Abstract
The canonical route from normal tissue to cancer occurs through sequential acquisition of somatic mutations. Many studies have constructed a linear genetic model for tumorigenesis using the genetic alterations associated with samples at different stages of neoplastic progression from cross-sectional data. The common interpretation of these models is that they reflect the temporal order within any given tumor. Linear genetic methods implicitly neglect genetic heterogeneity within a neoplasm; each neoplasm is assumed to consist of one dominant clone. We modeled neoplastic progression of colorectal cancer using an agent-based model of a colon crypt and found clonal heterogeneity within our simulated neoplasms, as observed in vivo. Just 7.3% of cells within neoplasms acquired mutations in the same order as the linear model. In 41% of the simulated neoplasms, no cells acquired mutations in the same order as the linear model. We obtained similarly poor results when comparing the temporal order to oncogenetic tree models inferred from cross-sectional data. However, when we reconstructed the cell lineage of mutations within a neoplasm using several biopsies, we found 99.7% cells within neoplasms acquired their mutations in an order consistent with the cell lineage mutational order. Thus, we find that using cross-sectional data to infer mutational order is misleading, while phylogenetic methods based on sampling intra-tumor heterogeneity accurately reconstructs the evolutionary history of tumors. Additionally, we find evidence that disruption of differentiation is likely the first lesion in progression for most cancers, and should be one of the few regularities of neoplastic progression across cancers.
Keywords: Evolution, heterogeneity, order
Introduction
Cancer is an evolutionary system that results from the accumulation of somatic mutations (1), resulting in cells that increase their fitness and divide faster, die less frequently, and no longer follow the rules dictated by their environments. New mutations in individual cells within a neoplasm can create subclones of genetically identical cells all sharing the same common ancestor, and these subclones then compete with each other for limited resources. Cancer cells are an evolving population of asexually-reproducing cells, and the rate and dynamics of adaptation in asexually-evolving populations have been studied experimentally and theoretically in evolutionary biology (reviewed in (2)).
A fundamental goal of research into carcinogenesis, with implications for cancer prevention, is to determine the order of mutations that occur in a neoplasm as it progresses from normal tissue to cancer. This has been explored in various cancer types, including breast cancer (3), lung cancer (4), and melanoma (5). In a canonical paper (6), the order of mutations for colorectal cancer was reconstructed from 172 colorectal tumor specimens (7). The specimens were classified according to tumor size and grade, and a small set of genetic alterations was characterized in these same samples. The order of mutations leading to colon cancer was constructed from these data by identifying the mutations whose frequency across tumors increased in conjunction with increases in the tumor size and grade (6). This analysis has been widely influential and used as a model for other cancer systems (4, 8, 9). Even at its conception, this model was not meant to provide a fixed roadmap for the accumulation of genetic alterations. In fact, in observations by the original authors of the linear genetic model, 2 of 7 tumors were inconsistent with the linear model, as determined through experiments in which the genetic alterations were identified in different stages within the same tumor. This was independently verified through experiments identifying the mutational state of colorectal tumors from over 100 patients (10). We analyzed the data from (10) and found that just 26.2% of the neoplasms had a mutational state consistent with the canonical genetic pathway inferred from cross-sectional data.
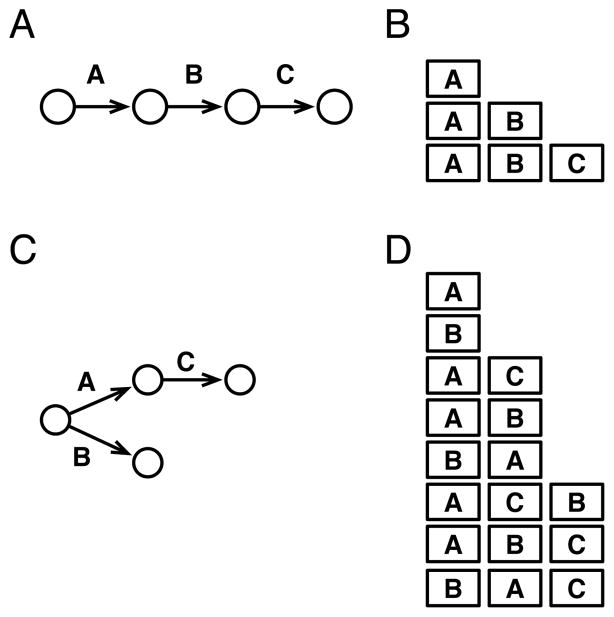
We begin by discussing two types of models of progression: the canonical path model and its extension, the tree model. A path model for a type of cancer is a linear sequence of mutations that must occur in order, beginning with wild-type (Fig. 1A and B). Because this is a path, each mutation has at most one following mutation, and a given mutation increases the chances of finding the next mutation. Oncogenetic tree models were developed to capture multiple independent mutational events, in effect enumerating several possible paths to cancer (11). Here, the mutational order can be a tree rather than a path (Fig. 1C and D). This is not an evolutionary model because the oncogenetic tree does not represent ancestral relationships within a neoplasm, but rather a summary of the observed co-occurrences of mutations across independent neoplasms. In this branching tree model, each mutation occurs just once in the tree, and can have multiple independently occurring following mutations. As before, the interpretation is that a given mutation increases the chances of obtaining the following mutation. A number of studies have constructed oncogenetic trees from cross-sectional data, including for renal cell carcinoma (12), endometrial adenocarcinoma (13), gastrointestinal stromal tumors (14), oral cancer (15), and colon cancer (16).
Fig. 1.
Illustration of path and oncogenetic tree mutational models inferred from cross-sectional data, and all possible temporal orders of clonal mutations that are consistent with the models. Each arrow between circles represents the acquisition of a new mutation in models inferred from cross-sectional data, and squares represent the accumulation of a new mutation in a clone during the evolution of a tumor. (A) The path model of carcinogenesis implies a linear order of sequential mutations from wild-type through A, B, and C, in order. (B) These are the temporal mutations that a cell lineage, or clone of cells, could acquire during evolution and still be consistent with the cross-sectional path model in A. All other sequences of mutations are inconsistent with the cross-sectional path model (e.g., B, C, AC, and BAC are inconsistent). (C) The oncogenetic tree model of carcinogenesis implies that all tumors begin as wild-type, and can next acquire either mutation A or B. Additionally, C can only occur at any point after mutation A has occurred. (D) All temporal mutations acquired by a clone that are consistent with the cross-sectional oncogenetic tree in C. Note that the order A, B, C is consistent because C occurs after A.
Here, we show that the temporal, or evolutionary, order of mutations acquired in the clones that survive over the lifetime of a neoplasm are not consistent with the path order or the oncogenetic tree order reconstructed from cross-sectional data. We explore why this is so and suggest an alternative approach by reconstructing the evolutionary history from samples within a tumor, rather than from across tumors.
Materials and Methods
Model Overview
See the Supplemental Methods for a detailed characterization of the model described in the standard (ODD) format for agent-based models (17). We focused the model and parameters on a colon crypt. Briefly, cells are represented as agents that can divide, mutate, differentiate, and die according to rules implemented stochastically using model parameters based on experimental literature. In principle, each cell in the model can be different from all others. Cells can acquire new mutations, expressed as changes in their phenotypes, when they divide, with probability given by the mutation rate. A mutation confers the loss of differentiation or one of the phenotypes of the hallmarks of cancer. The phenotypes and a summary of their model implementations are given in Table 1; detailed characterization of each hallmark of cancer and the loss of differentiation is given in the Supplemental Methods. Each phenotype can be in one of two states: normal or mutated. We chose the parameter values for both states based on known values for colon cancer whenever possible (Table S5). With high mutation rates, this often led to competition between clones of with similar fitness because multiple sub-clones evolved before any one sub-clone expanded to fill the crypt. The effect of most of the mutations on the phenotypes increases the fitness of a clone by allowing it to grow faster, divide less frequently, or self-renew. In principle, these phenotypes could be conferred through a variety of genetic alterations in the pathways that determine the phenotypes, but we do not model the pathways explicitly. By focusing on phenotypically related sets of genes, we have expanded the focus of this model from genes to sets of genes, similar to the expansion of cancer genes to cancer pathways (reviewed in (18)), without making a commitment to the particular genetic structure of the phenotypes. We assume that a mutation for each phenotype has an equal chance of occurring because the genotype-phenotype map is not well characterized. When a new mutation affecting a phenotype occurs in a cell, we assume that each mutation occurs at a different locus in one of the of genes responsible for the phenotype, in the spirit of the infinite alleles model (19). For each simulation, we recorded detailed information on mutations, cell lineages, and population size and composition over time.
Table 1.
Hallmarks of cancer phenotypes and their effects on the model when mutated.
| Phenotype | Model Effect |
|---|---|
| Insensitivity to antigrowth signals | Cells with this mutation have an increased probability for cell division |
| Self-sufficiency in growth signals | Cells with this mutation have an increased probability for cell division |
| Evasion of apoptosis | Cells with this mutation have a decreased probability of apoptosis |
| Limitless replicative potential | Mutation eliminates telomere loss during cell division. Cells with this mutation can divide more times than those without. |
| Sustained angiogenesis | Cells with this mutation increase the number of cells that can survive in a tumor. |
| Loss of differentiation | Cells with this mutation no longer differentiate at cell division. |
| Genome instability | Cells with this mutation have increased chances to acquire a new mutation. |
All cells in the model were initially wild-type cells with no mutant phenotypes as in a normal crypt; they were then allowed to divide, mutate, differentiate, and die. Simulations were stopped 1) when the number of cells crossed a cell population threshold 100 times greater than the target population size, 2) after 30 years, or 3) when all of the cells in the simulation eroded their telomeres to the point that they could no longer divide. The first two conditions correspond to the development of cancer and the presence of a benign neoplasm, respectively. Sensitivity analyses were conducted, in which we varied the parameter values across the range of realistic values and found to have little effect on the mean size of the largest clone or the mean diversity within the neoplasm (Table S3). Due to computational restrictions, our simulations were conducted in the realm of population sizes smaller than detectable clinically. However, when we varied the maximum possible size of the neoplasm, and thus the detection size of the tumor, there was little effect on the mean size of the largest clone or the heterogeneity. Thus, the conclusions we draw from smaller tumor sizes apply to larger tumor sizes. Accumulation of mutant phenotypes in a cell was low enough that cancer was a relatively rare event. Of the 10,002 simulations, 90 (0.9%) terminated in cancer and 4,639 (46.4%) were benign polyps.
Inferring the path model from cross-sectional data
In our analysis of the simulation data, we made the following assumptions: a tumor was detected as soon as it reached some size, a biopsy was taken immediately upon detection, and a mutation could be detected once it comprised 50% of the cells. We biopsied neoplasms at detection sizes ranging from 700 cells to 10,000 cells (the mean size of wild type cell populations were 772 cells, s.e.m. 7), and obtained the “genotype” for each biopsy (mutant phenotype status; see Fig. 2A and B for an example). For this analysis, a biopsy consisted of the entire tumor. From this, we constructed a matrix of the percent of biopsies having a particular mutation in a phenotype for each size threshold. This was used to identify the order in which the majority of tumors (> 50%) at a given size had a mutant allele. If multiple phenotypes reached 50% at the same size threshold, the mutant phenotype occurring in more of the biopsies was given precedence. Mutant phenotypes that did not occur in 50% of the samples in the final size biopsies were ordered by the percent of biopsies in which they occurred. For example, neither self-sufficiency in growth signals nor insensitivity to antigrowth signal mutations reached majority in 50% of the tumors, but self-sufficiency in growth signals was found in more biopsies and so we concluded that it occurred before insensitivity to antigrowth signals.
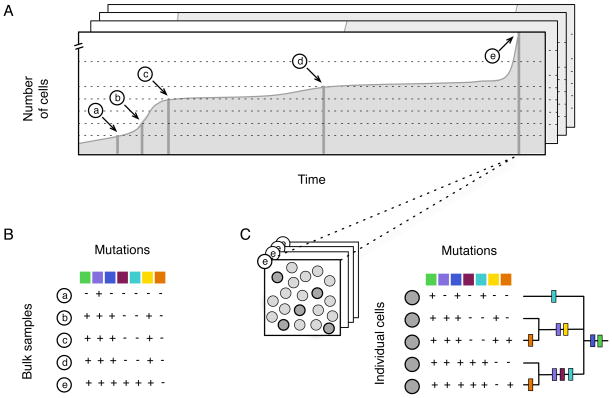
Fig. 2.
Explanation of sampling strategies using a representative simulation. Each simulation represents a single tumor and the order of mutations was inferred from the set of all tumors, using two alternative strategies. (A) The number of cells in this simulation increased over time until it became large enough to trigger detection of cancer (labeled as e). The cross-sectional path model was derived from sampling all tumors based on their size. Most cross-sectional studies take one biopsy per patient and categorize the tumors by size (and/or grade). To simulate this, we took biopsies of each tumor at pre-specified sizes (dashed lines) and then assayed the majority genotype for the biopsy (B). Data from each size class was summarized across all simulations to measure the frequency of mutations for each size class (see Fig. 4A). (C) The alternative to cross-sectional sampling is to reconstruct the cell lineages for each tumor. This was done using 5 randomly selected cells from the final timepoint e. During simulations, the cell lineages were recorded, since exact lineage relationships could be derived from detailed genetic data for each cell. The phenotypic effect of each mutation is represented by a different color, defined in Fig. 3B.
Inferring the oncogenetic tree model from cross-sectional data
We used the R Oncotree package (20) on the same biopsy data used to infer the path model. We estimated the confidence in this tree reconstruction by bootstrapping (98.0%, 1000 bootstrap samples).
Determining consistent temporal and cross-sectional orders
A temporal order was classified as consistent with a cross-sectional path order if it matched or was a prefix of the path order. A temporal order was classified as consistent with a cross-sectional oncogenetic tree order if the temporal mutations in the clone matched or was a prefix of any possible ordering of mutations along the branches of the oncogenetic tree (see Fig. 1C and D for example). When we analyzed the data from the Smith et al. study (10), we defined a biopsy as having a mutational state consistent with the cross-sectional path order if its genetic alterations matched, or matched a prefix, of the canonical order (APC; APC and K-ras; or APC, K-ras, and p53).
Obtaining the cell lineages from the genetic-dependency analysis
We sampled 5 or 10 cells for every tumor analyzed. Because we recorded the exact cell lineage relationship between all the clones in every tumor, we were able to obtain the exact cell lineage relationship between the sampled cells as in Fig. 2C. In our analyses, we assume perfect information and do not simulate experimental noise in either the cross-sectional or the cell lineage analyses. We assume that by assaying single crypts (21), cloning single cells (22) or, in the near future, sequencing single cells, enough genetic information would be available experimentally to accurately reconstruct the cell lineage. A temporal order is consistent with the cell lineage if the temporal mutations in the clone matched, or was a prefix of the phenotypic mutations along the branches of the tree, or if the prefix of a cell-lineage order matched the temporal order.
Results
To test if the temporal order and the order reconstructed from cross-sectional data are consistent, we need detailed data on the mutational order and timing for every clone in neoplasms that progress to cancer. It is difficult to obtain the data to do this experimentally. Thus, we constructed an agent-based model of cell evolution, in which we simulated the progression of normal cells to cancer cells computationally. An agent-based model is a discrete-time, stochastic, computational simulation technique in which cell behaviors (cell division, mutation, differentiation, and death) and traits (mutational states) are explicitly encoded in a computational model. Simulations of a population of cells are performed and the resulting system dynamics are recorded. Previous computational models of tumorigenesis (23, 24) explicitly model the phenotypes associated with the hallmarks of cancer (25). The proper functioning of differentiation is important in preventing somatic evolution and tumorigenesis (26). Thus, we have a created a computational model of cancer that incorporates ideas from previous models with a more realistic representation of differentiation.
First, we characterized the simulated neoplasms that progressed to cancer. We found that the necessary and sufficient mutant phenotypes for progression to cancer were loss of differentiation, evasion of apoptosis, and sustained angiogenesis (Fig. S1). We observed that tumors are heterogeneous and are comprised of cells from multiple different clones, which agrees with recent experimental evidence (27, 28). There were 327 (mean, s.e.m. = 17) clones generated over the lifetime of a tumor. When the tumor was large enough to be detected, it was made up of 256 distinct clones (mean, s.e.m. = 17), the largest clone of which comprised 67% (mean, s.e.m. = 2%) of the neoplasm. The Shannon Index of the tumors was 1.7 (mean, s.e.m. = 0.1). The temporal order of phenotypic mutations in the neoplasms that progressed to cancer was different between tumors; there were 50 different temporal orders found in the 91 cancers (Table S2). This agrees with recent experimental evidence suggesting that the same types of tumors can have different sets of mutations (29, 30).
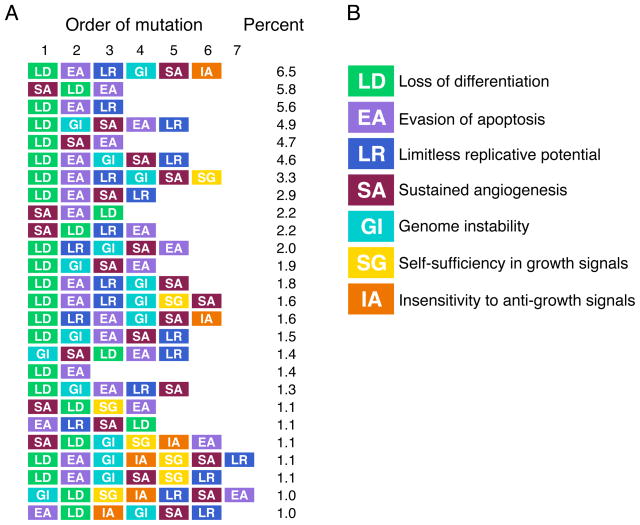
In order to characterize the actual, temporal order of mutations within a neoplasm, we extracted the temporal order of mutations for every cell that survived to become part of a cancer from the summary data. For our analyses, we focused on characterizing the ordering of mutant phenotypes. Of the 14,540* possible temporal paths to cancer, 381 were observed in cancer and 26 were found in more than 1% of the cells. Sixty-four percent of the cells had one of these 26 evolutionary paths (Fig. 3). Loss of differentiation was by far the most common first step in progression and occurred in 72% of the cells. For the subsequent mutations, there is at best a weak tendency for one mutation to occur as opposed to another (Fig. S2).
Fig. 3.
The most common temporal paths found in clones that survived to cancer. (A) Each of these common temporal paths comprise on average at least 1% of the neoplasm, and together these 26 paths account for 64% of the cells found in all the cancerous neoplasms. (B) Each mutation is represented by a different color: loss of differentiation (LD) is green, evasion of apoptosis (EA) is purple, limitless replicative potential (LR) is dark blue, sustained angiogenesis (SA) is red, genomic instability (GI) is light blue, self-sufficiency in growth signals (SG) is yellow, and insensitivity to anti-growth signals (IA) is orange.
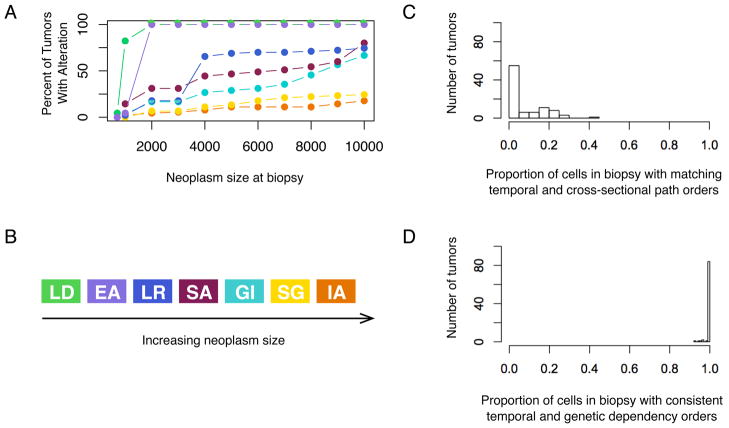
Next, we repeated the experiments to obtain the cross-sectional order of mutations, as was done in colon cancer (6), using our data. First, we obtained the mutational state of the simulated neoplasms at increasing tumor grades that we approximated by the neoplasm size, defined as the number of cells comprising a neoplasm. To do so, we biopsied neoplasms that progressed to cancer as they crossed various size thresholds and genotyped them for the presence of mutant alleles in the majority of cells. The frequency-based ordering of mutations inferred from this analysis was loss of differentiation, evasion of apoptosis, limitless replicative potential, sustained angiogenesis, genomic instability, self-sufficiency in growth signals, and insensitivity to antigrowth signals (Fig. 4A and B).
Fig. 4.
The temporal order of mutations in cancer clones rarely matches the path order from cross-sectional data, though the temporal order of clones matches the order inferred from the genetic-dependency analysis from intra-tumor data. (A) Plotting the percent of tumors with a given mutation at increasing neoplasm sizes can be used to infer (B) the cross-sectional path model of mutations. (C) However, the proportion of cells within any given simulated neoplasm whose temporal order is consistent with the cross-sectional path order tends to be low (mean = 7.3%, s.e.m. = 1.0%, n = 90). (D) The proportion of cells within any given simulation whose temporal order is consistent with the inferred order from the genetic-dependency analysis is high (mean = 99.7%, s.e.m. = 0.1%, n = 90). Each mutation is represented by a different color as given in Fig. 3B.
In order to quantify how well the temporal order matched the path order, we determined the percentage of tumor cells when cancer was detected whose evolutionary order over time matched the path order obtained from cross-sectional data. We found that most simulations had few cells in their tumors with consistent temporal and cross-sectional path orders (mean 7.3%, s.e.m. 1.0%; Fig. 4C). In fact, 41% of tumors had no cells with consistent orders and 68% had at most 1% of cells with consistent order. Thus, we saw that the order inferred from cross-sectional data was inconsistent with the temporal order of mutations for most cells. This is true across a range of parameter, or fitness, values for the cancer phenotypes, including telomere length, neoplasm size, mutation rate, and the probability of cell division. The sensitivity analyses are summarized in Table S3.
Then, we examined whether cells’ temporal orders were consistent with the order reconstructed from an oncogenetic tree model. We obtained the oncogenetic branching tree model from our cross-sectional cancer data (Fig. S3) and compared it to the temporal order from individual cells in cancers. We found that more temporal paths were consistent with the oncogenetic tree models than the simple path model, but most tumors still had relatively low fractions of temporal paths consistent with the oncogenetic tree (mean 11.5%; s.e.m. 2.2%; Fig. S3). Like for the path model comparison, 41% of tumors have no cells with consistent temporal orders to the reconstructed oncogenetic tree. However, the cells comprising the rest of the tumors are more consistent with the oncogenetic tree order; 54% of tumors have more than 1% of cells with consistent temporal orders. Like path models, these models do not fully specify the mutational spectrum observed in experimental cross-sectional studies of neoplasms. In one study in colon cancer, 11% of the neoplasms had combinations of mutations that were inconsistent with the oncogenetic tree identified (16). Of course, characteristics of neoplasms like mutation rate, number of cells, cell motility, and the relative fitness of new mutations affects the probability that a clone can reach fixation. In turn, this affects the percent of clones with consistent temporal and cross-sectional mutational orders.
We have shown that using cross-sectional methods to infer temporal order within tumors is often misleading. Now, we address whether using intra-tumor data to reconstruct the cell lineage, or genealogy of mutations, within individual tumors can better reflect the temporal order. To do so, we used a genetic-dependency analysis on a subsample of cells found in each final neoplasm. We sampled 5 cells per neoplasm and used these to reconstruct a cell lineage for each cancer as in Fig. 2C. We compared the temporal ordering of cells in the tumor at cancer to the same tumor’s genealogy and found that they were highly consistent (Fig. 4D; mean 99.7%, s.e.m. 0.1%). Increasing the number of cells sampled to 10 only slightly improved the mean consistency between the tumor’s genealogy and temporal order (from a mean of 99.7% to 99.9%, s.e.m. 0.1%). The appropriate number of cells to sample will depend on the evolutionary dynamics of the neoplasm (e.g., mutation rate, fitness effects of new mutations, neoplasm size, and cell motility). This approach has been previously implemented in pre-malignant tissues (31), malignant neoplasms (32, 33) and model systems (34). Of course, within-tumor cell lineage reconstruction could lead to difficulties teasing apart the order of some mutations when there aren’t enough different samples, or if key mutations occurred prior to recent selective sweeps that reached fixation. The former problem can be solved by obtaining more samples, while the latter can be solved by sampling from the same tumor at multiple time points. In either case, evolutionary methods can overcome the limitations of cross-sectional sampling.
Discussion
We have used our agent-based model to simulate neoplastic progression. This approach allowed us to record the cell lineage and population structure of neoplasms that progressed to cancer. We have shown that using cross-sectional data to infer the temporal order of mutations for all cells in a neoplasm rarely works; 41% of tumors had no clones with consistent temporal and cross-sectional orders. These results are robust and don’t depend on exactly how any one hallmark is implemented. Some of this mismatch between cross-sectional models and temporal orders can be due genetic instability and low clonal expansion rates within tumors. This prevents selective sweeps from reaching fixation, and thus neoplasms do not progress through discrete, homogenous mutational states as are assumed in path models. Additionally, clonal expansions may be transient during progression due to regression or competition with other clones. Using intra-tumor data to obtain the cell lineage for each tumor is a more accurate method of reconstructing the temporal order of mutations.
Path models implicitly assume that neoplasms pass through a series of selective sweeps, each of which homogenizes a tumor’s genotype. With our model, we have shown that there are many possible temporal paths to cancer and that, at detection, neoplasms are comprised of many clones. This heterogeneity has been observed experimentally (35–37). Exactly how much heterogeneity exists depends on the evolutionary parameters of the tumor, including the mutation rate, fitness advantage of the new mutations, and the level of cell motility within the neoplasm. Cancer cells are an evolving population of asexually-reproducing cells, and the rate and dynamics of adaptation in asexually-evolving populations has been studied experimentally and theoretically in evolutionary biology (reviewed in (2)). Homogenous and linear clonal evolution occurs when each clone outcompetes others and reaches fixation before the next mutation occurs. This tends to occur when mutations are infrequent, have strong selective advantages, and there is a high level of cell turnover in the neoplasm. In this case, the clear temporal path to cancer is simply the order of clonal selective sweeps and heterogeneity is likely to decrease as the neoplasm homogenizes, especially with low mutation rates. Clonal interference tends to occur when new mutant clones with relatively high fitness advantages occur frequently enough to interfere with fixation of other clones. Because there are many competing clones, determining the single temporal path to cancer for a neoplasm from its heterogeneous subclones becomes more complicated. Clonal interference, and the heterogeneity that accompanies it, can occur when mutation rates are high but the fitness effects of new mutations are small. Another condition generating exceptionally high intra-tumor heterogeneity occurs when mutation rates are high and there is geographical isolation of subpopulations of cells. It is currently unknown which of these clonal evolution dynamics are found in real tumors, though there is evidence for high mutation rates and small fitness effects in colon cancer (38), and evidence for small fitness effects in pancreatic cancer and glioblastoma multiforme (39). We observed dynamics consistent with both clonal evolution and clonal interference in our simulations, even within the same neoplasm (Fig. 5C). For example, we observed that heterogeneity decreased when a clone that acquired the loss of differentiation mutation fixed in the population, as might occur during a clonal selective sweep (Fig. 5B). Subsequently, heterogeneity increased as clonal interference dominated the rest of the neoplastic dynamics. In general, heterogeneity increased over the course of progression in the simulations, with occasional drops that were immediately followed by increases. Thus, one of the explanations for the low percent of clones within a tumor with matching temporal and path orders is that there is not a single evolutionary path for tumor. Instead, there are many clones, each of which can independently acquire genetic alterations as has been suggested in Barrett’s esophagus (21).
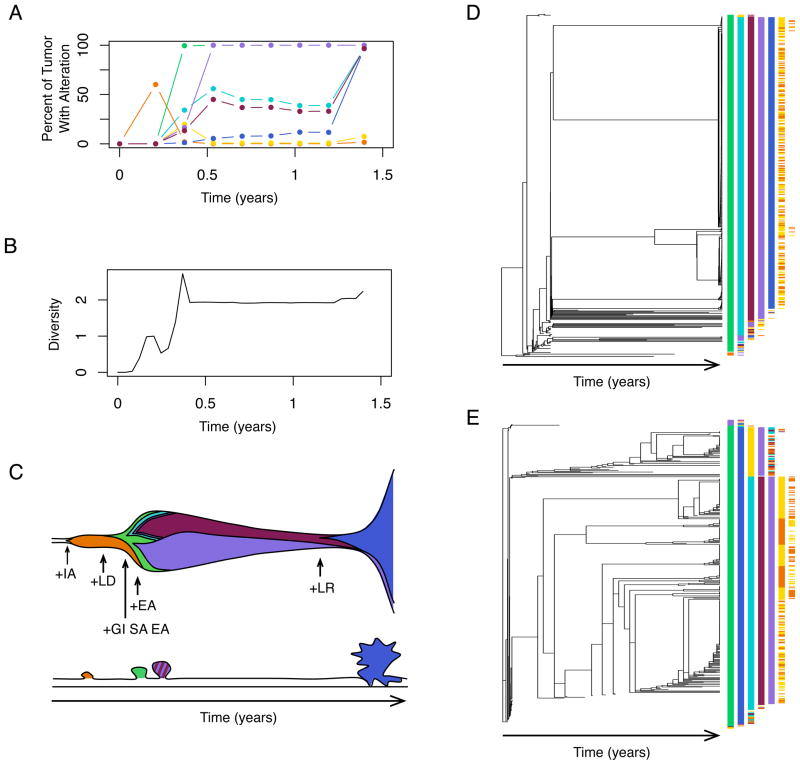
Fig. 5.
Details of a single simulation as it progresses to cancer. (A) Plot of the percent of cells within this neoplasm that contain a given mutation over time. Note that the IA reaches detection early in progression and regresses. (B) Plot of the Shannon index for diversity, or information entropy, over time for the simulation. (C) The top panel shows the clones, their mutational states, and their rough population sizes over time. The height is proportional to the population size of the neoplasm, and new mutations are indicated with an arrow. The bottom panel shows the type of neoplasm that would be identified at various points during progression from normal tissue to cancer, beginning with polyps and ending with cancer. (D) The genealogy, or cell lineage, for all of the clones that arose during the evolution of the neoplasm shows that a single evolutionary run doesn’t have a single evolutionary path. The temporal order of phenotypes is given at the tips of the genealogy. Because we are modeling phenotypes, the same set of phenotypic mutations can occur in clones that are unique by descent. Each new mutation for a phenotype is a new mutation in a gene or pathway conferring the phenotype. Thus, we have what looks like convergent evolution - there is phenotypic homogeneity, but it arose through different genetic alterations. Under these parameters, independent acquisition of hallmarks in different clones is common and leads to clonal interference and the suppression of clonal expansion for any one clone. Note that the most commonly-observed phenotypic order does not correspond to the cross-sectional path order given in Fig. 3B. (E) The genealogy, or cell lineage, for all of the clones that arose during the evolution of the neoplasm pictured in Fig. 2. Both neoplasms pictured here have relatively high genetic heterogeneity at cancer detection. As occurs here, genetic heterogeneity may lead to phenotypic homogeneity. Each mutation is represented by a different color as given in Fig. 3B.
Another reason the temporal order does not match the path order in cross-sectional studies is due to the detection of transient clones. Transient clones increase in size early in progression only to go extinct which may occur if a clone is outcompeted by another clone or if it fails to stabilize its telomeres. We observed both events (Fig. 5). For example, we observed a clone that acquired the insensitivity to antigrowth signals mutation early on which allowed the clone to expand to a detectable size. Then, a loss of differentiation mutation occurred independently in a wild-type cell. This new clone quickly expanded and drove the original clone extinct. Later in progression of this same neoplasm, a large clone eventually went extinct due to failure to stabilize its telomeres.
There is a further problem specifically with the construction of path models. Building path models requires the characterization of several different neoplasms at different stages. The stages are then ordered according to increasing size and grade, which is assumed to correspond to a single, linear order of changes during progression to cancer. This is how the in the path model in Fig. 4 was constructed. It may be an obvious point, but by basing the path model on mutations associated with increasing size and grade, we are identifying those mutations involved in increasing the neoplasm’s size and grade. That these mutations are involved in progression to cancer is an assumption (40). Thus, if histological grading does not reflect the necessary temporal sequence during progression, then studies based on that ordering will of course be invalid (41).
Previous work has identified other concerns with the cross-sectional approach. Using a probabilistic framework, Szabo and Yokovlev (42) showed that there are technical limitations in inferring the ordering of genetic events from frequency and correlation data, regardless if the cross-sectional order obtained was a path or a tree. In particular, small sample sizes, inherent undercounting of mutations associated with early tumor grades, and current methods that assume that the mutations are independent are problematic.
Oncogenetic tree models (11) accommodated more of the heterogeneity between tumors than path models because they do not impose a strict order on every mutation in a tumor. They also relax the assumption that the mutations that lead to neoplasms of increasing size and grade are the same mutations that lead to cancer. However, we have shown that even the oncogenetic tree order of mutations does not match the true evolutionary path. Oncogenetic tree models have already been extended. Distance-based methods have also been used to reconstruct oncogenetic tree models (43) and conjunctive Bayesian network models have used directed acyclic graphs to represent mutation ordering (44) and have been applied to cross-sectional data (45). These models still suffer from the weakness of cross-sectional data. Recently, a computational approach was developed to identify the most likely paths through a mutational network for colorectal cancer and glioblastoma using cross-sectional data; the authors found that not all evolutionary paths are accounted for in the mutational networks, perhaps due to heterogeneity of temporal orders within cancer types (46).
While understanding the dynamics of mutation accumulation has important implications for cancer prevention and risk stratification, it is difficult to reconstruct temporal order from cross-sectional data. A fundamental problem in the use of cross-sectional data to infer the temporal order of events is the assumption that the state of one tumor is informative for the history of a tumor in a different patient. Both our model and recent cancer resequencing efforts (30, 47, 48) show that there are likely many possible evolutionary paths to cancer, not just between types of cancers, but even within a given type of cancer. Each tumor is a unique evolutionary trajectory with occasional necessary and sufficient phenotypic mutations that can be acquired differently in different tissues. Further, tumors are populations of heterogeneous clones, each of which is evolving along a distinct path. Thus, identifying a single path or oncogenetic tree of mutational events is insufficient to describe this process. Reconstructing cell lineages within individual tumors should reveal the true temporal order of events for the different clones within a tumor (49).
Because cancer is an evolutionary process (50), we can use some of the powerful tools of evolutionary analyses to reveal the dynamics of cancer. We have shown that an evolutionary analysis applied to intra-tumor samples can overcome the limitations of cross-sectional analyses,. This can also resolve the conflicting results arising from analyses using cross-sectional data (10). Finally, evolutionary tools can help to reveal the dynamics of intra-tumor genetic heterogeneity that drives the process of tumor progression and therapeutic resistance.
Supplementary Material
Acknowledgments
Grant Support
This work was supported by the National Institutes of Health grants R03 CA137811, P01 CA91955, P30 CA010815, R01 CA119224 (C.C.M.), R01 CA140657 (K.S., J.W.P., C.C.M.), T32 HG000046 (K.S.), the Pew Charitable Trust (C.C.M.), the Martha W. Rodgers Charitable Trust (C.C.M.), a McLean Contributionship (C.C.M.), and the Landon AACR Innovator Award for Cancer Prevention (C.C.M.), and the American Cancer Society Research Scholar Grant 117209-RSG-09-163-01-CNE (C.C.M.).
Footnotes
This value can be calculated using , since there are 7 possible mutations and any subset of the 7 can be observed in a tumor.
References
- 1.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Sniegowski PD, Gerrish PJ. Beneficial mutations and the dynamics of adaptation in asexual populations. Philos Trans R Soc Lond, B, Biol Sci. 2010;365:1255–63. doi: 10.1098/rstb.2009.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sgroi DC. Preinvasive breast cancer. Annu Rev Pathol. 2010;5:193–221. doi: 10.1146/annurev.pathol.4.110807.092306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wistuba II, Mao L, Gazdar AF. Smoking molecular damage in bronchial epithelium. Oncogene. 2002;21:7298–306. doi: 10.1038/sj.onc.1205806. [DOI] [PubMed] [Google Scholar]
- 5.Balaban GB, Herlyn M, Clark WH, Nowell PC. Karyotypic evolution in human malignant melanoma. Cancer Genet Cytogenet. 1986;19:113–22. doi: 10.1016/0165-4608(86)90378-x. [DOI] [PubMed] [Google Scholar]
- 6.Fearon E, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 7.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 8.Bilke S, Chen Q-R, Westerman F, Schwab M, Catchpoole D, Khan J. Inferring a tumor progression model for neuroblastoma from genomic data. J Clin Oncol. 2005;23:7322–31. doi: 10.1200/JCO.2005.03.2821. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Bandyopadhyay N, Ranka S, Baudis M, Kahveci T. Inferring progression models for CGH data. Bioinformatics. 2009;25:2208–15. doi: 10.1093/bioinformatics/btp365. [DOI] [PubMed] [Google Scholar]
- 10.Smith G, Carey F, Beattie J, Wilkie M, Lightfoot T, Coxhead J, et al. Mutations in APC, Kirsten-ras, and p53--alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA. 2002;99:9433–9438. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desper R, Jiang F, Kallioniemi OP, Moch H, Papadimitriou CH, Schäffer AA. Inferring tree models for oncogenesis from comparative genome hybridization data. J Comput Biol. 1999;6:37–51. doi: 10.1089/cmb.1999.6.37. [DOI] [PubMed] [Google Scholar]
- 12.Jiang F, Desper R, Papadimitriou C, Schaffer A. Construction of evolutionary tree models for renal cell carcinoma from comparative genomic hybridization data. Cancer Res. 2000;60:6503–6509. [PubMed] [Google Scholar]
- 13.Chen L, Nordlander C, Behboudi A, Olsson B, Levan KK. Deriving evolutionary tree models of the oncogenesis of endometrial adenocarcinoma. Int J Cancer. 2007;120:292–6. doi: 10.1002/ijc.22165. [DOI] [PubMed] [Google Scholar]
- 14.Gunawan B, von Heydebreck A, Sander B, Schulten H-J, Haller F, Langer C, et al. An oncogenetic tree model in gastrointestinal stromal tumours (GISTs) identifies different pathways of cytogenetic evolution with prognostic implications. J Pathol. 2007;211:463–70. doi: 10.1002/path.2128. [DOI] [PubMed] [Google Scholar]
- 15.Pathare S, Schäffer AA, Beerenwinkel N, Mahimkar M. Construction of oncogenetic tree models reveals multiple pathways of oral cancer progression. Int J Cancer. 2009;124:2864–71. doi: 10.1002/ijc.24267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweeney C, Boucher KM, Samowitz WS, Wolff RK, Albertsen H, Curtin K, et al. Oncogenetic tree model of somatic mutations and DNA methylation in colon tumors. Genes Chromosomes Cancer. 2009;48:1–9. doi: 10.1002/gcc.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm V, Berger U, Bastiansen F, Eliassen S, Ginot V, Giske J, et al. A standard protocol for describing individual-based and agent-based models. Ecological Modelling. 2006;198:115–126. [Google Scholar]
- 18.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 19.Kimura M, Crow JF. The number of alleles that can be maintained in a finite population. Genetics. 1964;49:725–38. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabo A, Boucher K. Estimating an oncogenetic tree when false negatives and positives are present. Mathematical biosciences. 2002;176:219–36. doi: 10.1016/s0025-5564(02)00086-x. [DOI] [PubMed] [Google Scholar]
- 21.Leedham SJ, Preston SL, McDonald SAC, Elia G, Bhandari P, Poller D, et al. Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human Barrett’s oesophagus. Gut. 2008;57:1041–8. doi: 10.1136/gut.2007.143339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Notta F, Mullighan CG, Wang JCY, Poeppl A, Doulatov S, Phillips LA, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–7. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 23.Abbott R, Forrest S, Pienta K. Simulating the hallmarks of cancer. Artif Life. 2006;12:617–634. doi: 10.1162/artl.2006.12.4.617. [DOI] [PubMed] [Google Scholar]
- 24.Spencer S, Gerety R, Pienta K, Forrest S. Modeling Somatic Evolution in Tumorigenesis. PLoS Comput Biol. 2006;2:e108. doi: 10.1371/journal.pcbi.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg R. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 26.Pepper J, Sprouffske K, Maley C. Animal cell differentiation patterns suppress somatic evolution. PLoS Comput Biol. 2007;3:e250. doi: 10.1371/journal.pcbi.0030250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novelli MR, Williamson JA, Tomlinson IP, Elia G, Hodgson SV, Talbot IC, et al. Polyclonal origin of colonic adenomas in an XO/XY patient with FAP. Science. 1996;272:1187–90. doi: 10.1126/science.272.5265.1187. [DOI] [PubMed] [Google Scholar]
- 28.Park SY, Gönen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–44. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuukasjärvi T, Karhu R, Tanner M, Kähkönen M, Schäffer A, Nupponen N, et al. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res. 1997;57:1597–604. [PubMed] [Google Scholar]
- 30.Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 31.Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Galipeau PC, Paulson TG, et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet. 1999;22:106–9. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thirlwell C, Will OCC, Domingo E, Graham TA, McDonald SAC, Oukrif D, et al. Clonality assessment and clonal ordering of individual neoplastic crypts shows polyclonality of colorectal adenomas. Gastroenterology. 2010;138:1441–54. doi: 10.1053/j.gastro.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 33.Navin N, Krasnitz A, Rodgers L, Cook K, Meth J, Kendall J, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frumkin D, Wasserstrom A, Itzkovitz S, Stern T, Harmelin A, Eilam R, et al. Cell Lineage Analysis of a Mouse Tumor. Cancer Res. 2008;68:5924–5931. doi: 10.1158/0008-5472.CAN-07-6216. [DOI] [PubMed] [Google Scholar]
- 35.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–73. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 36.Tsao JL, Yatabe Y, Salovaara R, Järvinen HJ, Mecklin JP, Aaltonen LA, et al. Genetic reconstruction of individual colorectal tumor histories. Proc Natl Acad Sci USA. 2000;97:1236–41. doi: 10.1073/pnas.97.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.González-García I, Solé RV, Costa J. Metapopulation dynamics and spatial heterogeneity in cancer. Proc Natl Acad Sci USA. 2002;99:13085–13089. doi: 10.1073/pnas.202139299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beerenwinkel N, Antal T, Dingli D, Traulsen A, Kinzler KW, Velculescu VE, et al. Genetic progression and the waiting time to cancer. PLoS Comput Biol. 2007;3:e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazebnik Y. What are the hallmarks of cancer? Nat Rev Cancer. 2010;10:232–3. doi: 10.1038/nrc2827. [DOI] [PubMed] [Google Scholar]
- 41.Sontag L, Axelrod DE. Evaluation of pathways for progression of heterogeneous breast tumors. J Theor Biol. 2005;232:179–89. doi: 10.1016/j.jtbi.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 42.Szabo A, Yakovlev A. Preferred sequences of genetic events in carcinogenesis: quantitative aspects of the problem. Journal of Biological Systems. 2001;9:105–121. [Google Scholar]
- 43.Desper R, Jiang F, Kallioniemi O, Moch H. Distance-Based Reconstruction of Tree Models for Oncogenesis. Journal of Computational Biology. 2000;7:789–803. doi: 10.1089/10665270050514936. [DOI] [PubMed] [Google Scholar]
- 44.Beerenwinkel N, Eriksson N, Sturmfels B. Evolution on distributive lattices. J Theor Biol. 2006;242:409–20. doi: 10.1016/j.jtbi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Gerstung M, Baudis M, Moch H, Beerenwinkel N. Quantifying cancer progression with conjunctive Bayesian networks. Bioinformatics. 2009;25:2809–15. doi: 10.1093/bioinformatics/btp505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Attolini CS-O, Cheng Y-K, Beroukhim R, Getz G, Abdel-Wahab O, Levine RL, et al. A mathematical framework to determine the temporal sequence of somatic genetic events in cancer. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1009117107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campbell P, Yachida S, Mudie L, Stephens P, Pleasance E, Stebbings L, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navin NE, Hicks J. Tracing the tumor lineage. Mol Oncol. 2010;4:267–83. doi: 10.1016/j.molonc.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merlo L, Pepper J, Reid B, Maley C. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.