Abstract
T helper17 (Th17) cells are known to play a critical role in adaptive immune responses to several important extracellular pathogens. Additionally, Th17 cells are implicated in the pathogenesis of several autoimmune and inflammatory disorders as well as in cancer. Therefore, it is essential to understand the mechanisms that regulate Th17 differentiation. Notch signaling is known to be important at several stages of T cell development and differentiation. Here we report that Notch1 is activated in both mouse and human in-vitro polarized Th17 cells and blockade of Notch signaling significantly down-regulates the production of Th17 associated cytokines suggesting an intrinsic requirement for Notch during Th17 differentiation in both species. We also present evidence, using promoter reporter assays, knockdown studies as well as chromatin immunoprecipitation, that IL-17 and RORγt are direct transcriptional targets of Notch signaling in Th17 cells. Finally, in-vivo inhibition of Notch signaling reduced IL-17 production and Th17 mediated disease progression in experimental autoimmune encephalomyelitis, a mouse model of multiple sclerosis. Thus, this study highlights the importance of Notch signaling, in Th17 differentiation and indicates that selective targeted therapy against Notch may be an important tool to treat autoimmune disorders, including multiple sclerosis.
Introduction
The regulation of Th17 differentiation from naïve CD4+ T cells is an area of active investigation. Th17 cells and the pro-inflammatory cytokines (IL-17A, IL-17F, IL-21 and IL-22) produced by these cells, have been implicated in several autoimmune and inflammatory disorders (1, 2). The importance of this subset in autoimmune diseases was first recognized when mice lacking expression of the p19 subunit of IL-23, a cytokine involved in differentiation and expansion of Th17 cells, failed to develop certain autoimmune disorders (3, 4). The pathogenic role of IL-17 as well as Th17 cells has now been documented in numerous autoimmune diseases including multiple sclerosis (5), rheumatoid arthritis (6), psoriasis (7), Crohn’s disease (8) and systemic lupus erythematosus (9).
Several factors are known to affect Th17 differentiation including antigenic stimuli (10, 11), expression of particular transcription factors (12) and epigenetic changes in the IL-17 gene locus (13). The cytokine milieu leading to Th17 differentiation is the most carefully studied factor. In mice, it is known that IL-6 along with proinflammatory cytokines TGF-β and IL-21, promotes differentiation of naïve CD4+ T cells into the Th17 lineage (14). Manel et al (2008) have similarly shown that human Th17 differentiation requires exposure to low doses of TGFβ in concert with IL-1β, IL-6, IL-21 and/or IL-23 (15). In addition to the cytokine environment, transcription factors are important determinants of Th17 differentiation (14, 16, 17). The transcription factor retinoic acid receptor related orphan receptor γt (RORγt), in co-operation with RORα controls Th17 differentiation (18). Th17 differentiation also is regulated by histone-3 acetylation and H3Lys-4 methylation in both the IL-17A and the IL-17F promoters in a lineage dependent manner (13). Despite great progress in understanding the molecular mechanism of Th17 differentiation, the contribution of cell surface proteins found on CD4+ T cells is not well understood.
Notch proteins, are type 1 transmembrane proteins known to play a crucial role in cell fate determination in many cell lineages including early T cell development in the thymus (19). Four Notch receptors (Notch1, 2, 3 and 4) are found in mammals. In developing T cells, Notch1 has been reported to regulate αβ versus γδ T cell differentiation (20), T versus B cell fate determination (21) and CD4+ versus CD8+ T lineage decision (22). Notch1 is also present on naïve (23) and activated CD4+ T cells (24). Additionally, we and others have shown that Notch1 signaling is activated upon crosslinking of the T cell receptor (TCR) (24, 25).
Canonical Notch signaling is induced when one of the four mammalian Notch receptors (Notch1, 2, 3 or 4) encounter one of the five known ligands (Jagged 1, 2 or 3 or Delta like-1 or 2) on a neighboring cell. This interaction initiates a proteolytic cleavage of the transmembrane Notch peptide near the extracellular surface by an ADAM protease, which, in turn, induces a conformational change that allows access and cleavage of the Notch transmembrane domain by the γ-secretase complex. Cleavage of Notch receptors by γ-secretase results in the release of an intracellular Notch fragment (NIC), which rapidly translocates to the nucleus where it interacts with the DNA binding protein known as CSL (CBF-1, Suppressor of Hairless, Lag-1). In the absence of Notch signaling, CSL is bound to DNA in a complex with several repressor proteins. NIC translocation to the nucleus and binding to CSL results in disruption of the repressor complex followed by recruitment of several co-activator proteins resulting in the initiation of transcription of genes located downstream of Notch/CSL complexes (reviewed in (26, 27).
Notch is reported to play a critical role in Th1 (28, 29) and/or Th2 (30, 31) mediated immune responses. Data from several laboratories suggest that antigen presenting cells (APCs) expressing Delta like-4 (DLL-4) drive the differentiation of Th1 cells (32, 33) while APCs expressing Jagged1 promote differentiation of Th2 cells (23, 34)
In this study, we examined the role of Notch signaling in Th17 polarization. We used pharmacologic inhibitors as well as knockdown approaches to establish a role for Notch signaling in Th17 polarization. Promoter analysis and chromatin immunoprecipitation assays demonstrated regulation of both the IL-17 and RORγt promoters by Notch1. Lastly, we present in-vivo data demonstrating that inhibition of Notch signaling ameliorates the severity of EAE, a murine autoimmune disease that displays several characteristics of human multiple sclerosis. These data provide further understanding of the Th17 differentiation pathway and suggest opportunities for exploiting the Notch signaling pathway to treat Th17 mediated autoimmune disorders
Materials and Methods
Drugs and chemicals
γ-secretase inhibitors Compound E (Alexis Biochemicals, San Diego, CA), and ILCHO (25) were resuspended in DMSO and used in concentrations as indicated in figure legends.
Antibodies
For detection of human Notch 1, anti-Notch1 (C20) (Santa Cruz) and anti- activated Notch 1 (Rockland, Gilbertsville, PA) antibodies were used. Anti-Notch2 (Abcam, Cambridge MA), anti-Notch3 (M134) (Santa Cruz, CA) and anti-Notch4 (H225) (Santa Cruz, CA) were also used. For detection of mouse activated Notch1, anti-Notch 1 (ebioscience, mN1A clone) was used. β-actin antibody (Sigma Aldrich, St Louis) was used as a loading control.
Cell culture and mouse in-vitro polarization
For mouse in-vitro polarization assays, naïve CD4+ T cells were isolated from C57BL/6 (Jackson Laboratories) using the IMag magnetic system (BD Pharmingen, San Jose, CA), according to the manufacturer’s protocol. 2.5 ×106 cells/ml were pretreated in-vitro at 37°C for 30 min with 0.1% DMSO or with GSI (25 μM ILCHO or 4 μM Compound E) and then were plated onto 12 or 6 well plates precoated with 1μg/ml each of αCD3 and αCD28. To polarize CD4+ T cells to a Th17 phenotype 20 ng of IL-6 (R&D Systems), 5 ng of TGF-β (R&D Systems) and 10 μg of both anti IFNγ (BD Pharmingen) and anti IL-4 (BD Pharmingen) were used per ml of cells (35, 36). Cells were polarized for 24, 48 or 72 h. The activation supernatants were evaluated for IL-17A (BD Biosciences), IL-17F (R&D) and IL-21 (R&D) by ELISA. To study the effect of Notch inhibition on fully differentiated Th17 cells, naïve CD4+ T cells were differentiated towards the Th17 lineage for 4 days. These cells were then treated with either DMSO or ILCHO followed by culturing in αCD3-coated plates. After 24 hours, supernatants were collected and analyzed for IL-17 cytokine by ELISA (BD Biosciences).
Cell culture and human in-vitro polarization
Human in-vitro Th17 polarization was performed using a modified protocol from Manel et al (15). Naïve CD4+ T cells were purified from peripheral blood mononuclear cells by negative selection using MACS separation according to the manufacturer’s instructions (Miltenyi Biotech, Sunnyvale, CA) and were cultured in a 37°C at 5% CO2 in serum free X-VIVO 10 media (BioWhittaker, Maryland). Naïve cells (2 × 106 per ml) were plated in 24 well plates with beads coated with αCD3 and αCD28 (Dynabeads, Invitrogen, Norway) at a concentration of 1 bead per cell. Antibodies and cytokines were added at the time of plating at the following concentrations: 10 U/ml IL-2, 5 ng/ml TGF-β1, 10 ng/ml IL-6, 10 ng/ml IL-23, 10 ng/ml IL-21, 10 ng/ml IL-1β, anti IL-4 (10μg/ml) and anti IFNγ (10μg/ml). All antibodies and recombinant cytokines used in polarization were purchased from R&D systems, MN. IL-17A, IL-17F, IL-21 and IL-22 protein levels in the activation supernatants were quantified by ELISA (eBioscience, San Diego, CA). To evaluate the effect of Notch inhibition on differentiated Th17 cells, naïve CD4+ T cells were cultured in Th17 polarizing conditions for 4 days followed by treatment with either DMSO or ILCHO. Supernatants were collected after 24 hours and analyzed for IL-17 and IL-22 cytokines (eBioscience, San Diego, CA)
Cell lines and constructs
HEK 293T cells were obtained from the American Type Culture Collection (Manassas, VA) and cultured in DMEM medium (Mediatech, Inc, Manassas, VA) supplemented with 10% FBS (Cellgro, Mediatech, Manassa, VA), 2 mM glutamine, and 1 mM pyruvate. (Supplements are from Lonza, Walkersville, MD). The Notch1IC encoding plasmid construct was generated by cloning Notch 1IC cDNA into BamH1 and EcoRI sites of pcDNA3.0 (37).
Retroviral expression vector and transduction
The sequence encoding N1IC was subcloned into the retroviral vector LZRS and viral particles were produced as described previously (38). For transduction of virus, naïve human CD4+ T cells were isolated and stimulated with αCD3/αCD28-coated beads for 24h and transduced with retroviral supernatant in the presence of 8μg polybrene as described before (38).Transduced cells were then differentiated to Th0 or Th17 conditions. The cells were transduced again the following day with retroviral supernatants and cultured for an additional 48h.
Dual Luciferase assay
HEK 293 T cells were plated on 60 mm dishes and co-transfected with Notch1IC (1μg) expression vector constructs cloned into pcDNA 3.0 along with a human IL-17 (-1125bp) promoter luciferase construct (1μg) kindly provided by Dr Sarah Gaffen (University of Pittsburg) (39) and 0.1 μg Renilla luciferase construct as the internal transfection control. Luciferase assays (Dual Luciferase assay system, Promega, Madison, WI) were performed according to the manufacturer’s instructions.
Intracellular staining and cell surface staining
Mouse CD4+ T cells were polarized towards a Th17 phenotype as described above. After 72 hours, the cells were stimulated by adding 80 nM PMA (phorbol myristate acetate) and 2.5 μM ionomycin for 1h in addition to Brefeldin A for 5 hours. Cells were fixed and permeabilized using the BD Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s instructions. Fluorescent antibodies (anti mouse CD4-FITC, anti mouse IL-17A-APC and anti mouse IFNγ-PE) were obtained from BD Biosciences. Anti mouse Notch 1-PE was obtained from e-Bioscience. Cells were analyzed on a FACS LSR II (Becton Dickinson).
Real time PCR
Human naïve CD4+ T cells were polarized to Th17 as described above. RNA was extracted using the RNeasy mini kit (Qiagen, Valencia, CA). The RNA was then DNAase I-treated (Qiagen, Valencia, CA) and cDNA was synthesized using Superscript III (Invitrogen, Carlsbad, CA). Quantitative real time PCR was then performed using 18S rRNA to normalize following the 2-ΔΔCT method (40). The primer sequences used were: Notch1 (Forward): GTC AAC GCC GTA GAT GAC C, (Reverse): TTG TTA GCC CCG TTC TTC AG; RORγt (15) (Forward): TTT TCC GAG GAT GAG ATT GC, (Reverse): CTT TCC ACA TGC TGG CTA CA; 18SrRNA (Forward): GGC GCC CCC TCG ATG CTC TTA G, (Reverse): GCT CGG GCC TGC TTT GAA CAC TCT.
Mouse CD4+ T cells cultured as above were harvested and total RNA was isolated using the RNAqueous kit (Ambion, Austin, TX). Total RNA samples were subjected to treatment with DNase using the TURBO DNA-free kit (Ambion), cDNA was synthesized, and transcripts were amplified by quantitative real time polymerase (Q-PCR Stratagene MX3000P). Primer sequences for IL-17 were: (Forward): 5’-CTC CAG AAG GCC CTC AGA CTA C-3’, (Reverse): 5’-AGC TTT CCC TCC GCA TTG ACA CAG-3’ (41). RORγt (Forward): 5′-TTT GGA ACT GGC TTT CCA TC-3′, (Reverse): 5′-AAG ATC TGC AGC TTT TCC ACA-3′. The expression of each gene was normalized to the expression of β-actin by the 2-ΔΔCT method (40).
RNA- interference
To knock down the expression of Notch 1, CD4+ T cells were purified and nucleoporated with siRNA specific for Notch1 or scrambled siRNA (Santa Cruz Biotechnology, Santa Cruz, CA), using an Amaxa Nucleoporator system. Briefly, 5–10 × 106 CD4+ T cells were resuspended in 100 μl of nucleofector solution and transfected with 100 nM siRNA using the U-014 Amaxa nucleofector program (Lonza, Switzerland). After transfection, the cells were incubated for 6h at 37°C, and stimulated with αCD3/CD28 coated magnetic beads under Th17 polarizing conditions for 48 h.
MTS assay
Cytotoxicity assay was performed using CellTiter 96 AQueous one solution reagent (Promega) as per the manufacturer’s instructions.
Chromatin immunoprecipitation assay (ChIP)
ChIP assays (Upstate Cell Signaling Solutions) were performed using 1×107 naïve CD4+ T cells stimulated with αCD3/CD28-coated magnetic beads (1 bead/cell) under Th0 (no cytokines) or Th17 conditions and pretreated with DMSO or GSI (ILCH0) for 24 h. The following primers were used for quantitative as well as standard PCR. IL-17 primer sets were: 17 CSL1 (Forward): 5’-TTG ACC CAT AGC ATA GCA GC-3’, (Reverse):5’-TTC AGG GGT GAC ACC ATT TT-3’; 17CSL2 (Forward): 5’-GAA AAT CTC GTG TCT CTT GAA CC-3’, (Reverse): 5’-TTC CTC ACA GAT TCC TTG GC-3 ’; 17CSL3 (Forward): 5’-TTC CAC TTT CCA CTT CCC AC-3’, (Reverse): 5’-TTC CTC CCT GTC CTG CTC TA-3’; 17CSL4 (Forward): 5’-CAA TTG GGA AAA GCA AGC AT-3’, (Reverse): 5’-CCC TAC TGC CCC TCC TCT AC-3’. RORγt primer sets were: RCBF1 (Forward): 5’-ATC TCC AGC CTC AGC TTT GA-3’, (Reverse): 5’-GAT GCC CCT GTT TTC TTG AG-3’; RCBF2 (Forward): 5’-AGA GGG ACT CCT TGC CTC TC-3’, (Reverse): 5’-TCA AAG CTG AGG CTG GAG AT-3’. Antibodies used were rabbit anti Notch1 or normal rabbit IgG (Santa Cruz Biotechnology). Conditions for real-time PCR were 50°C 2min, 95°C 10min, 95°C 15sec, 60°C 1min (40 cycles); conditions for semiquantitative PCR were 95°C 5 min, 95°C 30s, 55°C 1min, 72°C 30s (35 cycles), 72°C 2min.
In-vivo GSI treatment
For the EAE experiments, 8–12 weeks old female SJL/J mice were purchased from Charles River Laboratory (Wilmington, MA). All mice were housed in the animal care facility at the University of Massachusetts, Amherst in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines. The GSI administered in-vivo was LY-411,575 (LY) formulated for two doses 5mg/kg and 2.5 mg/kg. Mice were fed 5mg/kg LY chow for four weeks. . They were then fed 2.5mg/kg LY chow for a week,immunized at that point, and after a total of two weeks on the 2.5mg/kg LYchow, they were returned to the 5mg/kg LY chow until the end of the experiment.
EAE Evaluation
EAE was induced by immunizing mice in the flank with 50μg PLP (139–151) (Invitrogen, Carlsbad, CA) supplemented with 400μg Mycobacterium tuberculosis H37RA (Difco, Detroit, MI). Pertussis toxin (Ptx; Sigma 200ng) was injected interaperitonially on the day of immunization. The progression and severity of disease was monitored and scored from 0–5 as follows: Score 0-no disease; 1-limp tail, 2-hind limb weakness; 3-hind limb paralysis; 4-hind and fore limb paralysis; 5-morbidity and death. Data is reported as the mean daily clinical score (42–44).
Mice were anesthetized and perfused through the left cardiac ventricle with PBS during the peak of disease (day 15 post immunization). Spinal cords and spleens were removed by dissection. Splenocytes were cultured at 37°C with medium alone or with different concentrations of PLP(139–151) antigen for 5 days. To prepare a single cell suspension, spinal cords were cut into pieces and the tissues were mashed and passed through a 70 μm mesh. Mononuclear cells were isolated over a Percoll gradient and were then cultured with PLP (139–151) antigen for 5 days. Cells from the spinal cord were stained for CD4+ and CD8+ T cells. IL-17 and IFNγ ELISAs were then performed on supernatants from all re-stimulated cells. Splenocytes were intracellularly stained for IL-17.
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 4.0. Unpaired t-test (α =0.05) were used when comparing two conditions.
Results
Gamma secretase inhibition (GSI) during murine Th17 polarization results in reduced Th17 associated cytokines production
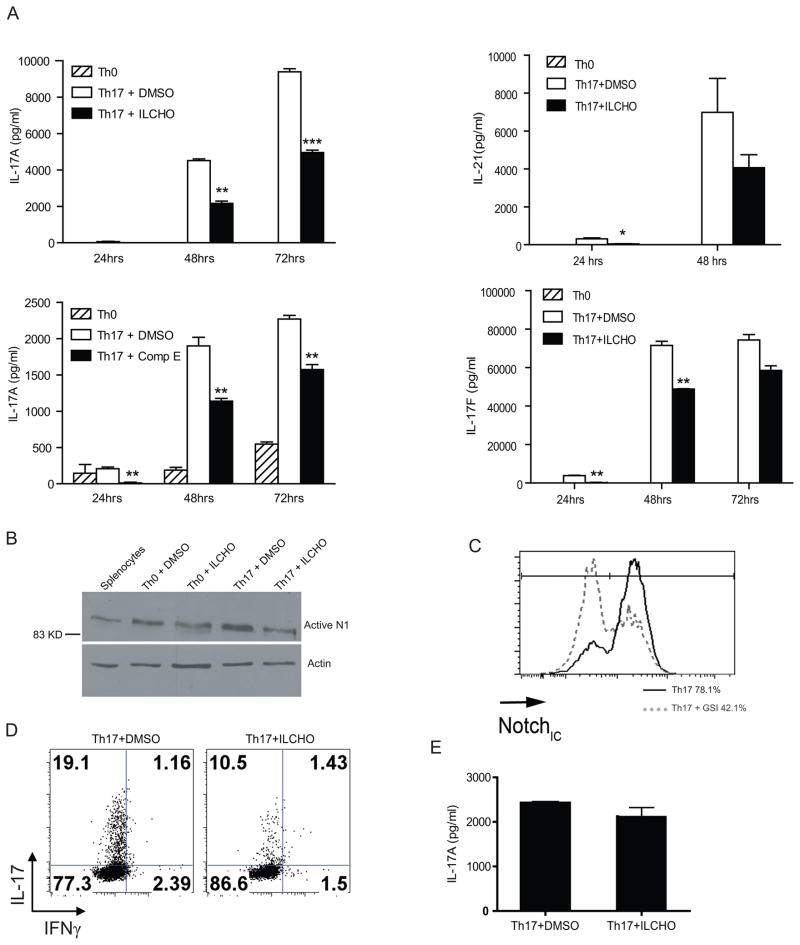
The effect of GSI on murine production of IL-17A, IL-17F and IL-21 was tested by treating Th17 polarized cells with ILCHO and Compound E, two chemically distinct GSIs that block γ-secretase by different mechanisms. ILCHO is a competitive peptide aldehyde inhibitor of γ-secretase activity that is thought to modify the active sites, while compound E is a non-peptide, non-transition state, non-competitive inhibitor of γ-secretase. Naive CD4+ T cells were isolated from spleens of 8–12 weeks old C57/BL6 mice, pre-treated with GSI or DMSO control for 30 min at 37°C and cultured in Th17 polarizing conditions for 24, 48 or 72 hours and IL-17A, IL-17F and IL-21 cytokine levels were assessed. The level of IL-17A produced by Th17 cells treated with GSI was significantly reduced in comparison to DMSO treated Th17 polarized cells (Fig.1A). Similarly, a reduction in IL-17F and IL-21 cytokine levels were observed after GSI treatment as compared to DMSO (Fig.1A). The observed cytokine profiles demonstrate that GSIs reduced Th17 associated cytokines from in-vitro differentiated Th17 cells. Interestingly we also observed that Notch1 is upregulated in Th17 polarized cells as compared to Th0 conditions (Fig.1B).
FIGURE 1.
Gamma secretase inhibitors (GSI) significantly down-regulate Th17 associated cytokine levels in murine Th17 in-vitro polarization assays. (A) ELISA for IL-17A, IL-17F and IL-21 in supernatants of activated CD4+ T cells from C57BL/6 mice. Cells were pretreated in-vitro with GSI (25μM ILCHO and 4μM Compound E) or with 0.1% DMSO (as a vehicle control) before 24, 48 and 72 hours culture in Th17 polarizing conditions. Cells were then lifted, recounted and cultured overnight. (B) Notch1 expression in cells pretreated with or without ILCHO was evaluated by immunoblotting using antibodies that recognized the cleaved active Notch1. Antibody specific for actin was used to control for loading. (C) Evaluation of Notch1 expression in cells pretreated with or without ILCHO by flow cytometry using antibodies specific for CD4+ cells and intracellular Notch 1 (Notch1IC). (D) Intracellular staining of IL-17 and IFNγ in Th17 differentiated cells treated with either DMSO or GSI. (E) Naïve CD4+ T cells were differentiated towads Th17 subset for 4 days followed by treatment with either DMSO or GSI. Supernatants were collected after 24 h and IL-17 A ELISA was performed. Data shown here represents one of at least three independent experiments done in triplicates.*p≤0.05, **p≤0.001, ***p≤0.0001.
Notch is a primary target of GSI in CD4+ T cells and to ensure that GSI was effective at reducing Notch1 activation, intracellular levels of Notch1 were assessed by immunoblot (Fig. 1B) and intracellular staining (Fig.1C). These data revealed that Notch1 protein expression was reduced in Th17 polarized murine CD4+ T cells treated with GSI.
In-order to determine the effect of GSI on IL-17A production on a per cell basis, intracellular staining of IL-17A was also performed in Th17 differentiated cells pre-treated with either DMSO or GSI (ILCHO). We observed a reduction in intracellular IL-17 levels in GSI treated Th17 cells as compared to DMSO (Fig.1D). Additionally, the effect of Notch inhibition on already differentiated Th17 cells was assessed. Naïve CD4+ T cells were cultured in Th17 polarizing conditions for 4 days followed by treatment with either DMSO or GSI. Interestingly no change in IL-17 levels were detected (Fig.1E).
The inhibition of gamma secretase during human Th17 polarization results in decreased Th17 associated cytokine levels
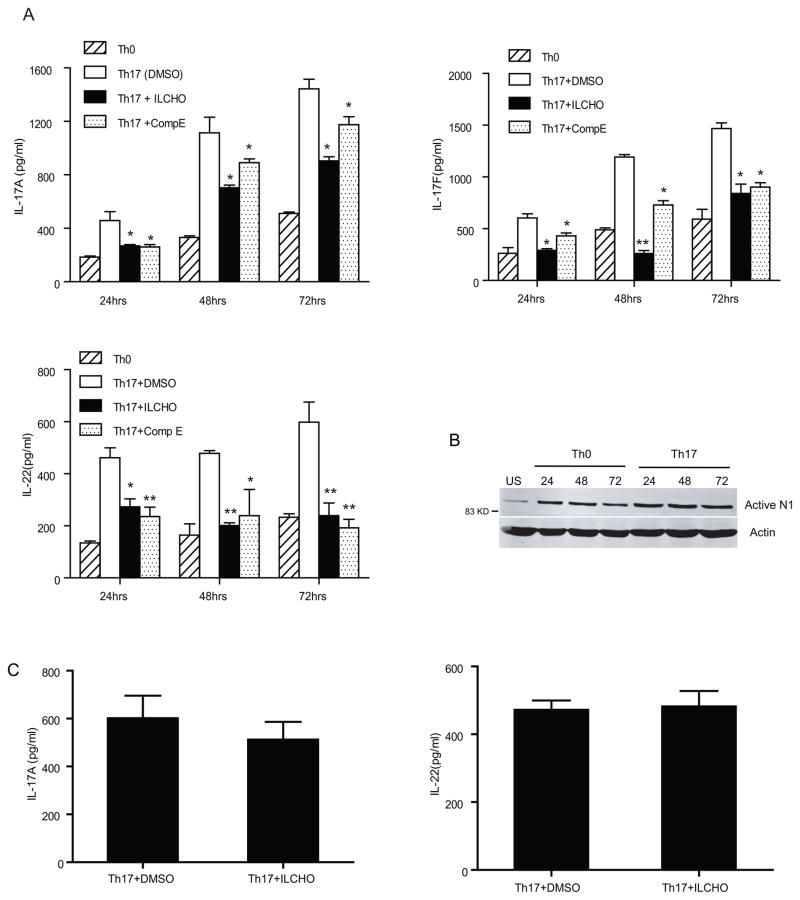
To determine whether Notch signaling also plays a role in human Th17 differentiation, we treated in-vitro human Th17 polarized cells with two different GSI (ILCHO and compound E). Naïve CD4+ T cells (CD4+CD45RA+) were purified from peripheral blood mononuclear cells, pretreated with either GSI or DMSO for 30 min and cultured in Th17 polarization conditions for 24, 48 and 72 h. IL-17A, IL-17 F and IL-22 secreted by human Th17 cells was significantly reduced in the presence of GSI compared to DMSO (Fig.2A). Surprisingly we did not detect significant levels of IL-21 in human in-vitro differentiated Th17 cells (data not shown). Consistent with the murine data, Th17 polarization of human CD4+ T cells resulted in increased levels of activated Notch1, compared to those activated under neutral conditions (Fig.2B). An MTS assay was performed to confirm the decrease in IL-17 secretion by GSI was not due to an effect on cell proliferation (data not shown). Next we assessed the effect of Notch inhibition in fully differentiated Th17 cells. Naïve CD4+ T cells were differentiated in Th17 conditions for 4 days followed by treatment with either DMSO or GSI. As seen in murine cells no change in IL-17 A and IL-22 cytokines were observed (Fig.1C). Taken together, data in Figures 1 and 2 show that treatment with GSI treatment blocks the differentiation of naïve CD4+ T cells into Th17 cells. Moreover treatment with GSI affects Th17 differentiation at earlier time points, but not in cells already committed to the Th17 lineage, suggesting a requirement for Notch signaling at early stages of Th17 differentiation.
FIGURE 2.
Gamma secretase inhibitors (GSIs) significantly reduce Th17 cytokine levels in human in-vitro Th17 polarization assays. (A) ELISA of IL-17A, IL-17F and IL-22 in supernatants of Th17 polarized naïve human CD4+ T cells treated with GSIs or DMSO as a vehicle control. Purified human CD4+ T cells were pretreated with GSIs (2μM ILCHO and 5μM Compound E) or DMSO as a vehicle control and then cultured in Th0 and Th17 polarizing conditions. Supernatants were collected at 24, 48 and 72 hours and were analyzed for IL-17A, IL-17F and IL-22. (B) Whole cell lysates were prepared from naive CD4+ T cells un-stimulated (US), or differentiated under Th0 and Th17 conditions and immunoblotted for interacellular active Notch1. β-actin was used to confirm equal loading. (C) Naïve CD4+ T cells were activated in-vitro under Th17 polarizing conditions for 4 days, followed by treatment with either DMSO or GSI. Supernatants were collected after 24 h and IL-17A and IL-22 ELISA were performed. Data shown here is representative of three independent experiments done in triplicates. *p≤0.05, ** ≤0.001.
Delivery of Notch1 siRNA to human naïve CD4+ T cells leads to decreased IL-17 secretion
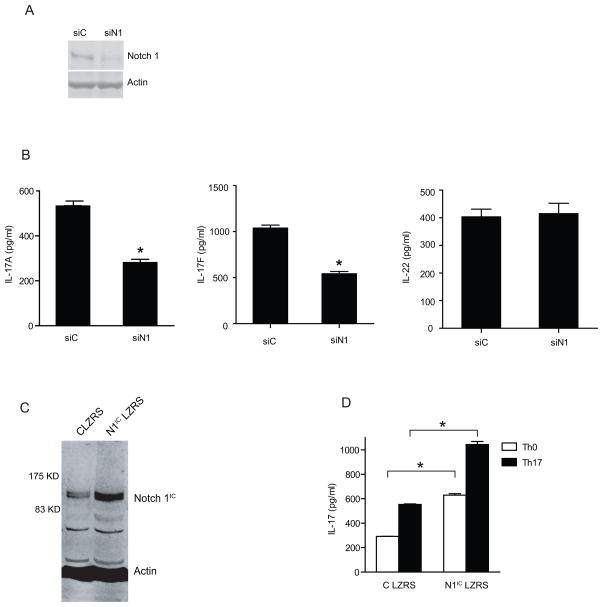
GSI blocks targets of γ-secretase including all members of the Notch family of proteins. To determine whether Notch1 is a functional target of GSIs during Th17 polarization, expression of Notch1 was reduced by delivery of siRNA to naïve CD4+T cells. Naïve CD4+ T cells were nucleoporated with Notch1 specific siRNA and subsequently polarized to the Th17 lineage and harvested 48 h after transfection. Western blot analysis of Notch1 protein and quantitative RT-PCR confirmed that Notch1 siRNA reduced the expression of Notch1 protein (Fig.3A) as well as mRNA (Supplementary Fig.2A). Western blot of Notch 2, 3 and 4 was also performed to confirm the specificity of Notch1 siRNA (Supplementary Fig.1). Notch1 knockdown significantly inhibited IL-17A and IL-17F production under Th17 polarizing conditions (Fig.3B). Surprisingly we did not observe a significant reduction in IL-22 production upon Notch1 knockdown (Fig.3B). An MTS assay was performed to check whether the reduction in IL-17 in Notch1 siRNA-treated cells was due to differential cell survival revealed no change between scrambled siRNA and Notch1 siRNA (Supplementary Fig.2B).
FIGURE 3.
Notch 1 controls human Th17 polarization. 1×107 purified human naïve CD4+ T cells were nucleoporated with Notch 1 specific siRNA or control siRNA. After transfection, the cells were cultured under Th17 skewing conditions and whole cell lysates and cDNA were prepared. (A) Immunoblot of Notch1 expression and β-actin (loading control). (B) ELISA of IL-17A, IL-17F and IL-22 were performed on the supernatants of naïve CD4+ T cells nucleoporated with control siRNA and Notch1 siRNA followed by in-vitro Th17 polarization. (C) Immunoblot of Notch1IC after transduction of naïve human CD4+ T cells with Notch1ICLZRS followed by Th17 differentiation. (D) ELISA of IL-17 performed after naïve CD4+T cells transduced with control LZRS and Notch IC LZRS followed by Th17 differentiation. The data is representative of three independent experiments done is triplicates. * p≤0.05.
The role of Notch1 in Th17 differentiation was confirmed by over-expressing activated Notch1 (intracellular domain of Notch1 cloned in the LZRS retroviral construct) in naïve human CD4+ T cells followed by Th17 polarization. An immunoblot for Notch1 confirmed over-expression (Fig.3C). Naïve CD4+ T cells overexpressing Notch1IC LZRS produced higher levels of IL-17 compared to control cells (Fig. 3D). Interestingly Notch1 overexpression also increased IL-17 secretion in cells activated under Th0 conditions.
Notch1 binds to the ROR gamma T promoter
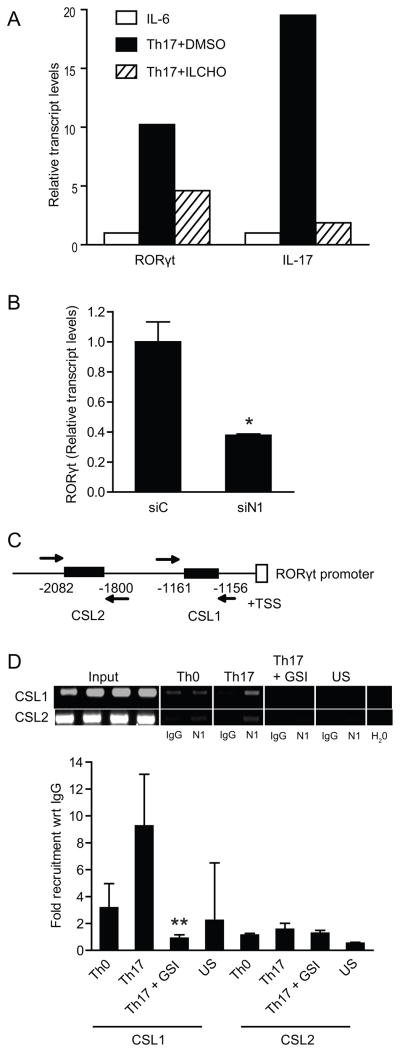
The orphan nuclear receptor RORγt is a key transcription factor that regulates the differentiation of the Th17 effector cell lineage. Thus, we explored whether Notch may regulate its expression. RNA was isolated from mouse CD4+ T cells polarized under Th17 conditions. cDNA was then synthesized to perform quantitative RT-PCR. RORγt mRNA expression was reduced by two-fold in Th17 polarized cells treated with GSI compared to DMSO-treated cells (Fig. 4A). To determine whether Notch1 influences human Th17 polarization by regulating RORγt expression, naïve human CD4+ T cells were purified, and nucleoporated with Notch1 specific siRNA, followed by culture under Th17 polarizing conditions. Quantitative RT-PCR of RORγt demonstrated that Notch1 knockdown resulted in decreased levels of RORγt transcripts (Fig. 4B). Taken together, these data indicate that Notch1 regulates the expression of RORγt. We then explored the possibility that Notch1 may directly regulate the human RORγt promoter. Analysis of this promoter revealed two potential CSL sites within the proximal 3kb promoter upstream of the RORγt transcriptional start site (Fig.4C). Chromatin immunoprecipitation analysis (ChIP) using an anti- Notch1 antibody was then performed to determine whether Notch1 binds directly to the RORγt promoter. The data presented in Figure 4D indicate that Notch1 binds directly to putative CSL binding sites in the human RORγt promoter (Fig.4D). In particular, Notch1 bound at the CSL1 site, which could be inhibited by treatment with GSI.
FIGURE 4.
Notch 1 regulates RORγt promoter activity. (A) In-vitro ILCHO treatment down-regulates RORγt and IL-17 mRNA expression. Total RNA was isolated from CD4+ T cells pretreated with 25μM ILCHO or DMSO as a vehicle control and cultured in Th17 polarizing conditions and analyzed by quantitative real time PCR. (B) Human naïve CD4+ T cells (1×107) were nucleoporated with Notch 1 specific siRNA or scrambled siRNA followed by in-vitro Th17 polarization. Cells were harvested and RORγt expression was determined by quantitative RT-PCR. Transcript abundance was normalized to 18s rRNA expression. (C) Schematic representation of putative CSL binding sites in human RORγt promoter. (D) Specific primers were used to amplify putative CSL binding sites on human RORγt promoter. ChIP assay was performed to determine recruitment of Notch1 on RORγt promoter. Data shown represents fold recruitment of Notch1 on CSL binding sites on human RORγt promoter with respect to control IgG normalized with 1% input DNA. Semiquantitative PCR was also performed using 2μl of DNA eluates using specific primers against CSL sites in RORγt promoter to confirm transcript size. Data represents mean± SD of three independent experiments done in triplicates. * p≤0.05, **p≤0.01.
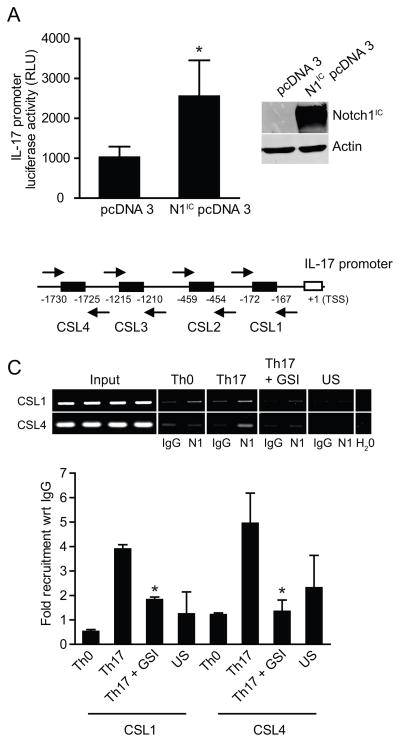
Notch 1 regulates IL-17 promoter activity
Since Notch has been reported to regulate and bind directly to the IFN-γ and IL-4 promoters, it is possible that Notch may also regulate the IL-17 promoter in addition to the RORγt promoter. Mouse CD4+ T cells were differentiated in-vitro towards the Th17 lineage in the presence of either DMSO or GSI. Transcript levels of IL-17 were reduced by 9 fold in GSI treated Th17 cells as detected by quantitative real time PCR (Fig.4A), suggesting Notch may directly regulate IL-17 promoter. Further, co-transfection of 293T cells with a human IL-17 promoter luciferase construct in combination with an activated Notch1 expression vector construct (N1IC) revealed that Notch1 expression significantly increased IL-17 promoter activity (Fig. 5A). This suggests that Notch1 regulates the IL-17 promoter. The human IL-17 promoter (3kb) upstream of the transcription start site (TSS) was therefore analyzed for putative CSL binding sites (Fig.5B). We found four putative CSL sites within this region (Fig. 5B). ChIP analysis of cells polarized under Th17 conditions, showed that Notch1 binds to putative CSL binding sites in the human IL-17 promoter, particularly CSL1 and 4 (Fig. 5C, 5D), but not CSL 2 and 3 (data not shown). The binding was inhibited by the pretreatment with GSI (Fig.5C). Thus Notch1 directly binds to both RORγt and IL-17 promoters and regulates Th17 differentiation.
FIGURE 5.
Notch1 regulates human IL-17 promoter activity. (A) 293 T cells were co-transfected with interacellular activated Notch expression vector construct (Notch1IC) along with a human IL-17 promoter construct cloned upstream of firefly luciferase gene. Luciferase assay was performed and data was normalized to renilla luciferase depicted as relative luciferase units (RLU). (B) Schematic representation of putative CSL binding sites in human IL-17 promoter. (C) ChIP assay was performed to determine the recruitment of Notch1 on human IL-17 promoter. Data shown represents fold recruitment of Notch1 on human IL-17 promoter with respect to isotype control IgG normalized to input DNA. Semiquantitative PCR was also performed (2μl of DNA eluates) using specific primers against different putative CSL binding sites in human IL-17 promoter to confirm transcript size. Data shown here represents the mean±SD of three independent experiments done in triplicates.*p≤0.05.
GSI ameliorates the severity of EAE–induced inflammation and Th17 differentiation in-vivo
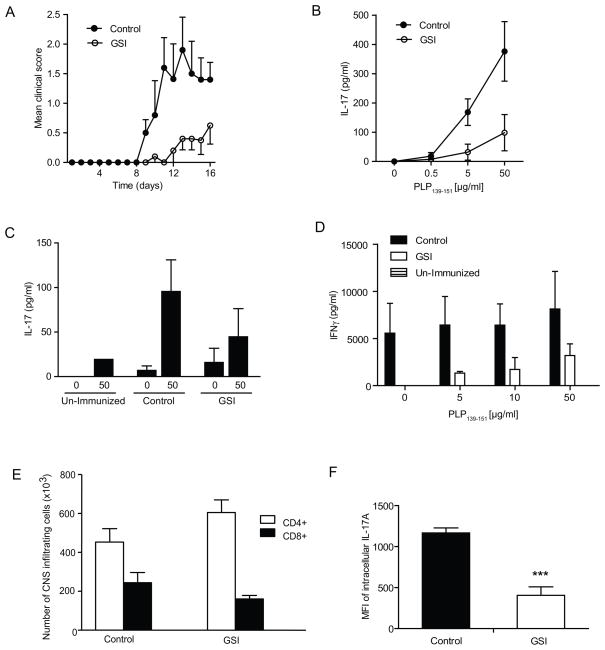
In-vitro experiments demonstrate that reducing Notch activation causes a significant decrease in IL-17 levels. To investigate if the in-vitro observations seen could be replicated in an in-vivo setting, an EAE model was used in this study. SJL/J mice were fed control chow or GSI (LY) chow. LY411,5745 is an orally active GSI, chemically similar to compound E. We have previously used LY411,575 incorporated into chow in mouse models of immune disorders and determined that doses between 2.5 and 5 mg/kg/day are safe and effective in reducing Notch activity systemically. Higher doses cause the well-known secretory diarrhea due to goblet cell metaplasia of the intestine. We have previously reported that GSI treatment ameliorates EAE progression (28). However, the role of Th17 cells in this model was not understood at the time, and in that report we explored exclusively Th1 responses. To induce EAE, mice were treated with PLP peptide emulsified in CFA and injected with pertussis toxin. The initial signs of EAE were observed eight days after immunization of the control group and ten days after immunization for the GSI treated group. At the peak of disease the clinical mean score for the control group was 2, while it was 0.8 for the GSI treated group (Fig.6A). Therefore GSI treatment significantly delayed the disease progression as well as reduced the severity of EAE symptoms, as previously shown (28).
FIGURE 6.
GSI treatment reduces EAE-induced inflammation and the development of PLP139–151-specific Th17 responses. (A) Clinical scores of SJL/J mice given GSI formulated chow, at 2.5mg/kg alternated with 5mg/kg for 4 weeks. Control mice were given regular chow. n=5 mice in each group. Results represent the mean disease score grouping each group. Splenocytes (B) and cells from the spinal cords (C) of EAE-induced mice were restimulated ex-vivo with PLP139–151 at increasing concentrations and cultured for 5 days. The restimulation supernatants were then analyzed for IL-17 by ELISA. (D) ELISA of IFNγ was performed on PLP139–151 restimulated splenocytes. (E) Total spinal cord cells were stained with CD4 and CD8 antibodies and analyzed by flow cytometry. (F) Intracellular staining of IL-17A in splenocytes of SJL/J mice fed with GSI or control chow. The data shown represent the mean fluorescent intensity of IL-17 in CD4+ T cells. ***p≤0.001
To determine whether Th17 responses were affected by GSI in-vivo,IL-17 levels in supernatents of peptide-stimulated splenocytes cultured from GSI- or control- treated mice were measured by ELISA.The GSI treated group showed significantly lower IL-17 levels than the control group (Fig.6B) Similarly, supernatants obtained from mononuclear cells isolated from spinal cords showed lower IL-17 levels in the GSI-treated mice than in the control group (Fig.6C). We also detected lower levels of IFNγ in peptide-restimulated splenocytes from GSI fed mice as compared to control mice (Fig.6D). To determine whether the effect of GSI mediated inhibition of IL-17 cytokines in-vivo is due to overall decrease in T cells number rather than Th17 cell differentiation, we determined the number of CD4+ and CD8+ T cells in spinal cord infiltrates. We found there were no significant differences in the number of cells infiltrating the spinal cord between the GSI or control chow fed mice (Fig.6E). Additionally, no significant differences in CD4+ and CD8+ cells were observed in the spleens of GSI-fed and control mice (data not shown). Indeed, we have maintained animals on GSI chow for as long as 6 months and not observed differences in CD4+ or CD8+ cell numbers (data not shown). This suggests that the decrease in IL-17 in the group of mice fed with GSI was not due to a difference in infiltrating cell numbers, but is more likely due to the effects of GSI on Th17 differentiation. Additionally we performed intracellular staining of IL-17 in CD4+ T cells of splenocytes treated in vivo and observed significant decrease in mean fluorescent intensity of IL-17 in GSI fed mice as compared to control mice (Fig.6F). Interestingly we did not observe a decrease in the percentage of CD4+ T cells producing IL-17, suggesting that inhibiting Notch signaling does not affect the number of CD4+ T cells producing IL-17 but rather their inherent ability to produce Th17 associated cytokines (Supplementary Fig.3).
Discussion
In this study, we addressed the role of Notch signaling in the development of a Th17 response in human and mouse CD4+ T cells. We employed several strategies to investigate the function of Notch in driving a Th17 response. Our data demonstrates that treatment with GSIs, compounds known to block γ-secretase function, also decreases Th17 differentiation and Th17 associated cytokines secretion. Additionally, we have shown that specific inhibition of Notch1 expression through the use of Notch1 siRNA abrogates IL-17 A and F production in polarized human Th17 cells. Surprisingly we do not observe a significant decrease in IL-22 cytokine levels upon Notch1 knockdown. Comparing our GSI data and specific Notch1 siRNA data, it may be possible that IL-22 is regulated by other downstream targets of γ secretase. Alternatively, IL-22 may be regulated by other Notch family members, particularly Notch 2 as reported before (45).
We also provide further insights into the role of Notch in Th17 induction by demonstrating that blockade of Notch, either through inhibition of γ-secretase or through siRNA mediated knockdown, results in reduced expression of RORγt, the transcription factor known to be required for effective induction of Th17 cells. These data, coupled with experiments showing Notch1 binding to both the RORγt and the IL-17 promoters suggest that Notch1 directly regulates the development of the Th17 subset of cells, at least in part, through the regulation of these two promoters. The biological consequences of Notch1 effects on Th17 development are highlighted in our in-vivo EAE experiments where GSI-mediated blockade of Notch activation results in reduced clinical disease as well as reduced levels of IL-17 produced by restimulated CD4+ T cells isolated from EAE-induced animals treated with GSI. Taken together, these data provide compelling evidence for a key role for Notch signaling in the development of an effective in-vitro and in-vivo Th17 response. We have also provide evidence that Notch signaling plays a role in early stages of Th17 differentiation as blocking Notch after 4 days of activation has no significant effect on Th17 associated cytokines in both mouse and human cells.
Notch1 has been implicated in the induction of both the Th1 and Th2 subsets of CD4+ T cells (28, 30, 31). Amsen et al suggested that different Notch ligands expressed on APCs drive differing T cell responses (23). In particular, this group provided evidence that DLL ligands preferentially drive a Th1 cell fate while Jagged ligands drive a Th2 fate. Lukacs and colleagues (10) recently revisited and expanded this observation and determined that DLL4 expression is induced on APCs by pathogen-associated signals and this ligand promotes expression of RORc and expansion of Th17 CD4+ T cells. The role of Notch signaling in mutually exclusive Th1, Th2 and Th17 differentiation may be mediated by different Notch ligands. Alternatively it may be due to upregulation of different Notch family members or by differential expression of the same Notch paralog (Notch1 being the most likely candidate). Notch signaling has been studied extensively but most experimental systems interrogate the conventional Notch signaling pathway, where activation of Notch leads to the production of intracellular Notch (NIC), which translocates to the nucleus and drives CSL-dependent transcription. More recent data indicate that activation of Notch also influences NF-κB signaling (46, 47) suggesting cross talk between these two signaling pathways in T cells. Additionally, evidence from several groups in a variety of vertebrate and invertebrate systems reveal a role for Notch in the cytosol and point toward a non-nuclear role for Notch in activation of cell survival pathways (48, 49). Therefore it is possible that different ligands activate different Notch signaling pathways, which, in turn, drive different outcomes that influence T helper differentiation and development. For example, the number of NotchIC molecules generated after activation and/or the duration of activation may dictate whether the canonical pathway or combinations of nuclear and cytoplasmic pathways are activated. Further experimentation is required to test this hypothesis.
In summary, in this report, we describe a role for Notch signaling in the development of both human and murine Th17 responses. A broad range of diseases require an active Th17 response, from multiple sclerosis to solid tumors. Our data suggest that Notch signaling inhibitors may act in-vivo at least by suppressing the Th17 response and may be useful in a variety of clinical situations where Th17 responses are required for disease pathogenesis.
Supplementary Material
Acknowledgments
We thank Dr Sarah Gaffen at University of Pittsburg for human IL-17 promoter luciferase construct. We thank Jared Klarquist for excellent technical support. We appreciate laboratory members for their support and criticism.
This work is NIH supported through AG025531 (LM and BAO), AI081179. (BAO and JA), Loyola University internal grant (BJN) and AR057643 (CLP).
Abbreviations used in this paper
- GSI
Gamma secretase inhibitors
- RORγt
Retinoic acid receptor related orphan receptor γ-t
- CSL
CBF-1, Suppressor of Hairless, Lag-1)
- ChIP
Chromatin immunoprecipitation
References
- 1.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 2.Bettelli E, Korn T, Kuchroo VK. Th17: the third member of the effector T cell trilogy. Curr Opin Immunol. 2007;19:652–657. doi: 10.1016/j.coi.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, Zurawski S, Wiekowski M, Lira SA, Gorman D, Kastelein RA, Sedgwick JD. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 4.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matusevicius D, Kivisakk P, He B, Kostulas N, Ozenci V, Fredrikson S, Link H. Interleukin-17 mRNA expression in blood and CSF mononuclear cells is augmented in multiple sclerosis. Mult Scler. 1999;5:101–104. doi: 10.1177/135245859900500206. [DOI] [PubMed] [Google Scholar]
- 6.Chabaud M, Durand JM, Buchs N, Fossiez F, Page G, Frappart L, Miossec P. Human interleukin-17: A T cell-derived proinflammatory cytokine produced by the rheumatoid synovium. Arthritis Rheum. 1999;42:963–970. doi: 10.1002/1529-0131(199905)42:5<963::AID-ANR15>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 7.Kagami S, Rizzo HL, Lee JJ, Koguchi Y, Blauvelt A. Circulating Th17, Th22, and Th1 Cells Are Increased in Psoriasis. J Invest Dermatol. 2009 doi: 10.1038/jid.2009.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakuraba A, Sato T, Kamada N, Kitazume M, Sugita A, Hibi T. Th1/Th17 immune response is induced by mesenteric lymph node dendritic cells in Crohn's disease. Gastroenterology. 2009;137:1736–1745. doi: 10.1053/j.gastro.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 9.Nalbandian A, Crispin JC, Tsokos GC. Interleukin-17 and systemic lupus erythematosus: current concepts. Clin Exp Immunol. 2009;157:209–215. doi: 10.1111/j.1365-2249.2009.03944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukherjee S, Schaller MA, Neupane R, Kunkel SL, Lukacs NW. Regulation of T cell activation by Notch ligand, DLL4, promotes IL-17 production and Rorc activation. J Immunol. 2009;182:7381–7388. doi: 10.4049/jimmunol.0804322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters M, Dudziak K, Stiehm M, Bufe A. T-cell polarization depends on concentration of the danger signal used to activate dendritic cells. Immunol Cell Biol. 2010 doi: 10.1038/icb.2010.3. [DOI] [PubMed] [Google Scholar]
- 12.Zhou L, Littman DR. Transcriptional regulatory networks in Th17 cell differentiation. Curr Opin Immunol. 2009;21:146–152. doi: 10.1016/j.coi.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 14.Ivanov, Zhou L, Littman DR. Transcriptional regulation of Th17 cell differentiation. Semin Immunol. 2007;19:409–417. doi: 10.1016/j.smim.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat Immunol. 2008;9:641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egwuagu CE. STAT3 in CD4+ T helper cell differentiation and inflammatory diseases. Cytokine. 2009;47:149–156. doi: 10.1016/j.cyto.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziegler SF, Buckner JH. FOXP3 and the regulation of Treg/Th17 differentiation. Microbes Infect. 2009;11:594–598. doi: 10.1016/j.micinf.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM, Dong C. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deftos ML, Bevan MJ. Notch signaling in T cell development. Curr Opin Immunol. 2000;12:166–172. doi: 10.1016/s0952-7915(99)00067-9. [DOI] [PubMed] [Google Scholar]
- 20.Garbe AI, von Boehmer H. TCR and Notch synergize in alphabeta versus gammadelta lineage choice. Trends Immunol. 2007;28:124–131. doi: 10.1016/j.it.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Radtke F, Wilson A, MacDonald HR. Notch signaling in T- and B-cell development. Curr Opin Immunol. 2004;16:174–179. doi: 10.1016/j.coi.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Germain RN. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol. 2002;2:309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 23.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 24.Adler SH, Chiffoleau E, Xu L, Dalton NM, Burg JM, Wells AD, Wolfe MS, Turka LA, Pear WS. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J Immunol. 2003;171:2896–2903. doi: 10.4049/jimmunol.171.6.2896. [DOI] [PubMed] [Google Scholar]
- 25.Palaga T, Miele L, Golde TE, Osborne BA. TCR-mediated Notch signaling regulates proliferation and IFN-gamma production in peripheral T cells. J Immunol. 2003;171:3019–3024. doi: 10.4049/jimmunol.171.6.3019. [DOI] [PubMed] [Google Scholar]
- 26.Osborne BA, Minter LM. Notch signalling during peripheral T-cell activation and differentiation. Nat Rev Immunol. 2007;7:64–75. doi: 10.1038/nri1998. [DOI] [PubMed] [Google Scholar]
- 27.Miele L. Notch signaling. Clin Cancer Res. 2006;12:1074–1079. doi: 10.1158/1078-0432.CCR-05-2570. [DOI] [PubMed] [Google Scholar]
- 28.Minter LM, Turley DM, Das P, Shin HM, Joshi I, Lawlor RG, Cho OH, Palaga T, Gottipati S, Telfer JC, Kostura L, Fauq AH, Simpson K, Such KA, Miele L, Golde TE, Miller SD, Osborne BA. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat Immunol. 2005;6:680–688. [PubMed] [Google Scholar]
- 29.Maekawa Y, Tsukumo S, Chiba S, Hirai H, Hayashi Y, Okada H, Kishihara K, Yasutomo K. Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity. 2003;19:549–559. doi: 10.1016/s1074-7613(03)00270-x. [DOI] [PubMed] [Google Scholar]
- 30.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, Flavell RA. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skokos D, Nussenzweig MC. CD8- DCs induce IL-12-independent Th1 differentiation through Delta 4 Notch-like ligand in response to bacterial LPS. J Exp Med. 2007;204:1525–1531. doi: 10.1084/jem.20062305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kassner N, Krueger M, Yagita H, Dzionek A, Hutloff A, Kroczek R, Scheffold A, Rutz S. Cutting edge: Plasmacytoid dendritic cells induce IL-10 production in T cells via the Delta-like-4/Notch axis. J Immunol. 2010;184:550–554. doi: 10.4049/jimmunol.0903152. [DOI] [PubMed] [Google Scholar]
- 34.Liotta F, Frosali F, Querci V, Mantei A, Fili L, Maggi L, Mazzinghi B, Angeli R, Ronconi E, Santarlasci V, Biagioli T, Lasagni L, Ballerini C, Parronchi P, Scheffold A, Cosmi L, Maggi E, Romagnani S, Annunziato F. Human immature myeloid dendritic cells trigger a TH2-polarizing program via Jagged-1/Notch interaction. J Allergy Clin Immunol. 2008;121:1000–1005. e1008. doi: 10.1016/j.jaci.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 35.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 36.Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 37.Hao L, Rizzo P, Osipo C, Pannuti A, Wyatt D, Cheung LW, Sonenshein G, Osborne BA, Miele L. Notch-1 activates estrogen receptor-alpha-dependent transcription via IKKalpha in breast cancer cells. Oncogene. 2010;29:201–213. doi: 10.1038/onc.2009.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curry CL, Reed LL, Nickoloff BJ, Miele L, Foreman KE. Notch-independent regulation of Hes-1 expression by c-Jun N-terminal kinase signaling in human endothelial cells. Lab Invest. 2006;86:842–852. doi: 10.1038/labinvest.3700442. [DOI] [PubMed] [Google Scholar]
- 39.Liu XK, Lin X, Gaffen SL. Crucial role for nuclear factor of activated T cells in T cell receptor-mediated regulation of human interleukin-17. J Biol Chem. 2004;279:52762–52771. doi: 10.1074/jbc.M405764200. [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Ivanov, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 42.Serada S, Fujimoto M, Mihara M, Koike N, Ohsugi Y, Nomura S, Yoshida H, Nishikawa T, Terabe F, Ohkawara T, Takahashi T, Ripley B, Kimura A, Kishimoto T, Naka T. IL-6 blockade inhibits the induction of myelin antigen-specific Th17 cells and Th1 cells in experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2008;105:9041–9046. doi: 10.1073/pnas.0802218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hofstetter HH, Grau C, Buttmann M, Forsthuber TG, Gaupp S, Toyka KV, Gold R. The PLPp-specific T-cell population promoted by pertussis toxin is characterized by high frequencies of IL-17-producing cells. Cytokine. 2007;40:35–43. doi: 10.1016/j.cyto.2007.07.192. [DOI] [PubMed] [Google Scholar]
- 44.Jurynczyk M, Jurewicz A, Bielecki B, Raine CS, Selmaj K. Overcoming failure to repair demyelination in EAE: gamma-secretase inhibition of Notch signaling. J Neurol Sci. 2008;265:5–11. doi: 10.1016/j.jns.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 45.Alam MS, Maekawa Y, Kitamura A, Tanigaki K, Yoshimoto T, Kishihara K, Yasutomo K. Notch signaling drives IL-22 secretion in CD4+ T cells by stimulating the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A. 2010;107:5943–5948. doi: 10.1073/pnas.0911755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, Golde TE, Sonenshein GE, Osborne BA. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. EMBO J. 2006;25:129–138. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellavia D, Campese AF, Alesse E, Vacca A, Felli MP, Balestri A, Stoppacciaro A, Tiveron C, Tatangelo L, Giovarelli M, Gaetano C, Ruco L, Hoffman ES, Hayday AC, Lendahl U, Frati L, Gulino A, Screpanti I. Constitutive activation of NF-kappaB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 2000;19:3337–3348. doi: 10.1093/emboj/19.13.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perumalsamy LR, Nagala M, Sarin A. Notch-activated signaling cascade interacts with mitochondrial remodeling proteins to regulate cell survival. Proc Natl Acad Sci U S A. 2010;107:6882–6887. doi: 10.1073/pnas.0910060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perumalsamy LR, Nagala M, Banerjee P, Sarin A. A hierarchical cascade activated by non-canonical Notch signaling and the mTOR-Rictor complex regulates neglect-induced death in mammalian cells. Cell Death Differ. 2009;16:879–889. doi: 10.1038/cdd.2009.20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.