tripeptide motif; they are likely important for the structure of the Ebov fusion loop. Mutation of Pro537 in the Ebov fusion loop, and its equivalent in ASLV Env, abrogated the fusion potential of their respective full-length glycoproteins (13, 44), perhaps due to a reduced depth of membrane penetration and/or an inability to form a membrane-destabilizing structure (43).
In conclusion, we report structures for a complete internal disulfide-bonded fusion loop, that of the Ebov GP, in a membrane environment. Our structures were determined at pH 7.0 and 5.5, which represent, respectively, fusion-inactive and fusion-active states. Our major finding is that low pH induces a major conformational change in the disulfide-bonded Ebov FL. The change involves a reorientation and compaction of hydrophobic residues through a hinge mechanism that redirects the hydrophobic surface at the tip of the loop. We propose that this change allows for tighter interaction with the target membrane and is therefore a requirement for optimal fusion by Ebov GP. Our findings set the stage for defining which residues of the fusion loop drive its conformational change and which engage the target membrane to promote fusion.
Materials and Methods
The Ebov FL was expressed with a cleavable His-tag in BL21 (DE3) E. coli cells and purified by nickel and gel filtration chromatography. Reducing agents were removed during the last purification steps, which resulted in a disulfide linked FL. Detailed descriptions and associated references for protein expression and purification, fluorescence, CD and NMR spectroscopy, and NMR structure determination are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by Grants R01 AI22470 (to J.M.W.), U54 AI57168 (to J.M.W.), and R37 AI30577 (to L.K.T.) from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The NMR and atomic coordinates, chemical shifts, and restraints reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2LCY and 2LCZ).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104760108/-/DCSupplemental.
References
- 1.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoenen T, Groseth A, Falzarano D, Feldmann H. Ebola virus: Unravelling pathogenesis to combat a deadly disease. Trends Mol Med. 2006;12:206–215. doi: 10.1016/j.molmed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Falzarano D, Geisbert TW, Feldmann H. Progress in filovirus vaccine development: Evaluating the potential for clinical use. Expert Rev Vaccines. 2011;10:63–77. doi: 10.1586/erv.10.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geisbert TW, et al. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: A proof-of-concept study. Lancet. 2010;375:1896–1905. doi: 10.1016/S0140-6736(10)60357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takada A, et al. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JE, Saphire EO. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 2009;4:621–635. doi: 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schornberg K, et al. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dube D, et al. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): Sequence and residues critical for host cell binding. J Virol. 2009;83:2883–2891. doi: 10.1128/JVI.01956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai AL, Li Y, Tamm LK. Interplay of protein and lipids in virus entry by membrane fusion. In: Tamm LK, editor. Protein-Lipid Interactions. Weinheim, Germany: Wiley-VCH; 2005. pp. 279–303. [Google Scholar]
- 13.Ito H, Watanabe S, Sanchez A, Whitt MA, Kawaoka Y. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J Virol. 1999;73:8907–8912. doi: 10.1128/jvi.73.10.8907-8912.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weissenhorn W, Carfí A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 16.Jeffers SA, Sanders DA, Sanchez A. Covalent modifications of the Ebola virus glycoprotein. J Virol. 2002;76:12463–12472. doi: 10.1128/JVI.76.24.12463-12472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallaher WR. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell. 1996;85:477–478. doi: 10.1016/s0092-8674(00)81248-9. [DOI] [PubMed] [Google Scholar]
- 18.Delos SE, White JM. Critical role for the cysteines flanking the internal fusion peptide of avian sarcoma/leukosis virus envelope glycoprotein. J Virol. 2000;74:9738–9741. doi: 10.1128/jvi.74.20.9738-9741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delos SE, et al. Cysteines flanking the internal fusion peptide are required for the avian sarcoma/leukosis virus glycoprotein to mediate the lipid mixing stage of fusion with high efficiency. J Virol. 2008;82:3131–3134. doi: 10.1128/JVI.02266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melder DC, Yin X, Delos SE, Federspiel MJ. A charged second-site mutation in the fusion peptide rescues replication of a mutant avian sarcoma and leukosis virus lacking critical cysteine residues flanking the internal fusion domain. J Virol. 2009;83:8575–8586. doi: 10.1128/JVI.00526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malashkevich VN, et al. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freitas MS, et al. Structure of the Ebola fusion peptide in a membrane-mimetic environment and the interaction with lipid rafts. J Biol Chem. 2007;282:27306–27314. doi: 10.1074/jbc.M611864200. [DOI] [PubMed] [Google Scholar]
- 23.Struck DK, Hoekstra D, Pagano RE. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 24.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, Blissard GW. Baculovirus GP64 disulfide bonds: The intermolecular disulfide bond of Autographa californica multicapsid nucleopolyhedrovirus GP64 is not essential for membrane fusion and virion budding. J Virol. 2010;84:8584–8595. doi: 10.1128/JVI.00264-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kielian M. Class II virus membrane fusion proteins. Virology. 2006;344:38–47. doi: 10.1016/j.virol.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 27.Han X, Bushweller JH, Cafiso DS, Tamm LK. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat Struct Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, et al. Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion. J Virol. 2005;79:12065–12076. doi: 10.1128/JVI.79.18.12065-12076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai AL, Tamm LK. Locking the kink in the influenza hemagglutinin fusion domain structure. J Biol Chem. 2007;282:23946–23956. doi: 10.1074/jbc.M704008200. [DOI] [PubMed] [Google Scholar]
- 30.Lai AL, Tamm LK. Shallow boomerang-shaped influenza hemagglutinin G13A mutant structure promotes leaky membrane fusion. J Biol Chem. 2010;285:37467–37475. doi: 10.1074/jbc.M110.153700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorieau JL, Louis JM, Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc Natl Acad Sci USA. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sainz B, Jr, Rausch JM, Gallaher WR, Garry RF, Wimley WC. Identification and characterization of the putative fusion peptide of the severe acute respiratory syndrome-associated coronavirus spike protein. J Virol. 2005;79:7195–7206. doi: 10.1128/JVI.79.11.7195-7206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamm LK, Han X. Viral fusion peptides: A tool set to disrupt and connect biological membranes. Biosci Rep. 2000;20:501–518. doi: 10.1023/a:1010406920417. [DOI] [PubMed] [Google Scholar]
- 34.Barry C, Key T, Haddad R, Duncan R. Features of a spatially constrained cystine loop in the p10 FAST protein ectodomain define a new class of viral fusion peptides. J Biol Chem. 2010;285:16424–16433. doi: 10.1074/jbc.M110.118232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 36.Melo MN, et al. Interaction of the Dengue virus fusion peptide with membranes assessed by NMR: The essential role of the envelope protein Trp101 for membrane fusion. J Mol Biol. 2009;392:736–746. doi: 10.1016/j.jmb.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun X, Belouzard S, Whittaker GR. Molecular architecture of the bipartite fusion loops of vesicular stomatitis virus glycoprotein G, a class III viral fusion protein. J Biol Chem. 2008;283:6418–6427. doi: 10.1074/jbc.M708955200. [DOI] [PubMed] [Google Scholar]
- 38.Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol. 2008;15:1024–1030. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- 39.Backovic M, Jardetzky TS, Longnecker R. Hydrophobic residues that form putative fusion loops of Epstein-Barr virus glycoprotein B are critical for fusion activity. J Virol. 2007;81:9596–9600. doi: 10.1128/JVI.00758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J Virol. 2007;81:4858–4865. doi: 10.1128/JVI.02755-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stampfer SD, Lou H, Cohen GH, Eisenberg RJ, Heldwein EE. Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1. J Virol. 2010;84:12924–12933. doi: 10.1128/JVI.01750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freitas MS, et al. Measuring the strength of interaction between the Ebola fusion peptide and lipid rafts: Implications for membrane fusion and virus infection. PLoS One. 2011;6:e15756. doi: 10.1371/journal.pone.0015756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gómara MJ, Mora P, Mingarro I, Nieva JL. Roles of a conserved proline in the internal fusion peptide of Ebola glycoprotein. FEBS Lett. 2004;569:261–266. doi: 10.1016/j.febslet.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Delos SE, Gilbert JM, White JM. The central proline of an internal viral fusion peptide serves two important roles. J Virol. 2000;74:1686–1693. doi: 10.1128/jvi.74.4.1686-1693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.