Abstract
DNA repair variants may play a potentially important role in an individual’s susceptibility to developing cancer. Numerous studies have reported the association between genetic single nucleotide polymorphisms (SNPs) in DNA repair genes and different types of hematologic cancers. However, to date, the effects of such SNPs on modulating Hodgkin Lymphoma (HL) risk have not yet been investigated. We hypothesized that gene-gene interaction between candidate genes in Direct Reversal, Nucleotide excision repair (NER), Base excision repair (BER) and Double strand break (DSB) pathways may contribute to susceptibility to HL. To test this hypothesis, we conducted a study on 200 HL cases and 220 controls to assess associations between HL risk and 21 functional SNPs in DNA repair genes. We evaluated potential gene-gene interactions and the association of multiple polymorphisms in a chromosome region using a multi-analytic strategy combining logistic regression, multi-factor dimensionality reduction and classification and regression tree approaches. We observed that, in combination, allelic variants in the XPC Ala499Val, NBN Glu185Gln, XRCC3 Thr241Me, XRCC1 Arg194Trp and XRCC1 399Gln polymorphisms modify the risk for developing HL. Moreover, the cumulative genetic risk score revealed a significant trend where the risk for developing HL increases as the number of adverse alleles in BER and DSB genes increase. These findings suggest that DNA repair variants in BER and DSB pathways may play an important role in the development of HL.
Introduction
Hodgkin’s lymphoma is an uncommon cancer of the lymphatic system. The American Cancer Society estimated that about 8,510 cases of HL were diagnosed in the United States in 2010 and 1,290 people will die from the disease (1). Due to remarkable treatment advances the death rate for this disease has now dropped by about 60%, however HL survivors report a higher rate of subsequent malignancies compared to other cancers. Recent studies indicate that HL patients have higher levels of overall genetic instability compared to others types of cancer which may potentially lead to unfavorable outcomes (2). DNA repair capacity is critical for maintaining genomic integrity and stability. In the general population, interindividual variability in DNA repair capacity has been reported, and an association between reduced DNA repair and susceptibility to a variety of hematological cancers, has been documented (3–5). Single nucleotide polymorphism (SNPs) in DNA repair genes is one mechanism that may lead to interindividual variations in repair capacity. Recent studies show that SNPs located within coding or regulating regions, lead to alterations in protein expression and in functional properties of the repair enzymes (6–7).
Epidemiological studies suggested associations between allelic variants in the different repair pathways and risk of hematologic cancers such as leukemias and lymphomas (8–12). Specifically, in the NER pathway, variants in the XPD, XPA, ERCC1, CCNH and ERCC5 genes have been associated with an increased risk (12–15). In the BER pathway, XRCC1 and PARP variants showed similar association (5, 16). In the post-replication repair pathway, XRCC3 Thr241Met polymorphism, a variant of XRCC3 gene, which is required for efficient repair of DSBs through the Homologous Replication Repair (HRR) pathway, was reported to be significantly associated with AML (17). Polymorphisms in MGMT, that participates in direct reversal of DNA damage was reported to contribute to development of NHL (18). However, to date, few studies investigated the role of DNA repair genetic polymorphisms as risk factors for development of HL. We have recently reported that the interaction between variants of XRCC1 Arg399Gln, XRCC3 Thr241Met and XPC Lys 939Gln polymorphisms may modify the risk of development HL (19). Given the multiple pathways involved in DNA repair, variants in the different pathways are likely to have a joint or additive effect on modulating HL risk and should therefore be evaluated simultaneously. In the current case control study, we evaluated the associations and interactions between 21 SNPs in 15 key genes in the different DNA repair pathways. Seven SNPs in 5 genes involved in the NER pathway, 6 SNPs in 5 genes involved in the BER pathway, 3 SNPs in 1 gene involved in the Direct Reversal pathway, and 5 SNPs involved in the HRR or NHEJ pathways were chosen for this study. These SNPs were selected from published association studies, a few were chosen from the National Institute of Environmental Health Science database (available at URL: http://egp.gs.washington.edu) on the basis of their location in the promoter, 5′-untranslated and coding regions of the genes and on the basis of commonality (minor allele frequencies>0.05).
Material and methods
Study subjects
The study population consisted of 200 newly diagnosed adult HL patients registered at M. D. Anderson Cancer Center between January 1987 and December 1992. The parent study that investigated the genotoxic effects of different chemotherapeutic treatment regimens for Hodgkin Lymphoma (20–22). Demographic information and the principal clinical characteristics (e.g. age at diagnosis, sex, race/ethnicity, family history of cancer, HL histologic subtypes, disease stage, and presence of B symptoms) were obtained through an interviewer-administered health risk questionnaires and medical records. Patients were excluded if they had HIV infection, or were unable to provide written informed consent (the participation rate for cases was 77%). The study required serial blood collection for cytogenetic studies and the main reason for non participation was the time commitment and constraints involved. The DNA used in our genotyping study was extracted from blood samples collected at baseline prior to intitiaion of therapy. Controls (n=220) were accrued through random-digit dialing (RRD) and were frequency matched to cases with regard to age (±5years) sex, and race/ethnicity. The RDD protocol that was used is based on the Waksberg procedure (23) stratified by the distribution of the cases by age (within five-year age, sex, and ethnic group). The Waksberg method involves a two-stage approach. First, telephone numbers are randomly generated using telephone tapes that exclude business and cell phone numbers, using equal probability simple random sampling. The second phase of the RDD consisted of using the first five digits of all residential phone numbers identified in the first stage as the “primary sampling unit.” Randomly generated two digit numbers were added to the primary sampling unit until four additional residences are contacted. This procedure was repeated for all residential numbers identified during the primary sampling. The secondary sampling can be shown to result in an equal probability sample of all residential numbers. Interclass correlation among respondents within the same primary sampling unit is minimized by contacting only one additional residence (denoted by the “k” by Waksberg) for each primary sampling unit during the secondary state of sampling. This procedure was used to identify and select eligible controls for all residences contacted during both stages of sampling. The telephone screener asked the ages of all individuals who reside in the household between 20 and 75, and then used the stratification tables to select in each strata based on the ratio of expected number of eligible individuals required for all strata. This process involved the generation of telephone numbers using equal probability simple random sampling and was used to identify and select eligible controls for all residences contacted. If more than one eligible individual resided at that number, one was randomly selected. The response rate for the initial call, after excluding 50% of the numbers dialed because of the numbers were businesses, caller ID screening, or fax machines, was 83%. An interviewer from M. D. Anderson then contacted the informant within 48 hours to explain the study, and ascertain willingness to participate and informed consent. In addition, a visit was set up for venipuncture. About 17% of the participants refuse us after they showed interested to the initial call. Ongoing studies in the department of epidemiology, had great success with the RDD method resulting in well-matched controls according to age, sex, and ethnicity. The research protocol was approved by the University of Texas, M.D. Anderson review board.
Genotyping assay
Candidate SNPs were selected from the best evidence from published studies, to represent more commonly occurring variants (minor allele frequencies, >5%) and to gain statistical power to detect interactions. Overall, a total of 21 potentially functional polymorphisms in 15 key genes were included, seven SNPs in 5 genes involved in the NER pathway, 6 SNPs in 5 genes involved in the BER pathway, 3 SNPs in 1 gene involved in the Direct Reversal pathway, and 5 SNPs involved in the HRR or NHEJ pathways were chosen for this study. These SNPs were selected from published association studies, a few were chosen from the National Institute of Environmental Health Science database (available at URL: http://egp.gs.washington.edu) on the basis of their location in the promoter, 5′-untranslated and coding regions of the genes and on the basis of commonality (minor allele frequencies>0.05). Genotyping was performed using Sequenom MassARRAY iPLEX7™ platform. Over 97% of the DNA samples were successfully genotyped for all selected SNP. Ten percent of the samples were randomly selected for repeat analysis as quality control for verification of genotyping procedures. Quality control measures included genotyping of internal positive control samples, the use of no template controls, and the replication of 40 study subjects for all SNPs (the concordance rates was of 100%).
Statistical analysis
Statistical analyses were completed using Stata 8.0 statistical software package (Stata Corp., College Station, TX). The Pearson χ2 test was used to test for differences in distribution between the cases and controls with regard to age, gender and race. Hardy-Weinberg equilibrium was tested in controls using the χ2 test. Multivariable logistic regression (MLR) analysis was used to estimate the associations between each genotype and risk of HL by computing the odds ratios (ORs) and 95% confidence intervals (CIs). A permutation test with M=1,000 replicates was used to reduce the potential for spurious findings due to multiple testing and to validate the results in our sample. Within each replicate, we chose a bootstrap sample of size N=N1+N2 such that N1=200 (total number of cases) and N2=220 (total number of controls) replacement from the original data (24). The random sample of the 200 cases was chosen separately from the random sample of 220 controls) to allow for any one participant to be chosen once, more than once, or not at all and to keep the ratio of cases to control constant. We then calculated the ORs for each replicate and constructed an empirical distribution for the ORs to calculate the empirical p-value for the observed OR.
Multi-factor-dimensionality reduction analysis (MDR)
MDR as used to identifying the genotype combinations associated with HL. MDR, is a nonparametric method developed by Ritchie et al (25) to detect and characterize high-order gene-gene interactions in studies with relatively small sample sizes. We conducted MDR using the dichotomous groupings of the polymorphisms. The procedure involves cross-validation where the entire dataset is divided into a training set (model building) and a test set (model testing) to estimate the accuracy rate of each k-level model. The k-level model that has the maximum cross-validation consistency (CVC) and maximum mean testing accuracy (TA) is selected as the final model. The TA measures how often individuals are correctly classified with respect to their case/control status and the CVC evaluate the consistency with which individuals are classified. The parsimonious interaction model that had the maximum mean TA and CVC was identified and included in multivariable logistic regression analysis to estimate the association between HL risk and MDR-identified combinations.
Cumulative genetic risk score analysis (CGRS)
We used the CGRS analysis for identifying the contribution of multiple SNPs and disease risk. The CGRS risk score was calculated for the genes of each pathway using MLR and for the SNP combinations identified through MDR. We calculated the CGRS by using a linear weighting of −1, 0, 1 for genotypes containing zero, one, or two risk alleles, respectively.
Classification and regression tree analysis (CART)
We conducted a CART analysis to identify subgroups of high- and low-risk based on stratified SNP profiles and explore possible gene-gene interactions between polymorphisms of DNA repair genes that may be identified through the tree (26). CART analysis was performed using the rpart package for the S-PLUS Analysis Software (version 8.1). CART produces a decision tree to identify subgroups of subjects with specific genotype combinations. Specifically, the recursive partitioning algorithm in rpart starts at the root node (with the entire data set) and determines all possible splits of the data set. The variable that produces the locally optimal split (based on the distribution of cases and controls in the two daughter nodes) is chosen as the first split. This process continues for each subsequent split of the data set until no optimal split is obtained or the terminal nodes reach a pre-specified minimum size (at least 20 subjects for each terminal node). The resulting tree is pruned back using an alternative pruning approach such that the Ps (corrected using Bonferonni test to control overall tree alpha level 0.05) for the terminal nodes are significant. Subgroups of individuals with differential risk associations were identified in the different nodes of the tree, indicating the presence of interactions. We also performed 10-fold cross-validation to ensure that the overall misclassification error rate did not exceed 50%.
Results
Study Subjects
The distribution of demographic characteristics in cases and controls are presented in Table 1. The mean age of the cases was 47.52 ± 13.37 yrs overall with 34% between the ages of 20–40 years and 66% between 40–70 years. Among the cases, 80% were histologically nodular sclerosis. Thirteen percent of the patients were stage I, 52% stage II, 23% stage III and 14% were stage IV. Twenty-six percent of the patients reported B symptoms. The Epstein-Barr virus status was unavailable on patients and controls.
Table 1.
Demographic Characteristics of the HL Cases and Controls
| Cases | Controls | ||||
|---|---|---|---|---|---|
| n | % | n | % | P value | |
| Subjects analyzed | 200 | 100 | 220 | 100 | |
| Age (mean ± SD) | |||||
| 47.52 ± 13.37 | 49.27 ± 15.19 | 0.21 | |||
| Age Range | |||||
| 20–40 | 68 | 34 | 72 | 33 | 0.78 |
| >40 | 132 | 66 | 148 | 67 | |
| Gender | |||||
| Female | 93 | 47 | 97 | 44 | 0.62 |
| Male | 107 | 53 | 123 | 56 | |
| Race | |||||
| white | 163 | 82 | 195 | 89 | 0.53 |
| others | 37 | 18 | 25 | 11 | |
| Smoking Status * | |||||
| Never | 95 | 51 | 120 | 55 | 0.45 |
| Ever | 92 | 49 | 100 | 45 | |
| Family History | |||||
| No | 119 | 60 | 99 | 45 | |
| Yes | 81 | 41 | 121 | 55 | 0.35 |
Smoking status was unknown in 13 of the HL patients
Frequency distribution of genotypes and associations with HL risk
The reference SNP (# rs), the single nucleotide changes and the chromosome position are presented in Table 2. The frequency distributions of the genotypes in the controls were tested for deviation from Hardy-Weinberg (HW) equilibrium. None of the genotypes showed significant deviation at the 5% alpha level. There was no significant difference in distribution frequency among the cases and controls for: all the 3 SNPs in the Direct Reversal pathways, 6/7 SNPs in NER, 4/6 SNPs in BER and 4/5 SNPs in HRR or NHEJ pathways. Significant differences in the frequency distributions among cases and controls were observed for XPC Ala499Val (P=0.01), XRCC1 Arg194Trp (P=0.01), XRCC1 Arg399Gln (P=0.04) and NBN Glu185Gln (P=0.04). The significant associations of the candidate SNPs with HL are shown in Table 3.
Table 2.
List of studied SNPs with chromosome location an reference number
| SNP | # rs | Nucleotide Change | SNP Region | Chromosome | |
|---|---|---|---|---|---|
| Nucleotide Excision Repair | |||||
| XPC codon 499 | 2228000 | C/T | Ex9−377C>T | 3 | |
| XPC codon 939 | 2228001 | C/A | Ex16+211C>A | 3 | |
| XPD codon 156 | 238406 | A/C | Ex6−10A>C | 19 | |
| XPD codon 751 | 13181 | A/C | Ex23+61A>C | 19 | |
| XPG codon 1104 | 17655 | G/C | Ex15−344G>C | 13 | |
| ERCC1 codon 504 | 3212986 | T/G | 196bp 3′ of STP T>G | 19 | |
| CCNH codon 270 | 2266690 | T/C | Ex7+49T>C | 5 | |
| Base Excision Repair | |||||
| XRCC1 codon 194 | 1799782 | C/T | Ex6−22C>T | 19 | |
| XRCC1 codon 399 | 25487 | A/G | Ex10−4A>G | 19 | |
| OGG1 codon 326 | 1052133 | C/G | Ex6−315C>G | 3 | |
| PARP1 codon 762 | 1136410 | T/C | Ex17+8T>C | 1 | |
| APEX 1 codon 148 | 3136819 | C/T | IVS4−61C>T | 14 | |
| POLB-10 | 3136794 | A/G | IVS11−235A>G | 8 | |
| Direct reversal of damage | |||||
| MGMT codon 143 | 2308321 | A/G | Ex7+13A>G | 10 | |
| MGMT codon 178 | 2308327 | A/G | Ex7+119A>G | 10 | |
| MGMT codon 84 | 12917 | C/T | Ex5−25C>T | 10 | |
| Others | |||||
| XRCC3 codon 241 | 861539 | C/T | Ex8−53C>T | 14 | |
| NBN codon 185 | 1805794 | G/C | Ex6−32G>C | 8 | |
| LIG1-01 codon 170 | 20580 | C/A | Ex7+44C>A | 19 | |
| LIG1-03 codon 114 | 20579 | C/T | Ex2−24C>T | 19 | |
| PARG codon 12 | 1801282 | C/G | Ex4−49C>G | 3 |
Table 3.
Allele frequency of significant genes and their associations with HD risk
| Cases | Controls | |||||||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | P value * | Adjusted OR [95% CI] | P value ** | ||
| Subjects analyzed | 200 | 100 | 220 | 100 | ||||
| Nucleotide Excision Repair | ||||||||
| XPC codon 499 | 0.01 | |||||||
| Ala/Ala | 92 | 48 | 137 | 63 | ||||
| Ala/Val | 90 | 47 | 71 | 33 | 1.77 [1.17–2.68] | 0.01 | ||
| Val/Val | 8 | 5 | 10 | 5 | 1.03 [0.38–2.84] | 0.95 | ||
| At least one Val | 98 | 52 | 81 | 38 | 1.68 [1.13–2.51] | 0.01 | ||
| ERCC1 codon 504 | 0.17 | |||||||
| Gln/Gln | 102 | 61 | 122 | 56 | ||||
| Gln/Lys | 52 | 31 | 84 | 39 | 0.57 [0.37–0.89] | 0.01 | ||
| Lys/Lys | 14 | 8 | 11 | 5 | 1.20 [0.51–2.80] | 0.68 | ||
| At least one Lys | 66 | 39 | 95 | 44 | 0.65 [0.43–0.97] | 0.04 | ||
| Base Excision Repair | ||||||||
| XRCC1 codon 194 | 0.01 | |||||||
| Arg/Arg | 183 | 93 | 184 | 84 | ||||
| Arg/Trp | 13 | 7 | 32 | 15 | 0.39 [0.20–0.79] | 0.01 | ||
| Trp/Trp | 0 | 0 | 2 | 1 | 0.00 [0.00–3.98] | 0.99 | ||
| At least one Trp | 13 | 7 | 34 | 16 | 0.37 [0.19–0.74] | 0.01 | ||
| XRCC1 codon 399 | 0.04 | |||||||
| Arg/Arg | 73 | 37 | 102 | 47 | ||||
| Arg/Gln | 106 | 53 | 89 | 41 | 1.77 [1.16–2.71] | 0.01 | ||
| Gln/Gln | 20 | 10 | 28 | 13 | 1.13 [0.58–2.18] | 0.72 | ||
| At least one Gln | 126 | 63 | 117 | 53 | 1.62 [1.08–2.43] | 0.02 | ||
| Other identified genes on DNA repair function | ||||||||
| NBN codon 185 | 0.04 | |||||||
| Glu/Glu | 82 | 42 | 103 | 47 | ||||
| Glu/Gln | 102 | 53 | 95 | 43 | 1.39 [0.93–2.10] | 0.11 | ||
| Gln/Gln | 9 | 5 | 22 | 10 | 0.49 [0.21–1.17] | 0.11 | ||
| At least one Gln | 111 | 58 | 117 | 53 | 1.23 [0.83–1.82] | 0.31 | ||
Two-sided χ2 test for the difference in the genotype or allele distributions between the cases and the controls
Adjusted for age, sex, race, and smoking status in logistic regression models
In the multivariate logistic regression analysis after adjustment for all covariates, XPC Ala499Val and XRCC1 Arg399Gln were significantly associated with HL risk as compared with wild type homozygote. The presence of the heterozygote Ala/Val genotype, in the NER gene XPC, was associated with a 77% increased risk of HL (OR=1.77 and 95% CI= 1.17–2.68) and the presence of at least one Val allele was associated with a 68% increase in risk (OR=1.68 and 95% CI=1.13–2.51). In addition, the presence of heterozygote Arg/Gln, in XRCC1 Arg399Gln, was associated with a 77% increased risk of HL (OR=1.77 and 95% CI= 1.16–2.71) and the presence of at least one XRCC1 Gln allele was associated with a 62% increased risk (OR=1.62 and 95% CI=1.08–2.43). On the other hand, a significantly protective effect was observed with at least one variant allele of ERCC1 Gln504Lys (OR=0.65 and 95% CI= 0.43–0.97) and XRCC1 Arg194Trp (OR=0.37 and 95% CI=0.19–0.74) as compared to homozygote wild type. Among the cases, no significant differences in genotype frequency were observed based on age at diagnosis, HL subtypes, the presence or absence of B symptoms, or family history of cancer.
MDR Analysis
Table 4 summarizes the best interaction models obtained from the MDR analysis for models involving one to four SNPs. The two-SNP interaction model involved XRCC1 Arg399Gln and XRCC3 Thr241Met (CVC=9.0 and TA=0.54 P=0.01), the three-SNP interaction model involved XRCC1 Arg399Gln, OGG1 Ser326Cys and NBN Glu185Gln (CVC=8.0 and TA=0.54, P=0.03) and the four-SNP interaction model involved XRCC1 Arg399Gln, OGG1 Ser326Cys, NBN Glu185Gln and XPC Ala499Val (CVC=8.0 and TA=0.54, P=0.02). All models with >2 SNPs had the same TA, but the two-SNP model had the highest CVC and is therefore our “best” multi-SNP model.
Table 4.
Multifactor dimensionality reduction model for gene-gene interactions
| No. of Factors | Best Candidate Model | TA * | CVC * | P value |
|---|---|---|---|---|
| 1 | XPC Ala499Val | 0.49 | 7.50 | 0.07 |
| 2 | XRCC1Arg399Gln, XRCC3Thr241Me | 0.54 | 9.00 | 0.01 |
| 3 | XRCC1Arg399Gln, OGG1 Ser326Cys and NBN Glu185Gln | 0.54 | 8.00 | 0.03 |
| 4 | XRCC1Arg399Gln, OGG1 Ser326Cys, NBN Glu185Gln and XPC Ala499Val | 0.54 | 8.00 | 0.02 |
TA indicates testing accuracy; CVC, cross validation consistency
CGRS and Hodgkin’s disease risk
Table 5 shows the effect of having more than one risk allele on modulating HL risk. Using the 0–2 risk alleles as the reference, we observed a 32% and 86% increased HL risk associated with having 3, or 4 combined variants in the BER pathway, respectively (P trend= 0.02). Using the genotypes involved in the MDR analysis in the two-SNP model, the OR were 1.14 (95% CI=0.69–1.89) and 3.67 (95% CI=1.70–7.91) in the presence of 3 or 4 combined variants, respectively.
Table 5.
Cumulative genetic risk score and HD risk
| Cases | Controls | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | P value | OR [95%CI] | P value | |||
| 191 | 100 | 215 | 100 | ||||||
| Nucleotide Excision Repair | |||||||||
| 0–2 | 26 | 17 | 44 | 21 | 0.48 | Referent | |||
| 3 | 42 | 28 | 57 | 27 | 1.25 | [0.67–2.34] | 0.49 | ||
| 4+ | 83 | 55 | 108 | 52 | 1.58 | [0.83–3.00] | 0.17 | ||
| Base Excision Repair | |||||||||
| 0–2 | 133 | 70 | 164 | 76 | 0.13 | Referent | |||
| 3 | 48 | 25 | 45 | 21 | 1.32 | [0.53–2.10] | 0.25 | ||
| 4 | 10 | 5 | 6 | 3 | 1.86 | [1.2–5.80] | 0.04 | ||
| Direct reversal of damage | |||||||||
| 0–1 | 151 | 82 | 177 | 82 | |||||
| 2 | 28 | 15 | 32 | 15 | 0.98 | [0.56–1.80] | 0.95 | ||
| 3 | 6 | 3 | 8 | 4 | 0.84 | [0.28–2.50] | 0.76 | ||
| Other identified genes on DNA repair function | |||||||||
| 0–2 | 94 | 53 | 115 | 53 | 0.61 | Referent | |||
| 3 | 57 | 32 | 76 | 35 | 0.92 | [0.60–1.42] | 0.70 | ||
| 4+ | 26 | 15 | 25 | 12 | 1.27 | [0.69–2.35] | 0.44 | ||
| MDR genes | |||||||||
| 0–2 | 82 | 58 | 144 | 69 | 0.02 | Referent | |||
| 3 | 35 | 25 | 54 | 26 | 1.14 | [0.69–1.89] | 0.62 | ||
| 4 | 24 | 17 | 11 | 5 | 3.67 | [1.70–7.91] | 0.01 | ||
| P trend=0.03 | |||||||||
Cases and controls were analyzed with adjustment for age, sex, race, and smoking status.
CART Analysis
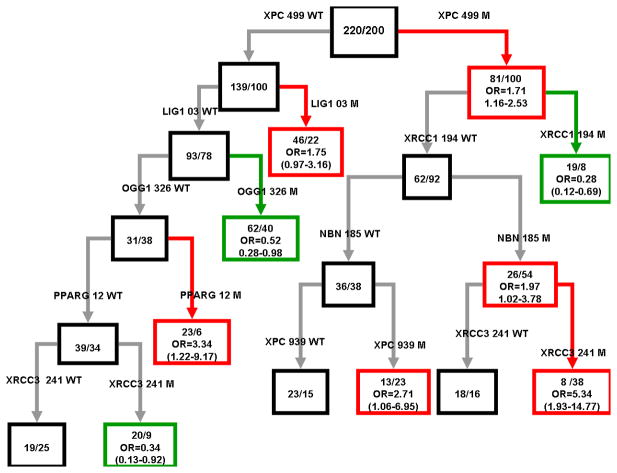
Figure 1 shows the subgroups of high and low-risk subjects and the higher-order interactions among polymorphisms detected by CART analysis such that a sister node serves as the reference group for each split. Consistent with the MDR analysis, one-locus model and the main effects observed by logistic-regression analyses, the initial split on the decision tree was XPC Ala499Val. Individuals with the combined polymorphisms of XPC Ala499Val (M), XRCC1 Arg194Trp (WT) and NBN Glu185Gln (M) exhibited an almost 2-foldincrease risk for HL as compared to controls. Furthermore, individuals who also carried the XRCC3 Thr241Met (M) polymorphism had a 5.3-fold increased risk of HL, suggesting the strong interaction between NBN Glu185Gln (M) and XRCC3 Thr241Me (M). At the same stratum, individuals with NBN Glu185Gln (WT) and XPC Lys939Gln (M) had a 2.7-fold of increase in risk to development HL. Moreover, individuals who carried the XPC Ala499Val (WT) with LIG1 Ala3Val (WT), OGG1 Ser326Cys (WT) and PARG Pro12Ala (WT) and XRCC3 Thr241Me (M) had a 66% risk reduction of HL.
Figure 1.
Nodes of the classification tree are formed by recursive splits of HL case/control status by SNPs. Within each black node, M/N is the number of controls/number of cases. Within each red (high risk) or green (low risk) node, M/N is the number of controls/number of cases and the OR (95% CI) is the odds of HD for a node with respect to its sister black node.
Discussion
In the current study, we confirmed our previous findings (19) and expanded the genes investigated and examined the role of polymorphisms in 15 DNA repair genes involved in the NER, BER, DSB and Direct Reversal repair pathways in modulating the risk of adult HL. Our results suggest that variations in DNA repair genes, particularly in combination, alter HL risk.
Polymorphisms in NER genes have been extensively studied for their associations with different cancers (27–29). In our study, only XPC of the 5 genes involved in the NER pathway was associated with HL risk. XPC encodes part of the XPC-HR23B complex, which is thought to play an early role in NER by initially detecting the DNA damage. In the single-locus analysis for NER pathway, the T allele of the XPC Ala499Val variant was associated with a 77% and 68% increased HL risk, for individuals who carried one and two copies of the variant allele. However, the XPC Lys939Gln variant showed no significant association with HL risk. These findings are in accordance with other studies that suggested that individuals who had 1 or 2 variant alleles of XPC Lys939Gln had higher DNA repair capacity (DRC) and individuals who had 1 or 2 variant alleles of XPC Ala499Val had lower DRC (30, 31). We observed that the interaction between XPC 939Gln(WT) and XPC499Val(M) led to a significant increase in risk of development of HL. Similar studies by Zhu et al. showed associations between the gene-gene interaction in these polymorphisms and risk for bladder cancer (32). Several investigators have observed similar effects associated with the heterozygote genotypes. A plausible explanation for such observation is that the variant allele may have a strong dominant effect that little difference between the effects of the variant homozygotes and heterozygotes state exists. However, due to the small numbers of the homozygotes, the effect is more likely subject to selection bias as compared to the heterozygotes that are present in a much larger number of observations. Simply the statistical power is insufficient to detect a real effect among homozygotes, this could potentially be overcome by larger studies (33,34).
A number of epidemiological studies suggested associations between polymorphisms in DNA repair and oxidative stress response genes with an increased risk for hematological malignancies. In particular, allelic variants in promoter regions of BER genes involved in the immunoglobulin (Ig) class switch recombination (CSR) and somatic hypermutation (SHM) events during the lymphomagenesis process (35). In the single-locus analysis for BER pathway, only XRCC1 gene was associated with HL risk. XRCC1 participates as a scaffolding intermediate and has multiple roles in repairing DNA base damage and single-strand DNA breaks, in the initial step of processing damaged DNA ends. The G allele of XRCC1 Arg399Gln variant was associated with a 62% increased HL risk and the variant T allele of XRCC1 Arg194Trp was associated with a 70% reduction in risk for HL. The scaffold protein XRCC1 plays a central role in BER and single strand break repair, coordinating the binding and activities of enzymes involved in the DNA damage recognition process (36). Recently, it has been suggested that the XRCC1 codon 399Gln allele may lead to diminished DNA repair proficiency compared with the Arg allele (37). XRCC1 Arg399Gln variant has been associated with decreased DNA repair capacity and subsequent accumulation of unrepaired DNA damage and increased cancer risk. Several studies (38, 39) suggested an association between the 399Gln allele and higher DNA adduct levels and higher sister chromatid exchange frequency since it is located within a well conserved region and encodes a non-conservative amino acid change. The XRCC1 Arg399Gln change occurs in the COOH-terminal side of the poly (ADP-ribose) polymerase-interacting domain and within an identified BRCT domain (40). Several studies link the BRCT domain to transcriptional regulation, DNA repair, apoptosis and growth/tumor suppression since the majority of p53 missense mutations in tumors are present within this central region (41, 42). Furthermore, with regard to the biologic significance, an association between the XRCC1 399Gln allele and p53 mutations has been suggested, with loss of the transcriptional activity of p53 and involvement in pathogenesis of HL and defective regulation of Reed-Sternberg cells (43, 44).
The XRCC1 Arg194Trp variant is a non-conservative amino acid substitution, but it is unclear whether this substitution affects the function of the protein. The amino acid substitutions reside in the linker region separating the DNA polymerase β domain from the poly (ADP-ribose) polymerase-interacting domain. This could alter the XRCC1 structure but not the function. Our findings are supported by epidemiologic studies that reported no or inverse associations with DNA repair capacity (4, 16, 45). We further observed that the interaction between XRCC1 399Gln (M) allele and XRCC1 194Trp (WT) allele led to a significant increase in risk of development of HL. Prior studies have shown that amino acid substitutions in the BRCT and the poly (ADP-ribose) polymerase domains could alter the functionality of XRCC1 and in turn lead to decrease repair (46, 47). Furthermore, when XRCC1 399Gln and XRCC1 194Trp variants were considered in combination with OGG1 Ser326Cys and PARP1 Val762Ala in the CGRS analysis by pathway, we observed a consistent increasing trend in the risk of developing HL with increasing number of risk alleles in the BER pathway. OGG1 repairs DNA by removing 8-oxo-dG which is a highly mutagenic oxidative DNA adduct while PARP1 has a role in repair of single-stranded DNA breaks. According to our findings, several reports have suggested that increased number of unfavorable genotypes in BER pathway might result in decreased DNA repair capacity and thus increased cancer risk (47, 48).
When evaluating the association between the candidate SNPs in DSB repair pathway and HL risk, no significant association between allelic variants of XRCC3 (encoding a protein that acts in the double strand break/homologous recombination repair pathway), NBN (encodes the nibrin protein that participates in double strand breaks repair) and LIG A (encoding a protein that plays a role in DNA replication and repair) and HL susceptibility. Nevertheless, we observed that interactions between NBN Glu185Gln and XRCC3 Thr241Me with XPC Ala499Val and XRCC1 Arg194Trp and XRCC1 399Gln polymorphisms led to a significant increase in HL risk. These findings are similar to reported studies suggesting that several polymorphisms in DNA repair genes may act together to contribute to cancer susceptibility (49–51). Perhaps the most significant finding in this study was the consistent association of XPC Ala499Val (M), NBN Glu185Gln (M), XRCC3 Thr241Me (M), XRCC1 Arg194Trp (WT) or/and XRCC1 399Gln (M), which were identified using different analytic approaches where together, MDR and CART complement each other in that they provide the user with a multi-faceted view of gene-gene interactions as well as stratified effects that can be further explored.
Overall, our findings support an association between SNPs that play key roles in the oxidative damage repair by BER and DSB pathways. The interactions between the pathways are supported by biologically plausible mechanisms where the NBN and XRCC3 proteins are involved in the homologous recombination repair (HRR) pathway, responsible for DNA double strand break repair (52). Inherited mutations in HRR genes have been recognized in familial cancer syndromes that involve an elevated lymphoma risk (53). The NBN protein is a member of theMRE11/RAD50/NBN (MRN complex) which participates in DSB detection and signaling. Several studies reported the association between variants of NBN gene with risk of developing solid tumors such as breast and prostate cancer (54, 55). It has been reported that mutations in NBN gene (including the Glu185Gln) cause genetic disorders (56, 57), some of which are characterized by increased risk of lymphoma. In addition, NBN Glu185Gln was reported to increase cancer risk in combination with variants of XRCC3 gene which is required for efficient repair of double strand breaks via HR, for correct chromosomal segregation as well as the repair of DNA cross links (58, 59). More recently, the association between mutations in XRCC3 and lymphomas risk was supported by Sale et al (60), which reported that XRCC3 could be involved in the SHM process and implicated in the recombination-dependent repair of the lesions in the Ig V gene.
In summary, our results suggest that allelic variants in BER and DSB genes may play an important role in the development of HL. A limitation of our study is the sample size that provided limited power for gene-gene interaction assessments; therefore, we cannot exclude the possibility that some of these findings may be due to chance, and thus should be interpreted with caution. Larger studies are needed to validate our findings and investigate the role of additional genes in the HL risk.
Acknowledgments
This study has been partially supported by the National Cancer Institute grants CA98549, CA129050, and NIEHS ES07784.
References
- 1.American Cancer Society. Cancer Facts and Figures 2008, American Cancer Society. Atlanta, GA: American Cancer Society; 2008. [Google Scholar]
- 2.Re D, Zander T, Diehl V, Wolf J. Genetic instability in Hodgkin’s lymphoma. Ann Oncol. 2002;13 (Suppl 1):19–22. doi: 10.1093/annonc/13.s1.19. [DOI] [PubMed] [Google Scholar]
- 3.Thoms KM, Baesecke J, Emmert B, Hermann J, Roedling T, Laspe P, Leibeling D, Truemper L, Emmert S. Functional DNA repair system analysis in haematopoietic progenitor cells using host cell reactivation. Scand J Clin Lab Invest. 2007;67(6):580–8. doi: 10.1080/00365510701230481. [DOI] [PubMed] [Google Scholar]
- 4.Holt SM, Scemama JL, Panayiotidis MI, Georgakilas AG. Compromised repair of clustered DNA damage in the human acute lymphoblastic leukemia MSH2-deficient NALM-6 cells. Mutat Res. 2009;674(1–2):123–30. doi: 10.1016/j.mrgentox.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Foroutan B, Ali Ruf A, Jerwood D, Anderson D. In vitro studies of DNA damage and its repair in cells from NHL patients with different p53 mutant protein status, resistant (p53(+)) and sensitive (p53(−)) to cancer chemotherapy. J Pharmacol Toxicol Methods. 2007;55(1):58–64. doi: 10.1016/j.vascn.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Ribas G, Gonzalez-Neira A, Salas A, et al. Evaluating Hap-Map SNP data transferability in a large-scale genotyping project involving 175 cancer-associated genes. Hum Genet. 2006;118:669–679. doi: 10.1007/s00439-005-0094-9. [DOI] [PubMed] [Google Scholar]
- 7.Hall J, Marcel V, Bolin C, et al. The associations of sequence variants in DNA-repair and cell-cycle genes with cancer risk: genotype-phenotype correlations. Biochem Soc Trans. 2009;37(Pt 3):527–33. doi: 10.1042/BST0370527. [DOI] [PubMed] [Google Scholar]
- 8.Seedhouse C, Faulkner R, Ashraf N, et al. Polymorphisms in genes involved in homologous recombination repair interact to increase the risk of developing acute myeloid leukemia. Clin Cancer Res. 2004;10(8):2675–80. doi: 10.1158/1078-0432.ccr-03-0372. [DOI] [PubMed] [Google Scholar]
- 9.Scrideli CA, Tone LG. Polymorphisms of DNA repair genes and susceptibility to acute childhood lymphoblastic leukemia. Leuk Res. 2007;31(6):759–63. doi: 10.1016/j.leukres.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Ganster C, Neesen J, Zehetmayer S, et al. DNA repair polymorphisms associated with cytogenetic subgroups in B-cell chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2009;48(9):760–7. doi: 10.1002/gcc.20680. [DOI] [PubMed] [Google Scholar]
- 11.Worrillow L, Roman E, Adamson PJ, et al. Polymorphisms in the nucleotide excision repair gene ERCC2/XPD and risk of non-Hodgkin lymphoma. Cancer Epidemiol. 2009;33(3–4):257–60. doi: 10.1016/j.canep.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Wang SS, Maurer MJ, Morton LM, et al. Polymorphisms in DNA repair and one-carbon metabolism genes and overall survival in diffuse large B-cell lymphoma and follicular lymphoma. Leukemia. 2009;23(3):596–602. doi: 10.1038/leu.2008.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monzo M, Brunet S, Urbano-Ispizua A, et al. Genomic polymorphisms provide prognostic information in intermediate-risk acute myeloblastic leukemia. Blood. 2006;107(12):4871–9. doi: 10.1182/blood-2005-08-3272. [DOI] [PubMed] [Google Scholar]
- 14.Batar B, Güven M, Bariş S, et al. DNA repair gene XPD and XRCC1 polymorphisms and the risk of childhood acute lymphoblastic leukemia. Leuk Res. 2009;33(6):759–63. doi: 10.1016/j.leukres.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Worrillow L, Roman E, Adamson PJ, et al. Polymorphisms in the nucleotide excision repair gene ERCC2/XPD and risk of non-Hodgkin lymphoma. Cancer Epidemiol. 2009;33(3–4):257–60. doi: 10.1016/j.canep.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Joseph T, Kusumakumary P, Chacko P, et al. DNA repair gene XRCC1 polymorphisms in childhood acute lymphoblastic leukemia. Cancer Lett. 2005;217(1):17–24. doi: 10.1016/j.canlet.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 17.Bhatla D, Gerbing RB, Alonzo TA, et al. DNA repair polymorphisms and outcome of chemotherapy for acute myelogenous leukemia: a report from the Children’s Oncology Group. Leukemia. 2008;22(2):265–72. doi: 10.1038/sj.leu.2405000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossi D, Gaidano G, Gloghini A, et al. Frequent aberrant promoter hypermethylation of O6-methylguanine-DNA methyltransferase and death-associated protein kinase genes in immunodeficiency-related lymphomas. Br J Haematol. 2003;123(3):475–8. doi: 10.1046/j.1365-2141.2003.04644.x. [DOI] [PubMed] [Google Scholar]
- 19.El-Zein RA, Monroy CM, Etzel CJ, et al. Genetic Polymorphisms in DNA Repair Genes as Modulators of Hodgkin Disease Risk. Cancer. 2009;115(8):1651–9. doi: 10.1002/cncr.24205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen P, Bailey NM, Hagemeister FB, Liang JC. Induction of chromosome breaks and sister chromatid exchanges in patients with Hodgkin’s disease by two combination chemotherapy regimens of different leukemogenic potential. Cancer Res. 1990;50(3):558–62. [PubMed] [Google Scholar]
- 21.Liang JC, Bailey NM, Gabriel GJ, Kattan MW, Wang RY, Hagemeister FB, Cabanillas FF, Fuller LM. A new chemotherapy regimen for treatment of Hodgkin’s disease associated with minimal genotoxicity. Leuk Lymphoma. 1993;9(6):503–8. doi: 10.3109/10428199309145757. [DOI] [PubMed] [Google Scholar]
- 22.Strom SS, Hess KR, Sigurdson AJ, Spitz MR, Liang JC. Evaluation of sister chromatid exchange and chromosome breaks in a cohort of untreated Hodgkin’s disease patients. Cancer Epidemiol Biomarkers Prev. 1997 Apr;6(4):291–3. [PubMed] [Google Scholar]
- 23.Hartge P, Brinton LA, Rosenthal JF, Cahill JI, Hoover RN, Waksberg J. Random digit dialing in selecting a population-based control group. Am J Epidemiol. 1984 Dec;120(6):825–33. doi: 10.1093/oxfordjournals.aje.a113955. [DOI] [PubMed] [Google Scholar]; Hartge, et al. Am J Epidemiol. 1984;120:825–833. doi: 10.1093/oxfordjournals.aje.a113955. [DOI] [PubMed] [Google Scholar]
- 24.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1984;AC-19:716–23. [Google Scholar]
- 25.Ritchie MD, Hahn LW, Roodi N, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–47. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breiman L, Friedman JN, Olsen RA. Classification and Regression Trees. Monterey: Wadsworth and Brooks/Cole; 1984. [Google Scholar]
- 27.Vogel U, Overvad K, Wallin H, et al. Combinations of polymorphisms in XPD, XPC and XPA in relation to risk of lung cancer. Cancer Lett. 2005;222:67–74. doi: 10.1016/j.canlet.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 28.Sanyal S, Festa F, Sakano S, et al. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25:729–734. doi: 10.1093/carcin/bgh058. [DOI] [PubMed] [Google Scholar]
- 29.Shen H, Sturgis EM, Khan SG, et al. An intronic Poly (AT) polymorphism of the DNA repair gene XPC and risk of squamous cell carcinoma of the head and neck: a case-control study. Cancer Res. 2001;61:3321–3325. [PubMed] [Google Scholar]
- 30.Zhu Y, Yang H, Chen Q, et al. Modulation of DNA damage/DNA repair capacity by XPC polymorphisms. DNA Repair (Amst) 2008;7(2):141–8. doi: 10.1016/j.dnarep.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vodicka P, Kumar R, Stetina R, et al. Genetic polymorphisms in DNA repair genes and possible links with DNA repair rates, chromosomal aberrations and single-strand breaks in DNA. Carcinogenesis. 2004;25(5):757–63. doi: 10.1093/carcin/bgh064. [DOI] [PubMed] [Google Scholar]
- 32.Zhu Y, Lai M, Yang H, et al. Genotypes, haplotypes and diplotypes of XPC and risk of bladder cancer. Carcinogenesis. 2007;28 (3):698–703. doi: 10.1093/carcin/bgl201. [DOI] [PubMed] [Google Scholar]
- 33.Chen D, Jin G, Wang Y, Wang H, Liu H, Liu Y, Fan W, Ma H, Miao R, Hu Z, Sun W, Qian J, Jin L, Wei Q, Shen H, Huang W, Lu D. Genetic variants in peroxisome proliferator-activated receptor-gamma gene are associated with risk of lung cancer in a Chinese population. Carcinogenesis. 2008;29(2):342–50. doi: 10.1093/carcin/bgm285. [DOI] [PubMed] [Google Scholar]
- 34.Ma H, Xu L, Yuan J, Shao M, Hu Z, Wang F, Wang Y, et al. Tagging single nucleotide polymorphisms in excision repair cross-complementing group 1 (ERCC1) and risk of primary lung cancer in a Chinese population. Pharmacogenet Genomics. 2007;17:417–423. doi: 10.1097/01.fpc.0000239975.77088.17. [DOI] [PubMed] [Google Scholar]
- 35.Schanz S, Castor D, Fischer F, et al. Interference of mismatch and base excision repair during the processing of adjacent U/G mispairs may play a key role in somatic hypermutation. Proc Natl Acad Sci U S A. 2009;106(14):5593–8. doi: 10.1073/pnas.0901726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caldecott KW, Aoufouchi S, Johnson P, et al. XRCC1 polypeptide interacts with DNA polymerase beta and possiblypoly (ADP-ribose) polymerase, and DNA ligase III is anovel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdel-Rahman SZ, El-Zein RA. The 399Gln polymorphism in the DNA repair gene XRCC1 modulates the genotoxic response induced in human lymphocytes by the tobacco-specific nitrosamine NNK. Cancer Lett. 2000;159:63–71. doi: 10.1016/s0304-3835(00)00532-2. [DOI] [PubMed] [Google Scholar]
- 38.Duell EJ, Holly EA, Bracci PM, et al. A population-based study of the Arg399Gln polymorphism in X-ray repair cross-complementing group 1 (XRCC1) and risk of pancreatic adenocarcinoma. Cancer Res. 2002;62(16):4630–6. [PubMed] [Google Scholar]
- 39.Matullo G, Palli D, Peluso M, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and 32P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 40.Dulic A, Bates PA, Zhang X, et al. BRCT domain interactions in the heterodimeric DNA repair protein XRCC1- DNA ligase III. Biochemistry. 2001;40:5906–5913. doi: 10.1021/bi002701e. [DOI] [PubMed] [Google Scholar]
- 41.Taylor RM, Thistlethwaite A, Caldecott KW. Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Mol Cell Biol. 2002;22:2556–2563. doi: 10.1128/MCB.22.8.2556-2563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feuerborn A, Moritz C, Von Bonin F, et al. Dysfunctional p53 deletion mutants in cell lines derived from Hodgkin’s lymphoma. Leuk Lymphoma. 2006;47:1932–1940. doi: 10.1080/10428190600667721. [DOI] [PubMed] [Google Scholar]
- 44.Kim LH, Peh SC, Poppema S. Expression of retinoblastoma protein and P16 proteins in classic Hodgkin lymphoma: relationship with expression of p53 and presence of Epstein-Barr virus in the regulation of cell growth and death. Hum Pathol. 2006;37:92–100. doi: 10.1016/j.humpath.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 45.Pakakasama S, Sirirat T, Kanchanachumpol S, et al. Genetic polymorphisms and haplotypes of DNA repair genes in childhood acute lymphoblastic leukemia. Pediatric Blood & Cancer. 2007;48:16–20. doi: 10.1002/pbc.20742. [DOI] [PubMed] [Google Scholar]
- 46.Vodicka P, Stetina R, Polakova V, et al. Association of DNA repair polymorphisms with DNA repair functional outcomes in healthy human subjects. Carcinogenesis. 2007;28(3):657–64. doi: 10.1093/carcin/bgl187. [DOI] [PubMed] [Google Scholar]
- 47.Huang M, Dinney CP, Lin X, et al. High-order interactions among genetic variants in DNA base excision repair pathway genes and smoking in bladder cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2007;16(1):84–91. doi: 10.1158/1055-9965.EPI-06-0712. [DOI] [PubMed] [Google Scholar]
- 48.Andrew AS, Karagas MR, Nelson HH, et al. DNA repair polymorphisms modify bladder cancer risk: a multi-factor analytic strategy. Hum Hered. 2008;65(2):105–18. doi: 10.1159/000108942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Ruyck K, Szaumkessel M, De Rudder I, et al. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res. 2007;631:101–110. doi: 10.1016/j.mrgentox.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, Scheurer ME, El-Zein R, et al. Association and interactions between DNA repair gene polymorphisms and adult glioma. Cancer Epidemiol Biomarkers Prev. 2009;18(1):204–14. doi: 10.1158/1055-9965.EPI-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Popanda O, Schattenberg T, Phong CT, et al. Specific combinations of DNA repair gene variants and increased risk for non-small cell lung cancer. Carcinogenesis. 2004;25(12):2433–41. doi: 10.1093/carcin/bgh264. [DOI] [PubMed] [Google Scholar]
- 52.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9(6):683–90. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 53.Puebla-Osorio N, Zhu C. DNA damage and repair during lymphoid development: antigen receptor diversity, genomic integrity and lymphomagenesis. Immunol Res. 2008;41(2):103–22. doi: 10.1007/s12026-008-8015-3. [DOI] [PubMed] [Google Scholar]
- 54.Bartkova J, Tommiska J, Oplustilova L, et al. Aberrations of the MRE11-RAD50-NBS1 DNA damage sensor complex in human breast cancer: MRE11 as a candidate familial cancer-predisposing gene. Mol Oncol. 2008;2(4):296–316. doi: 10.1016/j.molonc.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cybulski C, Górski B, Debniak T, et al. NBS1 is a prostate cancer susceptibility gene. Cancer Res. 2004;64(4):1215–9. doi: 10.1158/0008-5472.can-03-2502. [DOI] [PubMed] [Google Scholar]
- 56.Lu M, Lu J, Yang X, et al. Association between the NBS1 E185Q polymorphism and cancer risk: a meta-analysis. BMC Cancer. 2009;9:124. doi: 10.1186/1471-2407-9-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gładkowska-Dura M, Dzierzanowska-Fangrat K, Dura WT, et al. Unique morphological spectrum of lymphomas in Nijmegen breakage syndrome (NBS) patients with high frequency of consecutive lymphoma formation. J Pathol. 2008;216(3):337–44. doi: 10.1002/path.2418. [DOI] [PubMed] [Google Scholar]
- 58.Vineis P, Manuguerra M, Kavvoura FK, et al. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J Natl Cancer Inst. 2009;101(1):24–36. doi: 10.1093/jnci/djn437. [DOI] [PubMed] [Google Scholar]
- 59.Compton SA, Choi JH, Cesare AJ, et al. Xrcc3 and Nbs1 are required for the production of extrachromosomal telomeric circles in human alternative lengthening of telomere cells. Cancer Res. 2007;67(4):1513–9. doi: 10.1158/0008-5472.CAN-06-3672. [DOI] [PubMed] [Google Scholar]
- 60.Sale JE, Calandrini DM, Takata M, et al. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412(6850):921–6. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]