Abstract
Memory T cells survive throughout the lifetime of an individual and are protective upon recall. It is not clear how memory T cells can live so long. Here, we demonstrate that at the resolution of a viral infection, low levels of antigen are captured by B cells and presented to specific CD4+ memory T cells to render a state of unresponsiveness. We demonstrate in two systems that this process occurs naturally during the fall of antigen and is associated with a global gene expression program initiated with the clearance of antigen. Our study suggests that in the absence of antigen, a state of dormancy associated with low energy utilization and proliferation can help memory CD4+ T cells to survive nearly throughout the lifetime of mice. The dormant CD4+ memory T cells become activated by stimulatory signals generated by a subsequent infection. We propose that quiescence might be a mechanism necessary to regulate long-term survival of CD4 memory T cells and to prevent cross-reactivity to self, hence autoimmunity.
Keywords: B cell antigen presentation, Memory T cells, Anergy, Memory T cells survival, Microarray, BCR-Mediated Antigen Capture, CD4 memory T cells, Gene Regulation, Low-Dose Antigen, Memory Survival
The hallmarks of specific T cell immunity include proliferation, differentiation and generation of memory T cells. Memory helper T cell is an important component of immunological memory. Through production of cytokines CD4+ memory T cells activate dendritic cells (DCs) and help B cells as well as CD8+ T cells in the generation of robust secondary responses1-5. Understanding how memory T cells are generated and maintained, and what is required for their reactivation, is important in design of vaccines. Intense investigations over the past decade have advanced our knowledge of CD8+ T cell memory6. Among those are elegant studies reporting that different subpopulations of CD8+ memory T cells can differentiate from a single progenitor at the time of the first encounter with antigen7-10. It is generally accepted that CD8+ memory T cells are more efficient responders to antigens than naïve cells and are maintained at certain constant numbers throughout the life of an individual6, 11. Also despite a wealth of information regarding CD8+ memory T cell survival11-15, much less is known about the mechanism of long-term survival of memory CD4+ T cells16, 17.
Memory T cells are accepted to have lower antigenic threshold and be less dependent on the second signal for activation, although recent work supports an opposing view18-22. As such, memory T cells can potentially damage host tissues due to cross-reactivity, demanding a need for strictly regulating their reactivation to maintain self-tolerance. While activation-induced cell death and regulatory T cells are mechanisms evolved to regulate activated T cells23, not much is known as to how memory T cells are regulated. It is thought that memory T cells undergo homeostatic proliferation for long-term survival24,25, however, the precise molecular requirements for homeostatic proliferation in CD8+ and CD4+ T cells remain debatable. Also, repeated proliferation shortens the longevity of cells26, hence continuous proliferation might not serve memory T cells well in long-term survival27-29. Accordingly, the mechanism of memory T cell survival remains to be discovered.
We have shown in several antigenic systems including polyclonal HLA-DR1 transgenic mice30 that memory CD4+ T cells become hyporesponsive when stimulated with suboptimal doses of antigen31,32. Using peptides for immunization and challenge we have demonstrated that under such condition B cells are the APCs to induce this unresponsiveness in memory T cells33. B cells have the unique characteristics of carrying specific antigen receptors hence can potentially capture antigens at low levels. Coexistence of memory T cells and B cells carrying low levels of antigenic ligands might occur during the contraction phase and post resolution of an infection. In the present study, using CpG and Vaccinia virus expressing ovalbumin (VAC-OVA) as models of infection, we address how reduction in antigen level can trigger B cells to signal CD4+ memory T cells to become dormant and help their long-term survival.
Results
In vivo administration of suboptimal doses of OVA generates hyporesponsive memory CD4+ T cells
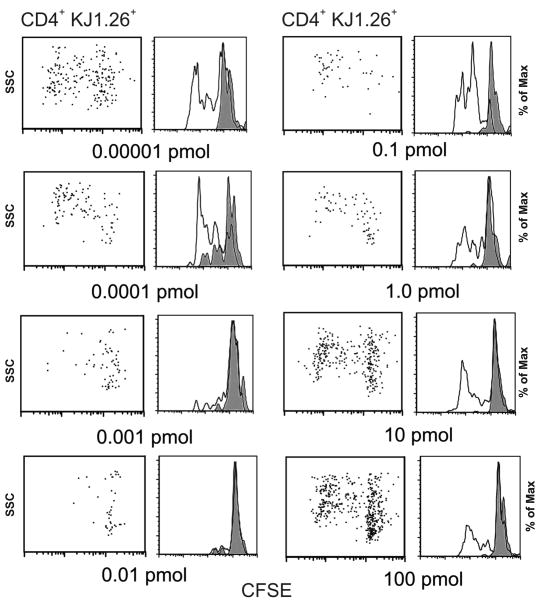
Previously, we have shown that B cells pulsed with cOVA323-339 peptide induce a state of anergy in specific memory CD4+ T cells33. Here, we asked whether suboptimal doses of OVA protein would induce hyporesponsive memory T cells. Using our established protocol for induction of unresponsive memory T cells in vivo 30, Balb/c mice transferred with 2 ×105 DO11.10 cells were immunized with cOVA323-339 in CFA to generate memory T cells. Once memory was established (5 weeks or later) multiple doses of OVA protein in IFA were administered and 8-12 days later, in different experiments, the state of memory T cell responsiveness was examined (Fig. 1). We found that majority of antigen-specific T cells that had encountered 0.001-0.01 pmol OVA in vivo had become unresponsive to their specific OVA323-339 peptide, while T cells from the mice immunized with doses below or above that range underwent several cycles of division as detected by CFSE dilution assay. These observations document that OVA protein at low doses generates hyporesponsive memory T cells.
Figure 1.
Suboptimal doses of OVA protein in IFA induces hyporesponsiveness in specific memory CD4+ T cells. Eight groups of mice (three mice per group) bearing memory CD4+ T cells (DO11.10 cells transferred and primed with cOVA323-339 peptide in CFA 5 weeks earlier) received increasing doses (0.00001 – 100 pmol) of OVA protein in IFA for induction of unresponsiveness. Ten days later, cells harvested from the draining lymph nodes were pooled, labeled with CFSE and re-challenged in vitro with cOVA323-339 peptide in triplicate wells for 72 h T cell proliferation was measured by CFSE dilution assay of antigen-specific CD4+ KJ1-26+ T cells. For CFSE dilution gating was done on DO11.10 cells that stained positive for CD4 and KJ1.26. Each panel depicts CFSE dilution of cells isolated from the eight groups of mice that received increasing doses of OVA protein in IFA. In vitro challenge with cOVA323-339 peptide: filled histogram, 0 μM; open histogram, 0.1 μM. Data shown represent one of three independent experiments. Cells from triplicate wells were pooled before staining for flow cytometry. FlowJo software was used for data analysis.
BCR dependent antigen uptake by B cells leads to hyporesponsive memory CD4+ T cells
Our previous experiments demonstrated that resting B cells can present OVA323-339 peptide to memory CD4+ T cells and render them tolerant33. To address if specific B cells might render memory T cells unresponsive, IgHelMD4 mice carrying transgenic B cells specific for Hen Egg Lysozyme (HEL) were used34. HEL was coupled to OVA (HEL-OVA) as previously described34 and was used as antigen. Unconjugated OVA served as the control antigen for comparison in parallel groups. Since IgHelMD4 mice are on B6 background, we switched to OT-II transgenic T cell (CD4+Vα2+Vβ5+) system specific for OVA323-339/I-Ab. Two hundred thousand OT-II T cells were transferred to B6 recipient mice and with subsequent immunization with OVA323-339 peptide in CFA to generate memory T cells. Purified B cells from IgHelMD4 mice were transferred to 16 groups (two sets of 8) of three mice each bearing OT-II memory cells. Each group received increasing doses of OVA (8 groups), or HEL-OVA in IFA (8 groups). Ten to twelve days later, cells from the draining lymph nodes were harvested and re-challenged with cOVA323-339 peptide in vitro. We found that while OVA at concentrations ranging between 0.01-1.0 pmol/IFA caused hyporesponsive CD4 memory T cells, HEL-OVA conjugate was effective at 1,000 to 10,000 folds lower doses as evidenced by IL-2 synthesis and cell proliferation (Fig. 2). These experiments suggest that B cells bearing specific receptors capture antigen more efficiently and present them to memory CD4+ T cells and render them hyporesponsive.
Figure 2.
BCR-mediated antigen uptake by B cells induces hyporesponsiveness in memory CD4+ T cells. B6 mice bearing memory OT-II (CD4+Vα2+Vβ5+) cells (OT-II cells transferred and primed with cOVA323-339 peptide in CFA 12 weeks earlier) were transferred intravenously with B cells from IgHelMD4 Tg mice. Eighteen hours later, the recipient mice were divided into eight groups of three and challenged with increasing doses (0.0001 – 1000 pmol) of ovalbumin in IFA (A and C), or increasing doses (0.00000001 – 10 pmol) of HEL coupled ovalbumin in IFA (B and D). Twelve days later, cells harvested from draining lymph nodes were pooled and re-challenged with cOVA323-339. T cell responses were measured by 3H-thymidine incorporation (A and B) and intracellular IL-2 (C and D) assays. Histograms underneath panels C and D summarize dot plot data above. Data shown represent one of two independent experiments. Bars represent mean ± standard deviation of triplicate cultures. There were three mice per group and cells from triplicate wells intended for cytokine measurement were pooled before measuring IL-2 synthesis. Analysis for cytokine synthesis was done on gated CD4+IL-2+ T cells.
During the fall of antigenic load, B cells induce hyporesponsive memory CD4+ T cells in vivo
CpG as an infection model
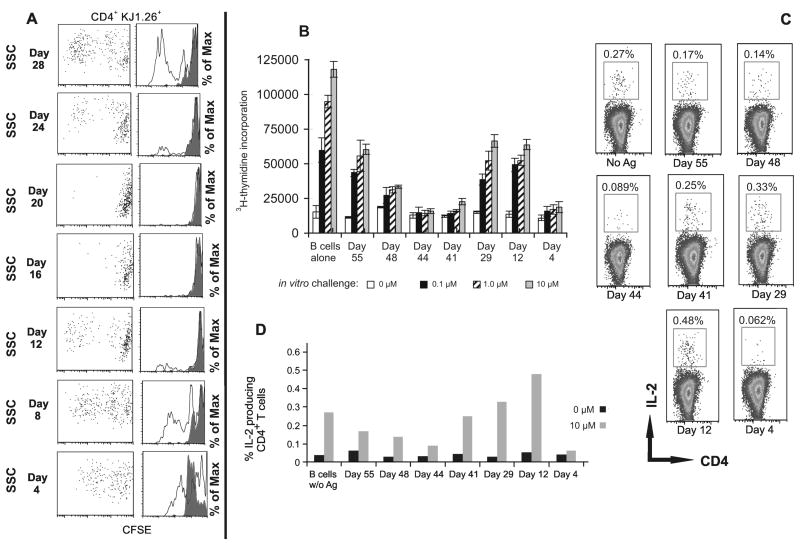
The above experiment showed that B cells capture OVA and generate hyporesponsive memory T cells. To examine if specific B cells might capture antigen during or after the resolution of an infection and present it to memory T cells, we used CpG as a mimic of an infection35. Normal BALB/c mice were injected with an immunogenic dose of OVA mixed with CpG, and their B cells were isolated at 4-day intervals and transferred to groups of recipient BALB/c mice harboring specific CD4+ (DO11.10) memory T cells. We found that B cells from mice harvested on day 16 or day 20 post-injected with OVA/CpG rendered memory CD4+ T cells hyporesponsive. B cells harvested before day 16 (days 4, 8 and 12), and after day 20 (days 24 and 28) were stimulatory and induced positive responses (Fig. 3A). This experiment suggested that the level of antigen presented by B cells during 16-20 days post CpG/OVA injection might be just the right amount for rendering CD4 memory T cells unresponsive.
Figure 3.
B cells during the fall of antigenic load induce hyporesponsive memory CD4+ T cells. (A) BALB/c mice bearing memory DO11.10 T cells per in previous figures, were transferred with purified B cells isolated from groups of BALB/c mice immunized with OVA protein/CpG in PBS at time intervals spanning 4–28 days as shown. The effects of B cell transfer on the proliferation of specific CD4+ T cells were determined by in vitro CFSE dilution assay of draining lymph node in response to an in vitro cOVA323-339 peptide challenge: filled histogram, 0 μM; open histogram, 0.1 μM. Data shown represent one of two independent experiments. In each experiment three mice per group were tested. For CFSE dilution, analysis was done on CD4+KJ1.26+ (DO11.10) gated cells. (B and C) B cells from IgHelMD4 transgenic mice immunized with HEL-OVA/ CpG for different time periods (day 5 – day 55) were transferred to parental wild type B6 mice bearing memory OT-II (CD4+Vα2+Vβ5+) T cells. Fourteen days later, cells from pooled draining lymph nodes were cultured in triplicate wells in the presence of cOVA323-339 peptide, and antigen specific CD4+ T cell proliferation was determined by 3H-thymidine incorporation in B, and intracellular IL-2 assays shown in C. Data shown represent one of two independent experiments. Bars represent mean ± standard deviation of triplicate cultures. There were three mice per group and cells from triplicate wells intended for cytokine measurement were pooled before measuring IL-2 synthesis. Analysis for cytokine synthesis was done on gated CD4+IL-2+ T cells.
To further test the role of BCR-mediated antigen uptake in this setting, groups of HEL-Ig BCR transgenic IgHelMD4 mice were injected with OVA-HEL in CpG. At different time intervals, B cells from those mice were isolated and injected into B6 mice harboring memory OT-II T cells. Eight-ten days later cells from those mice were tested for responsiveness in vitro. We found that B cells harvested from IgHelMD4 mice on days 41, 44, and 48 after injecting HEL-OVA caused reduced proliferation and IL-2 production in memory CD4+ T cells. As compared to B cells transferred on days 16 and 20 after immunization (Fig 3A), B cells with HEL specificity were more efficient in antigen capture and presentation (Fig. 3B-D). The response levels on day 4 post transfer were particularly low, likely because of antigen induced cell death since it is expected that at day 4 post immunization B cells would carry large amounts of antigen.
During the fall of antigenic load, B cells induce hyporesponsive memory CD4+ T cells in vivo
Vaccinia model
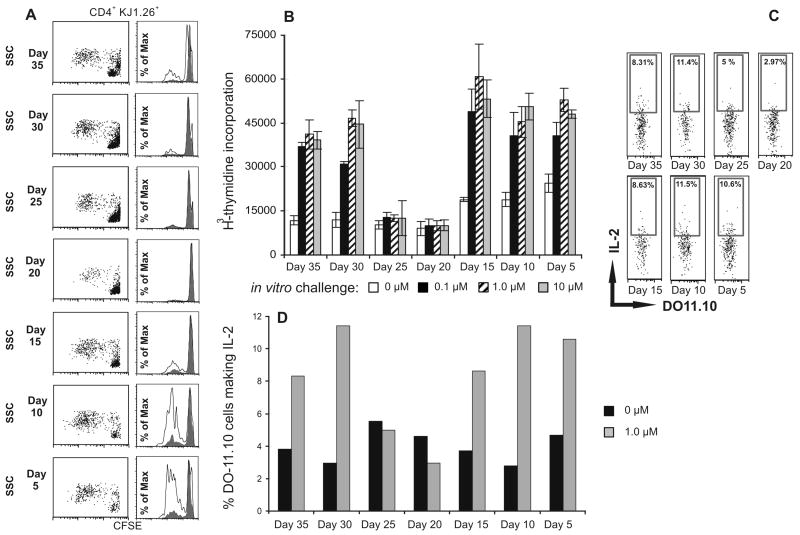
To test the role of B cells in a viral infection, BALB/c mice were infected with Vaccinia virus expressing OVA that stops replicating within 48-72 h post infection. Mice were divided into 7 groups of 3 and B cells were isolated from each group at 5-day intervals. B cells were transferred to recipient mice bearing DO11.10 memory T cells that were generated by immunization with cOVA323-339 peptide in CFA 4 months prior to the B cell transfer. Ten-twelve days later recipient mice were tested for T cell responses in vitro. We found that T cells from mice that had received B cells harvested on days 20 or 25 post Vaccinia infection responded poorly, whereas B cells harvested before day 20 (days 5, 10 and 15) and after day 25 (days 30 and 35) responded normally, as evidenced by the T cell proliferation and IL-2 production (Fig. 4). These observations indicate and enforce that antigen is efficiently captured by specific B cells and presented to memory T cells.
Figure 4.
B cells during the resolution of Vaccinia infection induce hyporesponsive memory CD4+ T cells in vivo. B cells from the mice, infected with Vaccinia-OVA for different time periods (day 5 – day 35), were transferred to mice bearing memory DO11.10 (CD4+KJ1.26+) T cells (day 66 post priming of T cells). Fourteen days later, cells pooled from the draining lymph nodes were cultured in the presence of peptide for 72 h, and antigen specific CD4+ T cell proliferation was determined by CFSE dilution as shown in (A), and 3H-thymidine incorporation shown in (B). In vitro challenge with cOVA323-339 peptide (A): filled histogram, 0 μM; open histogram, 1.0 μM. In parallel, T cell response was also measured by intracellular IL-2 synthesis (C) following 5 h stimulation with peptide. Data shown represent one of two independent experiments. Bars represent mean ± standard deviation of triplicate cultures. There were five mice per group and cells from triplicate wells intended for cytokine measurement were pooled before measuring IL-2 synthesis. Analysis for IL-2 synthesis was done on gated CD4+KJ1.26+ (DO11.10) T cells. Data shown represent one of two independent experiments.
Memory CD4+ T cell becomes hyporesponsive in the absence of experimental manipulations
CpG as mimic of infection
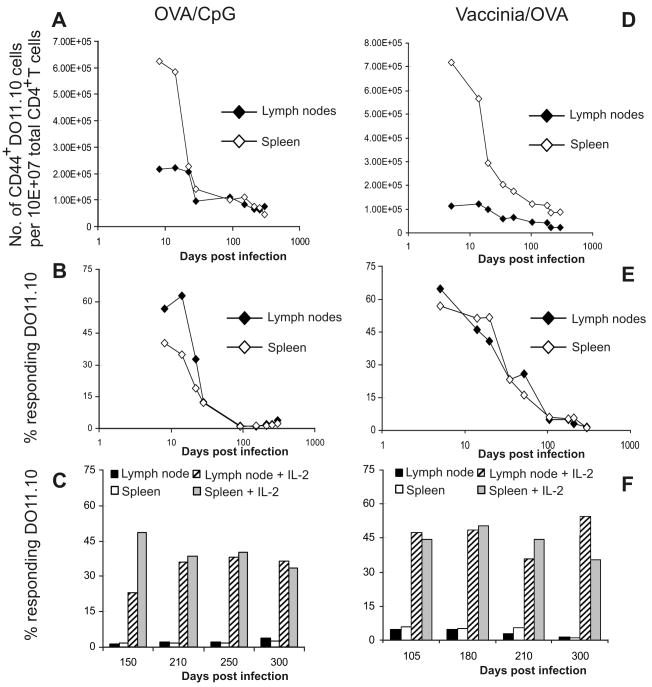
We thought if B cells from OVA or OVA-HEL immunized mice could render memory T cells unresponsive in recipient mice, they could do so in memory T cells developed in the same mouse. To test this idea, BALB/c mice were transferred with DO11.10 transgenic T cells and immunized with OVA protein mixed with CpG in PBS. On indicated days (Fig. 5A), cells from lymph nodes and spleens were harvested and stained for KJ1.26, CD4, CD44, CD25, CD45RB, and CD62L. We considered as memory cells those cells that stained high for CD44 and CD62L and low for CD25, CD45RB, and CD69. The numbers of KJ1.26+ CD4+ cells that were CD44hi per 107 CD4+ cells were plotted against time (Fig. 5). The number of cells staining for other markers was appropriately in accord with the level of staining expected for memory T cell. For example, when we plotted the number of cells staining low for CD69 per total CD4+ cells over time, the trend followed that of CD44hi, as shown in Fig. 5A. Antigen specific memory CD4+ T cells were counted and T cell responses to in vitro antigen challenge were measured. We found that CD4+ CD44hi memory T cells gradually declined in numbers over time in both spleen and lymph nodes, although greater decline was observed in the spleens compared to that in the lymph nodes (Fig. 5A). The percent of DO11.10 (KJ1.26+, CD4+ T) cells from lymph nodes or spleens that divided in response to cOVA323-339 -peptide challenge in vitro was plotted against time (Fig. 5B) and showed similar trend as the drop in the cell numbers shown in Fig. 5A. Thus, memory DO11.10 T cells lost their ability to respond to antigen gradually within 5 to 13 weeks post immunization. However, upon providing exogenous IL-2 along with antigen ex vivo, T cells became activated (Fig. 5C), or CpG plus antigen administered in vivo restored responses (Supplementary Fig. 1). These experiments suggest that majority of OVA-CpG induced memory CD4+ T cells become unresponsive to antigen by 3 months post infection naturally.
Figure 5.
Memory CD4+ T cell becomes hyporesponsive naturally during the resolution of Vaccinia infection or reduction of the immunizing antigen. Eighteen hours following adoptive transfer of DO11.10 cells (2.0 × 105/mouse) BALB/c mice were immunized with OVA and CpG in PBS (A-C), or were infected i.p. with Vaccinia virus expressing OVA (5 × 106 pfu) (D-F), and monitored for over 300 days. On indicated days post immunization, cells from spleens and the draining lymph nodes were harvested, and stained for CD4, KJ1.26, and memory markers CD44, CD25, CD45RB, CD62L, and CD69. In A and D, total numbers of CD4+, KJ1.26+ cells/107 counted cells, which also stained CD44hi, are plotted against days post immunization. Cells that stained high for CD44 also stained high for CD62L, but stained low for CD45RB, CD69, and CD25, all consistent with memory T cell markers. Similarly, cells from the spleens of those mice showed the same trends (data not shown). Proliferation of DO11.10 T cells was determined by CFSE dilution assay following cOVA323-339 peptide challenge for 72 h in vitro. Percent of DO11.10 T cells that proliferated to the in vitro peptide stimulation in the absence of exogenous IL-2 (B and E), or in the presence of exogenous IL-2 (C and F) was plotted against days post immunization. Data shown represent one of two independent experiments. For each time point three to five mice were tested.
Memory CD4+ T cell become hyporesponsive naturally
Vaccinia virus as a model infection
To verify reproducibility of the above phenomenon with a model virus, similar experiments as in Fig. 5A-C were designed with the difference that mice were infected with 5 × 106 pfu Vaccinia virus expressing OVA36. As described above, on indicated days, cells from lymph nodes and spleens were harvested and stained for KJ1.26, CD4, CD44, CD25, CD45RB, and CD62L. KJ1.26 and CD4 double positive cells that were CD44hi were considered memory T cells. We observed that again memory CD4+ T cells developed upon Vaccinia virus infection declined gradually (Fig. 5D) and lost ability to proliferate to antigen starting 8 weeks post-infection (Fig. 5E). After providing exogenous IL-2 along with OVA323-339 to the culture, T cells regained their ability to respond to antigen (Fig. 5F). Interestingly, unresponsiveness to antigen in OVA323-339 specific memory CD4+ T cells in Vaccinia infection developed slower than in CpG immunization, possibly suggesting a longer persistence of antigen due to Vaccinia infection versus CpG immunization.
Quiescent memory CD4+ T cells respond during re-infection
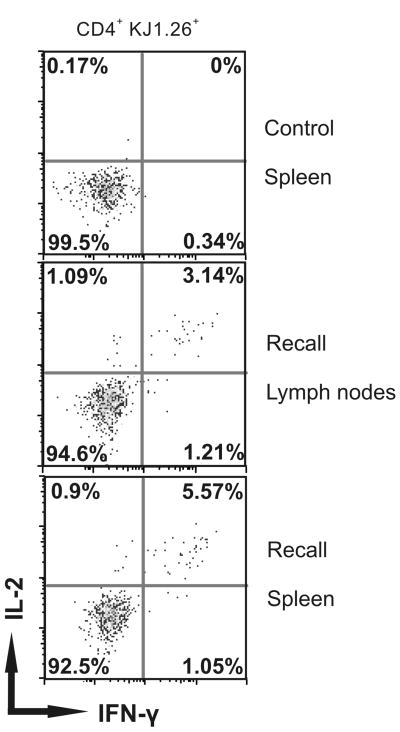
To demonstrate that unresponsive CD4+ memory T cells developed in mice infected originally with Vaccinia virus are functional upon a second encounter with the same infection, we challenged the mice with a second infection with Vaccinia-OVA 405 days (13.5 months) after the primary infection. Seven days later, mice were sacrificed, spleens and lymph nodes were removed, and the ability of DO11.10 T cells to make IL-2 and IFN-γ in response to 4 hours of cOVA323-339 peptide stimulation in vitro was measured. We found that nearly all of responding memory DO11.10 cells were polyfunctional, making both IL-2 and IFN-γ (5.57% of 6% total CD4+ cells making either IL-2 or IFN-γ (Fig. 6). In contrast, in mock-infected mice only 0.17% or 0.34 % of DO11.10 CD4+ cells made either IL-2 or IFN-γ respectively.
Figure 6.
Challenge with Vaccinia virus recalls cOVA323-339 specific responses in quiescent memory CD4+ T cells. Immediately following adoptive transfer of DO11.10 cells (2.0 × 105/mouse) BALB/c mice were infected i.p. with Vaccinia virus expressing OVA (5 × 106 pfu/mouse), and 13.5 months later mice were challenged with another inoculation of the same virus (5 × 106 pfu/mouse) i.p. Cells from lymph nodes and spleens were harvested, re-challenged in vitro with cOVA323-339, and T cell responses were measured by intracellular IL-2 and IFN-γ synthesis on CD4+ KJ1.26+ (DO11.10) gated cells. Control group represents mice without recall virus injection, while recall groups represent mice with a viral challenge 7 days earlier. Data shown represent one of two independent experiments. In each experiment three to five mice per group were tested.
Gene expression profile in long-term memory CD4+ T cells
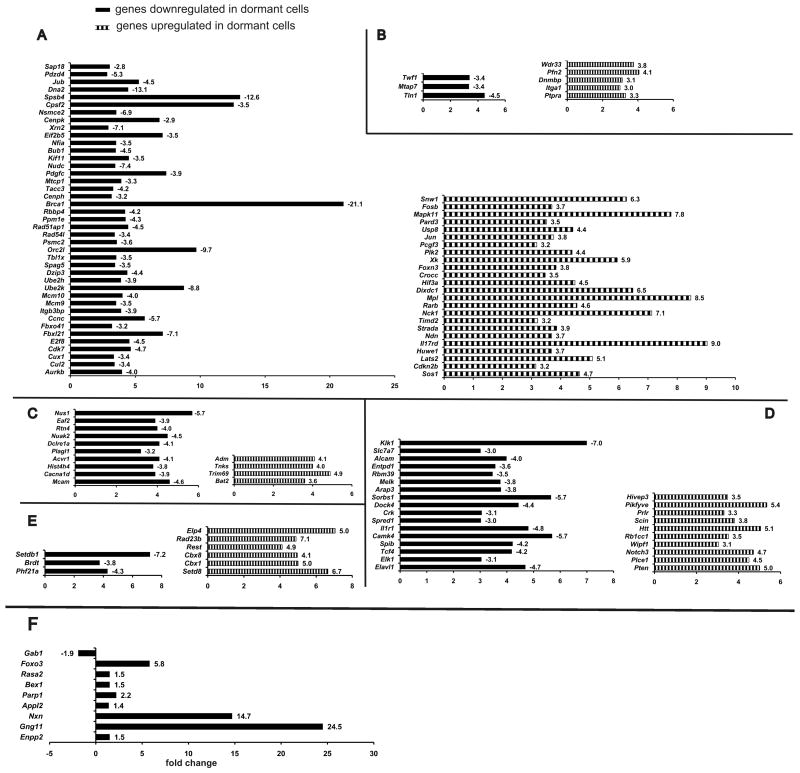
We performed gene chip analyses on memory CD4+ T cells isolated and FACS sorted from two groups of mice immunized with OVA-CpG, (hyporesponsive) or antigen-responsive memory T cells immunized with OVA323-339 emulsified in CFA (Fig. 7) nearly a year earlier. Differentially expressed genes were uploaded to and analyzed by DAVID database software. Under very stringent settings for data analysis gene clusters were assigned enrichment score by DAVID database. The most highly enriched clusters for genes downregulated in CpG immunized group appeared to contain genes responsible for cellular metabolism and regulation, cell cycle, and mitosis. The upregulated genes in the same cells belong to the clusters of intracellular organelles and regulation of cellular processes. Further evaluation of individual genes in those clusters revealed presence of specific groups of genes that were closely related functionally. Figure 7 shows the breakdown of gene clusters into proliferation and cell cycle (Fig. 7A), cytoskeletal rearrangement (Fig. 7B), apoptosis and cell survival (Fig. 7C), immune activation and suppression (Fig. 7D), and chromatin remodeling (Fig. 7E). Genes associated with chromatin remodeling were downregulated whereas genes belonging to the same cluster but with opposite known functions were upregulated in those cells. In memory T cells immunized with OVA-CpG, genes known to have a specific effect of preventing cells to progress from G1 to S phase were upregulated. Genes associated with promoting cellular cytoskeleton organization (including actin polymerization) were downregulated, whereas genes with the opposing function of preventing actin polymerization and cytoskeleton organization were upregulated. While apoptosis inducing genes were downregulated, cell-survival inducing genes were upregulated in hyporesponsive memory T cells. In light of the significance of regulation of the survival genes to the longevity of the memory T cells, we performed RT-PCR to re-assess the expression of some of the pro-survival genes. The results shown in Fig. 7F mostly confirmed the microarray findings indicating several genes that can cause cellular survival through synergistic functions. For example, Foxo3 and Nxn (nucleoredoxin, a member of a thioredoxin family) genes are known to become activated in oxidative stress and their specific actions together with β-catenin in repair of the oxidative damage in arrested cells and stopping apoptosis has been reported37-40. Gab1 is reported as the integrator of cell death versus cell survival signals in oxidative stress41,42; upregulated Parp1 gene has a protecting function from DNA damage by peroxides43. A high degree of upregulation of G-protein γ subunit Gng11 gene was observed, and although very little is known about this gene, it is reported to be a senescence-induced gene that directly responds to oxidative stress44. In the CpG-OVA immunized hyporesponsive group angiogenic factor adrenomedullin (ADM) was significantly upregulated although was not tested by RT-PCR. ADM has been reported to possibly rescue malignant cells from hypoxic cell death 45. A known marker of oxidative stress Nuak2 gene and its interaction partner Acvr146 were both upregulated in hyporesponsive group. The anti-apoptotic gene Bcl-2, however, showed only a slight 1.26-folds change in expression by RT-PCR. Pro-apoptotic genes, Eaf247 and Rtn4/Nogo-B48 and the gene for its receptor Nus149 were downregulated in hyporesponsive group. Apoptosis inducer Plagl150 was downregulated in hyporesponsive CD4 cells. Terf2 and Tnks, genes responsible for telomere protection and repair, were found to be upregulated in hyporesponsive OVA-CpG immunized mice51, 52. Taken together, these data are consistent with a picture of dormant CD4+ T cells accumulating peroxides during their long survival, yet because of the activation of genes that protect them from the peroxide damage, continue to survive.
Figure 7.
Gene expression and RT-PCR validation in quiescent memory CD4+ T cells by cRNA gene chip microarray. CD4+ KJ1.26+ T cells were extracted from spleens of 5 mice from each group (proliferation data shown in Supplementary Fig. 2) were pooled together, flow-sorted and their mRNA was isolated for determination of their global gene expression by cRNA microarray. Raw Affymetrix gene microarray expression data were statistically processed as described in Methods Section and filtered by signal strength. The resulting lists of up- and downregulated genes and functional clustering analysis performed by DAVID database software. (A) Proliferation and cell cycle, (B) Cytoskeletal rearrangement, (C) Apoptosis and cell survival, (D) Immune activation and suppression, (E) Chromatin remodeling. Line bars represent changes in gene expression in long-term memory CD4+ T cells, obtained by cell sorting from mice in dormant (CpG) group vs. non-dormant (CFA) group. F. Values for mRNA expression are normalized to four housekeeping genes. Fold differences in mRNA expression are shown as bars. mRNA expression pattern confirms the notion of oxidative stress in long-lived dormant memory CD4+ T cells and activation of protective mechanisms.
Homeostatic proliferation of long-lived CD4 memory T cells
The gene analyses experiments above suggested that the memory CD4 T cells have low metabolic rates and cease to become activated and/or proliferate, consistent with a resting state. To verify if the long-lived CD4 memory T cells underwent homeostatic proliferation, we used two groups of mice that had been immunized with Vaccinia OVA, or OVA in CFA 14 months, or 10 months prior to sacrifice, respectively. Mice were given BrdU in drinking water for 7 days. Spleens and lymph nodes were harvested and examined for surface markers and proliferation directly after harvest without further stimulation in culture. The analysis of surface marker expression in DO11.10 CD4 T cells in both groups showed that CD44+CD62L+ population was very small in the lymph nodes of OVA/CFA injected mice, while in Vaccinia-OVA injected mice those cells constituted more than half of the DO11.10 CD4 T cells, and more than half of them showed low uptake of BrdU, indicating that those cells were not proliferating. Additionally, expression of CD127 was noticeably higher in CD44+ cells from Vaccinia-OVA injected group. Comparison of the memory marker expression in CD4 DO11.10 cells in both groups showed noticeable difference; with over half of CD4 DO11.10 cells in Vaccinia-OVA injected group clearly showing predominant memory phenotype, CD44hiCD127hiCD62Lhi, as compared to only about 10% in OVA/CFA injected DO11.10 CD4 T cells. With respect to BrdU uptake, CD44hiCD127hiCD62Lhi cells from both groups picked up BrdU similarly as well. However, in OVA/CFA injected group over 72% of CD4 DO11.10 cells were CD44lo and proliferated actively at nearly 100%, as seen by high BrdU uptake. Interestingly, majority of long-lived memory CD4 T cells did not undergo homeostatic proliferation despite high expression levels of CD127.
Discussion
We here show that memory CD4+ T cells undergo a state of antigen unresponsiveness during the fall of antigen. We demonstrate that when antigen reaches to certain low levels, B cells capture antigen via their antigen receptors and induce unresponsiveness in CD4+ T cells. We provide evidence that long-lived quiescent memory T cells become activated upon re-infection with the virus or an in vivo challenge with antigen and a TLR-9 ligand, CpG. We suggest that this might be a mechanism adopted by memory CD4+ T cells for long-term survival in the absence of antigen.
We have previously shown that clonal CD4+ T cells become hyporesponsive by presentation of low levels of antigen31-33, 53. Furthermore, we have shown that different forms of peptide-MHC complexes including short-lived peptide–MHC and low densities of long-lived agonist peptides induce T cell unresponsiveness through engagement of ∼1000 TCR, as opposed to T cell activation that requires the engagement of over 4000 TCR31, 32, 53. Interestingly, only memory CD4+ T cells, and not activated or naïve CD4+ T cells were susceptible to become hyporesponsive upon encounter with low densities of peptide-MHC ligand. These observations led us to suggest that all forms of ligands that engage less than optimal numbers of TCR induce anergy or hyporesponsiveness in T cell clones or CD4+ memory T cells54. As such, the quantity of antigen or the density of peptide-MHC ligand presented to memory T cells can be regarded as a signal to activate or tolerize. In this study, one can associate high density of ligand with the presence of infection and low density of ligand with the resolution of infection.
Our original studies showing that low densities of agonist peptide induce anergy in CD4 memory T cells were established in HLA-DR1 transgenic mouse populated with heterogeneous CD4+ T cells as identified by HA306-318/DR1 tetramer staining31. A parallel comparison with TCR transgenic T cells adoptively transferred to recipient mice confirmed similar memory T cell sensitivity and responsiveness to the tolerogenic peptide treatments, giving us confidence that our experiments presented here are representative of the memory T cells in polyclonal systems55-58. Furthermore, the experiments presented here are performed with only 2.0×105 Tg T cells transferred into the recipient mice, which is five times lower than the original system34.
Here we provide strong evidence that the resolution of an infection as characterized by availability of low levels of antigen, would also associate with CD4+ memory T cells that are hyporesponsive. Indeed, our experiments provide direct evidence that B cells bearing specific antigen receptors are the APC that perform this task effectively. Using CpG or Vaccinia viral infection as models to study the immune response, we observed that purified B cells from the infected or immunized mice could induce hyporesponsive memory T cells. Those experiments showed that antigen capture was indeed 1,000-10,000 folds more efficient when OVA antigen was targeted to HEL-specific BCR transgenic B cells, providing a direct evidence for the role of BCR in capturing of antigen at very levels (10-5-10-8 pmol), making it highly unlikely for any other APC to capture the antigen.
Since transferring B cells from infected mice generated hyporesponsive memory CD4+ T cells in the recipient mice, it was possible that this state of rest could develop spontaneously in memory CD4+ T cells in mice that had recovered from infection. Indeed, we observed that memory T cells began to become hyporesponsive after the contraction phase in the absence of any external interference. When either Vaccinia-OVA, or OVA/CpG were used as mimics of infection, OVA specific CD4+ T cells contracted in numbers and became unresponsive to antigen once the effector phase ended. Several studies have already demonstrated that CD4+ T cells increase in number during the effector phase and decline over time after gaining memory characteristics following the resolution of infections, consistent with our observation11, 15, 59. Our findings are also in agreement with the reports that memory T cells adopt a resting state59 because of programmed metabolic switches that control glycolysis and/or fatty acid oxidation60-62. While those studies indicated that memory T cells are in resting state, we have demonstrated here that CD4+ memory T cells undergo a resting state initiated by B cells and triggered by certain low levels of antigen during the resolution of infection. It is noteworthy that low level of antigen presented by B cells is concurrent with low-level expression of danger signals as well. This resting state is a transient condition and may be reversed by antigen and IL-2, a condition that is met during the re-emergence of an infection due to inflammatory conditions. We demonstrate that a second viral infection even after nearly 14 months post infection is stimulatory to the memory populations that are otherwise fully unresponsive to a peptide alone challenge in vitro. Similarly, IL-2 plus peptide, or CpG plus antigen administered in vivo recalled vigorous responsiveness in quiescent memory CD4+ T cells.
Our microarray data, that was analyzed by the most stringent parameters set for gene clustering suggested that many genes from our final gene list appeared to have immediate interactions with each other. Quite remarkably, however, a number of differentially expressed genes that were excluded from the list, because of unclear immediate relevance to our data set, merged with the interactions in the gene map as functionally related to the genes of known functional importance. Altogether, differential expression of a significant number of genes that were connected to each other directly or through controlling “nodes” in our map support our findings that long-lived memory CD4+ T cells are dormant. These points strongly argue for the non-random nature of our microarray data.
Microarray data suggest that quiescent memory CD4+ T cells significantly reduce the expression of genes that induce cell proliferation and immune activation, while increasing the expression of genes that can protect cells from apoptosis and promote survival. The changes in profiles of genes belonging to the cluster of cytoskeletal rearrangement suggest that actin polymerization is prevented in rested memory T cells, confirming that the cells are not in activated but in quiescent state. The switch in gene expression corresponding to a state of quiescence may coincide with the disappearance of inflammation and depletion of circulating antigen, which appears to begin about 8 weeks from the onset of the viral entry in the infection models presented here. Recent reports indicate that memory T cells downregulate their activation genes and lower their metabolic activities59, 61, 62. Interestingly, our characterization of long-lived memory T cells after over a year post immunization, pointed to a remarkable observation. In Vaccinia-OVA injected mice, over half of CD4 DO11.10 cells showed high expression of memory markers, such as CD44, CD127, and CD62L from which more than half did not undergo homeostatic proliferation. In contrast, only about 10% of CD4 DO11.10 T cells from OVA emulsified in CFA injected mice showed high expression of CD44, CD62L and CD127. Majority of CD4 DO11.10 cells in the latter group expressed intermediate levels of CD127 and low levels of CD44 and proliferated well. If CD44 is to be considered a more representative memory marker, one can distinguish Vaccinia-OVA immunization as a better inducer of long-lived memory CD4 T cells as compared to OVA/CFA. Because of the 9 months time past OVA/CFA immunization, yet persistence of a clear population of KJ1.26 positive cells, one might propose that a new undefined population of memory CD4 cells that divide rapidly, yet are CD44- might have developed. Those findings are in agreement with the microarray data, indicating that injection of mice with Vaccinia-OVA led to fully developed CD4 memory T cells, whereas continuous release of antigen over a long period of time as expected in OVA-CFA63 did not efficiently induce long-lasting CD4 memory T cells. We like to suggest that OVA emulsified in CFA continues to be released and in a way mimics a chronic infection, whereas, Vaccinia-OVA infection clears in few days and leads to the generation of memory CD4 T cells that become fully rested and have their anti-stress genes turned on while being completely dormant. All these new findings document that perhaps dormant memory T cells survive longer and might be less harmful to self-tissues due to cross-reactivity.
Our findings highlight an important physiological process that takes place at the end of an infection, when, i) the antigenic load is reduced, and ii) memory T cells have developed and need to receive an inhibitory signal to cease proliferation and release of cytokines. Under such conditions, B cells bearing specific receptors for antigens are the natural choice for the immune system for capturing antigen at its lowest level and present it to the memory T cells for promoting a resting state. When the need arises to fight infection during challenge, the quiescent memory T cells get activated and exert their effector function. In all, our studies put forward a novel regulatory mechanism for CD4 memory T cells. Despite the general view that memory T cells are readily activated, our data reveal strict regulation on memory CD4+ T cells. First, lack of cell proliferation and reversibility by inflammatory cytokines fulfill the original definition of T cell clonal anergy64. Anergized memory T cells maintain low metabolic activity and cell cycle progression, criteria that would preserve cellular energy and might be the key mechanism in long-term survival. However, the critical requirement of anergized memory T cells for inflammatory cytokines for reactivation is another control mechanism of responding to danger65. Thus, memory T cells, while equipped to respond to antigens rapidly, also require second signals for the initiation of response, similar to naïve T cells. The need for an inflammation induced danger signal for the activation of memory T cells prevents them from self-reactivity.
Methods
Mice
All mice were purchased from the Jackson Laboratory and included five- to six-weeks old BALB/c, TCR Transgenic (Tg) mice (DO11.10 or OT-II) that express α/β T cell receptor recognizing chicken OVA323-339 in complex with I-Ad- or I-Ab in Balb/c or C57Bl/6 accordingly, and B cell receptor (IgHelMD4) transgenic mice on B6 background. IgHelMD4 mice were bred with non-transgenic B6 mice and the offspring heterozygous for BCR were used for the study. All mice were housed in the Johns Hopkins University animal facilities under virus-free conditions. All experiments were performed in accordance with protocols approved by the Animal Care and Use Committee of the Johns Hopkins University School of Medicine.
Protein, Peptides and Antibodies
Chicken OVA protein (Grade VI, [Sigma-Aldrich], OVA323-339 peptide (ISQAVHAAHAEINEAGR) was synthesized by Global-Peptides] at >90% purity. Fluorescently labeled antibodies to mouse CD4, CD25, CD44, CD45RB, CD62L, CD69, CD45R (B220), CD19, CD11c, IL-2, IFN-γ, antibody to OT-II TCR (Vα2 and Vβ5) were from [BD-Pharmingen]; antibody to clonotypic TCR (KJ1.26) specific for DO11.10 CD4 T cells was from Caltag Laboratories; MACs CD19 microbeads used for B cell purification were from Miltenyi Biotec.
Generation of memory CD4+ T cells
cOVA323-339 peptide specific transgenic CD4+ T cells (2.0 ×105 /cells per mouse in 100 μl of sterile PBS), prepared from pooled lymph nodes and spleens of DO11.10 or OT-II transgenic mice, were transferred i.v. into BALB/c or B6 recipients. Next day, mice were immunized subcutaneously with 15 nmol cOVA323-339 emulsified in CFA. For some experiments, mice were immunized subcutaneously with OVA protein (50 μg/mouse) and CpG (50 μg/mouse) in PBS, or were infected with 5×106 pfu Vaccinia-OVA/mouse intraperitoneally.
Induction of hyporesponsiveness in memory CD4+ T cells
Five to twelve week post-immunization with peptide/CFA, mice were injected subcutaneously with different concentrations of chicken OVA protein mixed with IFA to generate memory T cells. To test the role of BCR-mediated antigen presentation in anergy induction, IgHelMD4 Tg B cells were transferred to mice (3 × 106 B cells/mouse) bearing memory CD4+ T cells before injecting those mice with HEL-coupled OVA34 or OVA emulsified IFA. To test the role of B cells in anergy induction during the fall of antigen following resolution of infection, B cells from naive mice injected with OVA/CpG (in PBS) or infected with Vaccinia-OVA for different length of time were transferred to recipient mice (15 × 106 B cells/mouse) bearing memory T cells. Ten to fourteen days later, T cell responses in draining lymph nodes were measured in vitro to cOVA323-339 challenge by cell proliferation and intracellular IL-2 synthesis33.
Natural induction of hyporesponsiveness in memory CD4+ T cells: CpG or Vaccinia Virus as model infections
On indicated days following generation of memory DO11.10 cells by OVA/CpG immunization or Vaccinia-OVA infection, T cells from draining lymph nodes and spleens were stained for memory markers, or challenged in vitro with peptide. At some time points, T cell responses were measured after adding IL-2 to the culture.
Recall of quiescent memory CD4+ T cells during challenge
Fourteen months after generating memory T cells by Vaccinia-OVA infection, mice were challenged with the same virus (5 × 106 pfu/mouse). On day 7, cells from the draining lymph nodes and spleens were harvested, re-challenged in vitro with cOVA323-339, and T cell response to peptide was measured by intracellular IL-2 and IFN-γ synthesis.
GeneChip hybridization, microarray, and data analyses
Following 11 months post-immunization with OVA/CpG (for anergic T cells) or 9 months post-immunization with peptide/CFA (for non-anergic T cells), antigen specific (CD4+KJ1.26+) T cells were FACS sorted (FACSAria, Becton Dickinson) and total RNA was isolated by Qiagen RNAeasy kit. Hybridization of cRNA was done using Affymetrix mouse genearray chip (4302.0) containing 45,000 probes (for 35,000 genes) at the Johns Hopkins University School of Medicine Microarray Core Facility. Quality of the microarray samples was assessed with affyPLM and Affy. Robust Multiarray Analysis (RMA) expression measures66 were obtained as the gene expression signals with affyPLM. This probe level data processing includes a normalization procedure utilizing the quantile normalization method (Bolstad, 2003) to reduce the obscuring variation between microarrays, which might be introduced during the processes of sample preparation, manufacture, fluorescence labeling, hybridization and/or scanning.
Genes with signals in both samples ≤ 60 were filtered out prior to the downstream differential gene expression analysis. The Lognormal modeling of the bioconductor package EBarrays (www.bioconductor.org) was used to estimate the posterior probabilities of the differential expression of genes between the sample conditions67, 68. The control of 5% false discovery rate was taken to produce the differentially expressed gene lists. All computations were performed under R environment (http://www.r-project.org).
The lists of differentially expressed genes were submitted to The Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/summary.jsp)69, 70, and Functional Annotation Tool was used to calculate functional clustering of genes, separately for up- and downregulated groups of genes. Parameters for calculation were set as follows: Similarity Gene Overlap - the minimum number of annotation terms overlapped between two genes in order to be qualified for kappa calculation. The higher the value, the more meaningful are the results. This parameter was set to the highest possible value of 10. Similarity threshold - the minimum kappa values are considered biologically significant. The higher the setting, the higher is the quality of functional classification. Similarity Threshold was set to 0.85 (default value – 0.35). Multi-linkage Threshold - controls how seeding groups merge each other. The higher percentage gives sharper separation, i.e. more final functional groups with more tightly associated genes. Similarity Threshold was set to 0.85 (default value – 0.5). Output of the results from DAVID was obtained in the form of the annotated list of clustered groups of genes, assigned an enrichment score by Functional Annotation Tool. Pathway Commons (http://www.pathwaycommons.org) and Cytoscape (version 2.6.3, http://www.cytoscape.org)71 were used as tools to extract interaction information for genes, included in the final list of genes that we found to play significant roles in the biological processes of interest. Genes in this final list were thoroughly examined for their interactions with each other and with other genes, again using Cytoscape and Pathway Commons, by building the gene interaction map. Many genes from our final gene list appeared to have immediate interactions with each other. A detailed analysis of interactions for the remaining genes in our final list revealed a high degree of occurrence of particular genes in their interaction networks and placing those genes (“nodes”) in our gene interaction map provided connectivity with the genes already in the map.
RT-PCR
To perform RT-PCR, mRNA samples from CD4+ KJ1.26+ T cells were used to synthesize cDNA and RT2 qPCR Primer Assays (SABiosciences) were used to perform RT-PCR on ABI 7300 Real Time PCR system (Applied Biosciences). The comparison analysis was performed using Delta-Delta CT method72. The formula is 2ˆ-(delta-deltaCt+/-SD), where delta-deltaCt = (Ct _CpG - Ct _CFA) - (Ct _CpG Housekeeping Gene - Ct _CFA Housekeeping Gene), and CpG is the dormant cell group and CFA is the non-dormant cell group. For each housekeeping gene, Ct _CpG Housekeeping Gene - Ct _CFA Housekeeping Gene, was calculated and then average was calculated using the difference. The average was then used in the above formula in place of (Ct _CpG Housekeeping Gene - Ct _CFA Housekeeping Gene). The housekeeping genes used for normalization are beta-2 microglobulin (B2m), lactate dehydrogenase A-like 6B (Ldhal6b), TATA box binding protein (Tbp), transferrin receptor (Tfrc).
Homeostatic proliferation experiments
Two groups of BALB/c mice were adoptively transferred with 200,000 CD4 DO11.10 cells i.v into the tail vein, and immunized with either Vaccinia-OVA virus in PBS i.p., or cOVA323-339 in CFA subcutaneously at the base of tail. Ten months later for OVA/CFA group and 14 months later for Vaccinia-OVA group, mice were fed with BrdU in drinking water (0.8 mg/ml concentration), sacrificed and their draining inguinal lymph nodes extracted and pooled together for each group of mice. A multi-color flow cytometry analysis of cells from the lymph nodes was performed on LSR II (Becton Dickinson) instrument. Antibodies used were CD4-Pacific Blue, KJ1.26-PE, CD44-eFluor 605, CD62L-PerCp, CD127-FITC, CCR7-Alexa Fluor 700 and BrdU-APC.
Supplementary Material
Acknowledgments
We thank Drs. Drew Pardoll and Abdel Hamad for reading the manuscript and discussion, Phillipa Marrack, David Woodland, and Donna Farber for the discussion. This work was supported by R01GM53549 and R01AI063764 grants to SS-N.
Footnotes
A portion of this work was presented at the AAI meeting held at Miami, FL during May 18-22, 2007.
References
- 1.Bourgeois C, Rocha B, Tanchot C. A role for CD40 expression on CD8+ T cells in the generation of CD8+ T cell memory. Science. 2002;297:2060–2063. doi: 10.1126/science.1072615. [DOI] [PubMed] [Google Scholar]
- 2.Seder RA, Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat Immunol. 2003;4:835–842. doi: 10.1038/ni969. [DOI] [PubMed] [Google Scholar]
- 3.Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 4.Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–342. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 6.Antia R, Ganusov VV, Ahmed R. The role of models in understanding CD8+ T-cell memory. Nat Rev Immunol. 2005;5:101–111. doi: 10.1038/nri1550. [DOI] [PubMed] [Google Scholar]
- 7.Stemberger C, Huster KM, Koffler M, Anderl F, Schiemann M, Wagner H, et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27:985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Teixeiro E, Daniels MA, Hamilton SE, Schrum AG, Bragado R, Jameson SC, et al. Different T cell receptor signals determine CD8+ memory versus effector development. Science. 2009;323:502–505. doi: 10.1126/science.1163612. [DOI] [PubMed] [Google Scholar]
- 9.Bannard O, Kraman M, Fearon DT. Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science. 2009;323:505–509. doi: 10.1126/science.1166831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 11.Homann D, Teyton L, Oldstone MB. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nat Med. 2001;7:913–919. doi: 10.1038/90950. [DOI] [PubMed] [Google Scholar]
- 12.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson JM, MacLeod M, Marsden VS, Kappler JW, Marrack P. Not all CD4+ memory T cells are long lived. Immunol Rev. 2006;211:49–57. doi: 10.1111/j.0105-2896.2006.00383.x. [DOI] [PubMed] [Google Scholar]
- 14.Hammarlund E, Lewis MW, Hansen SG, Strelow LI, Nelson JA, Sexton GJ, et al. Duration of antiviral immunity after smallpox vaccination. Nat Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 15.Pepper M, Linehan JL, Pagan AJ, Zell T, Dileepan T, Cleary PP, et al. Different routes of bacterial infection induce long-lived TH1 memory cells and short-lived TH17 cells. Nat Immunol. 2010;11:83–89. doi: 10.1038/ni.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purton JF, Tan JT, Rubinstein MP, Kim DM, Sprent J, Surh CD. Antiviral CD4+ memory T cells are IL-15 dependent. J Exp Med. 2007;204:951–961. doi: 10.1084/jem.20061805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Leeuwen EM, Sprent J, Surh CD. Generation and maintenance of memory CD4(+) T Cells. Curr Opin Immunol. 2009 doi: 10.1016/j.coi.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macleod MK, Clambey ET, Kappler JW, Marrack P. CD4 memory T cells: What are they and what can they do? Semin Immunol. 2009 doi: 10.1016/j.smim.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachmann MF, Gallimore A, Linkert S, Cerundolo V, Lanzavecchia A, Kopf M, et al. Developmental regulation of Lck targeting to the CD8 coreceptor controls signaling in naive and memory T cells. J Exp Med. 1999;189:1521–1530. doi: 10.1084/jem.189.10.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crowe SR, Turner SJ, Miller SC, Roberts AD, Rappolo RA, Doherty PC, et al. Differential antigen presentation regulates the changing patterns of CD8+ T cell immunodominance in primary and secondary influenza virus infections. J Exp Med. 2003;198:399–410. doi: 10.1084/jem.20022151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Lin GH, McPherson AJ, Watts TH. Immune regulation by 4-1BB and 4-1BBL: complexities and challenges. Immunol Rev. 2009;229:192–215. doi: 10.1111/j.1600-065X.2009.00765.x. [DOI] [PubMed] [Google Scholar]
- 22.Zammit DJ, Cauley LS, Pham QM, Lefrancois L. Dendritic cells maximize the memory CD8 T cell response to infection. Immunity. 2005;22:561–570. doi: 10.1016/j.immuni.2005.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marrack P, Kappler J. Control of T cell viability. Annu Rev Immunol. 2004;22:765–787. doi: 10.1146/annurev.immunol.22.012703.104554. [DOI] [PubMed] [Google Scholar]
- 24.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 25.Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 26.Hathcock KS, Kaech SM, Ahmed R, Hodes RJ. Induction of telomerase activity and maintenance of telomere length in virus-specific effector and memory CD8+ T cells. J Immunol. 2003;170:147–152. doi: 10.4049/jimmunol.170.1.147. [DOI] [PubMed] [Google Scholar]
- 27.Seddon B, Tomlinson P, Zamoyska R. Interleukin 7 and T cell receptor signals regulate homeostasis of CD4 memory cells. Nat Immunol. 2003;4:680–686. doi: 10.1038/ni946. [DOI] [PubMed] [Google Scholar]
- 28.Zaph C, Uzonna J, Beverley SM, Scott P. Central memory T cells mediate long-term immunity to Leishmania major in the absence of persistent parasites. Nat Med. 2004;10:1104–1110. doi: 10.1038/nm1108. [DOI] [PubMed] [Google Scholar]
- 29.Wu CY, Kirman JR, Rotte MJ, Davey DF, Perfetto SP, Rhee EG, et al. Distinct lineages of T(H)1 cells have differential capacities for memory cell generation in vivo. Nat Immunol. 2002;3:852–858. doi: 10.1038/ni832. [DOI] [PubMed] [Google Scholar]
- 30.Rosloniec EF, Brand DD, Myers LK, Whittington KB, Gumanovskaya M, Zaller DM, et al. An HLA-DR1 transgene confers susceptibility to collagen-induced arthritis elicited with human type II collagen. J Exp Med. 1997;185:1113–1122. doi: 10.1084/jem.185.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirshahidi S, Huang CT, Sadegh-Nasseri S. Anergy in peripheral memory CD4(+) T cells induced by low avidity engagement of T cell receptor. J Exp Med. 2001;194:719–731. doi: 10.1084/jem.194.6.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirshahidi S, Ferris LC, Sadegh-Nasseri S. The magnitude of TCR engagement is a critical predictor of T cell anergy or activation. J Immunol. 2004;172:5346–5355. doi: 10.4049/jimmunol.172.9.5346. [DOI] [PubMed] [Google Scholar]
- 33.Dalai SK, Mirshahidi S, Morrot A, Zavala F, Sadegh-Nasseri S. Anergy in memory CD4+ T cells is induced by B cells. J Immunol. 2008;181:3221–3231. doi: 10.4049/jimmunol.181.5.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garside P, Ingulli E, Merica RR, Johnson JG, Noelle RJ, Jenkins MK. Visualization of specific B and T lymphocyte interactions in the lymph node. Science. 1998;281:96–99. doi: 10.1126/science.281.5373.96. [DOI] [PubMed] [Google Scholar]
- 35.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 36.Buller RM, Smith GL, Cremer K, Notkins AL, Moss B. Decreased virulence of recombinant vaccinia virus expression vectors is associated with a thymidine kinase-negative phenotype. Nature. 1985;317:813–815. doi: 10.1038/317813a0. [DOI] [PubMed] [Google Scholar]
- 37.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 38.Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ, Jr, DiStefano PS, et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 39.Funato Y, Michiue T, Asashima M, Miki H. The thioredoxin-related redox-regulating protein nucleoredoxin inhibits Wnt-beta-catenin signalling through dishevelled. Nat Cell Biol. 2006;8:501–508. doi: 10.1038/ncb1405. [DOI] [PubMed] [Google Scholar]
- 40.Korswagen HC. Regulation of the Wnt/beta-catenin pathway by redox signaling. Dev Cell. 2006;10:687–688. doi: 10.1016/j.devcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 41.Ren Y, Meng S, Mei L, Zhao ZJ, Jove R, Wu J. Roles of Gab1 and SHP2 in paxillin tyrosine dephosphorylation and Src activation in response to epidermal growth factor. J Biol Chem. 2004;279:8497–8505. doi: 10.1074/jbc.M312575200. [DOI] [PubMed] [Google Scholar]
- 42.Holgado-Madruga M, Wong AJ. Gab1 is an integrator of cell death versus cell survival signals in oxidative stress. Mol Cell Biol. 2003;23:4471–4484. doi: 10.1128/MCB.23.13.4471-4484.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hossain MN, Sakemura R, Fujii M, Ayusawa D. G-protein gamma subunit GNG11 strongly regulates cellular senescence. Biochem Biophys Res Commun. 2006;351:645–650. doi: 10.1016/j.bbrc.2006.10.112. [DOI] [PubMed] [Google Scholar]
- 45.Oehler MK, Norbury C, Hague S, Rees MC, Bicknell R. Adrenomedullin inhibits hypoxic cell death by upregulation of Bcl-2 in endometrial cancer cells: a possible promotion mechanism for tumour growth. Oncogene. 2001;20:2937–2945. doi: 10.1038/sj.onc.1204422. [DOI] [PubMed] [Google Scholar]
- 46.Lefebvre DL, Rosen CF. Regulation of SNARK activity in response to cellular stresses. Biochim Biophys Acta. 2005;1724:71–85. doi: 10.1016/j.bbagen.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 47.Su F, Pascal LE, Xiao W, Wang Z. Tumor suppressor U19/EAF2 regulates thrombospondin-1 expression via p53. Oncogene. 29:421–431. doi: 10.1038/onc.2009.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi B, Qi Y, Watari A, Yoshioka N, Inoue H, Minemoto Y, et al. Pro-apoptotic ASY/Nogo-B protein associates with ASYIP. J Cell Physiol. 2003;196:312–318. doi: 10.1002/jcp.10297. [DOI] [PubMed] [Google Scholar]
- 49.Harrison KD, Miao RQ, Fernandez-Hernando C, Suarez Y, Davalos A, Sessa WC. Nogo-B receptor stabilizes Niemann-Pick type C2 protein and regulates intracellular cholesterol trafficking. Cell Metab. 2009;10:208–218. doi: 10.1016/j.cmet.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bilanges B, Varrault A, Mazumdar A, Pantaloni C, Hoffmann A, Bockaert J, et al. Alternative splicing of the imprinted candidate tumor suppressor gene ZAC regulates its antiproliferative and DNA binding activities. Oncogene. 2001;20:1246–1253. doi: 10.1038/sj.onc.1204237. [DOI] [PubMed] [Google Scholar]
- 51.Donigian JR, de Lange T. The role of the poly(ADP-ribose) polymerase tankyrase1 in telomere length control by the TRF1 component of the shelterin complex. J Biol Chem. 2007;282:22662–22667. doi: 10.1074/jbc.M702620200. [DOI] [PubMed] [Google Scholar]
- 52.Gomez M, Wu J, Schreiber V, Dunlap J, Dantzer F, Wang Y, et al. PARP1 Is a TRF2-associated poly(ADP-ribose)polymerase and protects eroded telomeres. Mol Biol Cell. 2006;17:1686–1696. doi: 10.1091/mbc.E05-07-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korb LC, Mirshahidi S, Ramyar K, Sadighi Akha AA, Sadegh-Nasseri S. Induction of T cell anergy by low numbers of agonist ligands. J Immunol. 1999;162:6401–6409. [PubMed] [Google Scholar]
- 54.Sadegh-Nasseri S, Dalai SK, Korb Ferris LC, Mirshahidi S. Suboptimal engagement of the T-cell receptor by a variety of peptide-MHC ligands triggers T-cell anergy. Immunology. 2010;129:1–7. doi: 10.1111/j.1365-2567.2009.03206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hataye J, Moon JJ, Khoruts A, Reilly C, Jenkins MK. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science. 2006;312:114–116. doi: 10.1126/science.1124228. [DOI] [PubMed] [Google Scholar]
- 56.Badovinac VP, Haring JS, Harty JT. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity. 2007;26:827–841. doi: 10.1016/j.immuni.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ford ML, Koehn BH, Wagener ME, Jiang W, Gangappa S, Pearson TC, et al. Antigen-specific precursor frequency impacts T cell proliferation, differentiation, and requirement for costimulation. J Exp Med. 2007;204:299–309. doi: 10.1084/jem.20062319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marzo AL, Klonowski KD, Le Bon A, Borrow P, Tough DF, Lefrancois L. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat Immunol. 2005;6:793–799. doi: 10.1038/ni1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tokoyoda K, Zehentmeier S, Hegazy AN, Albrecht I, Grun JR, Lohning M, et al. Professional memory CD4+ T lymphocytes preferentially reside and rest in the bone marrow. Immunity. 2009;30:721–730. doi: 10.1016/j.immuni.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 60.Prlic M, Bevan MJ. Immunology: A metabolic switch to memory. Nature. 2009;460:41–42. doi: 10.1038/460041a. [DOI] [PubMed] [Google Scholar]
- 61.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, et al. mTOR regulates memory CD8 T cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lascelles AK, Eagleson G, Beh KJ, Watson DL. Significance of Freund's adjuvant/antigen injection granuloma in the maintenance of serum antibody response. Vet Immunol Immunopathol. 1989;22:15–27. doi: 10.1016/0165-2427(89)90160-8. [DOI] [PubMed] [Google Scholar]
- 64.Schwartz RH. T cell anergy. Annu Rev Immunol. 2003;21:305–334. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- 65.Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 66.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 67.Newton MA, Kendziorski CM, Richmond CS, Blattner FR, Tsui KW. On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J Comput Biol. 2001;8:37–52. doi: 10.1089/106652701300099074. [DOI] [PubMed] [Google Scholar]
- 68.Kendziorski CM, Zhang Y, Lan H, Attie AD. The efficiency of pooling mRNA in microarray experiments. Biostatistics. 2003;4:465–477. doi: 10.1093/biostatistics/4.3.465. [DOI] [PubMed] [Google Scholar]
- 69.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 70.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 71.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.