Abstract
PURPOSE
We investigated the feasibility of detecting aberrant DNA methylation of some novel and known genes in the serum of lung cancer patients.
EXPERIMENTAL DESIGN
To determine the analytical sensitivity, we examined the tumor and the matched serum DNA for aberrant methylation of fifteen gene promoters from 10 patients with primary lung tumors by using Quantitative methylation specific PCR. We then tested this 15 gene set to identify the more useful DNA methylation changes in the serum of a limited number of lung cancer patients and controls. In an independent set, we tested the six most promising genes (APC, CDH1, MGMT, DCC, RASSF1A and AIM) for further elucidation of the diagnostic application of this panel of markers.
RESULTS
Promoter hypermethylation of at least one of the genes studied was detected in all 10 lung primary tumors. In majority of cases, aberrant methylation in serum DNA was accompanied by methylation in the matched tumor samples. In the independent set, using a single gene that had 100% specificity (DCC), 35.5% (95% CI 25%, 47%) of the 76 lung cancer patients were correctly identified. For patients without methylated DCC, addition of a logistic regression score that was based on the five remaining genes improved sensitivity from 35.5% to 75% (95% CI: 64%, 84%) but decreased the specificity from 100% to 73% (95% CI:54%, 88%).
CONCLUSION
This approach needs to be evaluated in a larger test set to determine the role of this gene set in early detection and surveillance of lung cancer.
Keywords: DNA methylation/epigenetics, serum, lung cancer
INTRODUCTION
Lung cancer kills more people than breast, colon, and prostate cancers combined (1). Lung cancer remains the second most diagnosed cancer in the United States and the most common cause of cancer mortality, with an estimated 161 000 deaths in 2008, with 80% being non-small cell lung cancer (NSCLC) (2). Although the overall prognosis for patients with lung cancer is poor with a 5-year survival of <15%, patients diagnosed with early stage disease have a much more favorable prognosis. Patients with pathological Stages I and II disease have 5-year survivals of 57–67% and 38–55%, respectively (3, 4). Unfortunately, over half of patients with NSCLC present only after metastasis to lymph nodes or distant sites because of a lack of symptoms early in the disease (4, 5). Detection of lung cancer at earlier stages could potentially increase survival rates by 10–50 fold (6). Using chest X-ray and sputum cytology as screening techniques has proven ineffective in increasing patient survival (7, 8). Recently low dose spiral CT screening in high risk smokers was proven to find more lung tumors and to reduce mortality (3, 4). Identification of lung-cancer specific biomarkers combined with other non-invasive methods may allow for much needed further refinement of lung cancer screening to reduce mortality.
Methylation plays an important role in normal cells as well as in tumor development. In normal cells, it contributes to chromatin organization, silencing of transposable elements, X chromosome inactivation, tissue-specific expression and genetic imprinting (9–11). In addition to general hypomethylation of the genome, hypermethylation (further denoted as ‘methylation’), of CpG islands in gene promoter regions occurs in cancer cells (12). Methylation, one of the most common molecular alterations in human neoplasia refers to the addition of a methyl group to the cytosine ring of those cytosines that precede a guanosine (referred to as CpG dinucleotides) to form methyl cytosine (5-methylcytosine). CpG dinucleotides are found at increased frequency in the promoter region of many genes, and methylation in the promoter region is frequently associated with “gene silencing” (13). Several tumor suppressor genes (TSGs) contain CpG island in their promoters, and many of them show evidence of methylation silencing (9, 10). Aberrant promoter methylation may affect genes involved in cell-cycle control (p16INK4A, p15, Rb and p14) (14–16), DNA repair (MGMT and hMLH1) (10, 17), cell adhesion (H-Cadherin and CDH-1) (18, 19), signal transduction (RASSF1A) (20), apoptosis (DAPK and TMS1)(21) and cell differentiation (RARβ2) (17, 22). Studies in animals and in humans have demonstrated that these epigenetic changes are an early event in carcinogenesis and are present in the precursor lesions of a variety of cancers including breast (23, 24), lung (25, 26), colon (27) and endometrium (28).
The presence of abnormally high DNA concentrations in the sera and plasma of patients with various malignant diseases has been described (29–31). Recent publications have demonstrated the presence of promoter methylation in various bodily fluids including plasma, sputum and bronchoalveloar lavage DNA of lung cancer patients (32–36). Using Quantitative methylation specific PCR (QMSP) to test plasma DNA with a panel of 4 genes (KIF1A, DCC, RAR-β2 and NISCH) we were able to detect 73% of cancer cases with 71% specificity (37). This study evaluated the diagnostic potential of an extended panel of DNA methylation-based markers in pre-therapeutic sera of lung cancer patients. We decided to investigate these markers in patients who had not undergone any form of adjuvant systemic treatment. Of 15 markers tested in an evaluation set, six markers (APC, AIM1, DCC, CDH1, MGMT and RASSF1A) were tested in an independent set of serum samples from lung cancer patients.
MATERIALS AND METHODS
Patients
The gene evaluation set consisted of patients sera (n=17 to 25) collected before any therapeutic intervention and normal controls (n=15 to 614). The test set consists of 76 patients’ sera and sera from 30 age-matched controls. Selected demographics and histopathological characteristics of these 76 lung cancer patients are shown in Table 1. All patients subsequently underwent a biopsy and were confirmed to have lung cancer.
Table 1.
Demographic and clinicopathological parameters of lung cancer patients (n=76)
| na | |
|---|---|
| Gender | |
| Male | 40 |
| Female | 36 |
| Age | |
| ≥ 65 | 39 |
| < 65 | 37 |
| Race | |
| White | 52 |
| African American | 21 |
| Others | 3 |
| Histological type | |
| Adenocarcinoma | 36 |
| SSC | 26 |
| Others | 14 |
| Stage | |
| I | 41 |
| II | 17 |
| III | 11 |
| IV | 5 |
| Unknown | 2 |
| Tumor size | |
| ≥ 3cm | 41 |
| < 3cm | 29 |
| Unknown | 6 |
na, number of cases examined.
SCC, Squamous Cell carcinoma
Collection and Processing of Samples and DNA Preparation
We obtained samples of lung tumor tissue and serum from 10 patients with lung cancer who underwent curative surgery at The Johns Hopkins University School of Medicine. These patients were chosen consecutively on the basis of tissue availability. Tissue specimens were immediately snap-frozen in liquid nitrogen and stored at −80 °C. Frozen tissue was sectioned (12 μm thick), and every tenth section was stained with hematoxylin–eosin and histologically examined for the presence or absence of tumor cells as well as for tumor density. Only sections that showed more than 70%of neoplastic cells were used for DNA extraction.
Blood samples from all the cancer patients were drawn before any therapeutic intervention at at The Johns Hopkins University School of Medicine. The control serum samples consists of subjects enrolled in a community screening study for head and neck cancer approved by the Johns Hopkins institutional review board and through the early detection research networks (EDRN). The experimental protocol was approved by the Johns Hopkins Medical Institutions Institutional Review Board and informed consent was obtained from all enrolled subjects. All subjects were administered a confidential written survey of risk factors for upper aerodigestive tract malignancies, including alcohol and tobacco use as well as the presence of co-morbid illnesses. Smoking was defined as use of tobacco, chewable or smoked, for at least 1year continuously. Heavy alcohol use was defined as intake of more than two alcoholic drinks per day. For Head and Neck surveillance study, all individuals were called by phone once a year afterwards and interviewed to determine interval changes in tobacco and alcohol consumption and health history, including new cancer diagnosis. For the controls used in this study, we excluded those individuals presenting with premalignant or malignant lesions at head and neck area, past history of cancer regardless of site, those who were diagnosed of any cancer regardless of site during follow-up, and those not reachable by phone follow-up.
The blood was centrifuged at 2000 × g for 10 min at room temperature, and 1 ml aliquots of serum/plasma samples were stored at −80°C. DNA was obtained from primary tumor tissues and serum/plasma by digestion with 50 μg/mL proteinase K (Boehringer Mannheim, Germany) in the presence of 1% sodium dodecyl sulfate (SDS) at 48°C for 2 days, followed by phenol/chloroform extraction and ethanol precipitation and finally dissolved in 30 to 60 μL of LoTE (2.5 mmol/L EDTA and 10 mmol/L Tris-HCL).
Bisulfite Treatment
DNA extracted from primary tumors and serum DNA was subjected to bisulfite treatment, which converts unmethylated cytosine residues to uracil residues, as described previously (38), with minor modification. Briefly, 1 to 2 μg of genomic DNA from each sample was denatured with NaOH (final concentration, 0.2M) in a total volume of 20 μL for 20 minutes at 50 °C. The denatured DNA was diluted in 500 μL of a freshly prepared solution of 10 mM hydroquinone and 3 M sodium bisulfite and incubated for 3 hours at 70 °C. Bisulfite-modified DNA was purified using a Wizard DNA Clean-Up System (Promega), treated with 0.3 M NaOH for 10 minutes at room temperature, precipitated with ethanol, resuspended in 60μl– 120 μL of LoTE (2.5 mM EDTA,10 mM Tris–HCl [pH 8]), and stored at −80 °C.
Methylation Analysis
Bisulfite-modified DNA was used as a template for fluorescence-based real-time PCR, as previously described (39). Amplification reactions were carried out in duplicate or triplicate in a volume of 20 μL that contained 3 μL of bisulfite-modified DNA; 600 nM concentrations of forward and reverse primers; 200 nM probe; 5 U of platinum Taq polymerase (Invitrogen); 200 μM concentrations each of dATP, dCTP, and dGTP; 200 μM dTTP; and 5.5 mM MgCl2. Primers and probes were designed to specifically amplify the promoters of the fifteen genes of interest and the promoter of a reference gene, ACTB; primer and probe sequences and annealing temperatures are provided in Supplemental Table 1. Amplifications were carried out using the following profile: 95 °C for 3 minutes, followed by 50 cycles at 95 °C for 15 seconds and 60–62 °C for 1 minute. Amplification reactions were carried out in 384-well plates in a 7900 sequence detector (Perkin-Elmer Applied Biosystems) and were analyzed by a sequence detector system (SDS 2.2.1; Applied Biosystems). Each plate included patient DNA samples, positive (in vitro methylated leukocyte DNA) and negative (normal leukocyte DNA or DNA from a known unmethylated cell line) controls, and multiple water blanks. Leukocyte DNA from a healthy individual was methylated in vitro with excess SssI methyltransferase (New England Biolabs Inc., Beverly, MA) to generate completely methylated DNA, and serial dilutions (90–0.009 ng) of this DNA were used to construct a calibration curve for each plate. All samples were within the assay’s range of sensitivity and reproducibility based on amplification of internal reference standard (threshold cycle [CT] value for ACTB of ≤40). The relative level of methylated DNA for each gene in each sample was determined as a ratio of methylation specific PCR-amplified gene to ACTB (reference gene) and then multiplied by 1000 for easier tabulation (average value of triplicates of gene of interest divided by the average value of triplicates of ACTB × 1000). The samples were categorized as unmethylated or methylated based on the sensitivity of the assay.
Statistical Analysis
The major statistical endpoints in this study involved comparing normal and cancer methylation levels of six genes thought to be associated with lung cancer. The presence or absence of methylation was evaluated for an association with cancer using cross tabulations and chi-square or Fisher=s exact tests as appropriate. Continuous methylation levels were evaluated using logistic regression and receiver operating characteristic (ROC) curves. One gene with 100% specificity was identified in this group of patients. Among those patients without methylation of this gene, logistic regressions utilizing the remaining genes were performed. ROC curves were produced by combining the point of 100% specificity from the first step with the logistic results from the second step. Internal validation of the logistic regression models was done using an approximation to the leave one out jackknife procedure provided by the SAS classification table option.
Methylation values were visually compared using boxplots of the log transformed values. In these plots, the length of the box is the interquartile range (IQR) of the data and depicts the spread of the middle 50 percent of the observations. The median is displayed with a horizontal line inside of this box. The lines extending out from the box extend from the upper and lower quartiles to values defined as adjacent values. The adjacent values are the upper quartile plus 1.5 × IQR and the lower quartile minus 1.5 × IQR. Any value lying outside of this range is displayed with an open circle and can be considered an outlier.
Correlations of the methylation levels of genes were calculated with Spearman correlation coefficients. All p values are two-sided. Computations were performed using the Statistical Analysis System.
RESULTS
Methylation in primary tumor and matched serum
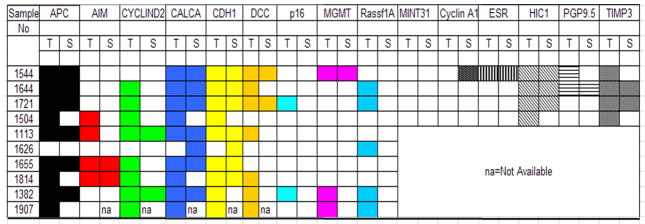
We initially measured the levels of promoter methylation for all the 15 genes (APC, AIM, CyclinD2, CALCA, CDH1, DCC, p16, MGMT, RASSF1, MINT31, CyclinA1, ESR1, HIC1, PGP9.5, TIMP3) in paired primary lung tumor and serum DNA samples from 10 lung cancer patients. Figure 1 summarizes the methylation profiles of each of the 15 genes for the 10 paired samples. In paired samples, the majority of genes methylated in serum DNA were always accompanied by methylation of tumor DNA, where as methylation of tumor DNA was not always accompanied by methylation of serum DNA. No aberrant methylation (i.e., hypermethylation) was detected in the serum of lung cancer patients who did not also have aberrant methylation of the same promoterin the corresponding tumor sample with one exception (sample number 1626). In general, relative methylation levels, which reflect the number of methylated alleles, were higher in tumor DNA than in serum DNA (Supplementary Figure 1). The frequency of methylation of all fifteen genes in primary tumors and the analytical sensitivity of the quantitative methylation–specific PCR assay are summarized in Table 2. Among the 10 primary tumor samples, the frequency of promoter methylation at each of the fifteen loci ranged from 0% (for MINT31, Cyclin A1) to 100% (for HIC1 and TIMP3), and the analytical sensitivity of the individual genes ranged from 0% to 75% (Table 2).
Figure 1.
Methylation pattern of primary tumor and matched serum DNA
Summary of methylation of pattern of 15 genes in corresponding tumor tissue (left) and serum DNA samples (right). Filled boxes represent samples that are methylated; white boxes represent samples devoid of methylation.
Table 2.
Sensitivity of the quantitative methylation-specific polymerase chain reaction assay in lung cancer detection in serum/plasma DNA
| Marker | Number of tumors with methylation/total number of tumors (%, 95% CI*) | No. of patients with methylation in serum or plasma/No. of patients with methylation in primary tumor (analytic sensitivity,† 95% CI) |
|---|---|---|
| APC | 9/10(90, 56 to 100) | 6/9(67, 30 to 93) |
| AIM | 4/10(40, 12 to 74) | 2/4(50, 7 to 93) |
| CYCLIND2 | 8/10(80, 44 to 97) | 2/8(25, 3 to 65) |
| CALCA | 9/10(90, 56 to 100) | 7/9(78, 40 to 97) |
| CDH1 | 9/10(90, 56 to 100) | 6/7(86, 42 to 100) |
| DCC | 7/10 (70, 35 to 93) | 2/6(33, 4 to 78) |
| P16 | 2/10(20, 3 to 56) | 0/2(0, 0 to 84) |
| MGMT | 3/10(30, 7 to 65) | 1/3(33, 1 to 91) |
| RASSF1A | 5/10(50, 19 to 81) | 0/5(0, 0 to 52) |
| MINT31 | 0/4(0, 0 to 60) | 0/0(0, 0 to 100) |
| CyclinA1 | 0/4(0, 0 to 60) | NA |
| ESR | 1/4(25, 1 to 81) | 1/1(100, 3 to 100) |
| HIC1 | 4/4(100, 40 to 100) | 3/4(75, 19 to 99) |
| PGP9.5 | 2/4(50, 7 to 93) | 1/2(50, 1 to 99) |
| TIMP3 | 4/4(100, 40 to 100) | 2/4(50, 7 to 93) |
CI = confidence interval.
The fraction of cases in which methylation of a marker was found in serum DNA for case patients who had confirmed methylation of the same marker in the tumor DNA. Analytical sensitivity were calculated only when tumor and paired DNA were available
Determination of the best performing genes for inclusion in a Gene Evaluation Set for serum DNA
In addition to determine the analytical sensitivity and clinical sensitivity as described above, to further characterize the best performers, we investigated 15 genes in the sera of 15 to 25 patients with lung cancer and 15 to 614 controls with no known neoplastic disease for the presence of aberrant methylation. The controls and cases used here for pre-selection of genes and were based on the availability of DNA from the samples. Due to limited amount of DNA we were not able to evaluate all the 15 genes in the same cohort of samples and we used different set of samples for pre-selection of genes. An overview of the frequency of methylation in this test set in serum samples is given in Table 3A. The most appropriate genes for our additional analyses were determined to be those that show high analytical sensitivity (unmethylated or very low frequency of methylation in serum samples from healthy controls but comparatively high frequency of methylation in serum samples from primary lung cancer patients) and a reported high frequency of lung cancer specific methylation. Based on the latter criteria, a total of six genes, namely APC, AIM1, CDH1, DCC, MGMT and RASSF1A, were selected for further analysis in an independent set of samples. Although we have more promising genes (like HIC1 and PGP9.5) in our evaluation set, we were not able to include all the interested genes in the same cohort of samples due to lack of required DNA from this cohort.
Table 3.
Frequency of methylation
| A. Fifteen genes in gene evaluation set
|
|||
|---|---|---|---|
| Number of methylation positive/number of total cases (Methylation positive %)
| |||
| Markers | Serum or plasma (Cancer) | Serum (Control) | Cutoff values |
| APC | 9/25(36%) | 2/30(7%) | 0 |
| AIM1 | 4/17(24%) | 1/15(7%) | 0 |
| CyclinD2 | 4/17(24%) | 3/35(9%) | 0 |
| CALCA | 12/17(71%) | 23/35(66%) | 0 |
| CDH1 | 10/17(59%) | 1/15(7%) | 0.3 |
| DCC | 3/17(18%) | 0/136(0%) | 0 |
| p16 | 0/25(0%) | 0/30(0%) | 0 |
| MGMT | 5/25(20%) | 1/30(3%) | 0 |
| Rassf1A | 2/25(8%) | 1/30(3%) | 0.1 |
| MINT31 | 0/20(0%) | 0/30(0%) | 0 |
| CyclinA1 | 2/20(10%) | 0/155(0%) | 0 |
| ESR | 1/20(5%) | 0/35(0%) | 0 |
| HIC1 | 10/20(50%) | 42/614(7%) | 0 |
| PGP 9.5 | 3/20(15%) | 9/318(3%) | 0 |
| TIMP3 | 5/20(25%) | 0/30(0%) | 1 |
| B. Six genes in gene independent set
|
|||||
|---|---|---|---|---|---|
| Number of methylation positive/number of total cases (Methylation positive %)
| |||||
| Markers | Serum or plasma (Cancer) | Serum (Control) | P | Sensitivity (%) | Specificity (%) |
| APC | 12/76(16%) | 3/30(10%) | 0.55 | 15.8 | 90 |
| AIM1 | 14/76(18%) | 1/30(3%) | 0.06 | 18.4 | 96.7 |
| CDH1 | 47/76(62%) | 9/30(30%) | 0.003 | 61.8 | 70 |
| DCC | 27/76(36%) | 0/30(0%) | 0.0002 | 35.5 | 100 |
| MGMT | 13/76(17%) | 1/30(3%) | 0.11 | 17.1 | 96.7 |
| Rassf1A | 6/76(8%) | 1/30(3%) | 0.67 | 7.9 | 96.7 |
| 1 of 6 markers | 64/76(84%) | 13/30(43%) | <.0001 | 84.2 | 56.7 |
Detection of lung cancer in an independent set of samples using the 6 gene panel
We tested quantitative analysis of promoter methylationin the serum DNA samples from 76 lung cancer patients and 30 age-matched control subjects. The demographic and clinical characteristics of the 76 lung cancer patients included in this study are summarized in Table 1. The study population was almost equal with males and females and had a median age of 65 years (interquartile range = 42–85 years). All lung cancer cases were eventually confirmed by standard pathology.
Among the serum samples from the 76 lung cancer patients, we detected aberrant methylation of the APC promoter in 12 samples (15.8 %, 95% confidence interval [CI] = 8.4 % to 26%), of the AIM1 promoter in 14 samples (18.4 %, 95 CI = 10.5% to 29.0%), of the CDH1 promoter in 47 samples (61.8%, 95% CI =50% to 73%), of the DCC promoter in 27 samples (35.5%, 95% CI = 25% to47%), of the MGMT promoter in 13 samples (17 %, 95% CI = 9.4% to 27.4%) and of the RassF1a promoter in 6 samples (7.9 %, 95% CI = 3.0% to 16.4%). The box plots in Figure 2 show the distribution of relative methylation values for each of the 6 genes of interest versus ACTB obtained by quantitative methylation–specific PCR using serum DNA from cancer patients and control subjects. These six genes were examined in the independent set to create a combined panel of methylation markers (Table 3B). Eighty-four percent (64 of 76) of patients with cancerous tumors showed methylation of at least one gene, whereas 13 of 30 (43%) of control subjects showed methylation (P = < 0.0001, Chi-Square).
Figure 2. Promoter methylation levels for the six markers in serum DNA from lung cancer patients (CA) and age-matched control subjects (N).
The quantity of methylated alleles of each gene was expressed as the ratio of the amount of polymerase chain reaction products amplified from the methylated gene to the amount amplified from the reference gene β-actin multiplied by 1000. Box plots show the middle 50% of the data, the line is the median, and the bars extend the median by 1.5 times the interquartile range.
Figure 3A depicts the combined two-stage algorithm that we used for disease classification. The ROC curves obtained by using the two-stage approach, which was based on 1 marker (DCC) with 100% specificity followed by logistic regression analysis on the remaining five markers, are shown in Figure 3B. Both curves have been corrected for over-fitting via internal validation. The DCC gene that had 100% specificity correctly identified 35.5% (95% CI 25%, 47%) of the 76 lung cancer patients (Figure 3B). For patients without methylated DCC, addition of a logistic regression score that was based on the five remaining genes improved sensitivity from 35.5% to 75% (95% CI: 64%, 84%) but decreased the specificity from 100% to 73% (54%, 88%) (Figure 3B). The detailed regression coefficients for the additional genes were also performed (data not shown). We then compared this overall ROC curve to those obtained by adding each of the five genes individually to the model (data not shown). As expected, we found that the individual genes performed less well as predictors than did the multivariable logistic score using the entire group.
Figure 3.

A. Two-stage algorithm for disease classification. Patients who were positive for promoter methylation of DCC were classified as having cancer. Those who were negative for promoter methylation of DCC were analyzed in a second stage, in which a logistic risk score was calculated based on adding additional markers (see text for details). B. ROC curve based on logistic scores using binary dichotomization of the genes at zero/nonzero methylation levels (Lighter line). ROC curve based on logistic scores using the actual log methylation levels (darker line). Both curves were corrected for overfitting via internal validation.
We also examined how the multivariable logistic score model performed for detecting stage I or stage II tumors. The sensitivity of the model increased with more advanced tumor stage, ranging from 73 % for stage I to 76.5 % of stage II tumors detectable by quantitative methylation–specific PCR with a specificity of 73% and 77% respectively Sample size did not permit this analysis for the stage III-IV subset.
Logistic regression was used to examine associations between clinicopathologic and demographic parameters (age ≥ 65 at diagnosis, tumor stage ≥ III, adeno cell type and tumor size ≥ 3cm) and the methylation status of the six genes chosen for testing in the final data set. CDH1 methylation was the only gene significantly associated with any of these factors. The presence of any CDH1 methylation decreased the probability of tumor size ≥ 3 cm, OR=0.22 (95% CI: 0.06, 0.74), p=0.01.
An indicator for the aberrant methylation of any one of the six genes investigated in serum DNA of lung cancer patients was also not associated with other clinical or demographic characteristics (data not shown). Methylation frequencies of 6 genes for different histological subtypes are shown in Supplementary Table 2.
Finally, we performed a correlation analysis for all pairs of markers (Supplementary Table 3). Promoter methylation of APC and CDH1 were often observed together and this was statistically significant: ρ=0.27, P=.01.
Discussion
This study confirms and extends previous observations that identification of serum DNA methylation in specific set of genesis a potentially useful approach to detect lung cancer patients. Serum DNA methylation was more frequently observed in patients with lung cancer than those with age-matched controls. Although the sensitivity for the diagnosis of lung cancer was only 35.5% when analyzed by a single gene (DCC), the high specificity (100%) indicates the usefulness of QMSP assay for lung cancer detection. For patients without methylated DCC, addition of a logistic regression score that was based on the five remaining genes improved sensitivity from 35.5% to 75% but decreased the specificity from 100% to 73%. Addition of gene/genes with reasonable sensitivity and very high specificity with DCC may allow higher diagnostic coverage while still proving 100% specificity. Of note, serum DNA methylation could be identified even in patients in the early stages of lung cancer, whereas conventional serum protein tumor markers were rarely elevated, indicating that this DNA-based method is more sensitive than protein-based method for diagnosis of lung cancer in early stage. In our previous study (37), DCC methylation was detected in 54% of cases with 100% specificity in evaluation set and fell to 26% in the independent set. In the present cohort DCC methylation was detected in 36% of cases, which compares favorably and suggests that about a third of lung cancer cases could be detected by DCC alone with high specificity.
In former studies, methylation in tumor tissues was detected in 25% to 41% for p16INK4a, 30% to 40% for RASSF1A, and 16% to 27% for MGMT (26). These results were consistent with our data in 10 tumor tissue samples. The frequency of detecting methylated genes in serum was about half to two thirds compared with that in tumor tissues. However, when we consider that tumor-derived DNA in blood is generally detectable in less than half of cancer patients (32), the frequency of methylation in serum DNA in our study may be quite reasonable. Laird reviewed the studies examining the methylation status of serum/plasma DNA in patients with various neoplasms and indicated that clinical sensitivity of DNA methylation was ~50% (40). Esteller et al. did methylation analysis in serum DNA from patients with non–small cell lung cancer for multiple genes and showed 33% to 80% clinical sensitivity by combination analysis of several genes (41).
Among various techniques used for methylation analysis, we adopted a simple method of QMSP analysis. The specificity of the primers and probes we used in this study has been verified using genomic sequencing and/or MSP. Recently, several studies showed improved detection rates of methylation status using a nested PCR approach or a quantitative real-time PCR technique (42, 43). The sensitivity of the Taqman method was reported to be 10-fold higher than conventional qualitative MSP and we feel that more sensitive technologies are very likely to reduce specificity (44).
Although promoter methylation was observed predominantly in lung cancer patients, several controls were methylation positive for one of the 5 genes (except DCC) in the independent test set. In methylation pattern analysis in primary tumors and paired serum samples, one lung cancer patient with serum DNA methylation for CALCA and CDH1 did not show the same alteration in the corresponding tumor tissue. We considered the following as possible explanation of these apparent false positive results. Firstly, the methylated serum DNA might be derived from undetected precancerous lesions in these cases. According to previous reports, aberrant promoter methylation is clearly detectable in precancerous lesions, such as dysplasia and nonmalignant lung tissues of patients with lung cancer (45, 46). In a prospective study Belinsky et al. reported that 3 genes in a panel of 6 were hypermethylated in the sputum of high risk individuals, resulting in a >6 fold risk of developing lung cancer within 18 months (47). Long term follow up of control subjects with methylated DNA of several genes in serum samples will elucidate the risk of developing lung cancer in these subjects. Secondly, aberrant methylation might be caused by environmental factors, such as smoking, yet may not necessarily lead to clonal expansion (25, 48). Finally, endogenous factors like the occurrence of other occult malignancies or age-related methylation could be the cause in the control group.
Hypermethylation of the APC and CDH1 promoters in serum DNA from lung cancer patients often occurred together. Given that hypermethylation of these genes is common in lung cancer, these associations could have occurred by chance alone and should be interpreted with caution. However, a possible interaction between these genes in lung cancer deserves further evaluation. CDH1 methylation occurred more often in patients with tumor sizes < 3 cm (20/24) 83% compared to patients with tumors ≥ 3 cm (24/46) 52%. p=0.01. It is well studied that CDH1 inactivation is related to invasion, metastasis and increased tumor size. So our finding of presence of any CDH1 methylation decreased the probability of tumor size ≥ 3 cm (OR=0.22 (95% CI: 0.06, 0.74), p=0.01) need to be further evaluated.
To consider the use of serum DNA methylation as a marker in lung cancer active screening, several issues must be considered. Certainly optimal specificity and sensitivity must be achieved before the approach can be used alone or in combination with imaging. It is tempting to combine methylation with promising methods such as low-dose spiral computed tomography (49, 50). Because one of the serious limitations of low-dose spiral computed tomography is its poor specificity, a combination with serum DNA methylation may overcome this limitation.
Serum DNA methylation is found in early-stage disease (73% and 77% in stage I and stage II respectively) and thus could be tested before invasive procedures after testing in a larger cohort and in a prospective setting. Although further evaluation is essential, the results in this study indicate the substantial usefulness of methylation marker for detection of lung cancer. Further studies are warranted to confirm the accuracy of the approach and search for the best combination of genes for methylation analysis. Moreover, it is important to investigate prospectively whether methylation-positive non-clinical cancer cases will ultimately develop malignancies in the near future.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
Identification of blood-based non-invasive or minimally invasive detection markers will improve the clinical management of NSCLC, the leading cause of cancer-related deaths in the United States and world-wide. From a panel of 15 cancer-specific methyalted genes we derived a 6-gene panel to identify NSCLC in blood by quantitative methylation specific PCR with high specificity. It would be envisaged that this simple, reliable and non-invasive blood test could aid the early detection of NSCLC in a high risk population, and could be used most effectively to direct imaging modalities with low specificity such as spiral CT. Such a test could therefore have a significant impact on the long term survival of these individuals. Moreover, these test can be develop to monitor the response to therapy and some of the methylated marker can be selected for targeted therapy in future.
Acknowledgments
This work was supported by National Cancer Institute Grant U01-CA84986, Oncomethylome Sciences, SA. The funding agencies had no role in the design of the study, data collection, analysis, interpretation of the results, preparation of the manuscript, or the decision to submit the manuscript for publication. Under a licensing agreement between Oncomethylome Sciences, SA and the Johns Hopkins University, D.S. is entitled to a share of royalty received by the University upon sales of diagnostic products described in this article. D.S. owns Oncomethylome Sciences, SA stock, which is subject to certain restrictions under University policy. D.S. is a paid consultant to Oncomethylome Sciences, SA and is a paid member of the company’s Scientific Advisory Board. The Johns Hopkins University in accordance with its conflict of interest policies is managing the terms of this agreement. Dr. Begum is supported by a Young Clinical Scientist Award from the Flight Attendant Medical Research Institute and Dr. Hoque is supported by the Young Investigator Award from the International Association for the Study of Lung Cancer. This paper/analysis is based on a web database application provided by Research Information Technology Systems (RITS): https://www.rits.onc.jhmi.edu/
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Mulshine JL, Sullivan DC. Clinical practice. Lung cancer screening. N Engl J Med. 2005;352:2714–20. doi: 10.1056/NEJMcp042630. [DOI] [PubMed] [Google Scholar]
- 3.Lu C, Soria JC, Tang X, et al. Prognostic factors in resected stage I non-small-cell lung cancer: a multivariate analysis of six molecular markers. J Clin Oncol. 2004;22:4575–83. doi: 10.1200/JCO.2004.01.091. [DOI] [PubMed] [Google Scholar]
- 4.Singhal S, Vachani A, Antin-Ozerkis D, Kaiser LR, Albelda SM. Prognostic implications of cell cycle, apoptosis, and angiogenesis biomarkers in non-small cell lung cancer: a review. Clin Cancer Res. 2005;11:3974–86. doi: 10.1158/1078-0432.CCR-04-2661. [DOI] [PubMed] [Google Scholar]
- 5.Wardwell NR, Massion PP. Novel strategies for the early detection and prevention of lung cancer. Semin Oncol. 2005;32:259–68. doi: 10.1053/j.seminoncol.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Wingo PA, Ries LA, Giovino GA, et al. Annual report to the nation on the status of cancer, 1973–1996, with a special section on lung cancer and tobacco smoking. J Natl Cancer Inst. 1999;91:675–90. doi: 10.1093/jnci/91.8.675. [DOI] [PubMed] [Google Scholar]
- 7.Marcus PM. Lung cancer screening: an update. J Clin Oncol. 2001;19:83S–86S. [PubMed] [Google Scholar]
- 8.Ellis JR, Gleeson FV. Lung cancer screening. Br J Radiol. 2001;74:478–85. doi: 10.1259/bjr.74.882.740478. [DOI] [PubMed] [Google Scholar]
- 9.Merlo A, Herman JG, Mao L, et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–92. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M, Levine R, Baylin SB, Ellenson LH, Herman JG. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413–7. doi: 10.1038/sj.onc.1202178. [DOI] [PubMed] [Google Scholar]
- 11.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 12.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–74. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 13.Leonhardt H, Cardoso MC. DNA methylation, nuclear structure, gene expression and cancer. J Cell Biochem Suppl. 2000;Suppl 35:78–83. doi: 10.1002/1097-4644(2000)79:35+<78::aid-jcb1129>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 14.Little M, Wainwright B. Methylation and p16: suppressing the suppressor. Nat Med. 1995;1:633–4. doi: 10.1038/nm0795-633. [DOI] [PubMed] [Google Scholar]
- 15.Baur AS, Shaw P, Burri N, et al. Frequent methylation silencing of p15(INK4b) (MTS2) and p16(INK4a) (MTS1) in B-cell and T-cell lymphomas. Blood. 1999;94:1773–81. [PubMed] [Google Scholar]
- 16.Greger V, Passarge E, Hopping W, Messmer E, Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet. 1989;83:155–8. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 17.Esteller M. Cancer epigenetics: DNA methylation and chromatin alterations in human cancer. Adv Exp Med Biol. 2003;532:39–49. doi: 10.1007/978-1-4615-0081-0_5. [DOI] [PubMed] [Google Scholar]
- 18.Yoshiura K, Kanai Y, Ochiai A, et al. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci U S A. 1995;92:7416–9. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato M, Mori Y, Sakurada A, Fujimura S, Horii A. The H-cadherin (CDH13) gene is inactivated in human lung cancer. Hum Genet. 1998;103:96–101. doi: 10.1007/s004390050790. [DOI] [PubMed] [Google Scholar]
- 20.Dammann R, Li C, Yoon JH, et al. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–9. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 21.Conway KE, McConnell BB, Bowring CE, et al. TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Res. 2000;60:6236–42. [PubMed] [Google Scholar]
- 22.Cirincione R, Lintas C, Conte D, et al. Methylation profile in tumor and sputum samples of lung cancer patients detected by spiral computed tomography: a nested case-control study. Int J Cancer. 2006;118:1248–53. doi: 10.1002/ijc.21473. [DOI] [PubMed] [Google Scholar]
- 23.Umbricht CB, Evron E, Gabrielson E, et al. Hypermethylation of 14-3-3 sigma (stratifin) is an early event in breast cancer. Oncogene. 2001;20:3348–53. doi: 10.1038/sj.onc.1204438. [DOI] [PubMed] [Google Scholar]
- 24.Evron E, Dooley WC, Umbricht CB, et al. Detection of breast cancer cells in ductal lavage fluid by methylation-specific PCR. Lancet. 2001;357:1335–6. doi: 10.1016/s0140-6736(00)04501-3. [DOI] [PubMed] [Google Scholar]
- 25.Belinsky SA, Nikula KJ, Palmisano WA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95:11891–6. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zochbauer-Muller S, Minna JD, Gazdar AF. Aberrant DNA methylation in lung cancer: biological and clinical implications. Oncologist. 2002;7:451–7. doi: 10.1634/theoncologist.7-5-451. [DOI] [PubMed] [Google Scholar]
- 27.Esteller M, Sparks A, Toyota M, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–71. [PubMed] [Google Scholar]
- 28.Tsuda H, Yamamoto K, Inoue T, Uchiyama I, Umesaki N. The role of p16-cyclin d/CDK-pRb pathway in the tumorigenesis of endometrioid-type endometrial carcinoma. Br J Cancer. 2000;82:675–82. doi: 10.1054/bjoc.1999.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sidransky D, Von Eschenbach A, Tsai YC, et al. Identification of p53 gene mutations in bladder cancers and urine samples. Science. 1991;252:706–9. doi: 10.1126/science.2024123. [DOI] [PubMed] [Google Scholar]
- 30.Ngan RK, Lau WH, Yip TT, et al. Remarkable application of serum EBV EBER-1 in monitoring response of nasopharyngeal cancer patients to salvage chemotherapy. Ann N Y Acad Sci. 2001;945:73–9. doi: 10.1111/j.1749-6632.2001.tb03866.x. [DOI] [PubMed] [Google Scholar]
- 31.Lo YM. Circulating nucleic acids in plasma and serum: an overview. Ann N Y Acad Sci. 2001;945:1–7. doi: 10.1111/j.1749-6632.2001.tb03858.x. [DOI] [PubMed] [Google Scholar]
- 32.Usadel H, Brabender J, Danenberg KD, et al. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer Res. 2002;62:371–5. [PubMed] [Google Scholar]
- 33.Topaloglu O, Hoque MO, Tokumaru Y, et al. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10:2284–8. doi: 10.1158/1078-0432.ccr-1111-3. [DOI] [PubMed] [Google Scholar]
- 34.Palmisano WA, Divine KK, Saccomanno G, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–8. [PubMed] [Google Scholar]
- 35.Belinsky SA, Palmisano WA, Gilliland FD, et al. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62:2370–7. [PubMed] [Google Scholar]
- 36.Ahrendt SA, Chow JT, Xu LH, et al. Molecular detection of tumor cells in bronchoalveolar lavage fluid from patients with early stage lung cancer. J Natl Cancer Inst. 1999;91:332–9. doi: 10.1093/jnci/91.4.332. [DOI] [PubMed] [Google Scholar]
- 37.Ostrow KL, Hoque MO, Loyo M, et al. Molecular analysis of plasma DNA for the early detection of lung cancer by quantitative methylation-specific PCR. Clin Cancer Res. 16:3463–72. doi: 10.1158/1078-0432.CCR-09-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoque MO, Feng Q, Toure P, et al. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol. 2006;24:4262–9. doi: 10.1200/JCO.2005.01.3516. [DOI] [PubMed] [Google Scholar]
- 39.Hoque MO, Begum S, Topaloglu O, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst. 2006;98:996–1004. doi: 10.1093/jnci/djj265. [DOI] [PubMed] [Google Scholar]
- 40.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–66. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 41.Esteller M, Sanchez-Cespedes M, Rosell R, et al. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59:67–70. [PubMed] [Google Scholar]
- 42.An Q, Liu Y, Gao Y, et al. Detection of p16 hypermethylation in circulating plasma DNA of non-small cell lung cancer patients. Cancer Lett. 2002;188:109–14. doi: 10.1016/s0304-3835(02)00496-2. [DOI] [PubMed] [Google Scholar]
- 43.Harden SV, Tokumaru Y, Westra WH, et al. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003;9:1370–5. [PubMed] [Google Scholar]
- 44.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zochbauer-Muller S, Fong KM, Virmani AK, et al. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–55. [PubMed] [Google Scholar]
- 46.Brabender J, Usadel H, Metzger R, et al. Quantitative O(6)-methylguanine DNA methyltransferase methylation analysis in curatively resected non-small cell lung cancer: associations with clinical outcome. Clin Cancer Res. 2003;9:223–7. [PubMed] [Google Scholar]
- 47.Belinsky SA, Liechty KC, Gentry FD, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–44. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 48.Tsou JA, Hagen JA, Carpenter CL, Laird-Offringa IA. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene. 2002;21:5450–61. doi: 10.1038/sj.onc.1205605. [DOI] [PubMed] [Google Scholar]
- 49.Henschke CI. Early lung cancer action project: overall design and findings from baseline screening. Cancer. 2000;89:2474–82. doi: 10.1002/1097-0142(20001201)89:11+<2474::aid-cncr26>3.3.co;2-u. [DOI] [PubMed] [Google Scholar]
- 50.Swensen SJ, Jett JR, Sloan JA, et al. Screening for lung cancer with low-dose spiral computed tomography. Am J Respir Crit Care Med. 2002;165:508–13. doi: 10.1164/ajrccm.165.4.2107006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.