Abstract
The level of TGF-β/bone morphogenetic protein (BMP) signaling through Smad is tightly regulated to ensure proper embryonic patterning and homeostasis. Here we show that Smad activation by TGF-β/BMP is blocked by a highly conserved phosphorylation event in the α-helix 1 region of Smad [T312 in Drosophila Smad1 (MAD)]. α-helix 1 phosphorylation reduces Smad interaction with TGF-β/BMP receptor kinase and affects all receptor-activated Smads except Smad3. Tissue culture and transgenic studies in Drosophila further demonstrate that the biological activity of MAD is repressed by T312 phosphorylation in vivo. Through RNAi screening of the kinome, we have identified Misshapen (Msn) and the mammalian orthologs TNIK, MINK1, and MAP4K4 as the kinases responsible for α-helix 1 phosphorylation. Targeted expression of an active form of Msn in the wing imaginal disk disrupted activation of endogenous MAD by Dpp and expression of the Dpp/MAD target gene. Msn kinases belong to the Ste20 kinase family that has been shown to act as MAP kinase kinase kinase kinase (MAP4K). Our findings thus reveal a function of Msn independent of its impact on MAP kinase cascades. This Smad inhibition mechanism by Msn likely has important implications for development and disease.
Keywords: myristoylation, TKV interaction
TGF-β cytokines are critical regulators of embryonic development and adult tissue homeostasis (1, 2). TGF-β signal transduction is mediated by the Smad transcription factors (3–5). Binding of the TGF-β ligands activates type I receptor kinase activity (ALK1-7 in mammals), which then directly phosphorylates the SXS motif at the C-termini of receptor-activated Smads (R-Smads), including Smad1/2/3/5/8 (6–9). After C-terminal phosphorylation, R-Smads associate with Smad4 and are transported by Msk/Imp7/8 into the nucleus (10). Thus, C-terminal phosphorylation of R-Smads is an obligatory step that switches on the TGF-β/bone morphogenetic protein (BMP) pathway. Among the TGF-β family members, TGF-β/activin/Nodal mainly induces C-terminal phosphorylation of Smad2/3, whereas BMPs activate Smad1/5/8. In the highly conserved C-terminal MH2 domains of Smads, the L3 loop contains sequence elements that distinguish between Smad1/5/8 and Smad2/3 and also confer specificity in the interaction of Smads with different ALKs (11–13). In the case of ALK1/2-mediated phosphorylation of Smad1/5/8 C-termini, additional sequences in the α-helix 1 region of these Smads play critical roles as well (14). Thus, both the L3 loop and α-helix 1 region of Smads are important for the recognition of and physical interaction with their upstream receptor kinases.
Cells must integrate other signal inputs in response to TGF-β/BMP. One important factor in the regulation of TGF-β/BMP signaling is the phosphorylation of Smads in the linker region by MAPKs, GSK3 kinase- or cyclin-dependent kinases (CDK2/4/8/9) (15–19). Linker phosphorylation induces the interaction of Smad with the E3 ubiquitin ligase Smurf1 or Nedd4L, and subsequent Smad degradation (18–20). TGF-β/BMP treatment induces Smad linker phosphorylation, but kinetically it lags behind Smad C-terminal phosphorylation. This led to the hypothesis that linker phosphorylation is a negative feedback mechanism for specifically removing activated Smads (18, 19). However, recent data also suggest that linker phosphorylation enhances the interaction of Smads with the transcription factor YAP and promotes Smad target gene expression (21). Although the exact outcome on linker phosphorylation in Smad is complex, these reports exemplify how cross-talking signals can modulate TGF-β/BMP signaling.
In this study, we identified a single phosphorylation event in the α-helix 1 region of Smads that effectively blocks Smad phosphorylation at the C terminus. Through RNAi screening of the kinome, we identified Msn and its mammalian orthologs, members of the Ste20 family, as the responsible kinases in Drosophila and mammalian cells. Functional assays in cell culture and Drosophila whole animals further demonstrated the ability of Msn kinases to suppress the biological functions of Smad.
Results
α-Helix 1 Phosphorylation of Smads Inhibits TGF-β/BMP Signal Transduction in Drosophila and Mammalian Cells.
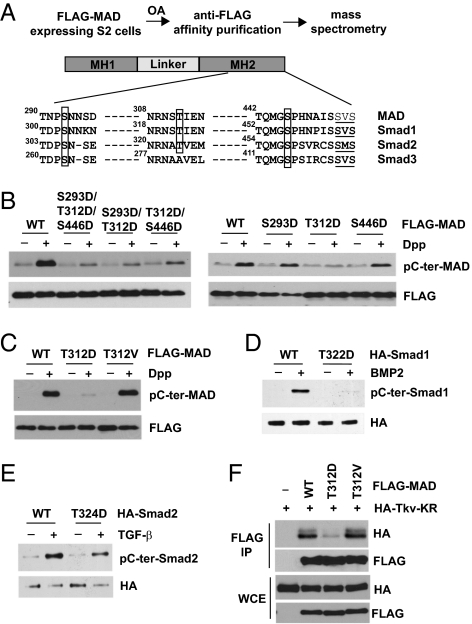
While studying the impact of phosphatases on TGF-β/BMP signaling, we found that depletion of endogenous PP2A in Drosophila wings resulted in greatly reduced C-terminal phosphorylation of MAD (Drosophila Smad1) in vivo (Fig. S1A). This suggested that a loss of PP2A function might induce a Smad inhibition mechanism. In the meantime, mass spectrometry analysis of MAD isolated from okadaic acid (OA; a PP2A inhibitor)-treated S2 cells revealed three phosphorylation sites in the MH2 domain: S293, T312, and S446 (Fig. 1A). These sites are highly conserved in mammalian Smads except for T312, which in Smad3 is an alanine instead (Fig. 1A).
Fig. 1.
Identification of a phosphorylation site in Smad that suppresses TGF-β/BMP-induced C-terminal phosphorylation. (A) Mass spectrometry identified phosphorylation sites in MAD. Drosophila and mammalian Smads sequences are aligned, with the newly identified phosphorylation sites boxed. The C-terminal SXS motifs that are phosphorylated in response to TGF-β/BMP are underlined. (B and C) Single, double, or triple mutants and WT MAD (all FLAG-tagged) were transfected into S2 cells. After Dpp treatment (1 nM, 1 h), FLAG-MAD was immunoprecipitated and analyzed for C-terminal phosphorylation. (D and E) WT and mutant mammalian Smad1 or Smad2 (HA-tagged) were transfected into COS1 cells. After treatment with BMP2 (100 ng/mL, 1 h) or TGF-β (100 pM, 1 h), HA-Smad1 or Smad2 was immunoprecipitated and analyzed for C-terminal phosphorylation. (F) WT and mutant MAD were expressed in S2 cells along with the catalytic mutant of Tkv. Whole cell extracts (WCE) were immunoprecipitated by anti-FLAG, and the bound proteins were evaluated using the indicated immunoblots.
We mutated these phosphorylation residues into aspartic acid and found that a single phosphomimetic mutation of T312 largely prevented MAD C-terminal phosphorylation in response to Dpp (Drosophila BMP2) (Fig. 1B). Importantly, mutating T312 to nonphosphorylatable valine (MADT312V) had no effect on C-terminal phosphorylation, suggesting that it is the phosphorylation status of T312 that critically determines whether MAD can be activated by Dpp (Fig. 1C). T312 and its neighboring residues are highly conserved in mammalian R-Smad, with Smad3 the only exception (Fig. 1A). Indeed, mutating the T312 equivalent in Smad1 (i.e., T322) into aspartic acid nearly abolished Smad1 C-terminal phosphorylation in response to BMP2 (Fig. 1D). A similar phosphomimetic mutation in Smad2 (i.e., T324D) also caused significantly decreased C-terminal phosphorylation in TGF-β–treated cells (Fig. 1E). Therefore, phosphorylation of the α-helix 1 residue appears to be a novel Smad inhibition mechanism in Drosophila and mammalian cells.
Interestingly, it has been suggested that amino acid residues adjacent to T312 are critical for recognition of Smad1 by the type I receptor kinase ALK1 (14). This raised the possibility that phosphorylation of T312 might affect the interaction of MAD with its upstream receptor kinase Thickvein (Tkv). In a coimmunoprecipitation experiment, MADT312D showed a much weaker interaction with Tkv compared with either WT MAD or MADT312V (Fig. 1F). Structural analyses have indicated that the strength of interaction between Smad and TGF-β/BMP receptor kinase is critical for the C-terminal phosphorylation of Smad (22). Thus, our observation here suggests a plausible mechanism by which T312 phosphorylation inhibits MAD activation in response to Dpp.
Biological Function of MAD Is Suppressed by T312 Phosphorylation.
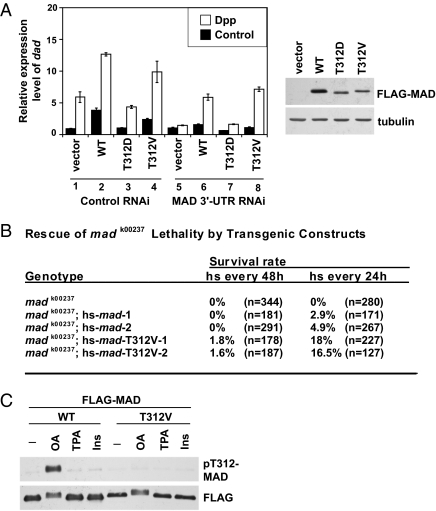
We further investigated whether the T312 phosphorylation of MAD suppresses MAD's biological activity. We examined transcriptional activation of dad, a typical Smad target gene from Drosophila to vertebrates. When endogenous MAD was knocked down in S2 cells by a dsRNA targeting its 3′-UTR, Dpp induction of dad was largely abolished (Fig. 2A; compare columns 1 and 5). Using exogenous mad cDNA to rescue the RNAi effect, we found that whereas both WT and a nonphosphorylatable mutant MADT312V fully restored the Dpp-induced dad transcription, MADT312D failed to rescue mad RNAi (Fig. 2A; compare columns 5 and 6–8). In cells with control RNAi, exogenous WT or MADT312V further elevated dad expression above the level achieved by endogenous MAD; in contrast, overexpression of MADT312D had no significant effect (Fig. 2A; compare columns 1 and 2–4). This suggests that phosphorylation of MAD on T312 inactivates the biological function of MAD.
Fig. 2.
α-helix 1 phosphorylation suppresses biological functions of MAD. (A) Stable S2 cell lines expressing WT and mutant MAD were treated with indicated RNAi to deplete endogenous MAD. After Dpp or vehicle (control) stimulation, the mRNA level of dad was measured by quantitative real-time PCR, with Rp49 as the standard (mean ± SD). (Right) The expression levels of MAD constructs are shown. (B) Heat shock–driven WT or mad T312V transgenes were tested for their ability to rescue the lethality caused by madk00237. Two kinds of heat shock treatments were used (e.g., once every 24 or 48 h). The percentages of progenies with straight wings are shown, which reflect rescue of the lethality associated with homozygous madk00237 (survival rate). A complete rescue is expected to result in 33.3% of the progenies having straight wings. n is the total number of curly-winged and straight-winged progenies scored in each case. (C) S2 cells expressing WT or FLAG-MADT312V were treated with OA (125 nM), TPA (5 μM), or insulin (300 nM) for 1 h. FLAG-MAD was immunoprecipitated and analyzed using the indicated immunoblots.
Is T312 phosphorylation an important regulatory input on MAD in vivo? Loss-of-function mad mutants, such as the madk00237 allele, are recessive lethal. We thus compared the ability of WT and MADT312V to rescue the lethality phenotype. Our reasoning for this is that if indeed T312 phosphorylation negatively regulates MAD, then MADT312V will be expected to be resistant to such inhibition and to exhibit a stronger biological activity than WT MAD. We generated transgenic flies with heat-shock–inducible mad transgene expression. Rearing the flies at 29 °C was insufficient to permit rescue of lethality by WT mad (Table S1). On a 37 °C heat shock, WT mad partially rescued the embryonic lethality of the madk00237 allele, and the rescue efficiency was increased with more frequent heat shocks (Fig. 2B). Such incomplete rescue might be attributed to heat shock not inducing adequate MAD expression in the right tissues with the right timing. Nevertheless, this assay allowed us to compare the activity in WT and mutant MAD. Indeed, the madT312V transgene was much more effective than the WT mad in rescuing the lethality phenotype (Fig. 2B). We obtained similar results using two independent transgenic lines of both WT and madT312V, and apparently WT and MADT312V were expressed at similar levels (Fig. S2A). This indicates that MADT312V has more potent biological activity than WT MAD in this assay, supporting the notion that T312 phosphorylation negatively regulates MAD in vivo.
We developed a phospho-T312–specific antibody, pT312-MAD (Fig. S2B). None of the common agents known to activate many kinases, including phorbol ester, insulin, camp, and calcium ionophore, caused any detectable phosphorylation of T312 (Fig. 2C). To date, OA is the only pharmacologic agent that we have tested that stimulated T312 phosphorylation (Fig. 2C). Perhaps OA treatment activates signaling events normally suppressed by PP2A, which culminate in Smad α-helix 1 phosphorylation. Therefore, in this study we used OA as a biochemical tool to trigger robust MAD phosphorylation on T312. Consistent with its ability to induce the inhibitory T312 phosphorylation, OA strongly inhibited MAD C-terminal phosphorylation in response to Dpp (Fig. S2C). This observation also agrees with the transgenic data shown in Fig. S1. However, it is important to point out that several studies have shown that PP2A physically interacts with and regulates TGF-β/BMP receptor kinases (23–25). Thus, our observations could be attributable to multiple impacts of PP2A on the receptor kinases, α-helix 1 of Smad, and possibly other components in the TGF-β/BMP pathway.
Msn Kinase Is Required for α-helix 1 Phosphorylation of Smad.
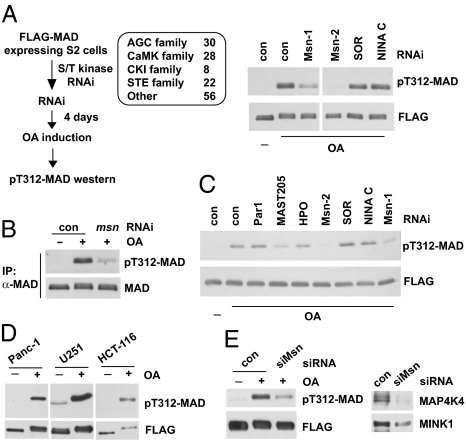
We screened a Drosophila Ser/Thr kinase RNAi library to search for the T312 kinase (Fig. 3A; Table S2 provides a complete list). The Ste20 kinase Msn emerged as a strong hit, and reduction of MAD T312 phosphorylation was observed with two nonoverlapping RNAi constructs against Msn (Fig. 3A). We used a MAD antibody to immunoprecipitate endogenous MAD and confirmed that T312 phosphorylation of endogenous MAD required Msn (Fig. 3B). Whole cell extract from OA-treated S2 cells was capable of phosphorylating purified recombinant MAD on T312 in vitro (Fig. 3C). Again, RNAi against Msn largely depleted such T312 kinase activity (Fig. 3C). Therefore, in two different assays, Msn appeared to be required for phosphorylation of MAD on T312.
Fig. 3.
RNAi screening of the kinome uncovered Msn as the Smad α-helix 1 kinase. (A) Indicated Ser/Thr kinase families were screened as described on the left; the immunoblots show that depletion of Msn (by two independent dsRNAs, Msn-1 and Msn-2) largely diminished T312 phosphorylation of MAD in OA-treated cells. SOR and NINA C are two other kinases that scored negative in the screening. (B) After msn RNAi and OA treatment, endogenous MAD was immunoprecipitated, and T312 phosphorylation was evaluated by immunoblot analysis. TrueBlot secondary antibodies were used to avoid detecting the IgG band. (C) In vitro kinase assays showing extracts from Msn-depleted cells (Msn-1 and Msn-2 RNAi) exhibited much weaker activity in phosphorylating T312 of MAD. Par1, MAST205, and HPO are other kinases that were tested in parallel. (D) T322 phosphorylation of Smad1 in several cancer cell lines. FLAG-Smad1 was immunoprecipitated after OA treatment to measure T322 phosphorylation. The pT312-MAD antibody recognizes T322 phosphorylation of Smad1 given the shared epitopes. (E) U251 cells expressing FLAG-Smad1 was transfected with control or siRNA that depleted both MAP4K4 and MINK1 (siMsn, Right). After OA treatment, FLAG-Smad1 was immunoprecipitated, and T322 phosphorylation was examined (Left).
Three Msn orthologs are annotated in the mammalian genome: TNIK, MINK1, and MAP4K4. In several human cancer cell lines that we have tested (Panc-1, HCT-116, and U251), OA treatment stimulated Smad1 phosphorylation at T322 (equivalent to T312 in MAD) (Fig. 3D). We tested a siRNA duplex that targets a nucleotide sequence highly conserved among all three mammalian Msn kinases. It significantly knocked down endogenous MAP4K4 and MINK1, and as a result, T322 phosphorylation of Smad1 in response to OA was significantly decreased (Fig. 3E). This result was validated by a different shRNA construct in a second cell line (e.g., Panc-1) (Fig. S3). Therefore, in mammalian cells, Smad1 phosphorylation in α-helix 1 also depends on Msn kinases.
Msn belongs to the Ste20 kinase family, in which the members share a conserved kinase domain but otherwise diverge substantially in sequences (26). Our finding here is the first indication that Msn may be a regulator of Smad signaling.
Msn Kinases Directly Phosphorylate α-Helix 1 of Smad.
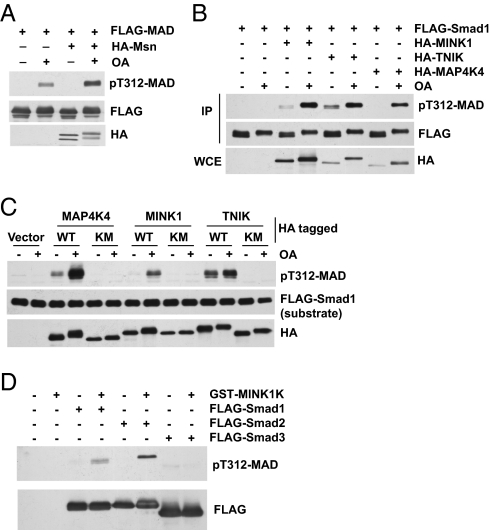
Overexpression of Msn alone did not result in detectable MAD T312 phosphorylation, although after OA treatment, cells with Msn overexpression exhibited higher levels of T312 phosphorylation (Fig. 4A). Therefore, Msn appears to be inactive in untreated S2 cells. In human 293T cells, OA alone did not cause detectable Smad1 T322 phosphorylation (Fig. 4B). Overexpression of TNIK, MINK1, or MAP4K4 caused weak Smad1 T322 phosphorylation; however, in all cases, OA strongly enhanced the ability of these kinases to phosphorylate Smad1 on T322 (Fig. 4B). We carried out an in vitro kinase assay using separately purified Msn kinases and FLAG-Smad1. The activities of Msn kinases, especially in the case of MAP4K4 and MINK1, were markedly stronger when purified from OA-treated cells (Fig. 4C). This suggests that these kinases are suppressed by PP2A in cells. Importantly, in all cases, mutation of the highly conserved K54 residue in the kinase domain completely abolished Smad1 phosphorylation in α-helix 1 (Fig. 4C), confirming that the Msn kinases themselves, not another copurified kinase, caused the Smad1 phosphorylation on T322.
Fig. 4.
Msn kinases directly phosphorylate α-helix 1 of Smad. (A) HA-Msn and FLAG-MAD were expressed in S2 cells. After OA stimulation, FLAG-MAD was immunoprecipitated and evaluated by immunoblot analysis. (B) 293T cells were transfected and treated with OA as indicated. FLAG-Smad1 was immoprecipitated, and phosphorylation on T322 was measured by immunoblot analysis. (C) IP-kinase assays. HA-tagged WT and catalytic mutants of MAP4K4, MINK1, and TNIK (KM, K54R mutation in all three cases) were purified from 293T cells with or without OA treatment as indicated. FLAG-Smad1 was affinity-purified separately and used as the substrate. (D) Recombinant MINK1 kinase domain (GST-MINK1K) purified from E. coli directly phosphorylated purified recombinant Smads in vitro.
Moreover, a GST fusion of the kinase domain of MINK1 produced in Escherichia coli phosphorylated α-helix 1 of Smad1 and Smad2, but not that of Smad3, in vitro (Fig. 4D). This result suggests that Msn kinases are perhaps kept inactive by an autoinhibitory mechanism, and that removing the rest of the molecule renders Msn constitutively active. More importantly, the result further confirms that Msn kinases can directly phosphorylate Smad in α-helix 1.
Msn Is Activated by Membrane Association.
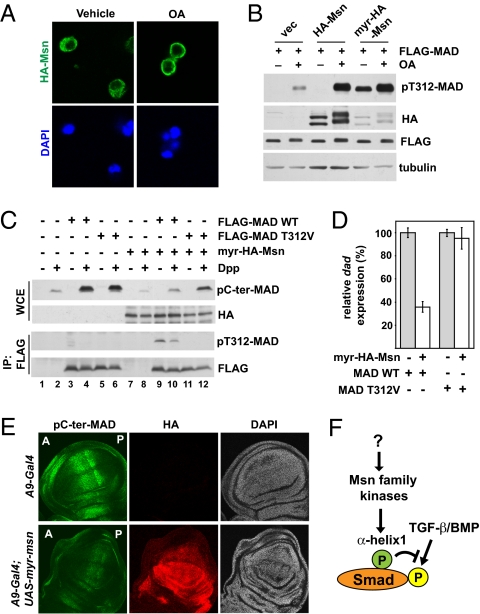
Very little is known about how Msn kinases are activated. Interestingly, Msn was present mostly in the cytoplasm, but after OA treatment, the bulk of Msn translocated to the cell membrane (Fig. 5A). This observation raised the possibility that membrane association might contribute to activation of the Msn kinase. We tested this idea by engineering a myristoylation sequence (MGNCLT) to the N terminus of Msn (myr-Msn). Indeed, myr-Msn was far more potent in causing phosphorylation of MAD on T312 without the need for OA, even though myr-Msn was expressed at a much lower level compared with WT Msn (Fig. 5B). It is important to determine whether the increase in MAD T312 phosphorylation was actually due to a gain in Msn catalytic activity or to another mechanism, such as physical proximity to MAD. In an IP kinase assay, myr-Msn exhibited much stronger catalytic activity than WT Msn, taking into consideration the much lower amount of myr-Msn (Fig. S4). Thus, membrane anchorage appears to be a critical step in activating the kinase function of Msn.
Fig. 5.
Msn is activated on membrane translocation and inhibits Dpp signaling in vivo. (A) Immunofluorescence staining showing the plasma membrane translocation of HA-Msn in S2 cells after OA (125 nM, 1 h) treatment. (B) HA-Msn with or without a myristoylation sequence was coexpressed with FLAG-MAD in S2 cells. FLAG-MAD was immunoprecipitated, and the level of T312 phosphorylation was examined. The effect of OA treatment is shown where indicated. (C) S2 cells were transiently transfected with indicated combinations of plasmids. After Dpp treatment, the whole cell extract (WCE), or the FLAG proteins (after IP) were analyzed using the indicated immunoblots. (D) The same cells from lanes 4, 6, 10, and 12 in (C) were analyzed by quantitative real-time PCR to measure the mRNA level of dad, using Rp49 as the standard (mean ± SD). (E) Third instar wing imaginal discs from the indicated transgenic flies were double-immunostained with pC-ter-MAD (green) and HA (red). The nuclei were stained by DAPI (blue). The orientation of the wing disk is denoted by “A” for anterior and “P” for posterior. (F) A model showing direct regulation of Smad by Msn kinases. The activity of Msn is regulated by as-yet unknown mechanisms.
With myr-Msn as a constitutively active kinase, we were able to specifically evaluate the functional impact of Msn on TGF-β/BMP signaling without the need for OA, which could activate multiple pathways. When myr-Msn was transiently transfected into S2 cells, the C-terminal phosphorylation of endogenous MAD in response to Dpp was decreased (Fig. 5C; compare lanes 2 and 8). We also cotransfected myr-Msn with WT MAD or MADT312V and specifically examined the exogenous MAD protein. Indeed, C-terminal phosphorylation of WT MAD was significantly suppressed by the cotransfected myr-Msn (Fig. 5C; compare lanes 4 and 10). Importantly, with the T312V mutation, such a reduction in C-terminal phosphorylation was significantly alleviated (Fig. 5C; compare lanes 6, 10, and 12). These data argue that Msn inhibits MAD C-terminal phosphorylation by phosphorylating T312. In parallel to the biochemical assays, we measured the transcription level of dad, a MAD downstream target gene. Overexpression of myr-Msn substantially decreased dad expression in Dpp-treated cells (Fig. 5D). In comparison, in cells expressing the MADT312V mutant, such inhibition by myr-Msn was largely abrogated (Fig. 5D). The results of these functional assays reinforce the notion that Msn suppresses MAD primarily through T312 phosphorylation.
Ectopic Msn Activity Suppresses MAD Activation in Vivo.
We asked whether ectopically activated Msn could disrupt Dpp activation of MAD in vivo. We generated transgenic flies containing UAS-driven myr-Msn (HA-tagged) and targeted its expression specifically to the wing imaginal disk using the A9-Gal4 driver. Myr-HA-Msn expression was greatest in the dorsal compartment of the wing discs, so we quantified the pC-ter-MAD signal in this segment to evaluate the level of MAD activation by Dpp (Fig. 5E; anti-HA staining). In the positive control experiment, A9-driven RNAi against mad strongly reduced pC-ter-MAD signal in the third instar wing disk (Figs. S5 A and B and S6A). Expression of myr-Msn also resulted in a greatly diminished pC-ter-MAD immunostaining signal compared with the negative control (Fig. 5E and Figs. S5B and S6A). As expected from the diminished pC-ter-MAD signal, the endogenous mRNA level of dad was significantly reduced in the wing discs with ectopic myr-Msn expression (Fig. S6B). These results are consistent with the observations in S2 cells and demonstrate in vivo that activated Msn is an inhibitor of MAD function.
Discussion
In this study, we uncovered a mechanism that keeps most R-Smads (i.e., Smad1/2/5/8) in an “off” state by phosphorylating a critical residue in the α-helix 1 domain (i.e., T312 in MAD, T322 in Smad1/5/8, and T324 in Smad2) (Fig. 5F). Therefore, Smad contains a built-in off switch that controls its availability to transduce TGF-β/BMP signals. Interestingly, the α-helix 1 phosphorylation site is conserved in all R-Smads except Smad3, indicating that the Msn kinases may selectively regulate physiological responses to BMPs and those TGF-β effects mediated by Smad2. Through an unbiased RNAi screening, we identified Msn kinase(s) as the key regulator of the phosphorylation status of α-helix 1 in Smad. Many Ste20 kinases, including Msn, have been suggested to function as MAP4Ks (MAP kinase kinase kinase kinase) upstream of MAP kinases, and here we uncovered an Msn function as a direct suppressor of the TGF-β/BMP pathway.
Msn kinases have been implicated in a number of biological processes. In development, Msn regulates dorsal closure in Drosophila and mesoderm patterning in mice. In these contexts, Msn has been postulated to act through the MAP kinase JNK, but the direct Msn targets relevant to these developmental processes remain unclear (27, 28). In mammalian cells, a recent RNAi screening led to the identification of MAP4K4 as an inhibitor of adipogenesis (29). In separate RNAi screenings, MAP4K4 was found to promote tumor cell motility (30), and MINK1 was identified as a mediator of Ras-induced cell growth arrest (31). More recently, TNIK was reported to be an activator of Wnt target genes by interacting with and phosphorylating the transcription factor TCF4 (32, 33). Therefore, the Msn kinases likely serve as critical signaling nodes that, in parallel to repressing Smad, may affect multiple pathways, giving rise to complex pathophysiological outcomes. Our finding that Smad is a substrate of Msn may provide insight into the biological functions of Msn kinases.
What is the biological context that activates Msn kinase activity? One clue is the fact that because OA activates Msn kinase activity, PP2A likely suppresses the hitherto unknown mechanism that activates Msn. However, given the broad impact of PP2A on many pathways, identifying the stimulus that turns on Msn remains difficult. Even though Msn was recently implicated in Wnt signaling (32, 33), we found no evidence that either canonical or noncanonical Wnt pathways activate Msn kinase function as measured by T312 phosphorylation of MAD/Smad1. Interestingly, previous reports have shown that oncogenic KRas activity causes increased MINK1 activity and expression (31), and that MAP4K4 expression is heightened in tumor cell lines and tumor tissues compared with their normal counterparts (34, 35). Those reports suggested the interesting possibility that Msn kinases might be involved in inhibiting the tumor-suppressing functions of TGF-β/BMP in various cancers. To understand the physiological context that stimulates Msn-mediated Smad inhibition, it is critical to elucidate the underlying molecular mechanism that activates Msn catalytic function. Interestingly, we found that Msn is activated when targeted to the cell membrane. This observation presents a stepping-stone to further dissect the upstream regulatory inputs imposed on Msn. The biological outcomes of TGF-β/BMP are critically dependent on the cellular context. Understanding the molecular control of Msn-mediated Smad inhibition will advance our knowledge of TGF-β/BMP integration with other signals during embryonic patterning, as well as in such diseases as cancer.
Materials and Methods
Tissue Culture, Transfection, and Cytokine Treatment.
Drosophila S2 cells were cultured in Schneider media with 10% FBS (Invitrogen) and transfected with Effectene (Qiagen). COS1, 293T, U251, and Panc-1 cells were cultured in DMEM with 10% FBS and transfected with Lipofectamine 2000 (Invitrogen). The experiments used Dpp (R&D Systems) at 1 nM, TGF-β (R&D Systems) at 100 pM, and BMP2 (R&D Systems) at 100 ng/mL.
Antibodies.
The pT312-MAD antibody was raised against the peptide CNRNS[pT]IENTRRHIG, and the sera were first absorbed by beads conjugated with the nonphospho-peptide and then affinity-purified using the phospho-peptide. Anti-MAD was described previously (36). pC-ter-Smad1, pC-ter-Smad2, and HA antibodies were purchased from Cell Signaling. Anti-FLAG was purchased from Sigma-Aldrich.
RNAi.
For RNAi screening of the Drosophila kinome, a stable S2 cell line expressing FLAG-MAD under the control of a metallothionine promoter (inducible by CuSO4) was transfected with dsRNA targeting each individual Ser/Thr kinase using DharmaFect 4 (Thermo Fisher). Four days later, cells were induced to express FLAG-MAD (0.1 mM CuSO4 for 2 h) and treated with OA (125 nM, 1 h). Cell lysates were analyzed by pT312-MAD immunoblotting. A total of 144 Ser/Thr kinases were screened. For siRNA against the mammalian Msn kinases, the targeted mRNA sequence is 5′-GACCAACUCUG-GCUUGUU-3′. For RNAi against the mad 3′-UTR, dsRNA was designed to target the mad mRNA sequence from nt 1770–2231.
In Vitro Kinase Assay.
HA-tagged Msn kinases were immunoprecipitated from 293T or S2 cells by anti-HA–conjugated beads (Roche). Cells were lysed in 20 mM Tris Cl (pH 7.4), 150 mM NaCl, 0.5% Nonidet P-40, and 2 mM DTT supplemented with protease inhibitors and phosphatase inhibitors (1 mM Na3VO4, 20 mM Na pyrophosphate, 20 mM NaF, and 20 mM β-glycerophosphate). The extracts were incubated with anti-HA beads (Roche) at 4 °C overnight. The beads were washed twice with the lysis buffer and once with the kinase reaction buffer [25 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 0.1% β-mercaptoethanol]. The beads were then incubated in the same kinase reaction buffer supplemented with 1 mM ATP, with affinity- purified FLAG-Smad1 as the substrate at 30 °C for 30 min. Purified GST-MINK1 kinase domain (Sigma-Aldrich) was used for in vitro kinase reactions at 10 μg/mL in a final volume of 10 μL. The kinase reaction buffer and incubation conditions are identical to those described above.
Identification of Phosphorylated Residues in MAD by Mass Spectrometry.
S2 cells expressing FLAG-MAD were treated with OA at 125 nM for 2 h. Whole cell extracts were prepared, and FLAG-MAD was immunoprecipitated as described above. The purified FLAG-MAD was resolved on 8% SDS/PAGE, and the protein band was excised and submitted for mass spectrometry analysis by the Taplin mass spectrometry facility at Harvard Medical School.
Drosophila Transgenic Studies.
Transgenic flies were generated that expressed WT or MADT312V under a heat shock-inducible promoter, and were crossed to the madk00237 mutant allele (recessive lethal, Bloomington Drosophila Stock Center) with a CyO (curly wing)-marked balancer. The resulting madk00237/CyO;hs-mad(WT or T312V)/TM3 lines were intercrossed. The fly strains were used for embryo deposition in regular fly food vials for 24 h and then heat-shocked in a 37 °C water bath for 45 min. Afterward, the heat shock was applied every day or every other day. The hatched adult flies were scored for the curly-wing or straight-wing phenotype. Straight-wing flies from such crosses indicate rescue of the madk00237 lethality phenotype.
cDNA encoding HA-tagged myr-Msn was cloned into the UASp-pPW vector and used to generate the transgenic line UAS-myr-msn. A9-Gal4 (Bloomington Drosophila Stock Center) was used for driving transgene expression in the wing disk. For in vivo RNAi against PP2A, the pp2a-1 and -2 strains were obtained from the Vienna Drosophila RNAi Center (transformants 4971 and 4972).
Immunofluorescence Staining of Third Instar Larvae Imaginal Discs.
The third instar larvae wing discs were dissected and fixed in 4% paraformaldehyde/PBS at room temperature for 30 min and permeabilized in 0.1% Triton X-100/PBS (2 × 30 min). The discs were blocked by incubation in 0.1% Triton X-100/PBS/0.5% BSA/2% normal horse serum for 2 h. Anti–phospho-SMAD1/5/8 (Cell Signaling), which also recognizes phosphor-MAD, was used at a 1:300 dilution, and anti-HA (Sigma-Aldrich; clone 6E2) was used at a 1:200 dilution for overnight incubation at 4 °C. After washing in 0.1% Triton X-100/PBS (4×), discs were incubated with Alexa Fluor 488 anti-rabbit and Alexa Fluor 633 anti-mouse (1:1,000 dilution for 2h), followed by washing in 0.1% Triton X-100/PBS and mounting in PBS/Vectashield (1:1) (Vector Laboratories). Confocal images were obtained with a Leica DM IRE2 microscope and analyzed using NIS-Elements v3.0 (Nikon). For each wing disk, a line (550 pixels long) that straddles the A/P border and perpendicular to the d-V axis is drawn across the wing pouch in the dorsal segment in which A9 targeting is the strongest (37). Using the intensity profile function of the program, the signal intensity of each pixel was measured. The program automatically draws 15 additional lines in parallel to the original line within a 32-pixel range, and an average value for each of the 550 pixels was calculated based on measurement of these parallel lines. For each transgenic line, multiple discs (20 for A9-Gal4, 26 for A9-Gal4;UAS-myr-HA-msn line 1 and 17 for line 2, and 16 for A9-Gal4;UAS-sh-mad) were analyzed in this way, and the mean ± SD for each pixel was plotted.
Quantitative Real-Time PCR Analysis.
Third instar wing discs were dissected and total RNA was isolated using the RNeasy Mini Kit (Qiagen). Reverse-transcription and SYBR-Green–based real-time PCR was performed as described previously using rp49 as the internal standard (10).
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center for the Drosophila strains, Dr. M. Czech for the MAP4K4 siRNA, and Drs. R. Davis, M. Czech, S. Newfeld, and Q. Xu for discussions and critical reading of the manuscript. This work was funded by grants from the National Institutes of Health (R01CA108509 to L.X. and DK83450 to Y.T.I.), a grant from the Worcester Foundation for Biomedical Research (to L.X.), and a Grant-in-Aid for Scientific Research (20590311) from the Japan Society for the Promotion of Science (to K.K.). Y.T.I. is a member of the UMass Diabetes and Endocrinology Center.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104128108/-/DCSupplemental.
References
- 1.Massagué J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 2.Wu MY, Hill CS. Tgf-β superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Attisano L, Wrana JL. Signal transduction by the TGF-β superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 4.Raftery LA, Sutherland DJ. TGF-β family signal transduction in Drosophila development: From Mad to Smads. Dev Biol. 1999;210:251–268. doi: 10.1006/dbio.1999.9282. [DOI] [PubMed] [Google Scholar]
- 5.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signaling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 6.ten Dijke P, et al. Activin receptor-like kinases: A novel subclass of cell-surface receptors with predicted serine/threonine kinase activity. Oncogene. 1993;8:2879–2887. [PubMed] [Google Scholar]
- 7.Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 8.Macías-Silva M, et al. MADR2 is a substrate of the TGFβ receptor, and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 9.Souchelnytskyi S, et al. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- 10.Xu L, et al. Msk is required for nuclear import of TGF-β/BMP-activated Smads. J Cell Biol. 2007;178:981–994. doi: 10.1083/jcb.200703106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo RS, Chen YG, Shi YG, Pavletich NP, Massagué J. The L3 loop: A structural motif determining specific interactions between SMAD proteins and TGF-β receptors. EMBO J. 1998;17:996–1005. doi: 10.1093/emboj/17.4.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Persson U, et al. The L45 loop in type I receptors for TGF-β family members is a critical determinant in specifying Smad isoform activation. FEBS Lett. 1998;434:83–87. doi: 10.1016/s0014-5793(98)00954-5. [DOI] [PubMed] [Google Scholar]
- 13.Chen YG, et al. Determinants of specificity in TGF-β signal transduction. Genes Dev. 1998;12:2144–2152. doi: 10.1101/gad.12.14.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YG, Massagué J. Smad1 recognition and activation by the ALK1 group of transforming growth factor-β family receptors. J Biol Chem. 1999;274:3672–3677. doi: 10.1074/jbc.274.6.3672. [DOI] [PubMed] [Google Scholar]
- 15.Matsuura I, et al. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 16.Kretzschmar M, Doody J, Massagué J. Opposing BMP and EGF signaling pathways converge on the TGF-β family mediator Smad1. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 17.Kretzschmar M, Doody J, Timokhina I, Massagué J. A mechanism of repression of TGFβ/Smad signaling by oncogenic Ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuentealba LC, et al. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131:980–993. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sapkota G, Alarcón C, Spagnoli FM, Brivanlou AH, Massagué J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Gao S, et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol Cell. 2009;36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alarcón C, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huse M, et al. The TGFβ receptor activation process: An inhibitor- to substrate-binding switch. Mol Cell. 2001;8:671–682. doi: 10.1016/s1097-2765(01)00332-x. [DOI] [PubMed] [Google Scholar]
- 23.Griswold-Prenner I, Kamibayashi C, Maruoka EM, Mumby MC, Derynck R. Physical and functional interactions between type I transforming growth factor β receptors and Bα, a WD-40 repeat subunit of phosphatase 2A. Mol Cell Biol. 1998;18:6595–6604. doi: 10.1128/mcb.18.11.6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petritsch C, Beug H, Balmain A, Oft M. TGF-β inhibits p70 S6 kinase via protein phosphatase 2A to induce G(1) arrest. Genes Dev. 2000;14:3093–3101. doi: 10.1101/gad.854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batut J, et al. Two highly related regulatory subunits of PP2A exert opposite effects on TGF-β/Activin/Nodal signaling. Development. 2008;135:2927–2937. doi: 10.1242/dev.020842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delpire E. The mammalian family of sterile 20p-like protein kinases. Pflugers Arch. 2009;458:953–967. doi: 10.1007/s00424-009-0674-y. [DOI] [PubMed] [Google Scholar]
- 27.Xue Y, et al. Mesodermal patterning defect in mice lacking the Ste20 NCK-interacting kinase (NIK) Development. 2001;128:1559–1572. doi: 10.1242/dev.128.9.1559. [DOI] [PubMed] [Google Scholar]
- 28.Su YC, Treisman JE, Skolnik EY. The Drosophila Ste20-related kinase Misshapen is required for embryonic dorsal closure and acts through a JNK MAPK module on an evolutionarily conserved signaling pathway. Genes Dev. 1998;12:2371–2380. doi: 10.1101/gad.12.15.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang X, et al. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPAR-γ, adipogenesis, and insulin-responsive hexose transport. Proc Natl Acad Sci USA. 2006;103:2087–2092. doi: 10.1073/pnas.0507660103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins CS, et al. A small interfering RNA screen for modulators of tumor cell motility identifies MAP4K4 as a promigratory kinase. Proc Natl Acad Sci USA. 2006;103:3775–3780. doi: 10.1073/pnas.0600040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicke B, et al. Involvement of MINK, a Ste20 family kinase, in Ras oncogene-induced growth arrest in human ovarian surface epithelial cells. Mol Cell. 2005;20:673–685. doi: 10.1016/j.molcel.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 32.Mahmoudi T, et al. The kinase TNIK is an essential activator of Wnt target genes. EMBO J. 2009;28:3329–3340. doi: 10.1038/emboj.2009.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shitashige M, et al. Traf2- and Nck-interacting kinase is essential for Wnt signaling and colorectal cancer growth. Cancer Res. 2010;70:5024–5033. doi: 10.1158/0008-5472.CAN-10-0306. [DOI] [PubMed] [Google Scholar]
- 34.Wright JH, et al. The STE20 kinase HGK is broadly expressed in human tumor cells and can modulate cellular transformation, invasion, and adhesion. Mol Cell Biol. 2003;23:2068–2082. doi: 10.1128/MCB.23.6.2068-2082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang JJ, et al. Expression of MAP4K4 is associated with worse prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin Cancer Res. 2008;14:7043–7049. doi: 10.1158/1078-0432.CCR-08-0381. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, Xu L. Specific nucleoporin requirement for Smad nuclear translocation. Mol Cell Biol. 2010;30:4022–4034. doi: 10.1128/MCB.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haerry TE, Khalsa O, O'Connor MB, Wharton KA. Synergistic signaling by two BMP ligands through the SAX and TKV receptors controls wing growth and patterning in. Drosophila. Development. 1998;125:3977–3987. doi: 10.1242/dev.125.20.3977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.