Abstract
Purpose
The type I Melanoma Antigen GEnes (MAGEs) are commonly expressed in cancers, fueling speculation that they may be therapeutic targets with oncogenic potential. They form complexes with RING domain proteins that have E3 ubiquitin ligase activity and promote p53 degradation. MAGE-A3 was detected in tumor specimens from patients with multiple myeloma and its expression correlated with higher frequencies of Ki-67+ malignant cells. In this report, we examine the mechanistic role of MAGE-A in promoting survival of proliferating multiple myeloma cells.
Experimental Design
The impact of MAGE-A3 expression on survival and proliferation in vivo was examined by immunohistochemical analysis in an independent set of tumor specimens segregated into two groups; newly diagnosed, untreated patients and patients who had relapsed after chemotherapy. The mechanisms of MAGE-A3 activity were investigated in vitro by silencing its expression by shRNA interference in myeloma cell lines and primary cells and assessing the resultant effects on proliferation and apoptosis.
Results
MAGE-A3 was detected in a significantly higher percentage of relapsed patients compared to newly diagnosed, establishing a novel correlation with progression of disease. Silencing of MAGE-A demonstrated that it was dispensable for cell cycling, but was required for survival of proliferating myeloma cells. Loss of MAGE-A led to apoptosis mediated by p53-dependent activation of pro-apoptotic Bax expression and by reduction of survivin expression through both p53-dependent and independent mechanisms.
Conclusions
These data support a role for MAGE-A in the pathogenesis and progression of multiple myeloma by inhibiting apoptosis in proliferating myeloma cells through two novel mechanisms.
Keywords: MAGE, Cancer-Testis Antigen, multiple myeloma, apoptosis, survivin
Introduction
The type I Melanoma Antigen GEnes (MAGE-A, B, and C) belong to the Cancer-Testis antigen (CTAg) group of tumor-associated genes. CTAg are expressed in a broad range of human cancers, but their normal expression is restricted to immunologically “privileged” tissues, including developing germ cells and trophoblastic tissue such as placenta (1). For these reasons, CTAg have been investigated as targets for therapeutic tumor vaccines (2). Their widespread expression in cancer also suggests that CTAg may have oncogenic activity and may be a novel class of therapeutic targets amenable to pharmacologic intervention. We previously reported that MAGE-A3 and CT7 (MAGE-C1) were detected in more than 70% of tumor specimens from patients with stage III (Durie-Salmon Staging) multiple myeloma (MM) (3), an incurable malignancy of plasma cells that is the second most common hematologic cancer (4). The patients in this study were a heterogenous group, including newly diagnosed, relapsed, and refractory cases. Expression of these type I MAGEs correlated with higher frequencies of Ki-67+ cells, a marker for proliferating cells, and with advanced stage of disease. The exceptionally high frequencies of expression and clinical correlations suggested that type I MAGEs play a role in the pathogenesis or progression of MM by promoting survival of proliferating cells.
Emerging data support the concept that type I MAGEs promote tumor cell survival. Type I MAGEs were shown to interact with the tumor suppressor p53 in sarcoma and non-small cell lung cancer cell lines, inhibiting p53 transactivation activity and conferring resistance to etoposide-induced apoptosis (5). In this setting, MAGE-A2 did not appear to regulate transcription of the p53 gene itself. However, Yang and colleagues reported that MAGE proteins interacted with the RING domain protein TRIM28/Kap1 and suppressed transcription of p53 in human and mouse melanoma cell lines, and that silencing of type I MAGEs also induced apoptosis (6). Silencing of MAGE-A3 or CT7 expression by small interfering (si)RNA transfection appeared to reduce viability in human myeloma cell lines (HMCL) (7), but the underlying mechanism was not explored. More recently, Doyle and colleagues used biochemical methods to demonstrate that MAGE/RING domain protein complexes function as E3 ubiquitin (Ub) ligases (8). Several MAGE proteins associated with TRIM28/Kap1, and the MAGE-A2 and -C2 complexes were capable of polyubiquitinylating p53, targeting it for proteasomal degradation. This report of a biochemical function for MAGE was a groundbreaking revelation; however, it remains to be determined whether this activity is responsible for the effects of type I MAGE on survival and if p53 is the sole or dominant target substrate in myeloma cells.
To investigate the role of type I MAGEs in the pathogenesis and progression of MM, we examined their expression by immunohistochemistry (IHC) in an independent set of samples from two critical clinical milestones; newly diagnosed, untreated patients and patients who had relapsed after chemotherapy. Our data revealed that both the number of patients expressing MAGE-A3 in their tumor cells and the percentage of MAGE-A3-positive tumor cells increased with progression of disease and correlated with higher frequencies of Ki-67+ myeloma cells. Silencing of MAGE-A in HMCL and primary patient cells did not impair cell cycle regulation, but it did result in apoptosis through p53-dependent activation of Bax and Bak genes and by repression of survivin through p53-dependent and independent mechanisms. We propose that MAGE-A plays a critical role in the survival of proliferating MM cells through regulation of anti-apoptotic mechanisms.
Materials and Methods
Patient samples
Specimens from patients diagnosed with multiple myeloma (9) were obtained at the Weill-Cornell Medical Center or New York University Clinical Cancer Center under Institutional Review Board-approved sample collection protocols (WCMC IRB #1000-422 and NYU IRB #06-523) in accordance with the Declaration of Helsinki.
Immunohistochemical Analysis
IHC of bone marrow biopsies was performed as described previously(10). Briefly, primary antibodies were: M3H67 (MAGE-A3), CT7-33 (CT7), CT10 (CT10), MA454 (MAGE-A1), 57B (MAGE-A4), and E978 (NY-ESO-1), all provided by the Ludwig Institute for Cancer Research. Slides were blinded to the pathologist in regards to newly diagnosed versus relapsed status. Images were acquired as JPEG format files with a Nikon Coolpix 990 digital camera. Minor adjustments of brightness, contrast, and color solely for the purpose of legibility were made with Adobe Photoshop 7.0 for Windows (Adobe Systems Inc., San Jose, CA).
Plasma cell proliferation index assay
PCPI was performed as previously described (11). To compute the plasma cell-specific proliferation index (PCPI), 500 cells with a positive membranous CD138 signal were counted; each of those cells was scored as proliferating (containing a brown Ki-67+ nucleus) or non-proliferating (containing a blue, counter-stained, Ki-67-negative nucleus). The PCPI was reported as a percentage.
Cell Lines
MM.1r and H929 were from ATCC (Manassas, VA). ARP-1 were provided by S. Chen-Kiang. Cells were cultivated at 37°C, 5% CO2 in R10 media (RMPI-1640 (Mediatech Cellgro, Manassas, VA), 10% heat-inactivated fetal bovine serum (Sigma-Aldrich, St. Louis, MO), 1% 1M HEPES buffer (Mediatech Cellgro) and 20 μg/mL gentamycin (Invitrogen Gibco, Carlsbad, CA).
Primary Myeloma Cells
Pt #1 cells were cultivated in X-VIVO 15 medium (Lonza Walkersville, Inc., Walkersville, MD) supplemented with 10% heat-inactivated pooled human AB serum (Omega Scientific, Inc, Tarzana, CA), 20 mcg/ml gentamicin, and 1 mcg/ml rhu IL-6 (R&D Systems, Minneapolis, MN) at 37°C, 5% CO2. These cells were greater than 90% CD138+ by flow cytometry. Experiments were performed on cells between passages 3 and 5 in tissue culture.
Genomic p53 sequencing
Genomic DNA was isolated from Pt #1 cells using the DNAeasy Blood and Tissue kit (Qiagen, Valencia, CA) according to manufacturers instructions. TP53 exons 2-11 were amplified and sequenced using the International Agency for Cancer Research (IARC) direct sequencing protocol: http://www-p53.iarc.fr/Download/TP53_DirectSequencing_IARC.pdf. Amplified exons were sequenced at the New York University Sequencing Core Facility.
Lentiviral shRNA transduction
Lentiviral shRNA construct particles targeting MAGE-A3 (shMA, TRCN0000128375 and TRCN0000129750, Sigma Aldrich), non-target (shNT) control (SHC002V) containing a scrambled sequence, and a construct encoding GFP (SHC003V) were used for these experiments. Briefly, cells were diluted to 105 cells/ml in R10 and incubated overnight at 37°C. Cells were then harvested and resuspended in R10 media without antibiotics supplemented with 8 μg/ml polybrene (hexadimethrine bromide, Sigma-Aldrich) and aliquoted into a 96-well round bottom plate at 30,000 cells per well. Lentivirus particles were added at a multiplicity-of-infection (MOI) of 5x for MM.1R and a MOI of 15x for ARP-1 and Pt #1. Untreated cells were included as additional negative controls. Cells were incubated for 18 hrs at 37°C, then the cells were washed three times with pre-warmed R10 media without antibiotics to remove excess virus and polybrene. Incubations were continued until the time points noted in individual experiments. Transduction efficiency typically exceeded 60% as measured at 24 and 48 hrs by flow cytometry for GFP fluorescence. For apoptosis analysis, cells were treated with 10 μM Quinoline-Val-Asp-Difluorophenoxymethylketone (Q-VD-OPh) pan-caspase inhibitor (R&D Systems) or DMSO vehicle control at 24 hrs, and annexin V staining was assessed at 48 hrs. For substrate-specific Ub assay, cells were treated with 40 nM MG132 (Calbiochem) or DMSO vehicle control eight hours prior to harvest.
BrdU labeling
Proliferation was analyzed with the BrdU flow kit from BD Biosciences (cat #559619) according to manufacturer's instructions.
Annexin V Staining
Lentivirus transduced and control cells were resuspended in a small volume (~100μl) of 1x binding buffer (BD Bioscience), and incubated with Annexin V-PE (BD Biosciences) and 5μl of 7-AAD (BD Biosciences) at room temperature, protected from light for 20 minutes. Untreated cells stained with annexin V only or 7-AAD only served as single color controls. Cells were then fixed in 100μl of 4% paraformaldehyde (Sigma-Aldrich) and analyzed by flow cytometry.
RNA isolation
RNA isolation was performed as previously described (3).
Quantitative RT-PCR
Semi-quantitative realtime PCR was performed as previously described (3). Taqman® primer sets (Applied Biosystems Inc., Foster City, CA) for the genes of interest in these experiments were: β-actin (Hs00181698_m1), cyclophilin B (Hx00168719_m1), p53 (Hs01034249_m1), p21 (Hs99999142_m1), cyclin B1 (Hs01030103_m1), p27 (Hs00153277_m1), survivin (Hs03063352_s1), MAGE-A3 (Hs00366532_m1), MAGE-A1 (Hs00607097_m1), CT7 (Hs00193821_m1), cyclin B2 (Hs00270424_m1), CDK1 (Hs00938777_m1), CDK2 (Hs01548894_m1), cyclin E2 (Hs01051894_m1), cyclin A2 (Hs00996789_g1). Relative expression was calculated against the geometric mean of the reference primers (β-actin and cyclophilin B) using the formula relative expression = 2^ –(ΔCt(sample) – ΔCt(reference)), where ΔCt = Ct(test) – Ct(reference control), the control is untreated cells, and Ct is the cycle threshold to reach a threshold fluorescence intensity of 0.2. Results were normalized to the shNT control in each set of samples and depicted as the mean of triplicate samples ± standard error of the mean.
Western Blotting
Protein lysates were prepared by freeze-thawing cells in lysis buffer (350mM NaCl, 20mM HEPES [pH7.9], 0.2% NP-40, 1mM MgCl2, 1mM DTT, 20% glycerol, 2mM sodium orthovanadate, 10mM beta-glycerol phosphate (all from Sigma-Aldrich), 1x protease inhibitor (Calbiochem, San Diego, CA)). Protein concentration was determined by Bradford assay (Bio-Rad, Herculaneum, CA) according to manufacturer's instructions. 10-20 μg of lysate was run on an SDS-PAGE gel and transferred to a PVDF membrane (Millipore, Billerica MA). Blocking and antibody dilutions were all done in 5% non-fat dry milk (NFDM) in TBS-T (10 mM Tris base, 150 mM NaCl, 0.01% (v/v) Tween 20, Sigma-Aldrich), with the exception of the pRb blots, for which 5% BSA + 0.4% NFDM in TBS-T was used for blocking, 1% BSA in TBS-T for primary antibody dilution and 1% BSA + 0.4% NFDM for secondary antibody dilution. Blots were visualized with Supersignal West Femto Substrate (Thermo Scientific).
Antibodies for these experiments were: p27 (cat #610242, BD Pharmingen, La Jolla, CA); Bcl-xl (2763), Bim (2819), Bid (2002), Mcl-1 (4572), Rb (9309), pRb (9308), p21 (2947, Cell Signaling Technology, Beverly, MA); Bcl-2 (M0887, Dako, Carpinteria, CA); Bax (sc-493), p53 (sc-126), MAGE-A (sc-20034) survivin (sc-17779, Santa Cruz Biotechnology, Inc., Santa Cruz, CA); Bak (AM03, EMD Biosciences, Gibbstown, NJ); β-actin (MS-1295-P0), goat anti-mouse IgG-HRP conjugate (1858413), goat anti-rabbit IgG-HRP conjugate (1858415, Thermo Scientific).
Immunoprecipitation
For substrate-specific ubiquitination analysis, 25 mcg lysate was incubated with 0.5 mcg p53-specific mAb or IgG isotype control on ice for one hour, then co-incubated with Protein A Sepharose beads (Pearse) that had been blocked for one hour with 10% BSA (Sigma-Aldrich) at room temperature. Immune complexes were tumbled overnight at 4°C, then the beads were washed four times with ice-cold RIPA buffer (0.05 % Tris pH 7.4, 0.15 M NaCl, 1% (w/v) Na Deoxycholate (Sigma-Aldrich), 1% (v/v) Triton X-100, 0.1% (w/v) SDS (MP Biomedical, Solon, OH). IP products were analyzed by western blot with p53 or ubiquitin (MMS-264R, Covance, Princeton, NJ) mAb.
Statistics and Data Presentation
Statistical analysis was performed with Excel 2008 for Mac (Microsoft Corporation, Redmond, WA) and Prism 5.0 (GraphPad Software, San Diego, CA) for Mac OSX (Apple Inc., Cupertino, CA). Figures were prepared with Prism 5.0, Adobe Photoshop CS3 and Illustrator CS3 (Adobe Systems Inc.) for Mac OSX (Apple Inc.). Densitometry of western blots was performed with ImageJ 1.44o (Wayne Rasband). Photo images were not altered in preparation except for the survivin western blot image for Pt #1 in fig. 5C, which was enhanced for contrast only.
Results
MAGE-A3 expression correlates with progression of disease in MM patients
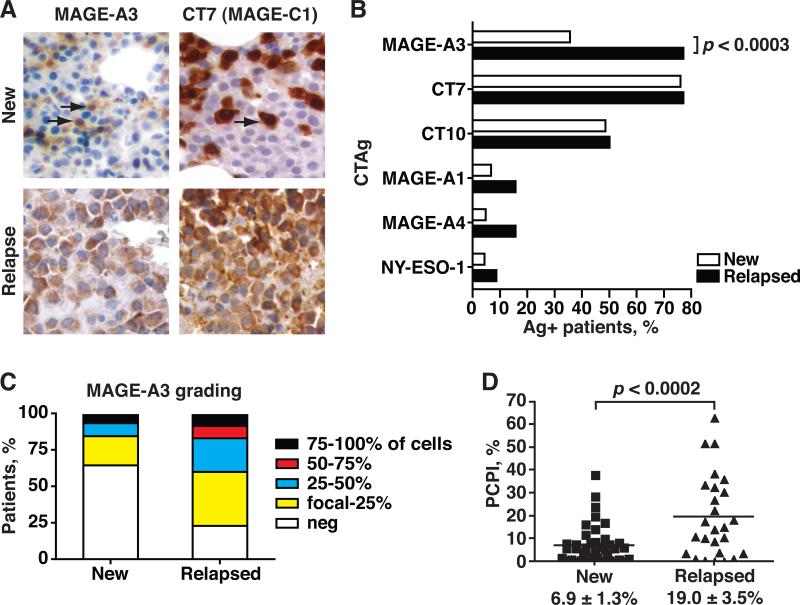
Expression of MAGE-A3 and other CTAg proteins was assessed by immunohistochemistry (IHC) in bone marrow biopsy specimens from two critical clinical milestones; newly diagnosed, untreated MM patients (n=46) and patients who relapsed after chemotherapy (n=35) (fig. 1A). Demographic data (age and sex distribution) and clinical characteristics were similar between the two groups and were representative of the general MM patient population (table 1). Karyotypic abnormalities were detected by either conventional cytogenetics or fluorescence in situ hybridization (FISH) in more than half of each group. High risk cytogenetic abnormalities, including chromosome 13 abnormalities, t(4;14), and complex cytogenetics, accounted for about one third of the tested patients in each group.
Figure 1. Immunohistochemical analysis of CTAg expression in newly diagnosed and relapsed myeloma patients.
A. Bone marrow biopsy sections were stained with a panel of mAb recognizing several CTAg (MAGE-A3 and CT7 shown). Representative sections are depicted. Brown chromophore indicates positive staining for CTAg (arrows). 20X magnification. B. Frequency of expression for each CTAg in new and relapsed patients were plotted and compared. C. Grading of MAGE-A3 expression (percent of MAGE-A3+ MM cells) was divided into quartiles (focal-25%, 25-50%, 50-74%, and 75-100%) and plotted by the percentage of patients in each quartile. D. Plasma Cell Proliferation Index was plotted for newly diagnosed and relapsed MM patient specimens. Mean PCPI ± standard error of the mean depicted. P-values calculated by Student's two-tailed t-test.
Table 1.
Demographic and staging data for multiple myeloma patients in this study.
| Newly diagnosed n=46 | Relapsed n=35 | |
|---|---|---|
| Age | 62.0 ± 1.5 yrs | 57.7 ± 1.5 yrs |
| Sex | M = 27, F = 19 | M = 20, F = 15 |
| Heavy chain isotype | IgG = 26 IgA = 10 IgM = 1 IgG + IgA = 1 Light chain only = 8 |
IgG = 18 IgA = 3 IgM = 1 IgD = 1 Light chain only = 7 Oligosecretory = 4 |
| Light chain isotype | Kappa = 31 Lambda = 15 |
Kappa = 23 Lambda = 11 Not done = 1 |
| Cytogenetics | Normal = 32 Del(20q) = 1 Inv(2) = 1 Inv(9) = 2 Y(-) = 2 Complex t(1;8); t(11;14); t(12;27) = 1 Not done = 7 |
Normal = 18 Complex = 7 Complex incl. t(5;13)(q31;q14) = 1 Inv(9) =1 Not done = 8 |
| FISH | Normal = 15 Del (13q14) = 4 Complex incl. del(13q14) = 7 t(11;14) = 4 t(4;14) = 1 Trisomy/ tetrasomy 11 = 5 Trisomy 11/ trisomy 17 = 1 Complex = 1 Not done = 8 |
Normal = 19 t(11;14) =2 Del(13) = 2 Trisomy 11 = 1 Trisomy 11 + del(13q14) = 1 Complex = 1 Not done = 9 |
| Serum β2-microglobulin | 4.8 ± 0.6 mg/ dL | 4.7 ± 0.7 mg/ dL |
| Serum albumin | 3.6 ± 0.1 mg/ dL | 3.5 ± 0.1 mg/ dL |
| Hemoglobin | 10.4 ± 0.4 gm/ dL | 10.2 ± 0.3 gm/ dL |
| Serum creatinine | 1.5 ± 0.2 mg/ dL | 2.2 ± 0.4 mg/ dL |
The frequency of MAGE-A3 expression was significantly higher in relapsed (77.1%) versus newly diagnosed patients (35.6%, p<0.0003, fig. 1B), and the grading of MAGE-A3 expression (percentage of MAGE-A3-positive MM cells) also increased in the relapsed patients (fig. 1C). In contrast, although CT7 (MAGE-C1) and CT10 (MAGE-C2) were also commonly detected, their frequencies of expression were not increased in relapsed patients (fig. 1B), although the grading of CT7 expression appeared to increase with progression of disease (suppl. fig. 1A). Other CTAgs (MAGE-A1, MAGE-A4, and NY-ESO-1) were detected in less than 20% of patients in either group. Proliferation in new and relapsed specimens was examined by Plasma Cell Proliferation Index (PCPI), defined as the percentage of myeloma cells expressing both the plasma cell marker CD138 and the proliferation marker Ki-67 (11). As expected, the average PCPI was significantly higher in relapsed patients (19.0 ± 3.5%) compared to newly diagnosed (6.9 ± 1.3%, p < 0.0002, fig. 1D)(12). Therefore, although CT7 and MAGE-A3 were the predominant type I MAGEs expressed in MM, MAGE-A3 was distinguished by unique correlations with progression of disease and higher frequencies of Ki-67+ myeloma cells.
Loss of MAGE-A does not impair cell cycling in HMCL and primary myeloma cells
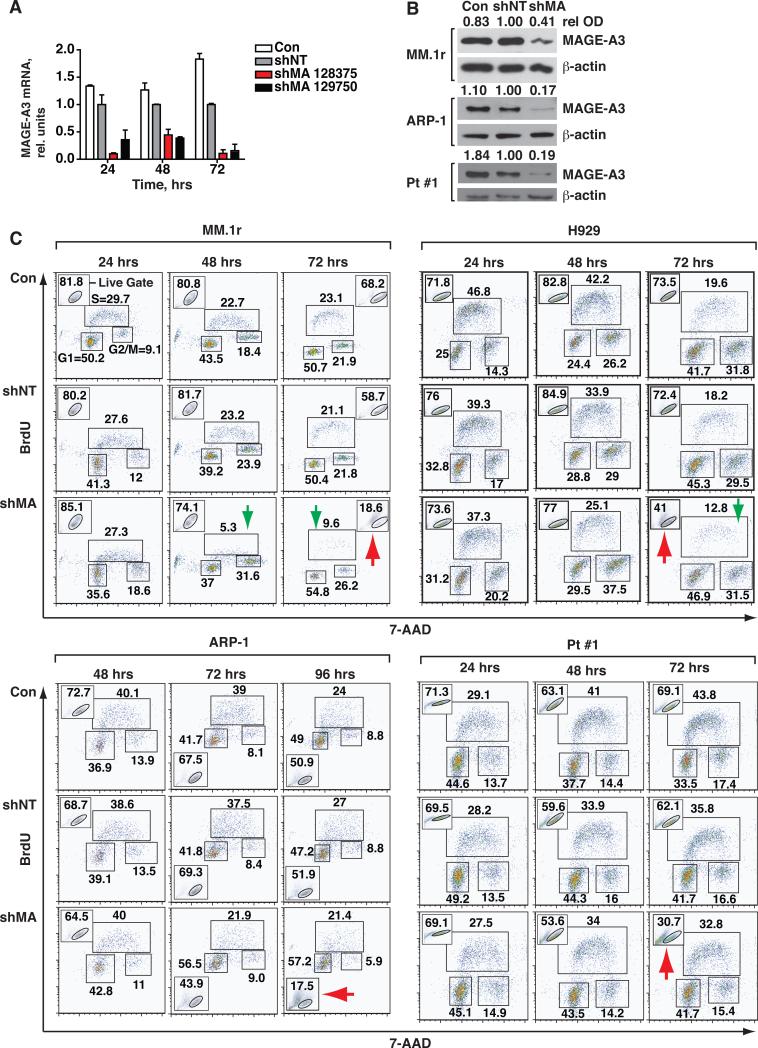
To investigate the mechanisms by which MAGE-A3 contributes to progression in MM, we silenced its expression in the HMCL MM.1r and H929, both of which express wt p53, and in ARP-1, which bears homozygous deletions of the TP53 loci on chromosome 17. We also used polyclonal primary cells (Pt #1) that had a monoallelic deletion of TP53 by fluorescence in situ hybridization, and sequencing of the other TP53 allele revealed a loss-of-function mutation in the DNA-binding region of exon 7, rendering them p53-null (suppl. fig. 2A, B). MAGE-A3 was silenced with targeted lentiviral shRNA constructs (suppl. fig. 1B) that demonstrated high transduction efficiency and low background toxicity. Two distinct constructs (shMA 129750 and 128375) silenced MAGE-A3 mRNA and protein expression compared to non-target shRNA-transduced (shNT) and untreated negative control (Con) cells (fig. 2A and B). Subsequent experiments were performed with shMA 128375. This construct also silenced MAGE-A1 (suppl. fig. 1C), most likely due to the high degree of sequence conservation among the MAGE-A family (13, 14). CT7 mRNA levels in ARP-1 and Pt #1 cells were not affected by shMA 128375 (suppl. fig. 1D). Since the potential activity of MAGE-A1 and other -A family members could not be formally excluded in these shRNA experiments, the target genes will be referred to collectively as “MAGE-A.” However, it should be noted that MAGE-A1 was detected in less than 20% of patient samples by IHC (fig. 1B).
Figure 2. Silencing of MAGE-A does not directly affect proliferation in MM cells.
MM.1r and ARP-1 HMCL and Pt #1 cells were transduced with MAGE-A-targeted (shMA 128375 and 129750) or non-target (shNT) shRNA lentiviral particles and samples were harvested at the indicated time points. A. MAGE-A3 mRNA in MM.1r cells was assessed by semi-quantitative real time (q) RT-PCR. Results were normalized to shNT control at each time point. Error bars depict standard error of the mean for triplicates of each sample. Control (Con), untreated negative control cells. B. HMCL and Pt #1 cells were treated with shMA 128375 or controls and MAGE-A3 protein expression was assessed by western blot at 48 hrs. Relative optical density (rel OD) was normalized to matched β-actin and expressed as a fraction of shNT control. C. shMA-transduced and control cells were pulsed for 30 minutes with 10 mM BrdU at the indicated time points. Proliferation was assessed by intracellular staining for BrdU and 7-AAD, followed by flow cytometry. Acquisition populations were gated on viable cells in forward vs. side scatter plots (Live Gate, insets) and the gated events plotted by 7-AAD (total DNA content) vs. FITC (BrdU incorporation) fluorescence, resulting in segregation into the G1, S, and G2/M populations as labeled in the first panel.
MAGE-A may contribute to myeloma progression by promoting cell cycling, cell survival, or both. To investigate the role of MAGE-A in cell cycle regulation, we labeled replicating DNA in MAGE-A-silenced and control cells with 5-bromo-2-deoxyuridine (BrdU) for 30 minutes. BrdU uptake and total DNA content per cell were then assessed by flow cytometry. Short pulses of BrdU provided a “snapshot” of cells in S phase at a given time without skewing the distribution of cells in each phase of the cycle due to death. Silencing of MAGE-A led to a drastic decrease in the percentage of live cells by 72 hrs in MM1.r, H929, and Pt #1 cells and by 96 hrs in ARP-1 cells (fig. 2C, red arrows). Of note, the reduction of viable MM1.r and H929 cells was accompanied by a decrease in the percentage of S phase cells (fig. 2C, green arrows), but this selective loss of S phase cells was not observed in ARP-1 or Pt #1 cells.
The loss of MM.1r and H929 cells in S phase was not accompanied by an increase in G1 cells (fig. 2D), suggesting that the loss was due to cell death rather than a block in progression through the G1 cell cycle checkpoints. Corroborating this observation, silencing of MAGE-A in MM.1r did not perturb the ratio of cyclin-dependent kinase (CDK)4/CDK6-specific phosphorylation of Rb to total Rb compared to controls at 48 hrs (suppl. fig. 3A), indicating that cell cycle progression through mid-G1 was intact. Total Rb protein was reduced in MAGE-A-silenced cells, possibly reflecting the decrease in overall cell viability. Thus, although dysregulation of G1 progression via overexpression of CDK4 or CDK6 is central to loss of cell cycle control in myeloma (13), these data confirmed that MAGE-A is not required for cell cycle progression through G1. The CDK inhibitors p21Cip1 and p27Kip1 can act in both G1 and G2/M to mediate cell cycle arrest. The expression of p21 and p27 proteins also remained unchanged upon silencing of MAGE-A, despite an increase in p21 mRNA (suppl. fig. 3B-C). Collectively, these data demonstrated that MAGE-A is required to maintain myeloma cell survival, particularly survival of S phase cells in the presence of p53, but is dispensable for cell cycling.
Silencing of MAGE-A triggers the intrinsic apoptosis pathway in proliferating myeloma cells
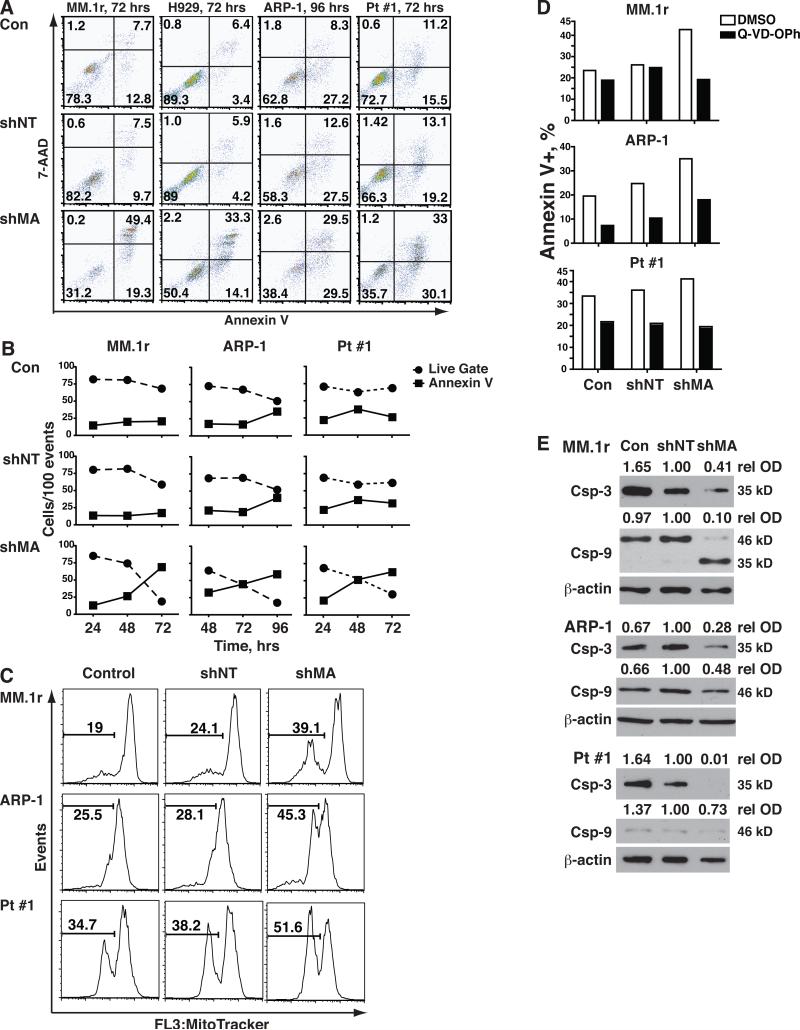
This led us to investigate the mechanisms by which silencing MAGE-A induced cell death. We first assessed apoptotic versus necrotic cell death in this system. Silencing of MAGE-A with shMA 128375 (fig. 3A) or 129750 (suppl. fig. 1E) in HMCL and Pt #1 cells was associated with an increased number of apoptotic cells (annexin V-positive) in the total acquisition population, compared to controls. Analysis of the kinetics of cell death demonstrated an inverse correlation between live cells and annexin V-positive cells (fig. 3B), indicating that the loss of viable cells was due to apoptosis. Furthermore, apoptosis in MAGE-A-silenced HMCL and primary cells was characterized by increased mitochondrial depolarization (decreased MitoTracker Red® staining) (fig. 3C) suggesting that it triggered the intrinsic apoptosis pathway. Apoptosis in MAGE-A-silenced cells was reversed by co-incubation with the pan-caspase inhibitor Quinoline-Val-Asp-Difluorophenoxymethylketone (Q-VD-OPh) (fig. 4D), indicating that it was caspase-dependent. In fact, silencing of MAGE-A lead to cleavage of caspase-3 (fig. 3E). Caspase-9 cleavage/activation was also evident after MAGE-A silencing in p53+/+ MM.1r but not in p53-/- ARP-1, and very little caspase-9 was detected in p53mut/- Pt #1 cells. These data demonstrated that MAGE-A inhibits intrinsic apoptosis in both p53+/+ and p53-null myeloma cells.
Figure 3. Silencing of MAGE-A results in activation of intrinsic apoptosis.
HMCL and Pt #1 cells were transduced shMA or controls as previously described and apoptosis was assessed by staining with annexin V. A. Total (ungated) acquisition populations at 72 or 96 hrs were plotted by Annexin V-PE (apoptosis) vs. 7-AAD (necrosis) fluorescence. B. The percentages of viable cells (Live Gate as described in fig. 2, solid circles) and annexin V+ cells in the ungated acquisition population (solid squares) were plotted over time for the MAGE-A-silenced and control groups. C. HMCL and Pt#1 cells were stained with MitoTracker Red®, which is fixed in mitochondria with intact membrane polarization, and analyzed by flow cytometry. Total (ungated) events were plotted by MitoTracker Red® fluorescence and mitochondrial depolarization was assessed by decreased fluorescence (bars). E. Lentivirus-transduced cells were incubated with 10 μM Q-VD-OPh or DMSO vehicle control for 72 hrs and apoptosis was assessed by staining with annexin V. D. Caspase-3 and -9 were analyzed by western blot in at 48 hrs for MM.1r and Pt #1 and 72 hrs for ARP-1 cells.
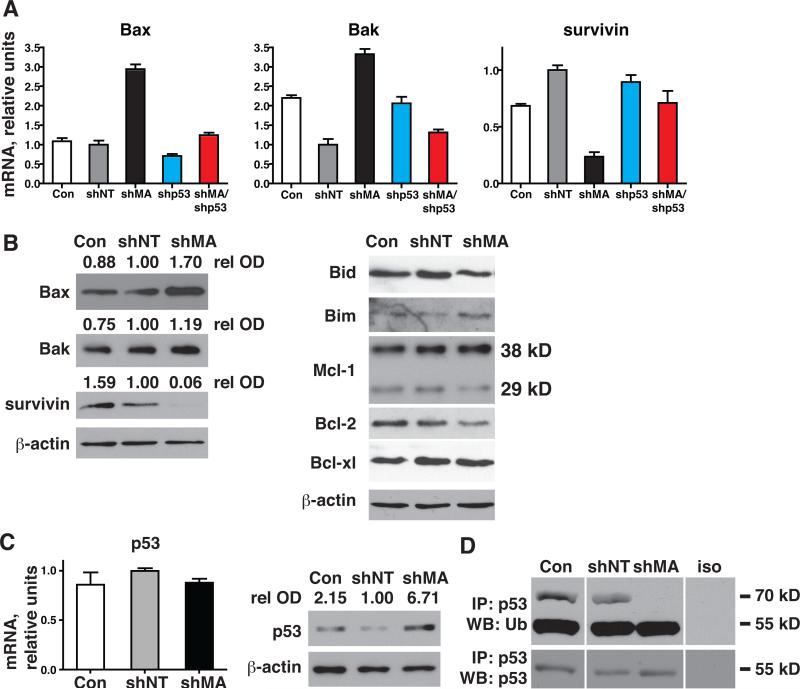
Figure 4. Silencing of MAGE-A induces p53-dependent Bax/Bak expression and repression of survivin.
MM.1r cells were transduced with MAGE-A-targeted (shMA) or p53-targeted (shp53) shRNA lentiviral constructs, both targeted constructs, or controls as indicated and cells were harvested at 48 hrs. A. Bax, Bak, and survivin mRNA expression was assessed by qRT-PCR. B. Protein expression of Bcl-2 proteins and survivin was assessed by western blot. C. p53 mRNA expression was assessed by qRT-PCR and protein expression by western blot. D. Cells were incubated with 40 nM MG132 prior to harvest and lysates were immunoprecipitated (IP) with p53-specific mAb or isotype control mu IgG (iso). Ubiquitinylation of p53 was assessed by western blot.
MAGE-A regulates Bax and survivin through p53-dependent mechanisms
Having demonstrated that MAGE-A antagonized intrinsic apoptosis in MM.1r, we investigated expression of pro- and anti-apoptotic Bcl-2 genes in these cells. MAGE-A silencing in MM.1r lead to increased mRNA expression of pro-apoptotic Bax and Bak (Fig. 4A). Increased expression of Bax protein was also evident, but Bak protein was not significantly changed under these conditions (fig 4B). Other Bcl-2 proteins (Bid, Bim, Mcl-1, Bcl-2, and Bcl-xl) similarly demonstrated very little changes compared to controls. Bax is a target of p53 transcriptional activation, so we next examined p53 expression. Silencing of MAGE-A in MM.1r increased p53 protein without affecting mRNA expression (fig. 4C). MAGE-A post-translational regulation of p53 was confirmed by substrate-specific Ub assay, which demonstrated loss of Ub-p53 (70 kD band, Fig. 4D) after silencing of MAGE-A compared to controls. Silencing of p53 by shRNA lentiviral transduction in addition to MAGE-A in MM.1r reverted Bax and Bak mRNA to control levels, indicating that MAGE-A repressed their expression by inhibiting p53 transactivation (fig. 4B). We then examined survivin, a multifunctional protein that plays critical roles in survival and cell division in normal and cancer cells (15, 16). It can be transcriptionally repressed by p53 (17, 18). Silencing of MAGE-A in MM.1r resulted in a marked reduction in survivin mRNA and protein (Fig. 4A and B). Survivin mRNA was partly restored by silencing of p53 in addition to MAGE-A. Collectively, these findings demonstrated that MAGE-A inhibits Bax transcription through inhibition of p53 and promotes survivin expression in part through p53-dependent mechanisms.
MAGE-A promotes expression of survivin by p53-independent mechanisms
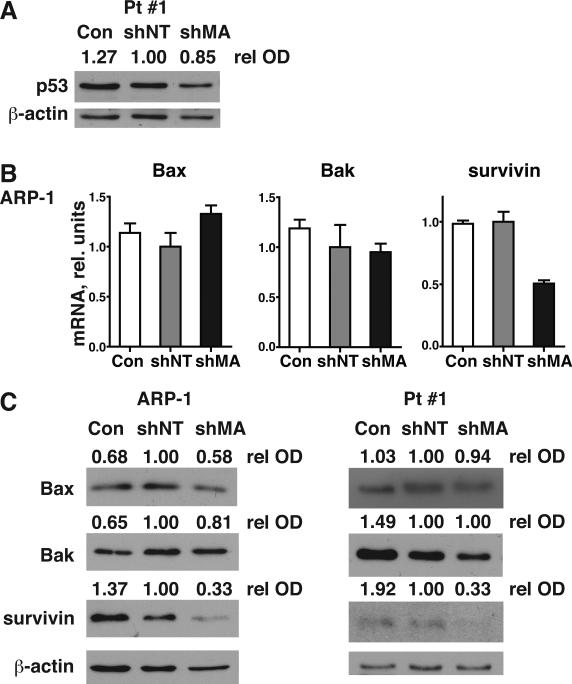
Since silencing of MAGE-A also induced apoptosis in p53-null ARP-1 and Pt #1, we investigated p53-independent mechanisms of MAGE-A inhibition of apoptosis by examined expression of Bax, Bak, and survivin in these cells. MAGE-A silencing did not stabilize mutant p53 in Pt #1 cells (fig. 5A). Bax and Bak mRNA or protein did not increase upon silencing of MAGE-A (fig. 5B and C). In contrast, MAGE-A silencing resulted in marked decreases in survivin protein. The expression of survivin mRNA paralleled that of protein in ARP-1 cells. These data indicate that MAGE-A also promoted survivin protein expression through p53-independent mechanisms in myeloma cells.
Figure 5. Silencing of MAGE-A results in repression of survivin in the absence of p53.
ARP-1 (p53-/-) and Pt #1 (p53mut/-) cells were transduced with MAGE-A-targeted (shMA) shRNA lentiviral constructs or controls as indicated and cells were harvested at 48 hrs (Pt #1) or 72 hrs (ARP-1). A. p53 protein in Pt #1 cells was assessed by western blot. B. Bax, Bak, and survivin mRNA expression was assessed by qRT-PCR. C. Protein expression of Bax, Bak, and survivin was assessed by western blot.
Discussion
These results demonstrated that MAGE-A inhibited apoptosis in proliferating MM cells through at least two novel mechanisms: inhibition of p53-dependent upregulation of Bax and maintenance of survivin expression by p53-dependent and independent mechanisms. These mechanisms were active in cell lines and primary MM cells. Furthermore, MAGE-A3 expression was unique among the CTAg characterized in that it was specifically correlated with progression of disease. Identification of these apoptotic mechanisms provides long-sought insight into the role of MAGE-A in the pathogenesis and progression of MM and validates them as functional therapeutic targets in this disease.
Silencing of MAGE-A resulted in loss of p53 ubiquitinylation and stabilization of p53 protein, increased expression of Bax, and apoptosis in p53+/+ MM.1r cells. Progression through G1 and entry into S phase appeared intact even as the majority of cells became apoptotic. Therefore, apoptosis in the S phase population of MM.1r cells was likely a consequence of p53-dependent up-regulation of Bax after loss of MAGE-A, since disproportionate S phase apoptosis was not observed in p53-null ARP-1 or Pt #1 cells. These results strongly suggest that MAGE-A is an antagonist of p53-dependent pro-apoptotic transcriptional regulation in MM cells.
Survivin was down-regulated in HMCL and primary MM cells after MAGE-A silencing regardless of p53 status. This is the first evidence of a functional link between type I MAGE and survivin and suggests that survivin is a principle mediator of MAGE-dependent resistance to apoptosis in MM. Interestingly, high levels of survivin mRNA in primary MM specimens from newly diagnosed patients were correlated with poor prognosis (19). Survivin expression is at its lowest in G1 and increases throughout the cycle, peaking in G2/M where it regulates the mitotic spindle checkpoint (15). Therefore, it is unlikely that loss of survivin made a significant contribution to S phase apoptosis observed in MM.1r cells after MAGE-A silencing.
These results support a model in which MAGE-A promotes survival in proliferating MM cells through at least two distinct mechanisms that may act at different phases of the cycle: Bax in S phase and survivin in G2/M. This model also reconciles the correlation between MAGE-A3 and increases in the frequency of proliferating cells in patient samples as measured by Ki-67 expression (3, 20), despite the lack of evidence for a direct effect on cell cycle progression in vitro. In a heterogeneous tumor mass, cells expressing MAGE-A3 are protected from apoptosis as they transit through the cell cycle, effectively increasing the percentage of MAGE-A3+/Ki-67+ cells. Therefore, the correlations between type I MAGE, progression of disease, and proliferation in primary MM specimens are a consequence of survival of proliferating cells.
The lentiviral shRNA constructs in these experiments efficiently silenced MAGE-A1 as well as MAGE-A3, which is likely a reflection of the extensive sequence homology shared by this family (13). However, MAGE-A1 was only detected in a minority of samples in this study and in our previous set of patients (3), suggesting that MAGE-A3 plays a dominant role in vivo. Several MAGE-A family members and MAGE-C2 complex with TRIM28/Kap1 to form E3 ubiquitin ligases, and p53 is one of their targets for proteasomal degradation (8). The results presented here indicate this activity reduces p53 protein in myeloma cells and decreases apoptosis. However, since apoptosis and down-regulation of survivin were also observed after MAGE-A silencing in cells that lacked functional p53, it is likely that non-p53 substrates of MAGEA3/TRIM28 Ub ligase activity contribute to inhibition of apoptosis by regulating survival factors such as survivin.
Silencing of CT7 by siRNA transfection was reported to decrease viability in MM cell lines (7). In our hands, silencing of MAGE-A by lentiviral shRNA transduction in ARP-1 and Pt #1 did not affect CT7 expression, but expression of CT7 in the absence of MAGE-A was not sufficient to prevent apoptosis. This discrepancy may be due to differences in RNA interference efficiency and greater background cell death with siRNA transfection. Although CT7 is commonly expressed in MM and its expression appears to be an early event in MM pathogenesis (3, 21), it is unknown whether it has a similar spectrum of activity as MAGE-A family members. Further investigation is needed to illuminate the role of CT7 in MM pathogenesis.
These data indicate that MAGE-A is critical for the survival of proliferating MM cells and therefore a promising therapeutic target in this disease. This strategy is already under investigation in the form of MAGE-A3 tumor vaccines (22). Further investigation of the mechanisms of MAGE-A3 inhibition of p53 and promotion of survivin expression in MM may reveal novel therapeutic targets that will be amenable to pharmacologic agents. These may be combined with vaccines in multi-targeted, MAGE-specific therapy designed to induce apoptosis in proliferating MM cells or make them more susceptible to chemotherapy-induced apoptosis, resulting in durable remission of this currently incurable disease. Proof of principle in MM may lead to broader application in other cancers that express type I MAGE. These include lung, breast, prostate and skin cancers, among the leading causes of cancer death.
Statement of Translational Relevance.
Multiple myeloma is an incurable blood cancer. MAGE-A3 is a cancer-associated gene that is commonly expressed in multiple myeloma and correlates with higher frequencies of proliferating malignant cells. The results presented here show that expression of MAGE-A3 in patient specimens also correlated to relapse of multiple myeloma, suggesting a pathogenic role in progression of disease. Silencing of MAGE-A3 in myeloma cell lines and patient cells resulted in activation of intrinsic apoptosis due to p53-dependent expression of Bax and survivin by p53-dependent and independent mechanisms. MAGE-A3 appeared to contribute to progression of disease by promoting resistance to apoptosis in proliferating multiple myeloma cells. Therefore, MAGE-A3 is a promising therapeutic target in multiple myeloma, and strategies targeting it, such as MAGE-A3-specific tumor vaccines currently in clinical trials, may result in myeloma cell apoptosis and prevent relapse.
Supplementary Material
Acknowledgments
VCCA is supported by scholarship 200024/2009-4 from Conselho Nacional de Desenvovimento Cientifico e Tecnologico (CNPq) of Brazil. HJC is supported by K01-CA115917 from the NIH/NCI and a grant from the Multiple Myeloma Working Group Initiative of the Cancer Research Institute.
Footnotes
The authors have no relevant conflicts of interest to declare
The authors wish to thank Wayne Austin and Hasina Outtz for their technical contributions.
References
- 1.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 2.Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 3.Jungbluth AA, Ely S, DiLiberto M, et al. The cancer-testis antigens CT7 (MAGE-C1) and MAGE-A3/6 are commonly expressed in multiple myeloma and correlate with plasma-cell proliferation. Blood. 2005;106:167–74. doi: 10.1182/blood-2004-12-4931. [DOI] [PubMed] [Google Scholar]
- 4.Kyle RA, Rajkumar SV. Multiple myeloma. Blood. 2008;111:2962–72. doi: 10.1182/blood-2007-10-078022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monte M, Simonatto M, Peche LY, et al. MAGE-A tumor antigens target p53 transactivation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc Natl Acad Sci U S A. 2006;103:11160–5. doi: 10.1073/pnas.0510834103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang B, O'Herrin SM, Wu J, et al. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res. 2007;67:9954–62. doi: 10.1158/0008-5472.CAN-07-1478. [DOI] [PubMed] [Google Scholar]
- 7.Atanackovic D, Hildebrandt Y, Jadczak A, et al. Cancer-testis antigens MAGE-C1/CT7 and MAGE-A3 promote the survival of multiple myeloma cells. Haematologica. 2009 doi: 10.3324/haematol.2009.014464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Molecular cell. 2010;39:963–74. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol. 2003;121:749–57. [PubMed] [Google Scholar]
- 10.Jungbluth AA, Chen YT, Stockert E, et al. Immunohistochemical analysis of NY-ESO-1 antigen expression in normal and malignant human tissues. Int J Cancer. 2001;92:856–60. doi: 10.1002/ijc.1282. [DOI] [PubMed] [Google Scholar]
- 11.Ely SA, Chadburn A, Dayton CM, Cesarman E, Knowles DM. Telomerase activity in B-cell non-Hodgkin lymphoma. Cancer. 2000;89:445–52. doi: 10.1002/1097-0142(20000715)89:2<445::aid-cncr33>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 12.Kumar SK, Therneau TM, Gertz MA, et al. Clinical course of patients with relapsed multiple myeloma. Mayo Clinic proceedings. 2004;79:867–74. doi: 10.4065/79.7.867. [DOI] [PubMed] [Google Scholar]
- 13.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–51. [PubMed] [Google Scholar]
- 14.De Plaen E, Arden K, Traversari C, et al. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics. 1994;40:360–9. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 15.Li F, Ambrosini G, Chu EY, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–4. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 16.Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000–5. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–57. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 18.Mirza A, McGuirk M, Hockenberry TN, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–22. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- 19.Romagnoli M, Seveno C, Wuilleme-Toumi S, et al. The imbalance between Survivin and Bim mediates tumour growth and correlates with poor survival in patients with multiple myeloma. Br J Haematol. 2009;145:180–9. doi: 10.1111/j.1365-2141.2009.07608.x. [DOI] [PubMed] [Google Scholar]
- 20.Chng WJ, Kumar S, Vanwier S, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67:2982–9. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- 21.Atanackovic D, Arfsten J, Cao Y, et al. Cancer-testis antigens are commonly expressed in multiple myeloma and induce systemic immunity following allogeneic stem cell transplantation. Blood. 2006 doi: 10.1182/blood-2006-04-014480. [DOI] [PubMed] [Google Scholar]
- 22.Brichard VG, Lejeune D. GSK's antigen-specific cancer immunotherapy programme: pilot results leading to Phase III clinical development. Vaccine. 2007;25(Suppl 2):B61–71. doi: 10.1016/j.vaccine.2007.06.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.