Abstract
Covalent modifications of histones, such as acetylation, methylation and ubiquitination, are central for regulation of gene expression. Heterochromatic gene silencing, for example, is associated with hypoacetylation, methylation and demethylation, and deubiquitination of specific amino acid residues in histone molecules. Many of these changes can be effected by histone-modifying repressor complexes that include histone lysine demethylases, such as KDM1 in animals and KDM1C in plants. However, whereas KDM1-containing repressor complexes have been implicated in histone demethylation, methylation and deacetylation, whether or not they can also mediate histone deubiquitination remains unknown. We identify an Arabidopsis otubain-like deubiquitinase OTLD1 which directly interacts with the Arabidopsis KDM1C in planta, and use one target gene to exemplify that both OTLD1 and KDM1C are involved in transcriptional gene repression via histone deubiquitination and demethylation. We also show that OTLD1 binds plant chromatin and has enzymatic histone deubiquitinase activity, specific for the H2B histone. Thus, we suggest that, during gene repression, lysine demethylases can directly interact and function in a protein complex with histone deubiquitinases.
Keywords: otubain superfamily, repression of gene expression, bimolecular fluorescence complementation
Covalent modifications of histones are critical for eukaryotic DNA metabolism, determining structure and gene expression activity of the corresponding chromosomal region (1–5). They govern chromatin folding, which in turn determines cell fate. Proteins containing the hallmark SWIRM (Swi3p, Rsc8p, Moira) (6) and polyamine oxidase (PAO) domains, including histone lysine demethylase LSD1/KDM1 in animals (7) and LDL1/KDM1C in plants (8), are frequently found in chromatin-associated transcriptional repressor enzymatic complexes (9) and play a key role in regulation of gene expression (10) during such diverse developmental events as acquisition of neuron-specific traits in mammals (11, 12) and determination of flower timing in plants (8). One of the major effects of KDM1-containing complexes is transcriptional gene silencing through posttranslational modifications of the core histones. Silenced heterochromatic loci are distinguished by general histone hypoacetylation, methylation on lysines 9 (K9) and 27 (K27) (1, 13, 14) and demethylation on lysine 4 (K4) (7) and lysine 36 (15) of histone H3, and recently described deubiquitination of histones H2A and H2B in animals and yeast (16–19) and H2B in plants (20). Although KDM1-containing repressor complexes have been implicated in histone methylation, demethylation, and deacetylation, whether or not they can also mediate histone deubiquitination is enigmatic.
Monoubiquitination of histone H2B, in both animals and plants, is typically associated with transcriptional activation, and amino acid sequences adjacent to the H2B ubiquitination sites are remarkably conserved between various eukaryotes (20). For instance, ubiquitinated H2B is enriched around transcriptionally active sequences in bovine thymus, chicken erythrocytes, and Tetrahymena macronuclei (21). In Drosophila, the USP7 deubiquitinase catalyzes ubiquitin removal from H2B and contributes to epigenetic silencing of homeotic genes (22). It is thought that ubiquitination mechanistically disrupts histone–DNA interaction, due to the large size of the ubiquitin moiety, and thus contributes to euchromatic features of the surrounding chromatin (20, 23). In plants, similarly to other eukaryotes, monoubiquitination of histone H2B lysine 143 (K143) (23) has been shown to associate with actively transcribed genes. For example, the Arabidopsis ubiquitin-conjugating enzymes UBC1/2 monoubiquitinate H2B, leading to up-regulation of the FLC flower timing gene (24, 25). Furthermore, UBC1 and -2 interact with histone monoubiquitinating enzymes HUB1 and -2, which also contribute to derepression of FLC and inhibit transition from vegetative to reproductive phases in the plant life cycle (24, 25).
Conversely, deubiquitination of H2B results in transcriptional silencing. Mutations in Arabidopsis H2B deubiquitinase UBP26/SUP32, which appears to be vital for heterochromatin formation, release heterochromatic silencing of transgenes and transposons (20). Also, a T-DNA insertion resulting in UBP26/SUP32 loss-of-function leads to up-regulation of PHE1, a MADS-box gene expressed in developing siliques (26). Another Arabidopsis ubiquitin-specific protease, UBP14, which may be directly involved in the ubiquitin/26S proteasome pathway, is essential for embryonic development (27).
Otubains are recently discovered ubiquitin proteases that belong to a superfamily of OTU (ovarian tumor)-like proteins, are conserved primarily among eukaryotes and viruses (28), and show no homology to other deubiquitinating enzymes (29). Otubains also differ from other deubiquitinases in their substrate specificity by cleaving only isopeptide bonds (29, 30). Originally described in animal systems, OTU-like proteins, including otubains, are involved in many ubiquitin-related cellular processes. Little is known about otubain-like ubiquitin proteases in plants. Only a single otubain-like protein, with as yet uncharacterized biological activity and function, was shown to be expressed during early somatic embryogenesis of pine trees (31).
Here, we describe an Arabidopsis otubain-like deubiquitinase OTLD1. We show that OTLD1 directly interacts with the Arabidopsis KDM1C in vivo, and use one target gene common to OTLD1 and KDM1C to exemplify that both of them are involved in transcriptional gene repression via histone deubiquitination and demethylation. We also show that OTLD1 binds plant chromatin and has a histone deubiquitinase activity in vitro.
Results
KDM1C Directly Interacts with OTLD1.
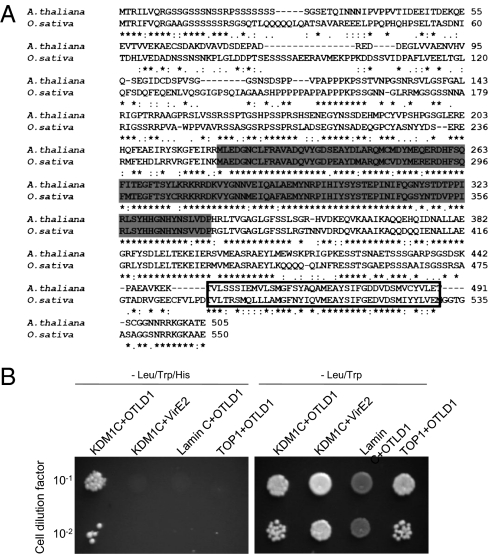
Previously, we have conducted a yeast two-hybrid screen for KDM1C-interacting proteins (8). Here, using this approach, we identified an otubain-like histone deubiquitinase, designated OTLD1, and encoded by the Arabidopsis At2g27350 gene. OTLD1 contains a highly conserved N-terminal isopeptidase OTU domain, found in deubiquitinating enzymes (28, 29), and a C-terminal UBA (ubiquitin-associated) domain, commonly found in proteins involved in ubiquitin-related cell signaling and transcriptional processes (32) (Fig. 1A). Although both OTU and UBA domains each are found in many different eukaryotic proteins, the combination of these two domains is distinctly unique to OTLD1. In Arabidopsis, OTLD1 is a single-copy gene, and its homologs are found in diverse plant species, including rice (Oryza sativa, NP_001052752) (Fig. 1A), corn (Zea mays, ACG29802), grape (Vitis vinifera, CAO48705), and moss (Physcomitrella patens, XP_00178178).
Fig. 1.
OTLD1 is an otubain-like protein that interacts with KDM1C. (A) Sequence alignment and domain structure of A. thaliana OTLD1 and its homolog from rice (O. sativa). Conserved OTU and UBA domains are indicated by shaded and boxed residues, respectively. (B) Specific interaction of OTLD1 with KDM1C in the two-hybrid system. (Left) Growth in the absence of histidine, tryptophan, and leucine. (Right) Growth in the absence of tryptophan and leucine. Growth in histidine-deficient medium represents selective conditions for protein–protein interactions.
We cloned the full-length cDNA of OTLD1 and confirmed its interaction with KDM1C in the yeast two-hybrid system (Fig. 1B). This interaction was specific because it was not observed with an unrelated Agrobacterium VirE2 protein or with the known nonspecific two-hybrid activators lamin C and topoisomerase I (33, 34) (Fig. 1B, Left). Under the nonselective conditions, i.e., in the presence of histidine, all combinations of the tested proteins resulted in the efficient cell growth (Fig. 1B, Right).
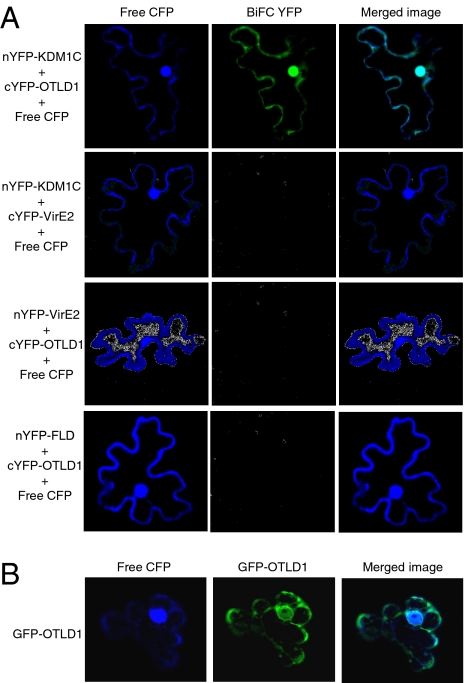
The KDM1C–OTLD1 interaction was then examined in planta using bimolecular fluorescence complementation (BiFC) (35–38) in a heterologous, Nicotiana benthamiana system. Fig. 2A shows that KDM1C and OTLD1 interacted with each other in planta, producing the BiFC signal. Negative control experiments coexpressing KDM1C and VirE2 or OTLD1 and VirE2 failed to reconstitute the YFP fluorescence. To demonstrate further the specificity of the KDM1C–OTLD1 interaction in the BiFC assay, we coexpressed OTLD1 with a KDM1C homolog, the Arabidopsis FLD histone demethylase (39). No BiFC signal was detected in these experiments (Fig. 2A), substantiating the specificity of the OTLD1–KDM1C interaction as detected by BiFC.
Fig. 2.
Specific interaction between OTLD1 and KDM1C and OTLD1 subcellular localization in planta. (A) The BiFC assay for OTLD1–KDM1C interaction in planta. (B) Nucleocytoplasmic localization of OTLD1 in plant cells. GFP or YFP signal is in green, CFP signal is in blue, and overlapping GFP/YFP and CFP signals are in blue-green. All images are single confocal sections.
Interestingly, the KDM1C–OTLD1 BiFC signal not only accumulated in the cell nucleus, but was also observed in the cytoplasm, colocalizing with coexpressed free CFP (Fig. 2A), which is known to label both cellular compartments (8). This pattern is consistent with the subcellular localization of OTLD1 itself (Fig. 2B), suggesting that OTLD1 might be involved in both nuclear and cytoplasmic metabolic pathways. On the other hand, KDM1C accumulates in the cell nucleus (40); however, because both proteins initially are produced in the cytoplasm, they should have cytoplasmic pools, and the cytoplasmic BiFC signal may reflect detection of this cytoplasmic pool of KDM1C.
OTLD1 Binds Plant Chromatin and Deubiquitinates Histone H2B in Vitro.
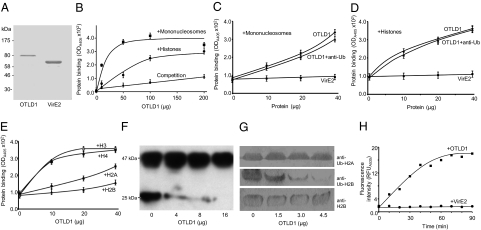
If OTLD1 is involved in gene regulation by histone deubiquitination, it may directly interact with the chromatin. Indeed, purified OTLD1 (Fig. 3A) efficiently bound purified plant nucleosomes as well as total purified bovine histones in a concentration-dependent and saturable manner (Fig. 3B). The specificity of the OTLD1–histone interaction was confirmed by competition experiments, in which increasing concentrations of free histones efficiently inhibited binding (Fig. 3B). On the other hand, neither binding to nucleosomes nor to histones was observed with an unrelated Agrobacterium protein VirE2 (Fig. 3 C and D, respectively). To examine whether binding of OTLD1 to nucleosomes or histones is affected by ubiquitination, we blocked the ubiquitin residues with antiubiquitin antibodies. Fig. 3 shows that the presence of these antibodies had virtually no effect on binding of OTLD1 to nucleosomes (Fig. 3C) or histones (Fig. 3D), suggesting that these interactions do not involve exposed ubiquitin moieties. We then compared binding of OTLD1 to each of the core histones, H2A, H2B, H3, and H4. Fig. 3E shows that, whereas the association with H2B was barely detectible, binding to H3 and H4 was significantly more efficient and saturable. The binding of OTLD1 to H2A occurred only with relatively low affinity (Fig. 3E).
Fig. 3.
OTLD1 binds nucleosomes and histones and has a histone H2B deubiquitinase activity. (A) SDS/PAGE analysis of purified OTLD1 and VirE2. Molecular mass markers in thousands of daltons are indicated on the Left. (B) Binding of OTLD1 to nucleosomes and purified histones. Competition curve indicates competitive inhibition of OTLD1 binding to immobilized histones by free histones. (C) Effects of antiubiquitin (Ub) antibody on OTLD1 binding to nucleosomes. (D) Effects of anti-Ub antibody on OTLD1 binding to purified histones. VirE2 represents negative control. (E) OTLD1 preferentially binds H3 and H4. All data represent average values of three independent experiments with indicated SD. (F) Deubiquitination of a total histone preparation by OTLD1. (G) Deubiquitination of monoubiquitinated H2B, but not H2A, by OTLD1. Each panel represents an immunoblot analysis of a separate membrane, done in parallel. (H) Kinetic analysis of the OTLD1 deubiquitinase activity. VirE2 represents negative control.
Next, we directly tested whether OTLD1 possesses the histone deubiquitinase enzymatic activity. To this end, purified OTLD1 was incubated with purified total bovine histones, and the degree of histone ubiquitination was estimated by Western blot analysis. Fig. 3F shows that the input preparation contained monoubiquitinated histones (Lower) as well as slower migrating, potentially polyubiquitinated, protein species (Upper). Incubation in the presence of increasing concentrations of OTLD1 gradually decreased the amount of monoubiquitinated histones until they were completely deubiquitinated at the highest tested concentration of OTLD1 (Fig. 3F). Histone deubiquitination by OTLD1 occurred on the H2B. Fig. 3G shows that, whereas OTLD1 did not affect the amounts of monoubiquitinated H2A, it substantially reduced the content of monoubiquitinated H2B in the histone preparation. This was due to deubiquitination, rather than proteolysis, because OTLD1 did not affect the total amounts of H2B with the electrophoretic mobility similar to that of the monoubiquitinated H2B (Fig. 3G). Thus, OTLD1 is an enzymatically active histone deubiquitinase specific for the monoubiquitinated histone H2B. Note that, because the major histone species ubiquitinated in plants is H2B (20, 23, 41), we did not test the OTLD1 deubiquitinase activity against H3 and H4.
Finally, we performed kinetic analysis of the OTLD1 activity using a commercially available fluorimetric assay. Fig. 3H shows that OTLD1 exhibited typical saturation kinetics, on the basis of which we calculated its apparent specific activity as 0.01 mU/mg (or 0.17 pkatal). No deubiquitinase activity was observed with an unrelated protein VirE2 (Fig. 3H).
KDM1C–OTLD1 Complex Represses Gene Expression via Histone Deubiquitination.
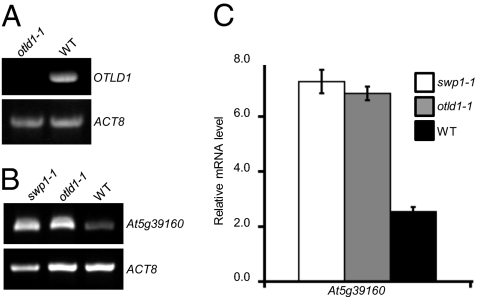
That KDM1C and OTLD1 can function in complex implies that they can act on the same target gene and, because KDM1C promotes heterochromatin formation (8, 40), that the effect of OTLD1 could also be heterochromatinization. Our earlier microarray analysis (8) identified several genes targeted by KDM1C and derepressed in a KDM1C mutant, swp1-1, out of which we selected At5g39160 that was also up-regulated in an OTLD1 mutant, otld1-1, and analyzed it in detail. First, we demonstrated that the homozygous otld1-1 line, which carried the mutagenic T-DNA insertion within the OTLD1 exon at position coding for the amino acid residue 402 (SALK_028707), between the OTU and UBA domains, is indeed a null mutant and expresses no OTLD1 RNA (Fig. 4A). The absence of KDM1C transcript in the swp1-1 line has already been reported (8). Then, we compared expression of At5g39160 between the wild-type plants and swp1-1 and otld1-1 mutant plants by RT-PCR. Fig. 4B shows that the level of expression of the At5g39160 gene was substantially increased in swp1-1 as well as in otld1-1 mutants. Control experiments (Fig. 4B), using constitutively expressed actin (ACT8), confirmed equal input of RNA and reaction efficiency. Finally, the At5g39160 derepression in swp1-1 and otld1-1 was measured by quantitative real-time PCR (qPCR) (Fig. 4C), showing that the expression of this gene was increased approximately threefold in both mutants. Thus, consistent with their possible function in the same repressor complex, both KDM1C and OTLD1 repress the At5g39160 target gene, potentially fine tuning its expression similarly to other KDM1C-containing repressors (8, 40, 42). Alternatively, KDM1C and OTLD1 may affect At5g39160 indirectly, through regulation of other genes whose protein products then act—also via histone demethylation and deubiquitination—on At5g39160.
Fig. 4.
OTLD1 and KDM1C target and repress the At5g39160 gene. (A) RT-PCR detection of the OTLD1 mrna-specific product in the otld1-1 and wild-type (WT) plants. (B) RT-PCR analysis of At5g39160 derepression in the swp1-1, otld1-1, and WT plants. Constitutively expressed ACT8 was used as internal control. (C) qPCR analysis of At5g39160 derepression in the swp1-1, otld1-1, and WT plants. The data represent average values of three independent experiments with indicated SD.
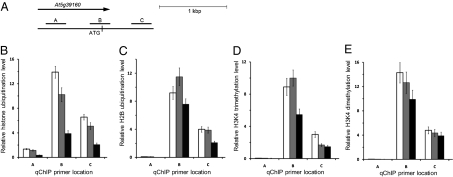
Next, we used quantitative chromatin immunoprecipitation (qChIP) to examine whether the repressor activity of KDM1C/OTLD1 involves histone deubiquitination. Because the regulatory elements of At5g39160 associated with histone modifications have not been defined, this analysis was performed on several sequences that surround the translation initiation site of this gene (Fig. 5A). Using antiubiquitin antibody, we showed that, in the At5g39160 chromatin, histone ubiquitination was most pronounced in the regions B and C (Fig. 5B). Importantly, not only the otld1-1 mutant, but also the swp1-1 plants, exhibited hyperubiquitination of the target chromatin, both showing a 2.5- to 4-fold increase in histone ubiquitination relatively to the wild-type plants (Fig. 5B). Consistent with the ability of OTLD1 to deubiquitinate H2B (Fig. 3), antiubiquityl H2B antibody detected hyperubiquitination of H2B in both otld1-1 and swp1-1 plants, with the highest signal observed for region B (Fig. 5C); the signal differences between total histone ubiquitination and that of H2B most likely are due to the relatively low titer of the monoclonal antiubiquityl H2B antibody compared with the polyclonal anti-H2B antibody. No differences in the total H2B levels were detected in all tested plant lines.
Fig. 5.
Repression of the At5g39160 gene is mediated via histone deubiquitination and demethylation. (A) Location of sequence regions A–C in At5g3916, used for qChIP analyses. (B) Relative levels of total histone ubiquitination. (C) Relative levels of H2B ubiquitination. (D) Relative levels of H3K4 trimethylation. (E) Relative levels of H3K4 dimethylation. White, gray, and black bars indicate the swp1-1, otld1-1, and wild-type plants, respectively. All data represent average values of three independent experiments with indicated SD.
KDM1C is a member of the histone lysine demethylase family (43); thus, we examined potential changes in major chromatin methylation marks, such as lysines 4, 9, 27, and 36 of H3 (H3K4, H3K9, H3K27, and H3K36, respectively). Our qChIP analysis showed that only H3K4 methylation—which is diagnostic of active chromatin, frequently associated with histone ubiquitination (20, 24, 25) and targeted by the human KDM1 (7)—was affected in the At5g39160 chromatin of the swp1-1 plants (Fig. 5 D and E), whereas no changes in other H3 methylation marks could be detected. The increase in both tri- and dimethylated H3K4 was observed; this H3K4 hypermethylation was consistent, albeit less dramatic, i.e., 30–50% of the wild-type plants, than the increase in ubiquitination, and detected mostly in region B (Fig. 5 D and E). Comparable levels of H3K4 hypermetylation in the At5g39160 chromatin were observed also in the otld1-1 mutant (Fig. 5 D and E). Thus, OTLD1 and KDM1C affected both their direct and reciprocal histone modifications, i.e., the absence of OTLD1 not only reduced the target gene chromatin deubiquitination, but also brought about its H3K4 hypermethylation, whereas the absence of the KDM1C increased not only H3K4 di- and trimethylation, but also elevated ubiquitination of the target chromatin.
Discussion
OTLD1 is the first example of an otubain-like histone deubiquitinase involved in chromatin modification and regulation of plant gene expression. To date only one histone deubiquitinase UBP26/SUP32, with no homology to OTLD1, has been identified in plants (20). OTLD1 also represents the first eukaryotic histone deubiquitinase shown to associate directly and function together with the KDM1-type lysine demethylases. Because the presence of only OTLD1 or KDM1C in the swp1-1 or otld1-1 mutants, respectively, was insufficient to effect their respective histone modifications, the functions of these repressors are most likely interdependent. Taken together with the observations that OTLD1 and KDM1C directly interact with each other in vivo and affect at least one common target gene, these results suggest that both proteins may function within the same corepressor complex that regulates gene expression through chromatin modification. In this complex, OTLD1, due to its affinity toward nucleosomes, may bind directly to and deubiquitinate the target gene chromatin.
OTLD1, which carries the unique combination of OTU–UBA domains, may be specific to plants because we did not find its full-sequence homologs in nonplant species databases. On the other hand, that OTLD1 homologs are encoded by both dicots and monocots suggests that this protein architecture has evolved before the dicot/monocot divergence. Because histone deubiquitination plays an important role in gene regulation, the OTLD1 function most likely is backed up by other deubiquitinases. Indeed, the homozygous otld1-1 mutant lines did not display apparent phenotypic changes, suggesting functional redundancy with additional deubiquitinating enzymes. Similarly, knockdown of otubain expression in HeLa cells had little effect on the pattern of ubiquitinated proteins (29).
Unlike mammalian cells, which ubiquitinate both H2A and H2B (19), the primary ubiquitinated histone found in plants is H2B (20, 23, 41), and, consistent with its potentially plant-specific function, OTLD1 deubiquitinated only H2B within the mixture of ubiquitinated mammalian H2B and H2A. Interestingly, the binding affinity of OTLD1 to core histones differs from its enzymatic specificity toward the same substrates. OTLD1 associated preferentially with H3 and H4, and to a lesser extent with H2A, but not with H2B, whereas it deubiquitinated only H2B. Thus, OTLD1 most likely recognizes and modifies the target chromatin at different histone molecules within the nucleosome. This is consistent with the nucleosome topology where portions of the H3 and H4 molecules are located in close proximity to the H2B ubiquitination site (19). Similar strategy of a protein modifier binding to one component of the target protein complex and modifying another component of the same complex has been reported, for example, for the E2 enzyme, which binds to the RING (U-Box) and A20 finger-type E3 ligase–target protein complexes via E3, but transfers ubiquitin to the target protein and not to E3 (44).
How histone deubiquitination may lead to heterochromatin formation and target gene repression is not yet understood, although several possible mechanisms have been proposed (19). For example, the removal of bulky ubiquitin residues may directly affect chromatin folding and result in its denser packaging, restricting access of the transcription machinery to the DNA. Or, histone deubiquitination may affect chromatin structure indirectly, through its impact on other covalent modifications. Indeed, monoubiquitination of H2B is linked with H3K4 methylation in plants (20, 25) as well as in yeast (45–47) and animals (48). This last scenario is consistent with our data, indicating that OTLD1 functions as part of KDM1C-containing repressor complexes that deubiquitinate H2B and induce hypermethylation of H3K4 in their target chromatin. Alternatively, KDM1C and OTLD1 may cooperate with each other to protect the existing heterochromatin from H2B ubiquitination and H3K4 methylation. Indeed, a Drosophila KDM1 homolog SU(VAR)3-3 in concert with its associated histone deacetylase and methyltransferase, albeit not deubiquitinase, is thought to exert a protective effect against expanding H3K4 methylation of heterochromatin (49).
Materials and Methods
Plants.
The T-DNA insertion mutants swp1-1 (8) and otld1-1 [SALK_142477 and SALK_028707, respectively, obtained from the Arabidopsis Biological Resource Center (ABRC)] and wild-type plants were derived from the Col-0 ecotype of Arabidopsis thaliana. We used the unsilenced heterozygous otld1-1 stock to analyze segregation of the kanamycin resistance marker contained in the mutagenic T-DNA; the resulting segregation ratio, 102:29 seedlings, is consistent with T-DNA insertion into a single segregating site. We then crossed the heterozygous otld1-1 plants to homozygocity, whereas the homozygous swp1-1 line was produced earlier (8). The homozygocity of both mutant lines was verified using the KDM1C- or OTLD1-specific primer pairs 5′GTTTTGGCGAGGCAACTTGGT3′/5′CCCATCTGGAACAGAGGGCTT3′ and 5′tgcttgacttgttctgtgttggaa3′/5′cacgtggacaaggaacaggtg3′, respectively, and the T-DNA left border-specific reverse primer 5′GCGTGGACCGCTTGCTGCAACT3′ as described (50) (http://signal.salk.edu/cgi-bin/tdnaexpress). Notably, the otld1-1 mutant was indistinguishable from the wild-type plants in its overall morphology and development; thus, the knockout of OTLD1 most likely did not interfere with essential plant cellular functions. For RNA extractions and ChIP analysis, plants were grown on Gamborg's B5 (Sigma)/0.1% sucrose medium in an environment-controlled chamber at 22–24 °C and maintained under long-day conditions of 16 h of white light (70–80 μmol photons m−2 s−1) and 8 h of darkness.
Preparation of Recombinant OTLD1.
The full-length OTLD1 cDNA (ABRC stock U24564) was amplified using the primer pair 5′CTAGCTAGCATGACTCGGATTTTGGTTCAAAG3′/5′GTTAGCGGCCGCTTCCGTGGCTTTGCCTTTGC3′, cloned into the NheI/NotI sites of pET21a (Novagen), and verified by DNA sequencing. The VirE2 coding sequence was subcloned as a HindIII/XhoI fragment of pET3b::virE2 (51, 52) into the same sites of pET28b(+) (Novagen). The recombinant His-tagged OTLD1 and VirE2 proteins were expressed in BL21(DE3) Escherichia coli strain (Novagen) as described (51, 53), and purified on Ni-NTA resin (Qiagen). This procedure resulted in >95% pure protein preparations as determined by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 3A). Note that the electrophoretic mobility of OTLD1 was higher than its predicted molecular mass, most likely due to posttranslational modifications (54).
OTLD1 Binding to Nucleosomes and Histones.
Maxisorb plates (Nunc) were incubated overnight at 4 °C with 200 μL of carbonate buffer (50 mM Na2CO3/50 mM NaHCO3, pH 9.6) containing 0.2 mg/mL of plant mononucleosomes purified from cauliflower florets as described (55), purified total bovine histones (Sigma; H9250), purified recombinant human H2B, H2A, H3.3, an H3 variant conserved in all eukaryotes, from yeast to humans (56), and H4 (New England Biolabs; M2505S, M2502S, M2507S, and M2504S, respectively), or BSA (Sigma-Aldrich; A7906). After a wash in PBS pH 7.4, the plates were blocked by incubation with 200 μL/well of 4% BSA in PBS for 2 h at room temperature. Then, the increasing concentrations of purified His-tagged OTLD1 or VirE2 in 4% BSA/PBS were added to the wells and incubated for 1–2 h at room temperature, followed by a wash in PBS and addition of 1:500 dilution of rabbit anti-His antibody (Bethyl Laboratories; A190-114A). After 1-h incubation and a wash in PBS, secondary antirabbit IgG conjugated to alkaline phosphatase (AP) (1:500 dilution; Sigma; A3687) was added, and the incubation continued for 1 h at room temperature. OTLD1 binding was detected colorimetrically at 405 nm after addition of the AP substrate p-nitrophenyl phosphate (PNPP) in diethanolamine buffer. For competitive inhibition of OTLD1 binding, the OTLD1-containing binding solution was supplemented with 2 mg/mL of free bovine histones. For blocking the ubiquitin residues, antiubiquitin antibody (1:100 dilution; Santa Cruz Biotechnology; sc-8017) was added to the wells and incubated for 1 h before addition of OTLD1.
In Vitro Histone Deubiquitination.
The purified OTLD1 was incubated with 20 μg of total purified bovine histones in 50 mM Tris-HCl (pH 8.0), 50 mM NaCl for 3–4 h at room temperature. For Western blot analysis, samples were resolved by 6–12% gradient SDS/PAGE, electrotransferred onto a NitroPlus membrane (Micron Separations), blocked with 4% milk and probed with the following antibodies: antiubiquitin antibody conjugated to horse radish peroxidase (HRP) (1:1,000 dilution; Santa Cruz Biotechnology; sc-8017 HRP), or anti-H2B (1:1,000 dilution; Santa Cruz Biotechnology; sc-10808), antiubiquityl H2B (1:1,000 dilution; Medimabs; MM0029), or antiubiquityl H2A (1:1,000 dilution; Upstate/Millipore; 05–678) antibodies followed by alkaline phosphatase (AP)-conjugated goat anti-rabbit antibody (1:2,000 dilution; Jackson ImmunoResearch). After additional washing, proteins recognized by the antibodies were visualized using either a chemiluminescent HRP substrate (Millipore) or chromogenic AP staining with bromochloroindolyl phosphate (BCIP) and nitro blue tetrazolium (NBT).
For quantification of the OTLD1 deubiquitinase activity, we used the DUB-Detector kit with a fluorescent universal ubiquitin substrate (Active Motif) according to the manufacturer's instructions. For calculation of the OTLD1-specific activity, one enzyme unit was defined as the amount of OTLD1 that catalyzes the conversion of 1 mmole of substrate per minute.
Yeast Two-Hybrid Assay.
The KDM1C cDNA was cloned into the SalI/PstI sites of a LexA plasmid pSTT91 (TRP1+; ref. 57) and the OTLD1 cDNAs was cloned into the EcoRI/PstI sites of pGAD424 (LEU2+; Clontech). The Arabidopsis cDNA library and the virE2 gene in pGAD424 as well as human lamin C and topoisomerase I in pSTT91 were described previously (58–60). Protein interaction indicated by histidine prototrophy was assayed in the Saccharomyces cerevisiae strain L40 (34) (61) by growing cells for 3 d at 30 °C on a leucine-, tryptophan- and histidine-deficient medium in the presence of 10 mM of 3-amino-1,2,4-triazole (3-AT).
BiFC, Subcellular Localization, and Microbombardment.
For BiFC, the KDM1C or FLD cDNAs were inserted into the XhoI/KpnI or SacI/KpnI sites, respectively, of pSAT4-nEYFP-C1 (GenBank accession no. DQ168994), and OTLD1 cDNA was inserted into the BglII/EcoRI sites of pSAT1-cEYFP-C1(B) (GenBank accession number DQ168996) (62). The virE2 gene was cloned into the BglII/BamHI sites of pSAT4-nEYFPC1 and pSAT1-cEYFP-C1(B). For subcellular localization, the OTLD1 cDNA was fused to GFP in pSAT6-EGFP-C1 (GenBank accession no. AY818377) (62). Free CFP was expressed from SAT6-ECFP-C1 (GenBank accession no. AY818374 (62). Tested constructs were mixed in a weight ratio of 2:2:1 for BiFC or 1:1 for subcellular localization studies, adsorbed onto 10 mg of 1-μm gold particles (Bio-Rad) and bombarded at 80–110 psi into the leaf epidermis of greenhouse-grown N. benthamiana using a Helios gene gun (PDS-1000/He; Bio-Rad) as described (63). After incubation for 24–48 h at 22–24 °C, the bombarded tissues were viewed under a Zeiss LSM 5 Pascal confocal laser scanning microscope.
RT-PCR and qPCR.
For RT-PCR, total RNA from 2-wk-old seedlings was isolated using TRI-reagent (Molecular Research Center) and treated with RNase-free DNase (DNA-free kit; Ambion). cDNA was synthesized using ProtoScript First Strand cDNA synthesis kit (New England Biolabs) with Oligo-dT primers and ∼500 ng of the DNA-free RNA for each sample. Equal amounts of the RT reaction products were PCR amplified using primer pairs specific for the OTLD1 cDNA (5′AGCTGTGGACAGTGATGAACCTGC3′/5′CATATACTTGATCTGCAACAGCTCG3′; to determine that this is a null mutant, we designed them to span an intron upstream of the mutagenic T-DNA sequence), ACTIN8 (Act8) (5′GTCTGTGACAATGGTACTGG3′/5′CCTGCTTCATCATACTCTGC3′), and At5g39160 (5′TGATCCAAGTCCACTCCAAG3′/5′ACCTGAAAATGGATCATTCC3′). These primers were designed to amplify across introns to discriminate between cDNA and potential residual genomic DNA contaminants. The absence of such contamination was also demonstrated by control PCR reactions performed without RT. The PCR products were analyzed by agarose gel electrophoresis.
qPCR was performed using the same cDNA preparations in Light Cycler 480 Real-Time PCR system (Roche) with SYBR-green I Master mix (Roche) and the primer pairs specific for Act8 (5′TGTATGTTGCCATTCAAGCTGTTC3′/5′GAAACCCTCGTAGATAGGCACAGTG3′) and At5g39160 (5′AATGCTTATGATCCAAGTCCACTCC3′/5′AATCTTTTGCATCGACTCGCTTCGG3′). Relative abundance of the At5g39160 mRNA-specific product was normalized to the amount of the product specific for Act8, which represented an internal control of a constitutively expressed gene.
qChIP.
ChIP was performed as described (8, 64). Briefly, 2-wk-old seedlings were cross-linked with formaldehyde, chromatin was isolated, shared by sonication, and immunoprecipitated with 5–6 μg of antiubiquitin (Santa Cruz Biotechnology; sc-8017), antiubiquityl H2B (Medimabs; MM0029), anti-H3 (Upstate/Millipore; 06–755), antidimethyl H3K4 (Upstate/Millipore; 07–030), or antitrimethyl H3K4 (Upstate/Millipore; 07–473) antibodies. The cross-links were heat reversed at 65 °C, DNA purified on spin columns (Zymogen), and qPCR was performed with primer pairs specific for Act2/7 promoter regions (64) or for regions A–C within At5g39160: (region A) 5′TTTGGTCCCTCCCAAATACATG3′/5′AAACATAAATTATAACTTACGTTC3′, (region B) 5′AATATGACTTATATGCTCTTGTCC3′/5′GGATCATAAGCATTGACAAAGG3′, and (region C) 5′GGAAAATTCCAGCAGTTGCTTTTGC3′/5′TGGACCCAAACTTGGCCTGAAGTTC3′. Relative abundance of the products specific for ubiquitinated At5g39160 regions was normalized to the amount of the Act2/7-specific product.
Acknowledgments
The work in our laboratory is supported by grants from the National Science Foundation, US Department of Agriculture's National Institute of Food and Agriculture, National Institutes of Health, and Binational Agricultural Research and Development (V.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Kadonaga JT. Eukaryotic transcription: An interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Carrozza MJ, Utley RT, Workman JL, Côté J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 5.Goodrich J, Tweedie S. Remembrance of things past: Chromatin remodeling in plant development. Annu Rev Cell Dev Biol. 2002;18:707–746. doi: 10.1146/annurev.cellbio.18.040202.114836. [DOI] [PubMed] [Google Scholar]
- 6.Aravind L, Iyer LM. The SWIRM domain: A conserved module found in chromosomal proteins points to novel chromatin-modifying activities. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-research0039. research0039.0031–0039.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Krichevsky A, et al. C2H2 zinc finger-SET histone methyltransferase is a plant-specific chromatin modifier. Dev Biol. 2007;303:259–269. doi: 10.1016/j.ydbio.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krichevsky A, Kozlovsky SV, Gutgarts H, Citovsky V. Arabidopsis co-repressor complexes containing polyamine oxidase-like proteins and plant-specific histone methyltransferases. Plant Signal Behav. 2007;2:174–177. doi: 10.4161/psb.2.3.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- 11.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Lunyak VV, Prefontaine GG, Rosenfeld MG. REST and peace for the neuronal-specific transcriptional program. Ann N Y Acad Sci. 2004;1014:110–120. doi: 10.1196/annals.1294.011. [DOI] [PubMed] [Google Scholar]
- 13.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 14.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 15.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 16.Vissers JH, Nicassio F, van Lohuizen M, Di Fiore PP, Citterio E. The many faces of ubiquitinated histone H2A: Insights from the DUBs. Cell Div. 2008;3:8. doi: 10.1186/1747-1028-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weake VM, Workman JL. Histone ubiquitination: Triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Henry KW, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003;17:2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- 20.Sridhar VV, et al. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447:735–738. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- 21.Nickel BE, Allis CD, Davie JR. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry. 1989;28:958–963. doi: 10.1021/bi00429a006. [DOI] [PubMed] [Google Scholar]
- 22.van der Knaap JA, et al. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Zhang K, Sridhar VV, Zhu J, Kapoor A, Zhu JK. Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS ONE. 2007;2:e1210. doi: 10.1371/journal.pone.0001210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu X, Jiang D, Wang Y, Bachmair A, He Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 2009;57:522–533. doi: 10.1111/j.1365-313X.2008.03709.x. [DOI] [PubMed] [Google Scholar]
- 25.Cao Y, Dai Y, Cui S, Ma L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell. 2008;20:2586–2602. doi: 10.1105/tpc.108.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo M, et al. UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis. Genetics. 2008;180:229–236. doi: 10.1534/genetics.108.091736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doelling JH, Yan N, Kurepa J, Walker J, Vierstra RD. The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J. 2001;27:393–405. doi: 10.1046/j.1365-313x.2001.01106.x. [DOI] [PubMed] [Google Scholar]
- 28.Makarova KS, Aravind L, Koonin EV. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem Sci. 2000;25:50–52. doi: 10.1016/s0968-0004(99)01530-3. [DOI] [PubMed] [Google Scholar]
- 29.Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: A new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4:517–522. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komander D, Barford D. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem J. 2008;409:77–85. doi: 10.1042/BJ20071399. [DOI] [PubMed] [Google Scholar]
- 31.Aquea F, Gutiérrez F, Medina C, Arce-Johnson P. A novel Otubain-like cysteine protease gene is preferentially expressed during somatic embryogenesis in Pinus radiata. Mol Biol Rep. 2008;35:567–573. doi: 10.1007/s11033-007-9124-0. [DOI] [PubMed] [Google Scholar]
- 32.Hofmann K, Bucher P. The UBA domain: A sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- 33.Bartel P, Chien CT, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 34.Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 36.Citovsky V, et al. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol. 2006;362:1120–1131. doi: 10.1016/j.jmb.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 37.Citovsky V, Gafni Y, Tzfira T. Localizing protein-protein interactions by bimolecular fluorescence complementation in planta. Methods. 2008;45:196–206. doi: 10.1016/j.ymeth.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 39.He Y, Michaels SD, Amasino RM. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003;302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- 40.Krichevsky A, Zaltsman A, Kozlovsky SV, Tian GW, Citovsky V. Regulation of root elongation by histone acetylation in Arabidopsis. J Mol Biol. 2009;385:45–50. doi: 10.1016/j.jmb.2008.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergmüller E, Gehrig PM, Gruissem W. Characterization of post-translational modifications of histone H2B-variants isolated from Arabidopsis thaliana. J Proteome Res. 2007;6:3655–3668. doi: 10.1021/pr0702159. [DOI] [PubMed] [Google Scholar]
- 42.Jiang D, Yang W, He Y, Amasino RM. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell. 2007;19:2975–2987. doi: 10.1105/tpc.107.052373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, Ma H. Evolutionary history of histone demethylase families: Distinct evolutionary patterns suggest functional divergence. BMC Evol Biol. 2008;8:294. doi: 10.1186/1471-2148-8-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 45.Dover J, et al. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 46.Lee JS, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 47.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 48.Zhu B, et al. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 49.Rudolph T, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 50.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 51.Citovsky V, Wong ML, Zambryski PC. Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: Implications for the T-DNA transfer process. Proc Natl Acad Sci USA. 1989;86:1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Citovsky V, DE Vos G, Zambryski PC. Single-stranded DNA binding protein encoded by the virE locus of Agrobacterium tumefaciens. Science. 1988;240:501–504. doi: 10.1126/science.240.4851.501. [DOI] [PubMed] [Google Scholar]
- 53.Lacroix B, Loyter A, Citovsky V. Association of the Agrobacterium T-DNA-protein complex with plant nucleosomes. Proc Natl Acad Sci USA. 2008;105:15429–15434. doi: 10.1073/pnas.0805641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iakoucheva LM, et al. Aberrant mobility phenomena of the DNA repair protein XPA. Protein Sci. 2001;10:1353–1362. doi: 10.1110/ps.ps.40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bowler C, et al. Chromatin techniques for plant cells. Plant J. 2004;39:776–789. doi: 10.1111/j.1365-313X.2004.02169.x. [DOI] [PubMed] [Google Scholar]
- 56.Gabrielli F, et al. Histone complements of human tissues, carcinomas, and carcinoma-derived cell lines. Mol Cell Biochem. 1984;65:57–66. [PubMed] [Google Scholar]
- 57.Sutton A, et al. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol Cell Biol. 2001;21:3514–3522. doi: 10.1128/MCB.21.10.3514-3522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ballas N, Citovsky V. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc Natl Acad Sci USA. 1997;94:10723–10728. doi: 10.1073/pnas.94.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001;20:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tzfira T, Vaidya M, Citovsky V. Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis VIP1 gene. Proc Natl Acad Sci USA. 2002;99:10435–10440. doi: 10.1073/pnas.162304099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.SenGupta DJ, et al. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tzfira T, et al. pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol. 2005;57:503–516. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- 63.Ueki S, Lacroix B, Krichevsky A, Lazarowitz SG, Citovsky V. Functional transient genetic transformation of Arabidopsis leaves by biolistic bombardment. Nat Protoc. 2009;4:71–77. doi: 10.1038/nprot.2008.217. [DOI] [PubMed] [Google Scholar]
- 64.Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]