Abstract
Unlike BCR and secreted immunoglobulin, TCR expression is not currently thought to occur in a bivalent form. The conventional monovalent model of TCR/CD3 is supported by published studies of complexes solubilized in the detergent digitonin, in which bivalency was not observed. We have revisited the issue of TCR valency by examining complexes isolated from primary αβ T cells after solubilization in digitonin. Using immunoprecipitation followed by flow cytometry (IP-FCM), we unexpectedly observed TCR/CD3 complexes that contained two TCRs per complex. Standard anti-TCR antibodies, being bivalent themselves, tended to bind with double occupancy to bivalent TCRs; this property masked the presence of the second TCR per complex in certain Ab binding assays, which may partially explain why previous data did not reveal these bivalent complexes. We also found that the prevalence of bivalency among fully assembled, mature TCR/CD3 complexes was sufficient to impact the functional performance of immunoprecipitated TCRs in binding antigenic peptide/MHC-Ig fusion proteins. Both TCR positions per bivalent complex required an antigen-specific TCR in order to effect optimal binding to these soluble ligands. Therefore, we conclude that in primary T cells, TCR/CD3 complexes can be found that are physically and functionally bivalent. The expression of bivalent TCR/CD3 complexes has implications regarding potential mechanisms by which antigen may trigger signaling. It also suggests the possibility that the potential for bivalent expression could represent a general feature of antigen receptors.
Introduction
TCR is highly related to BCR in terms of evolutionary pedigree, gene structure, recombinase-dependent gene rearrangement during development, protein domain organization, and expression within multiprotein signaling complexes (1). However, one major structural difference between these two receptors is that whereas transmembrane BCR and secreted Ab are at least bivalent, current models suggest that TCR is not. As a result, most paradigms of T cell activation predict that low affinity binding of peptide/MHC (pMHC) to monvalent αβ TCR represents the decisive molecular event of antigen recognition, the initial interaction that culminates in TCR aggregation and T cell signaling (2).
Because TCR/CD3 is expressed only in a transmembrane complex with no naturally secreted form, its valency has been studied via biochemical analyses involving immunoprecipitation (IP) and other methods. The general format of the definitive IP experiment has been to examine T cells that express two different TCRs, allowing IP of one TCR to be followed by Western blotting for the second TCR to test for their inclusion in shared complexes. Three groups reported that there was little to no co-association between TCRs under these conditions (3–5). Importantly, the detergent digitonin was used in all of those studies, since digitonin is known to maintain TCR/CD3 associations while excluding extraneous proteins from the complex (6). Due to this property, digitonin has been used to solubilize the αβ TCR/CD3 complex, and define its subunit constituency and stoichiometry as αβγε2ζ2 (7).
The possibility that TCR/CD3 might be bi- or polyvalent is a controversial idea that is not new (8, 9), though it has been supported by few studies. Using the same strategy described above, two groups reported co-association by IP of two different TCRs when solubilized in Brij-family detergents (10, 11), although it is known that Brij lysates fail to separate TCR/CD3 from extraneous membrane proteins (12, 13). Still, these groups reported Förster resonance energy transfer (FRET) between fluorescent Ab-labeled surface TCR (10), and concatemeric expression of heterogeneous numbers of TCR observed via electron microscopy and blue native polyacrylamide gel electrophoresis (BN-PAGE) (11). Therefore, it has been proposed that digitonin-solubilized complexes are monovalent (7), with higher orders of concatemeric complexes detectable by alternative methods that avoid complete membrane solubilization (14). Notably, no published data has previously provided empirical evidence for specific bivalency, in either digitonin-solubilized TCR/CD3, or putative higher-order concatemers of heterogeneous copy number.
We have revisited the issue of αβ TCR valency by using IP-FCM, a sensitive technique for analyzing the subunit constituency of native multiprotein complexes (15–19). Primary T cells provided the source of TCR/CD3 complexes, which were solubilized in digitonin, a condition previously used to define TCR/CD3 valency. The present data support a model wherein a significant proportion of TCR/CD3 complexes display bivalency, their prevalence being sufficient to impact the outcome of a functional antigen binding assay. Additionally, understanding the conditions that govern detection of both TCRs in these bivalent complexes allows a plausible explanation to be suggested as to why they may not have been readily detectable in previous experimental systems. These observations evoke the speculation that the potential for bivalent expression could represent a general feature of the antigen receptors that mediate adaptive immunity.
Materials and Methods
Mice
BALB/c and C56BL/6 (B6) were purchased from the Jackson Laboratory. DO11.10 (BALB/c) (20), DO11.10/RAG20, 2C, OT1, and 2C × OT1 (F1) mice were bred and maintained in our animal facilities, and all mice were used between 6–16 weeks of age. Animal procedures were in accordance with IACUC regulations at Mayo Clinic, University of Pennsylvania, and University Hospital-Basel.
Antibodies
Purified mAbs were obtained from hybridoma supernatant: B20.1 (anti-Vα2); MR9-4 (anti-Vβ5); 1B2 (anti-2C TCR); 145-2C11 (anti-CD3ε); 37.51 (anti-CD28); H57–597 (anti-TCRβ); H146 (anti-CD3ζ); B21.14 (anti-Vα8). Purified AF6–120.1 (anti-I-Ab) was purchased from BD Pharmingen. PE-conjugated mAbs purchased from BD Pharmingen included: G155–178 (Mouse Ig); R35–95 (Rat Ig); A19-3 (Hamster Ig); 30-F11 (anti-CD45); 53-2.1 (anti-Thy1.2); H129 (anti-CD4); 53.6.7 (anti-CD8α); 53.5.8 (anti-CD8β); AMS-32.1 (anti-H-2Kd); B20.1 (anti-Vα2); MR9-4 (anti-Vβ5); 145-2C11 (anti-CD3ε); H57–597 (anti-TCRβ); B21.14 (anti-Vα8). PE-conjugated KJ126 (anti-DO11.10 αβ TCR) was purchased from Caltag. H2-Ld-Ig fusion protein and associated reagents, including PE-conjugated secondary mAb, were purchased from BD Pharmingen. Where indicated, H2-Ld-Ig was loaded with exogenous p2Ca or QL9 peptides following manufacturer instructions.
Fabs
Fab fragments were prepared using papain digestion as previously described (21). Following cleavage, Protein A-sepharose beads (Pierce) were used to bind and remove Fc fragments from the digest. Fabs were then de-salted and passed over a Resource Q anion exchange column (GE Healthcare). Homogeneous eluate fractions were subjected to SEC using a Superdex 200 10/300 GL column (GE Healthcare) to isolate fractions corresponding to ~50 kDa. Final fractions were coupled to IP-FCM beads or FITC, and those capable of binding specific antigen from lysates were used in experiments.
T cells
Unless otherwise stated, whole splenocyte and/or lymph node cells were used as the source of T cells without further isolation. Where noted, cells were stained with anti-Vα2-FITC, anti-Vα8-PE, and anti-Thy1.2-APC, and specific cell subsets were isolated by FACS sorting using FACSVantage or FACSAria cytometers (BD). Sorted cells were cultured in vitro in a 1:1 mixture of Aim V (Gibco) and RPMI (+10% Cosmic Calf serum, HyClone Laboratories), with 1:40 addition of tissue culture supernatant from X63Ag8–653.IL2 cells that secrete recombinant murine IL-2 (22). To remove the mAbs used for FACS sorting, and to expand the cell numbers, sorted cells were stimulated in vitro with plate-bound anti-CD3ε and anti-CD28 mAbs for 3 days, followed by re-plating in mAb-free wells where expansion occurred for 9–11 days prior to lysate preparation.
Lysates
Cells were lysed in buffer containing 50 mM Tris, 150 mM NaCl, freshly added protease inhibitors (P2714 and A8456, Sigma-Aldrich), and 1% Digitonin (ultra-pure grade, Calbiochem), except where an alternative non-ionic detergent is specified (saponin, Sigma-Aldrich). Lysis was performed at a concentration of up to 200 × 106 cells/mL of lysis buffer. Nucleii and cellular debris were discarded by centriguation such that post-nuclear lysates were used in all experiments.
IP-FCM
The method and protocol have been previously described in detail (17, 18, 23). Briefly, IP Abs or Fabs were covalently coupled to polystyrene latex beads. These IP beads were incubated with lysates from which protein complexes were captured. Subsequently, beads were washed, probed with PE- or FITC-conjugated probe Abs, and analyzed by FCM. Fluorescence data was analyzed using FlowJo (Treestar, Inc.) or CFLow (Accuri, Inc.) software, and is displayed in raw format without smoothing or scaling except as noted in Figure 5. Although the minimum number of acquired events suggested for commercial bead-based flow cytometry applications is 25, we acquired 500–3500 bead events for all samples. The y-axis for all histograms is “bead count”, which is omitted from figure display. In some experiments, the geometric mean fluorescence intensity (gMFI) was compared to fluorescent bead standards to convert semi-quantitative data into estimated number of PE molecules (#PE) per bead, as described previously (17, 18). PE-conjugated mAbs used in these experiments were verified to possess PE:mAb ratio = 1:1 by at least one of two means: (i) communication with commercial departments of Technical Service; (ii) in-house FPLC fractionation and purification of the 1:1 conjugation product (data not shown).
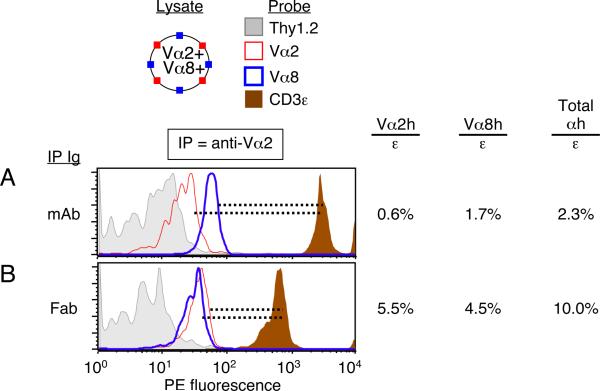
Figure 5. From dual-TCR T cells, IP with anti-Vα2 Fab improves detection of both Vα2+ and Vα8+ TCRs present in shared complexes.
These data are associated with the experiment shown in Figure 4. To improve the legibility of the four overlain histograms, FCM plots were smoothed and scaled. (A) This data is identical to that shown in Fig. 4B, left, with the addition of the anti-Vα2-PE probe. The anti-Vα2 mAb IP captured complexes from which relatively few free Vα2+ and Vα8+ TCRs were detected (Vα2h/ε = 0.6%; Vα8h/ε = 1.7%). (B) Capture of TCR/CD3 complexes with anti-Vα2 Fab-beads improved the detection of free Vα2+TCRs (Vα2h/ε = 5.5%), and free Vα8+ TCRs (Vα8h/ε = 4.5%). Data in both panels represent one of three experiments.
Statistics
Where indicated, Student's t-tests were performed using Microsoft Excel software for duplicate data samples within a representative experiment. For summary fluorescence data, either geometric mean fluorescence intensity (gMFI) or median values are displayed with standard error bars.
Results
IP-FCM assesses specific inclusion of subunits and exclusion of extraneous proteins in TCR/CD3
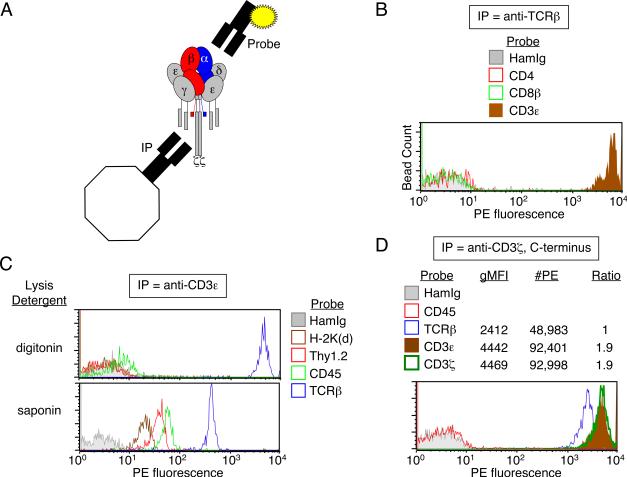
In IP-FCM, immunoprecipitating mAbs are covalently coupled to polystyrene latex beads that are used to capture a specific protein (primary analyte) in native conformation from cell lysates. The beads are probed with a fluorochrome-conjugated mAb that may be specific for another protein (secondary analyte) in shared complexes with the primary analyte (Fig. 1A). We performed IP-FCM analysis of TCR/CD3 isolated from primary BALB/c T cells after lysis in digitonin. Complexes were immunoprecipitated with H57 anti-TCRβ mAb-coupled microbeads, separated into parallel samples, and probed with a PE-conjugated mAb specific for the extracellular domain of either CD4, CD8β, or CD3ε (see Materials and Methods and associated references). We found that whereas the known subunit CD3ε was clearly found in complexes together with TCRβ, the functionally relevant coreceptors CD4 and CD8 were not physically associated (Fig. 1B). Conversely, IP of digitonin-solubilized complexes with 2C11 anti-CD3ε mAb confirmed specific inclusion of CD3ε in complexes with TCRβ; however, neither the highly expressed membrane proteins Thy1.2 and CD45, nor the potential TCR ligand H-2Kd were found in digitonin-solubilized TCR/CD3 complexes (Fig. 1C, top). In contrast to digitonin, the detergent saponin did not fully solubilize cell membranes (24), as observed by failure to fully separate TCR/CD3 from extraneous membrane proteins such as H-2Kd, Thy1.2, and CD45 (Fig. 1C, bottom). These data confirm that IP-FCM analysis involving digitonin-solubilized complexes allows TCR/CD3 to be isolated and detected with high stringency and specificity. To estimate TCR/CD3 subunit stoichiometry, we performed IP-FCM analysis on digitoninsolubilized TCR/CD3 from primary BALB/c T cells by capturing complexes with H146, a mAb that binds a C-terminal epitope of the cytoplasmic domain of CD3ζ (25). Captured complexes were probed in parallel with PE-conjugated mAbs (PE:mAb conjugation ratio = 1:1) specific for the extracellular domains of TCRβ or CD3ε, or an N-terminal epitope of the intracellular domain of CD3ζ, to minimize possible steric interference between the paired capture and detection mAbs. Since TCR/CD3 is composed of the associated dimers αβ/εγ/εδ/ζζ, we found that IPFCM confirmed the conventionally accepted dimer ratios (Fig. 1B, β:ε:ζ = 1:2:2) that were previously observed by experiments involving IP (4) and BN-PAGE (11). Thus, TCR/CD3 complexes analyzed by IP-FCM appear to contain the expected subunits in their expected relative quantities.
Figure 1. IP-FCM assesses specific inclusion of subunits and exclusion of extraneous proteins in TCR/CD3.
(A) Schematic representation of TCR/CD3 associated subunits captured by immunoprecipitating (IP) beads and probed with a PE-conjugated probe mAb. (B) BALB/c T cells were lysed in 1% digitonin, and TCR/CD3 complexes were immunoprecipitated with H57 anti-TCRβ mAb and probed with PE-conjugated probe mAbs. The CD3ε probe shows that many complexes were captured on the beads, but not the functionally related CD4 or CD8 coreceptors. The relative number of beads whose fluorescence is measured is denoted as “Bead Count”, a label that is omitted from histogram y-axes hereafter. (C) BALB/c T cells were lysed in either 1% digitonin, which maintains TCR/CD3 complex integrity while separating it from extraneous membrane proteins, or 1% saponin, which does not display these properties but preserves “membrane chunks”. TCR/CD3 complexes were immunoprecipitated with anti-CD3ε and probed with various PE-conjugated probe mAbs. The TCRβ probe shows plentiful capture of complexes from the digitonin lysates, with exclusion of MHC class I (H-2Kd), and the highly expressed plasma membrane proteins, Thy1.2 and CD45. However, in saponin, these extraneous proteins are present on the beads together with TCR/CD3. (D) BALB/c T cell lysates were incubated with H146 mAb-coupled beads specific for the cytoplasmic C-terminus of CD3ζ. Captured TCR/CD3 complexes were probed in parallel samples with PE-conjugated probe mAbs that were either specific for TCR/CD3 subunits or control proteins. Following published methods (17), the geometric mean fluorescence intensity (gMFI) was compared to fluorescent bead standards to convert semi-quantitative data into estimated number of PE molecules (#PE) per bead (17, 18). Either gMFI or #PE readily allowed calculation of apparent subunit ratios, which displayed the expected TCR/CD3 stoichiometry. Data in all panels represent one of at least two experiments.
Observation of TCR/CD3 complexes containing two β-chains
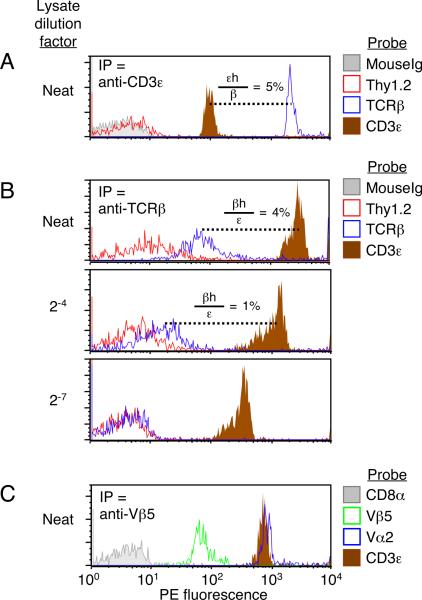
By using the same mAb for both IP and probe, it was possible to observe the inclusion of multiple copies of identical subunits within complexes. Thus, when complexes were captured using anti-CD3ε mAb and probed with a PE-conjugated version of the same mAb, a positive assay signal was obtained (Fig. 2A, brown histogram). Since each TCR/CD3 complex contains more than one copy of CD3ε (26), one must be required for binding the complex to the anti-CD3ε IP bead, while another may be free for detection by a probe mAb. We refer to plural subunits that could be detected under these conditions with the notation `h' (`hanging', e.g., εh). Because detection of εh was relatively low compared to the total number of complexes captured on beads (Fig. 2A, εh/β = 5%), we reasoned that most ε epitopes in the captured complexes were either occupied by IP mAbs on the beads or otherwise sterically inaccessible to probes.
Figure 2. Multiple copies of identical TCR/CD3 subunits in shared complexes detected by IP-FCM.
(A–B) TCR/CD3 complexes were captured from BALB/c T cell lysates that were either undiluted (neat) or diluted in excess lysis buffer by the factor indicated. (A) The 2C11 anti-CD3ε mAb was used for both IP and probe. CD3ε subunits that could be probed (and thus did not mediate binding of the complex to the IP bead) are referred to as `hanging' (εh), and their relative detection is reported as the ratio of εh to β-chain gMFI (107/2340 = 5%). Thy1.2 is a highly expressed surface T cell marker whose absence from the captured complexes indicates adequate membrane solubilization and TCR/CD3 isolation. (B) H57 anti-TCRβ mAb was used for both IP and probe in the analysis of captured TCR/CD3 complexes. As in (A), β-chains that could be probed under these conditions are `βh'. The ratio of detectable βh to CD3ε is calculated from gMFI (110/2838 = 4%; 15/1076 = 1%). (C) TCR/CD3 complexes from OT1 TCR transgenic T cell lysates were captured with anti-Vβ5 and probed in parallel with PE-conjugated probe mAbs. MR9-4 anti-Vβ5 mAb was used for both IP and probe. Data in all panels represent one of at least two experiments.
A similar detection pattern occurred when the H57 anti-TCRβ mAb was used as both capture and probe reagent (Fig. 2B, blue histograms), despite the expectation that only one β-chain would be expressed in each digitonin-solubilized TCR/CD3 complex. The H57 anti-TCRβ mAb binds all mouse TCRβ-chains through an epitope in the β constant domain, and the mAb:epitope interaction has been crystallized and shown to involve a unique epitope present in a single copy per β-chain (27). Therefore, we considered the possibility that at least some TCR/CD3 complexes contain more than one β-chain, and we referred to putative plural β-chains that could be probed under these conditions as βh. Detection of βh required the capture of TCR/CD3 and was titratable, since dilution of the lysate during IP reduced βh detection (Fig. 2B). Additionally, observation of βh was not limited to the H57 mAb or its epitope, because TCRs from OT1 TCR transgenic T cells displayed the same pattern when the MR9-4 anti-Vβ5 mAb was used as both capture and probe reagent (Fig. 2C). Therefore, as with CD3ε multiplicity (Fig. 2A), observation of βh suggested that digitonin-solubilized TCR/CD3 complexes can contain more than one β-chain.
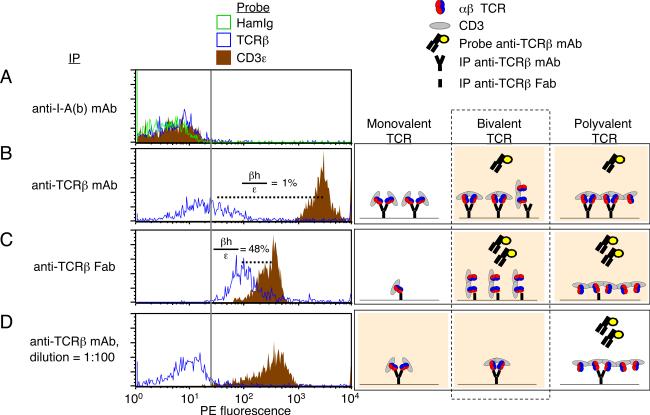
To further investigate this possibility, we tested the hypothesis that IP mAbs on the bead influence detection of multiple β-chains per complex. Either intact bivalent H57 anti-TCRβ mAb, or monovalent Fab fragments derived therefrom, were covalently coupled to beads for use in IP-FCM. Whereas negative control IP beads failed to capture TCR/CD3 complexes (Fig. 3A), IP with anti-TCRβ mAb captured many complexes with a low degree of detectable free β-chains (Fig. 3B, βh/ε = 1%). In contrast, IP with H57 Fab captured fewer complexes, but these displayed significantly enhanced detection of free β-chains (Fig. 3C, βh/ε = 48%). This result indicated that there were complexes containing a β-chain copy number that was >1, because at least one β-chain mediated capture of a complex onto the Fab-beads, while at least one additional β-chain remained accessible to subsequent probes.
Figure 3. TCR/CD3 complexes containing two β-chains.
BALB/c T cell lysates were subjected to IP-FCM analysis to ascertain whether 1, >1, or >2 β-chains were present in complexes. Histograms extending beyond the vertical gray line are considered to be above background. (A) Negative control IP with anti-I-Ab mAb did not capture TCR/CD3 complexes. (B) IP with standard, bivalent anti-TCRβ mAb captured many TCR/CD3 complexes, including a low level of free β-chains when probed with the same mAb clone (H57) that had been used to IP. (βh/ε = 28.3/2831 = 1%). (C) IP with monovalent Fab fragments of anti-TCRβ (H57) allowed capture of complexes with increased detectable free β-chains when probed with PE-conjugated anti-TCRβ (H57). (βh/ε = 132/280 = 48%). (D) Dilution of the IP mAb on the beads (100-fold) resulted in capture of fewer complexes, but did not facilitate the detection of free β-chains. (Right) Possible outcomes are illustrated, as TCR/CD3 complexes are captured and probed with the H57 anti-TCRβ Abs. Those outcomes compatible with the data are displayed with peach-colored background. Inclusion of exactly two TCRs in bivalent complexes is consistent with all data considered together (dashed rectangle). Data in all panels represent one of at least two experiments.
It remained possible that the β-chain copy number in these complexes was >2, if multiple epitopes within a complex were occupied cooperatively between adjacent IP mAbs on a bead. If correct, then increasing the distance between mAbs on the IP-beads would be predicted to diminish adjacent cooperativity, and allow the capture of complexes with free, detectable β-chains, in a similar pattern to that observed with Fab-beads. However, this proved not to be the case. We tested the idea, increasing the average spacing between IP mAbs on beads by coupling 100-fold less H57 anti-TCRβ mAb. These IP beads captured fewer complexes, and lost detection of free β-chains (Fig. 3D) when compared with Fab-beads (Fig. 3C). Therefore, the copy number of TCR β-chains in complexes that possessed >1 was not >2, meaning it equaled exactly two. Notably, Figure 3 panels C and D displayed roughly similar quantities of complexes per bead (CD3ε probe, brown histograms), indicating that these samples were “capture-matched” (analogous to protein loading controls that show equal content between lanes on a gel or Western blot). But free β-chains were only available for detection by probes if the complexes were captured by monovalent Fab fragments instead of bivalent mAbs. Thus, we conclude that when bivalent anti-β-chain mAbs are used to immunoprecipitate digitoninsolubilized complexes bearing two β-chains, both β-chain epitopes tend to be occupied by the two mAb binding sites. IP using Fab fragments allows capture of complexes in which a second TCR remains freely detectable.
Observation of TCR/CD3 complexes containing two α-chains
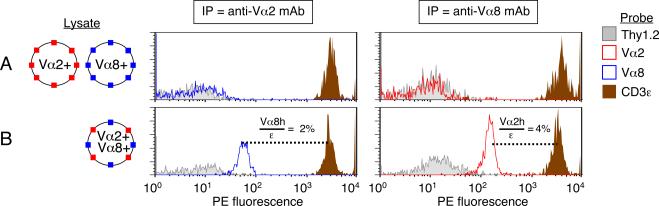
To determine whether digitonin-solubilized complexes could be observed that contained two copies of TCRα, we followed the classic strategy as previously discussed, utilizing T cells that express two different TCR α-chains to determine whether each would co-IP with the other. Experimental manipulation of TCR expression was avoided, however, by focusing on the small percentage of wild-type T cells that naturally express two different α-chains (28). Rare BALB/c Vα2+ Vα8+ dual-TCR T cells were FACS-purified and expanded, as well as Vα2+ Vα8- T cells and Vα2- Vα8+ T cells. A single lysate was prepared of Vα2+ Vα8- cells and Vα2- Vα8+ cells that had been mixed together in equal numbers, and separately a lysate of Vα2+ Vα8+ dual-TCR cells was prepared for analysis by IP-FCM. We found no evidence for shared complexes containing both Vα2 and Vα8 when each TCR type had been expressed in separate cells (Fig. 4A). Thus, the detergent, lysis, and other conditions of these experiments do not induce TCR co-association per se. In contrast, lysates from dual-TCR T cells revealed that Vα8+ TCRs co-IP with anti-Vα2, and conversely Vα2+ TCRs co-IP with anti-Vα8 (Fig. 4B).
Figure 4. TCR/CD3 complexes containing two α-chains in dual-TCR-expressing T cells.
BALB/c T cells were FACS-sorted that displayed the following TCR expression patterns: (i) Vα2+ Vα8−, (ii) Vα2− Vα8+, (iii) Vα2+ Vα8+. Each cell type was stimulated with plate-bound anti-CD3ε + anti-CD28 mAbs for 72h in vitro, followed by 9–11 days of expansion in the absence of further CD3 engagement but presence of exogenous IL-2. (A) Single-TCR expressing blasts were mixed 1:1 and then lysed, while (B) dual-TCR-expressing cells were also lysed. TCR/CD3 complexes were captured with either anti-Vα2 mAb (left) or anti-Vα8 mAb (right). Following IP, samples were probed in parallel with PE-conjugated mAbs. Calculations: Vα8h/ε = (55.6/3289) = 2%; Vα2h/ε = (149/3452)= 4%. Data in all panels represent one of at least two experiments.
When compared to complexes captured by the bivalent anti-Vα2 mAb, its Fab fragment permitted detection of greater quantities of both free Vα2+ and free Vα8+ TCRs (Fig. 5; anti-Vα8 Fab did not function as an IP capture reagent, data not shown). We conclude that primary αβ T cells express TCR/CD3 complexes that contain two α-chains. Because IP with anti-Vα2 Fab improved detection of both Vα2h and Vα8h, we also conclude that in dual-TCR cells, complexes can be expressed as either homo-bivalent (two copies of Vα2) or hetero-bivalent (one copy each of Vα2 and Vα8).
Inclusion of properly folded αβ heterodimers and a second TCR in shared complexes
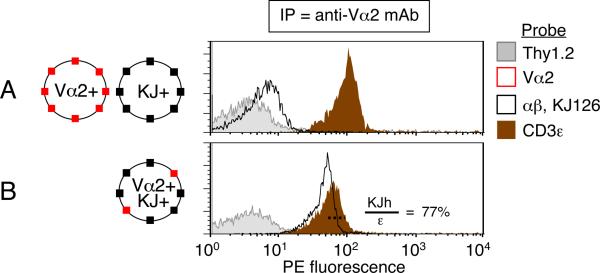
We sought direct evidence to determine whether a properly folded αβ TCR heterodimer can be present in a shared complex with a second TCR. We used DO11.10 TCR transgenic mice (20), which, on RAG+ background, generate endogenously re-arranged Vα2+ TCRs co-expressed together with the transgenic TCR (29). Importantly, the DO11.10 TCR can be probed with KJ126 mAb, which only binds if the transgenic Vα13 and Vβ8.2 subunits are both present and properly folded (30). In this system, Vα2+ KJ126+ dual-TCR T cells represent ~10% of the T cells, the KJ126+ TCR being expressed in considerable excess over the endogenous Vα2+ TCR (29). We wished to determine whether capture of the minor TCR (Vα2+) would reveal its inclusion in shared complexes with the major, properly folded αβ heterodimer (KJ126+). For control samples, Vα2+ 3bbm74 TCR transgenic T cells (31) were mixed with a 9-fold excess of DO11.10/RAG20 T cells, which express only the KJ126+ TCR. Lysates of the mixed cells or DO11.10 (RAG+) cells were prepared and analyzed by IP-FCM. We found that co-expression of Vα2+ TCRs in an excess of KJ126+ TCRs revealed the presence of shared complexes including the two TCR types (Fig. 6; KJh/ε = 76.9%, where KJh is free KJ126+ TCR). We conclude that properly folded αβ heterodimers can be expressed in TCR/CD3 complexes that include a second TCR.
Figure 6. Complexes containing both KJ126+ αβ heterodimer and Vα2+ TCRs.
(A) Vα2+ 3bbm74 transgenic T cells were mixed together with 9-fold excess of DO11.10/RAG20 T cells prior to lysis. This lysate was compared with that of (B) DO11.10 (RAG2+) T cells in which ~10% of T cells co-express endogenously re-arranged Vα2+ TCR together with the DO11.10 clonotype transgenic TCR (Vα13+ Vβ8.2+, detected by mAb clone KJ126, `KJ'). The less expressed TCR (Vα2) was immunoprecipitated and captured complexes were analyzed by IPFCM using the indicated probes. Calculation: KJh/ε = 44.1/58.3 = 77%. Data in both panels represent one of at least two experiments.
Functional TCR bivalency contributes to optimal binding of antigenic pMHC-Ig fusion protein ligands
We examined whether bivalency in digitonin-solubilized TCR/CD3 complexes can influence binding to pMHC ligands. Here, the strategy was to capture fully assembled TCR/CD3 complexes onto beads, and subsequently assess binding to pMHC ligands under varying conditions of TCR bivalency (Fig. 7A). Because assessment of TCR/CD3 complexes required their isolation from digitonin lysates, relatively high-affinity pMHC ligands would be needed since no coreceptors would be present with the isolated TCRs to aid in pMHC binding (32–34).
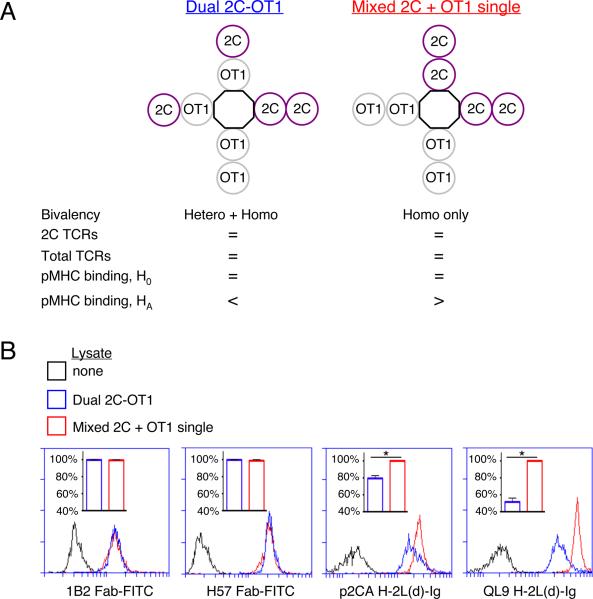
Figure 7. Functional bivalency of TCR/CD3 complexes contributes to optimal binding of antigenic pMHC-Ig fusion protein ligands.
Cells from 2C and OT1 transgenic mice were mixed 1:1 and then lysed, while dual-TCR-expressing cells from 2C × OT1 (F1) mice were also lysed. From the mixed or dual-TCR lysates, TCR/CD3 complexes were captured on beads coupled with H146 anti-CD3ζ mAb. Following IP, samples were probed in parallel with fluorochrome-conjugated reagents and assessed by FCM as indicated. (A) Experimental strategy: from both lysates, equal numbers of 2C and total TCRs were captured onto IP-beads; thus, 2C TCRs would be present in both homo- and hetero-bivalent complexes on beads from the dual-TCR lysate, but only in homo-bivalent complexes on beads from the mixed lysate. The null hypothesis (H0) was that expression of 2C TCRs in hetero-bivalent complexes has no impact on pMHC-Ig:TCR binding, while the alternate hypothesis (HA) was that hetero-bivalent complexes would show decreased pMHC-Ig binding. (B) Equal capture of 2C and total TCRs from both lysates was confirmed (via 1B2-Fab-FITC and H57-Fab-FITC staining, respectively). However, 2C TCRs from the mixed lysate bound p2Ca/H-2Ld and QL9/H-2Ld MHC-Ig fusion protein ligands to a greater degree than did 2C TCRs from the dual-TCR lysate (H0 rejected, one-tailed Student's t-tests, *p=0.04 p2Ca, *p=0.02 QL9). Data in all panels represent one of two experiments.
2C, OT1, and 2C × OT1 (F1) TCR transgenic mice were used as sources of TCR/CD3 for the following reasons: (i) the F1 mice were previously shown to co-express both TCRs (35); (ii) the 2C αβ heterodimer can be detected by the anti-clonotype mAb 1B2 (36); (iii) relatively high-affinity, CD8-independent, allogeneic pMHC complexes (p2Ca/H-2Ld and QL9/H-2Ld) specific for the 2C TCR have been previously characterized (37); (iv) in the presence of pMHC ligands specific for 2C, the OT1 TCR could serve as a physically present but functionally inert TCR in this experiment. A single lysate was prepared of 2C+ OT1− cells and 2C− OT1+ cells that had been mixed together in equal numbers, while separately a lysate of 2C+ OT1+ dual-TCR cells was prepared. As learned from Figures 4 and 6, lysate from mixed cells was considered a source of homo-bivalent complexes, while lysate from dual-TCR cells was a source of both homo- and hetero-bivalent complexes (Fig. 7A). After lysis, complexes were captured using H146 anti-CD3ζ IP-beads, since capturing complexes via the C-terminus of the cytoplasmic domain of CD3ζ would not be predicted to sterically impede pMHC:TCR binding. Also, because it is the last subunit added during assembly (4), ζ-chain was captured as a means of including only fully assembled TCR/CD3 complexes in the experiment.
We found that an equal number of 2C and total TCRs could be captured from each lysate, as confirmed by 1B2 Fab and H57 anti-TCRβ Fab probes, respectively (Fig. 7A–B). Next, because it was previously shown that soluble pMHC ligands must themselves be at least bivalent in order to stimulate T cell activation (38), we used bivalent MHC-Ig fusion proteins as probes for the capture-matched, digitonin-solubilized TCR/CD3 complexes. We observed that 2C TCR binding to either p2Ca/H-2Ld or QL9/H-2Ld MHC-Ig ligands was inhibited for 2C TCRs originating from dual-TCR cells compared with single-TCR cells (Fig. 7B). Because the number of captured TCRs was equal between these two experimental groups, the data indicate that arrangement of TCRs in hetero-bivalent complexes inhibited binding to the MHC-Ig fusion proteins. Thus, these data suggest that the property of bivalency is sufficiently prevalent among the population of TCR/CD3 complexes to impact the outcome of this assay of model antigen-binding. Additionally, both TCR positions in bivalent complexes must be occupied by antigen-specific TCRs in order to effect optimal binding to these soluble pMHC-Ig ligands.
Discussion
We present evidence that complexes containing two TCRs and possessing functional bivalency can be found among digitonin-solubilized αβ TCR/CD3 complexes from primary T cells. IP with anti-TCR Fab fragments significantly increased visualization of a second TCR in shared complexes when compared to IP with intact mAbs. Several strategies focused on the critical issue of determining whether the bivalent complexes could be considered mature, properly folded TCR/CD3, rather than misfolded or incompletely assembled, intermediate complexes. First, much of the data relied on ex-vivo, physiologic systems that do not involve over-expression, and thus should reflect normal subunit interactions in the complex (Figs. 1–5). Second, the Abs used for both IP and probe were conformation-dependent and do not bind denatured epitopes, implying that subunits were only detected if they were properly folded. Third, a properly folded and assembled αβ heterodimer was shown to be included in complexes with a second co-expressed endogenous α-chain (Fig. 6). Finally, we showed that the fully assembled TCR/CD3 pool, defined as containing CD3ζ, contains complexes that display functional bivalency (Fig. 7).
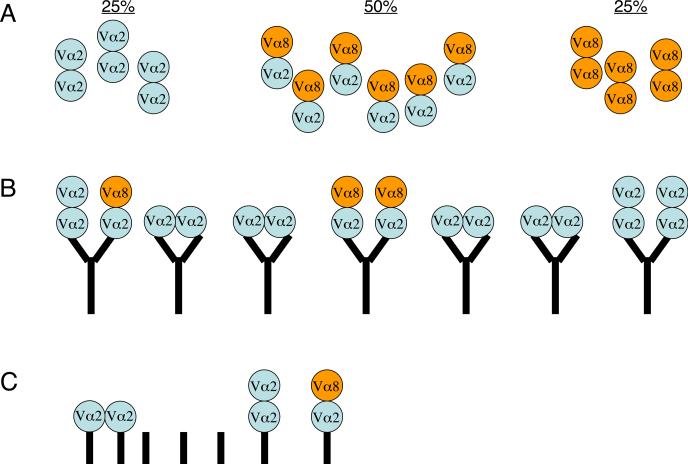
The fact that anti-TCR mAbs can bind with double occupancy to bivalent TCR/CD3 suggests a reason why these complexes may not have been apparent in previous studies. A T cell which expresses two different α chains (e.g., Vα2 and Vα8) and a single β-chain (Vβ*) would express three different kinds of bivalent complexes, Vα2:Vβ*/ Vα2:Vβ*, Vα2:Vβ*/ Vα8:Vβ*, and Vα8:Vβ*/ Vα8:Vβ* (Fig. 8). An IP mAb specific for one of the TCRs, such as anti-Vα2, is predicted to preferentially capture homo-bivalent Vα2+ Vα2+ complexes, while the hetero-bivalent Vα2+ Vα8+ complexes are at a competitive disadvantage for binding to the IP mAb, since the latter can only bind one site per mAb. The net result is that Vα8+ TCRs are under-represented in such an IP. We observed that the presence of a second TCR per bivalent complex was uncovered when monovalent Fab fragments were used to IP (Figs. 3, 5). Therefore, we propose that the tendency of bivalent mAbs to bind to bivalent TCRs with double occupancy may have contributed to the previous interpretation of past data where bivalent TCR/CD3 complexes were not detected.
Figure 8. Model of bivalent IP mAbs binding with double occupancy to bivalent TCRs.
Assay bias can occur when using bivalent mAbs to assess inclusion of two different co-expressed TCRs in shared complexes (such as those containing Vα2 and Vα8; all other TCR/CD3 subunits not depicted). For this example, two assumptions are made: (i) expression of the two TCR species is equal; (ii) since two different Vα chains can be detected in single complexes, then the sequence(s) mediating bivalency involve constant, non-variable domains. (A) Lysates of T cells expressing both Vα2+ and Vα8+ TCRs could contain 3 populations of bivalent receptors that might freely associate with the following ratios: Vα2+ Vα2+ (25%), Vα2+ Vα8+ (50%), Vα8+ Vα8+ (25%). (B) IP mAbs specific for one TCR (here, Vα2) would capture large numbers of Vα2+ Vα2+ homo-bivalent complexes, because this TCR species would bind with highest affinity to the bivalent mAbs with double occupancy on the beads. At a low level, some Vα2+ Vα2+ complexes would bind with single-occupancy, allowing their detection (mAb-bound complexes, far right). Due to competitive disadvantage, hetero-bivalent Vα2+ Vα8+ complexes would rarely be captured and detected on beads, although this occurs to detectable levels by IP-FCM (mAb-bound complexes, left and middle). The net result is that bivalent IP mAbs selectively capture homo-bivalent TCRs that match the specificity of the IP mAb, underestimating the levels of the second TCR species in bivalent complexes, and resulting in the previously held interpretation that bivalent complexes are not expressed. (C) Capturing TCRs with anti-TCR Fab fragments would minimize the assay effects described in (B), although some double-occupancy binding would still be possible if two Fabs attached to a bead can bind TCR cooperatively, or if the Fabs have any natural tendency to spontaneously dimerize noncovalently. Never the less, Fabs will allow single-occupancy binding to a greater extent than expected for bivalent mAbs, and this property allows increased visualization of complexes containing two TCRs in the present work.
Although we have shown that bivalent TCR/CD3 are present among digitonin-solubilized complexes, we do not know the proportion of complexes they represent. One focus of future experiments must be to develop other methods that would permit a quantitative estimate of the prevalence of bivalency among all TCR/CD3 complexes. However, despite this current technical limitation, the data in Figure 7 imply that the proportion of bivalent complexes is sufficiently high to impact the outcome of the pMHC-Ig fusion protein binding assay performed. Thus, based on the assumption that this assay reflects a true potential for functional impact, we speculate that it is likely that a biologically significant number of TCR/CD3 complexes display bivalency.
Which motifs might specifically interact to compose a bivalent complex are currently unknown. Even in the standard monovalent model of TCR/CD3, the interactions between subunit dimers (αβ/εγ/εδ/ζζ) that compose the multiprotein complex are not fully characterized, and as a consequence it is not known where the various dimers are situated relative to each other. Most schematics of TCR/CD3 follow the model integrated by Sun et al. (39), with TCR in the middle of CD3 subunits, compatible with the proposed interaction of ionizable amino acids between subunit transmembrane domains (7). However, other data supports the possibility that the relative subunit positions may be arranged differently, with αβ closer to one extreme instead of in the middle of the complex (40). The inability to observe binding between engineered extracellular domains of subunit dimer pairs has left the issue empirically unresolved by crystallography to date. Thus, how the various subunit dimers interact to compose the complex remains a significant outstanding question, which is not simplified by the proposal that a proportion of complexes may be bivalent. However, our observations argue that bivalency in these complexes is `closed', mediated by inteaction(s) without an open end, because concatemerization is not observed in the digitonin-solubilized complex.
Higher-scale oligomerization or concentration is possible beyond the digitonin-solubilized complexes on which we have focused (14, 41). Using size-exclusion chromatography, sedimentation-velocity ultracentrifugation, surface plasmon resonance, and other methods, several groups have shown that some CD3-free TCR αβ ectodomains can oligomerize when bound by agonist pMHC (42–44). In the context of full TCR/CD3 expression, a recent study identified sequences in the extracellular constant domain of TCRα required for optimal TCR accumulation at the principal site of pMHC contact (40). Therefore, it is an interesting possibility that antigenic engagement might induce association of formerly separate complexes into higher-order concatemers/oligomers through TCR motifs.
Bivalent complexes might impact T cell function by enhancing the strength with which these receptors could bind to pMHC ligands (Fig. 7). At least some pMHC ligands have been described to be expressed as a “dimer of dimers” (45–48), meaning that two pMHC are expressed in co-association. In such complexes, if both peptides are identical, then a bivalent TCR:pMHC interaction could occur 2:2, with predicted enhanced binding strength over 1:1 or 1:2 interactions. Other data has shown that two co-associated pMHC can enhance T cell activation even if only one of the two peptides is antigenic (49–51). Thus, a pseudo-dimer model of T cell activation was proposed that requires engagement of one TCR by antigenic pMHC to recruit a second TCR that would somehow contribute to signaling upon interaction with the second non-antigenic pMHC. A model of TCR expression that includes bivalent complexes now suggests that a second TCR is already present in these complexes, and therefore would not require recruitment in order to interact with a second pMHC. Finally, it is possible that many pMHC complexes may be initially expressed in single-copy (52), but are effectively joined into pairs by bivalent TCR engagement to initiate the extensive crosslinking and concentration that will generate microclusters and the immunological synapse (53). In summary, under several different possible scenarios of pMHC copy number and peptide specificity, it can be speculated that expression of bivalent TCRs might contribute to T cell triggering by providing a mechanism to enhance antigen binding strength over that expected of a monovalent complex.
In conclusion, bivalent TCRs can be observed among the TCR/CD3 complexes isolated from primary T cells. The glycoprotein sequences mediating this bivalency and its precise functional consequences to T cell signaling are a focus of future work.
Acknowledgements
For helpful discussions, we thank Mark Daniels, Gennaro De Libero, Mike Dustin, Nick Gascoigne, Michel Mallaun, Dieter Naeher, Larry Stern, and Emma Teixeiro. For review of the manuscript, we thank Rachel Gibbons and Virginia Shapiro. For reagents and technical advice, we thank Ursula Gunthert (DO11.10 mice), Bernard Malissen (B21.14 hybridoma), and Larry Pease (2C and OT1 mice). For other technical help we thank Tessa Davis, Verena Jäggin, Chris Parks, Robert Stiles, and the Mayo Flow Cytometry and Optical Morphology (FCOM) core facility.
This work was supported by the Sangstadt-American Society of Transplantation Basic Science Fellowship; Ruth L. Kirschstein National Research Service Award from National Cancer Institute, National Institutes of Health (NIH) (A.G.S.); the Mayo Foundation (A.G.S. and D.G.); Ramón y Cajal program and project PR3/04-12454 from Universidad Complutense; Dirección General de Universidades e Investigación and Universidad Complutense de Madrid program, 920631; Instituto de Salud Carlos III, grant PI060057 (D.G.); NIH grants AI41521 and AI43620 (L.A.T.); Swiss National Science Foundation; Hoffmann- La Roche, Ltd; Novartis, AG (E.P.).
Abbreviations
- B6
C57BL/6
- FCM
flow cytometry
- gMFI
geometric mean fluorescence intensity
- IP
immunoprecipitation
- IP-FCM
immunoprecipitation detected by flow cytometry
- pMHC
peptide/MHC
References
- 1.Oltz EM. Regulation of antigen receptor gene assembly in lymphocytes. Immunol Res. 2001;23:121–133. doi: 10.1385/IR:23:2-3:121. [DOI] [PubMed] [Google Scholar]
- 2.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Punt JA, Roberts JL, Kearse KP, Singer A. Stoichiometry of the T cell antigen receptor (TCR) complex: each TCR/CD3 complex contains one TCR alpha, one TCR beta, and two CD3 epsilon chains. J Exp Med. 1994;180:587–593. doi: 10.1084/jem.180.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan Q, Gollapudi AS, Dave VP. Biochemical evidence for the presence of a single CD3delta and CD3gamma chain in the surface T cell receptor/CD3 complex. J Biol Chem. 2004;279:51068–51074. doi: 10.1074/jbc.M406145200. [DOI] [PubMed] [Google Scholar]
- 6.Call ME, Pyrdol J, Wucherpfennig KW. Stoichiometry of the T-cell receptor-CD3 complex and key intermediates assembled in the endoplasmic reticulum. Embo J. 2004;23:2348–2357. doi: 10.1038/sj.emboj.7600245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Call ME, Wucherpfennig KW. The T cell receptor: critical role of the membrane environment in receptor assembly and function. Annu Rev Immunol. 2005;23:101–125. doi: 10.1146/annurev.immunol.23.021704.115625. [DOI] [PubMed] [Google Scholar]
- 8.Terhorst C, Exley M, Franco R, Hall C, Kang J, Mueller B, Sancho J, She J, Wileman T. Coupling of T-cell activation with T-cell receptor assembly. Year Immunol. 1993;7:1–24. [PubMed] [Google Scholar]
- 9.Jacobs H. Pre-TCR/CD3 and TCR/CD3 complexes: decamers with differential signalling properties? Immunol Today. 1997;18:565–569. [PubMed] [Google Scholar]
- 10.Fernandez-Miguel G, Alarcon B, Iglesias A, Bluethmann H, Alvarez-Mon M, Sanz E, de la Hera A. Multivalent structure of an alphabetaT cell receptor. Proc Natl Acad Sci U S A. 1999;96:1547–1552. doi: 10.1073/pnas.96.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schamel WW, Arechaga I, Risueno RM, van Santen HM, Cabezas P, Risco C, Valpuesta JM, Alarcon B. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J Exp Med. 2005;202:493–503. doi: 10.1084/jem.20042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beyers AD, Spruyt LL, Williams AF. Molecular associations between the T-lymphocyte antigen receptor complex and the surface antigens CD2, CD4, or CD8 and CD5. Proc Natl Acad Sci U S A. 1992;89:2945–2949. doi: 10.1073/pnas.89.7.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerny J, Stockinger H, Horejsi V. Noncovalent associations of T lymphocyte surface proteins. Eur J Immunol. 1996;26:2335–2343. doi: 10.1002/eji.1830261010. [DOI] [PubMed] [Google Scholar]
- 14.Alarcon B, Swamy M, van Santen HM, Schamel WW. T-cell antigen-receptor stoichiometry: pre-clustering for sensitivity. EMBO Rep. 2006;7:490–495. doi: 10.1038/sj.embor.7400682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teixeiro E, Daniels MA, Hausmann B, Schrum AG, Naeher D, Luescher I, Thome M, Bragado R, Palmer E. T cell division and death are segregated by mutation of TCRbeta chain constant domains. Immunity. 2004;21:515–526. doi: 10.1016/j.immuni.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Gil D, Schrum AG, Alarcon B, Palmer E. T cell receptor engagement by peptide-MHC ligands induces a conformational change in the CD3 complex of thymocytes. J Exp Med. 2005;201:517–522. doi: 10.1084/jem.20042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schrum AG, Gil D, Dopfer EP, Wiest DL, Turka LA, Schamel WW, Palmer E. High-sensitivity detection and quantitative analysis of native protein-protein interactions and multiprotein complexes by flow cytometry. Sci STKE. 2007;2007:12. doi: 10.1126/stke.3892007pl2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schrum AG. Visualization of multiprotein complexes by flow cytometry. Curr Protoc Immunol. 2009;Chapter 5(Unit5):9. doi: 10.1002/0471142735.im0509s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teixeiro E, Daniels MA, Hamilton SE, Schrum AG, Bragado R, Jameson SC, Palmer E. Different T Cell Receptor Signals Determine CD8+ Memory Versus Effector Development. Science. 2009;323:502–505. doi: 10.1126/science.1163612. [DOI] [PubMed] [Google Scholar]
- 20.Murphy KM, Heimberger AB, Loh DY. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science. 1990;250:1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 21.Andrew SM, Titus JA. Fragmentation of immunoglobulin G. Curr Protoc Immunol. 2001;Chapter 2(Unit 2):8. doi: 10.1002/0471142735.im0208s21. [DOI] [PubMed] [Google Scholar]
- 22.Karasuyama H, Melchers F. Establishment of mouse cell lines which constitutively secrete large quantities of interleukin 2, 3, 4 or 5, using modified cDNA expression vectors. Eur J Immunol. 1988;18:97–104. doi: 10.1002/eji.1830180115. [DOI] [PubMed] [Google Scholar]
- 23.Davis TR, Schrum AG. IP-FCM: Immunoprecipitation Detected by Flow Cytometry. J Vis Exp. 2010 doi: 10.3791/2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montixi C, Langlet C, Bernard AM, Thimonier J, Dubois C, Wurbel MA, Chauvin JP, Pierres M, He HT. Engagement of T cell receptor triggers its recruitment to low-density detergent-insoluble membrane domains. Embo J. 1998;17:5334–5348. doi: 10.1093/emboj/17.18.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rozdzial MM, Kubo RT, Turner SL, Finkel TH. Developmental regulation of the TCR zeta-chain. Differential expression and tyrosine phosphorylation of the TCR zeta-chain in resting immature and mature T lymphocytes. J Immunol. 1994;153:1563–1580. [PubMed] [Google Scholar]
- 26.de la Hera A, Muller U, Olsson C, Isaaz S, Tunnacliffe A. Structure of the T cell antigen receptor (TCR): two CD3 epsilon subunits in a functional TCR/CD3 complex. J Exp Med. 1991;173:7–17. doi: 10.1084/jem.173.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Lim K, Smolyar A, Teng M, Liu J, Tse AG, Liu J, Hussey RE, Chishti Y, Thomson CT, Sweet RM, Nathenson SG, Chang HC, Sacchettini JC, Reinherz EL. Atomic structure of an alphabeta T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody. Embo J. 1998;17:10–26. doi: 10.1093/emboj/17.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padovan E, Casorati G, Dellabona P, Meyer S, Brockhaus M, Lanzavecchia A. Expression of two T cell receptor alpha chains: dual receptor T cells. Science. 1993;262:422–424. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- 29.Schrum AG, Turka LA. The proliferative capacity of individual naive CD4(+) T cells is amplified by prolonged T cell antigen receptor triggering. J Exp Med. 2002;196:793–803. doi: 10.1084/jem.20020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yague J, White J, Coleclough C, Kappler J, Palmer E, Marrack P. The T cell receptor: the alpha and beta chains define idiotype, and antigen and MHC specificity. Cell. 1985;42:81–87. doi: 10.1016/s0092-8674(85)80103-3. [DOI] [PubMed] [Google Scholar]
- 31.Backstrom BT, Muller U, Hausmann B, Palmer E. Positive selection through a motif in the alphabeta T cell receptor. Science. 1998;281:835–838. doi: 10.1126/science.281.5378.835. [DOI] [PubMed] [Google Scholar]
- 32.Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Hollander GA, Gascoigne NR, Palmer E. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 33.Naeher D, Daniels MA, Hausmann B, Guillaume P, Luescher I, Palmer E. A constant affinity threshold for T cell tolerance. J Exp Med. 2007;204:2553–2559. doi: 10.1084/jem.20070254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gil D, Schrum AG, Daniels MA, Palmer E. A role for CD8 in the developmental tuning of antigen recognition and CD3 conformational change. J Immunol. 2008;180:3900–3909. doi: 10.4049/jimmunol.180.6.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniels MA, Schober SL, Hogquist KA, Jameson SC. Cutting edge: a test of the dominant negative signal model for TCR antagonism. J Immunol. 1999;162:3761–3764. [PubMed] [Google Scholar]
- 36.Kranz DM, Tonegawa S, Eisen HN. Attachment of an anti-receptor antibody to non-target cells renders them susceptible to lysis by a clone of cytotoxic T lymphocytes. Proc Natl Acad Sci U S A. 1984;81:7922–7926. doi: 10.1073/pnas.81.24.7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speir JA, Garcia KC, Brunmark A, Degano M, Peterson PA, Teyton L, Wilson IA. Structural basis of 2C TCR allorecognition of H-2Ld peptide complexes. Immunity. 1998;8:553–562. doi: 10.1016/s1074-7613(00)80560-9. [DOI] [PubMed] [Google Scholar]
- 38.Cochran JR, Cameron TO, Stern LJ. The relationship of MHC-peptide binding and T cell activation probed using chemically defined MHC class II oligomers. Immunity. 2000;12:241–250. doi: 10.1016/s1074-7613(00)80177-6. [DOI] [PubMed] [Google Scholar]
- 39.Sun ZY, Kim ST, Kim IC, Fahmy A, Reinherz EL, Wagner G. Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling. Proc Natl Acad Sci U S A. 2004;101:16867–16872. doi: 10.1073/pnas.0407576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhns MS, Girvin AT, Klein LO, Chen R, Jensen KD, Newell EW, Huppa JB, Lillemeier BF, Huse M, Chien YH, Garcia KC, Davis MM. Evidence for a functional sidedness to the alphabetaTCR. Proc Natl Acad Sci U S A. 2010;107:5094–5099. doi: 10.1073/pnas.1000925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reich Z, Boniface JJ, Lyons DS, Borochov N, Wachtel EJ, Davis MM. Ligand-specific oligomerization of T-cell receptor molecules. Nature. 1997;387:617–620. doi: 10.1038/42500. [DOI] [PubMed] [Google Scholar]
- 43.Alam SM, Davies GM, Lin CM, Zal T, Nasholds W, Jameson SC, Hogquist KA, Gascoigne NR, Travers PJ. Qualitative and quantitative differences in T cell receptor binding of agonist and antagonist ligands. Immunity. 1999;10:227–237. doi: 10.1016/s1074-7613(00)80023-0. [DOI] [PubMed] [Google Scholar]
- 44.Deng L, Langley RJ, Brown PH, Xu G, Teng L, Wang Q, Gonzales MI, Callender GG, Nishimura MI, Topalian SL, Mariuzza RA. Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor. Nat Immunol. 2007;8:398–408. doi: 10.1038/ni1447. [DOI] [PubMed] [Google Scholar]
- 45.Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 46.Schafer PH, Pierce SK, Jardetzky TS. The structure of MHC class II: a role for dimer of dimers. Semin Immunol. 1995;7:389–398. doi: 10.1006/smim.1995.0043. [DOI] [PubMed] [Google Scholar]
- 47.Cherry RJ, Wilson KM, Triantafilou K, Toole PO, Morrison IE, Smith PR, Fernandez N. Detection of dimers of dimers of human leukocyte antigen (HLA)-DR on the surface of living cells by single-particle fluorescence imaging. J Cell Biol. 1998;140:71–79. doi: 10.1083/jcb.140.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Unternaehrer JJ, Chow A, Pypaert M, Inaba K, Mellman I. The tetraspanin CD9 mediates lateral association of MHC class II molecules on the dendritic cell surface. Proc Natl Acad Sci U S A. 2007;104:234–239. doi: 10.1073/pnas.0609665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krogsgaard M, Li QJ, Sumen C, Huppa JB, Huse M, Davis MM. Agonist/endogenous peptide-MHC heterodimers drive T cell activation and sensitivity. Nature. 2005;434:238–243. doi: 10.1038/nature03391. [DOI] [PubMed] [Google Scholar]
- 50.Yachi PP, Ampudia J, Gascoigne NR, Zal T. Nonstimulatory peptides contribute to antigen-induced CD8-T cell receptor interaction at the immunological synapse. Nat Immunol. 2005;6:785–792. doi: 10.1038/ni1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cebecauer M, Guillaume P, Mark S, Michielin O, Boucheron N, Bezard M, Meyer BH, Segura JM, Vogel H, Luescher IF. CD8+ cytotoxic T lymphocyte activation by soluble major histocompatibility complex-peptide dimers. J Biol Chem. 2005;280:23820–23828. doi: 10.1074/jbc.M500654200. [DOI] [PubMed] [Google Scholar]
- 52.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 53.Fooksman DR, Vardhana S, Vasiliver-Shamis G, Liese J, Blair DA, Waite J, Sacristan C, Victora GD, Zanin-Zhorov A, Dustin ML. Functional anatomy of T cell activation and synapse formation. Annu Rev Immunol. 2010;28:79–105. doi: 10.1146/annurev-immunol-030409-101308. [DOI] [PMC free article] [PubMed] [Google Scholar]