Abstract
Pigment epithelium-derived factor (PEDF) is a broad-spectrum angiogenesis inhibitor that displays potent anti-metastatic activity in multiple tumor types. We have previously shown that PEDF prevents primary tumor growth and metastatic spread of human melanoma in mouse experimental models. Consistent with these observations, PEDF expression is lost at the late stages of melanoma progression, allowing melanoma cells to become angiogenic, migratory and invasive. PEDF’s ability to modify the interplay between the host and tumor tissues strongly supports its use as a therapeutic agent for the treatment of metastatic melanoma. However, transition to the clinic requires a more detailed knowledge of the molecular mechanisms underpinning PEDF’s activity. In this study we describe changes in the gene expression profile of A375 human melanoma cells induced by PEDF over-expression. PEDF modulated diverse categories of genes known to be involved in angiogenesis and migration. It downregulated cytokines like interleukin 8 and extracellular matrix proteins like collagen IV, while it upregulated fibronectin. Multiple transcripts previously described as contributing to the acquisition of malignant phenotype by melanoma were also diminished by PEDF over-expression, among which we validated galectin 3 and jagged 1. Additionally, PEDF downregulated S100β and melanoma inhibitory activity (MIA), which are widely used in the pathological diagnosis of melanoma. Interestingly, PEDF increased the expression of melanophilin and decreased rab27A, which are relevant targets for melanosome transport; suggesting that PEDF could directly impinge on melanocytic lineage-specific processes. Our study identifies new molecular targets and signaling pathways that may potentially contribute to determine PEDF’s ability to restrict the aggressiveness of A375 human melanoma cells.
Keywords: angiogenesis, PEDF, melanoma, gene expression profile
Introduction
Angiogenesis, the formation of new blood vessels from pre-existing ones [1], is an essential process for tumor growth maintenance, dissemination and metastasis growth [2]. Angiogenesis is regulated by the dynamic balance between two general types of regulators, inducers (pro-angiogenic) and inhibitors (anti-angiogenic). In normal tissues, vascular quiescence is maintained due to a predominance of inhibitors, whose blocking action is dominant over a wide array of inducers [3, 4]. However, during tumor progression, the angiogenic balance is inverted by genetic alterations as well as epigenetic regulation which determine a predominance of inducers over inhibitors [5]. Strict dependency of tumor growth and dissemination on sustained angiogenesis prompted the design of new therapeutic strategies for cancer treatment based on angiogenesis blockade, either by using neutralizing antibodies to relevant angiogenic factors or to their signaling receptors. Despite major impact of these strategies in preclinical models [6], both preclinical and clinical studies show that when a single angiogenic factor is targeted, the tumors acquire resistance to therapy, most likely due to production of alternative angiogenesis inducers [7]. In order to overcome these limitations a number of alternative approaches are currently being tested [8–11]. One such strategy involves restoration of endogenous angiogenesis inhibitors, whose expression is lost during malignant progression and which block the action of multiple inducers [11]. Treatment of xenografted tumors with endogenous anti-angiogenic factors efficiently halts primary tumor growth and significantly impairs metastasis [12–14]. However, translating endogenous inhibitors of angiogenesis into the clinic requires deeper knowledge of their biological effects and underlying molecular mediators.
Pigment epithelium-derived factor (PEDF) is a 50 kDa secreted glycoprotein and a non-functional member of the serine protease inhibitor (Serpin) superfamily [15, 16]. PEDF plays a critical role in maintaining vascular quiescence and controls the angiogenic switch in multiple tissues [17, 18]. Potent anti-tumor and anti-metastatic effects of PEDF were demonstrated in mouse experimental models using a variety of cancer cell lines, and an inverse correlation between PEDF levels and aggressiveness was also shown for different types of cancers [19]. We have previously described PEDF’s anti-tumor and anti-metastatic effects in melanoma [19, 20], a type of cancer that displays a relevant angiogenic activity [21, 22]. PEDF ability to inhibit melanoma growth and metastasis is based on its dual action on endothelial and tumor cells. In both cell types, PEDF can cause apoptosis and impair migratory and invasive capacity. Recent reports have identified non-overlapping epitopes on PEDF molecule responsible for its diverse biological actions [23–25]. We have also found that PEDF is expressed at high levels in normal skin melanocytes, and that the loss of PEDF expression enables the switch of melanoma cells from proliferative to invasive phenotype highlighting the relevance of this factor in the control of melanoma progression [26]. Thus we propose that restoration of PEDF expression by delivery of recombinant PEDF or short PEDF-derived peptides could be employed as a therapeutic strategy in advanced melanoma, which is a especially aggressive cancer refractory to currently available drugs [27]. Using this multifaceted factor, which targets diverse biological processes critical for cancer progression, may serve double purpose by obliterating the vasculature and switching off their ability to invade, leading to reduced metastatic spread of melanoma cells [19]. Despite the interesting biological properties described for PEDF, a detailed understanding of its molecular mechanisms remains elusive.
In this study we identify novel candidate factors and pathways that could mediate the effects of PEDF on A375 human melanoma cells. Using global gene expression profiling analysis, we have found that PEDF over-expression in A375 cells mostly downregulated distinct sets of genes, which fall into several functional categories including the control of angiogenesis and several other functions required for melanoma progression; while it increased expression of some relevant transcripts such as fibronectin and melanophilin. Identified targets provide molecular insight into PEDF’s actions leading to reduced aggressiveness of A375 melanoma cells.
Methods
Cell culture
A375 human melanoma cell line, originally established in culture from a lymph node metastasis of a melanoma patient [28, 29], was kindly provided by Dr Vidal-Vanaclocha (Universidad del Pais Vasco, Bilbao, Spain). UCD-Mel-N human melanoma cell line was originally derived from a patient with multiple primary melanoma tumors, and generously provided by Dr. E. Medrano [30]. C8161 human melanoma cell line was initially isolated from an abdominal wall metastasis of a cutaneous melanoma patient, and generously provided by Dr. M. Hendrix [31]. All melanoma cell lines were grown in DMEM (Gibco, New York, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco), 2 mM glutamine (Gibco), 50 μg/ml streptomycin (Gibco), 50 U/ml penicillin (Gibco) and 2.5 μg/ml fungizone (Sigma, St Louis, MO, USA).
Generation of PEDF over-expressing cell lines
A375 melanoma cells were seeded into 21 cm2 plates and 24 h later they were transfected with 10.8 μg pCEP4-PEDF plasmid (provided by Dr Bouck, Northwestern University, Chicago, IL, USA) or control pCEP4 plasmid and 21.6 μl jetPEI (Polyplus Transfection, Illkirch, France) for 4 h. Pooled transfected cells (herein after referred to as A375-pCEP4-PEDF Pool and A375-pCEP4 Pool) were selected using 300 μg/ml hygromycin B (Sigma) for 2 weeks and used for the gene expression profile study. In an independent transfection a number of isolated clones were selected as described above and used for the validation of target genes identified using the pools.
Further validation of selected target genes was carried out by transient over-expression using lentivirus. The lentiviral construct encoding the full-length human PEDF cDNA in the prrl.CMV.EGFP.wpre.SIN lentiviral vector (kindly provided by A Bernad, Centro Nacional de Investigaciones Cardiovasculares, Madrid, Spain) has been previously reported [26]. Lentiviral particles were produced as described earlier [28]. Transduction of A375 melanoma cells was performed by incubating cells with lentiviruses at a multiplicity of infection (MOI) of 10 in the presence of 8 μg/ml polybrene (Sigma) for 8 h, typically yielding more than 95% transduced (GFP-positive) cells. After 72 h, PEDF over-expression was assessed in transient studies. PEDF-over-expressing UCD-Mel-N and C8161 melanoma cell lines (UCD-PEDF, C8161-PEDF) and their respective controls (UCD-GFP, C8161-GFP) have been described previously [20, 26].
Western blot
Conditioned media (CM) were prepared and analyzed as described earlier [32], and 1.5 μg CM were loaded per lane. PEDF monoclonal antibody (Chemicon, Temecula, CA, USA) was diluted 1:1200.
IL8 ELISA
Secreted IL8 protein levels in 48 h CM were quantified using Human CXCL8/IL8 Quantikine ELISA Kit (R&D) following manufacturer’s instructions.
Reporter assays
We used pGL3-basic reporter plasmids (Promega, Madison, WI, USA) containing the Firefly luciferase gene (Luc) under the control of the −2000 bp fragment of the human IL8 gene promoter (pGL-2000-IL8-Luc, provided by Dr Raingeaud, INSERM, Chatenay-Malabry, France) [33]; and the pRL-SV40 plasmid (Promega, Madison, WI, USA) containing the Renilla reniformes luciferase gene (Renilla) under the control of the SV40 virus promoter as a control to correct for the efficiency of transfection. 5×104 cells were seeded onto 2 cm2 wells 24 h before transfection, each condition in triplicate. Then, cells were incubated with 1 μl Lipofectamine 2000 (Sigma), 300 ng pGL-2000-IL8-Luc and 20 ng pRL-SV40 plasmids in 50 μl OptiMEM (Gibco) per well for 4 h, and medium was changed. Next day, cells were incubated with 10% FBS or 10 ng/ml tumor necrosis alpha (TNFα) in serum-free medium for 24 h, and then plates were frozen at −80°C. Analysis of Luc and Renilla activity was performed using the Dual Luciferase Reporter Assay System (Promega) and a Lumat LB9507 luminometer (Berthold Technologies, Bad Wildbad, Germany). The Luc activity was then normalized to Renilla activity.
RNA extraction and quantitative RT-PCR
Total RNA was extracted using TRIzol (Molecular Research Center Inc., Cincinnati, OH, USA) and was retro-transcribed to cDNA using High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA). See Supplementary Methods and Supplementary Table 1 for TaqMan (Applied Biosystems) and Universal Probe Library (UPL) (Roche, Basel, Switzerland) probes and oligonucleotides used. The quantitative PCR reaction was performed in an ABI Prism 7900 HT thermal cycler (Applied Biosystems). Thermal cycling conditions consisted of a denaturing step at 95°C for 10 min, 40 cycles of denaturing at 95°C for 15 s and annealing and elongation at 60°C for 60 s. Each condition was assessed in triplicate. Relative mRNA levels were determined using the comparative CT method.
Microarray hybridization and data analysis
Global gene expression profiles of A375-pCEP4 Pool and A375-pCEP4-PEDF Pool cells were determined using whole genome oligonucleotide GeneChip Human Genome-133 plus 2.0 microarrays (Affymetrix, Santa Clara, CA). Microarrays were hybridized in duplicate with RNAs from two independent experiments. RNA integrity was determined using a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA). Biotinylated cRNA was synthesized from total RNA using the 3′ Amplification One-cycle Target labeling kit (Affymetrix). Briefly, 2 μg of RNA were reverse transcribed to produce first strand cDNA using an oligodeoxythymidylic acid 24 primer with a T7 RNA polymerase promoter site added to the 5′ end. After second strand synthesis, in vitro transcription was performed using T7 RNA polymerase and biotinylated nucleotides, to produce biotin-labeled cRNA. Ten μg cRNA was fragmented at 95° C for 35 min into 35–200 bases in length. Fragmented cRNAs were hybridized to microarrays at 45°C for 16 h in an oven at 60 rpm.
Each microarray was washed and stained in the Affymetrix Fluidics Station 450 following the standard protocols developed by Affymetrix. Microarrays were scanned in an Affymetrix GeneChip® Scanner 3000 7G. Normalized gene expression values were obtained using GCRMA algorithm [34]. To determine statistically significant changes in each individual comparison, we used Microarray Suite (MAS 5.0) comparison analysis from Affymetrix (Statistical Algorithm Reference Guide). In this analysis a change algorithm generates a Change p-value and an associated Change (Increase, Decrease or NoChange). Probe sets were considered as regulated when there was a significant Change (Increase or Decrease) and at least a two-fold upregulation or downregulation in A375-pCEP4-PEDF compared to A375-pCEP4 cells in both independent hybridizations (regulated probe sets are listed in Supplementary Table 2). Heat maps of regulated genes were generated using Multiexperimental Viewer software (Microarray Software Suite, http://www.tm4.org). Most regulated genes were classified according to their function into biological process categories from the Gene Ontology database (http://www.geneontology.org/). This classification was further completed with new “melanoma progression-related” categories according to information from Gene database (NCBI) and published studies in PubMed.
Invasion assay
Cell invasion was evaluated using cell Invasion Assay Kit (Chemicon) following the manufacturer’s instructions. In brief, cells were serum-starved 16 h prior to seeding, and 10% FBS was used as chemoattractant. After 24 h, non-invaded cells were wiped off using a cotton swab, and the filters were stained with Diff Quik (Dade Behring, Newark, DE, USA). Invaded cells were counted at ten areas of maximum invasion under a light microscope at 40× magnification. Average ± standard deviation (SD) values shown are representative of at least three independent experiments.
Statistical analysis
All statistical analyses were performed using GraphPad Instat (GraphPad Software, San Diego, CA, USA). P values ≤ 0.05 were considered as significant.
Results
Functional grouping of genes modulated by PEDF in A375 human melanoma cells
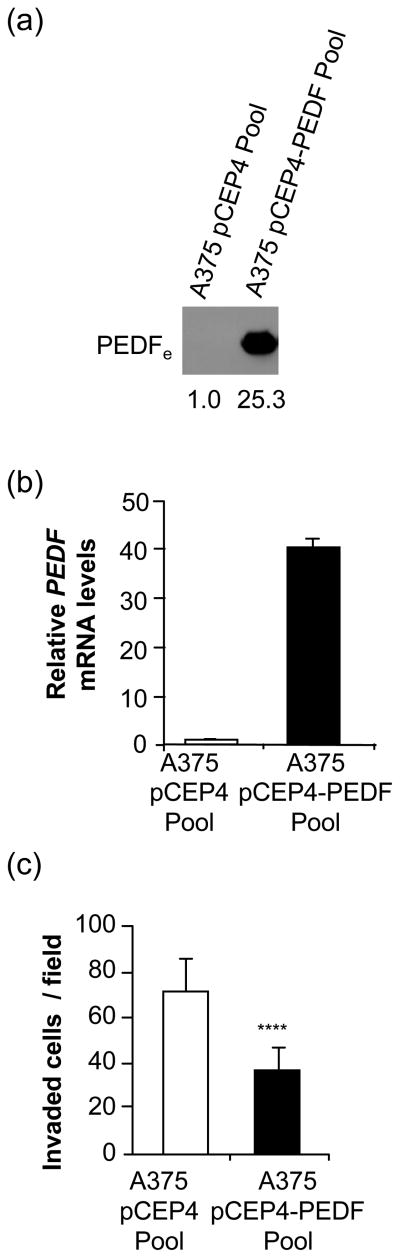
To gain insight into the molecular mechanisms underlying effects of PEDF on melanoma, we performed genome-wide analysis of the changes in gene expression induced by PEDF over-expression in A375 human melanoma cell line. A375 cells expressed very low endogenous levels of PEDF compared to other human melanoma cell lines tested [26], and PEDF over-expression significantly reduced its metastatic potential in mouse models [20]. We generated a pool of A375 melanoma cells stably expressing high levels of PEDF (A375-pCEP4-PEDF Pool) and a pool of empty vector-transfected A375 cells (A375-pCEP4 Pool). PEDF expression was assessed by Western blot of conditioned medium (CM) (Fig. 1a) and quantitative RT-PCR (Fig. 1b). PEDF over-expression reduced the invasive ability of A375 cells (Fig. 1c), in agreement with previously described results in other human melanoma cell lines [20, 26, 35].
Fig. 1.
Characterization of PEDF-over-expressing A375 cell lines. (a) Western blot analysis of secreted extracellular PEDF (PEDFe) protein in 48 h conditioned medium (CM) from control (A375-pCEP4 Pool) and PEDF-over-expressing (A375-pCEP4-PEDF Pool) cells. Numbers below blot show densitometry values normalized to A375-pCEP4 Pool expression. (b) Quantitative RT-PCR analysis of PEDF mRNA levels in A375-pCEP4 Pool and A375-pCEP4-PEDF Pool cells. PEDF mRNA levels are shown relative to A375-pCEP4 Pool after normalization to GAPDH. Bars represent average ± standard deviation (SD). (c) Invasion assay of A375-pCEP4 Pool and A375-pCEP4-PEDF Pool cells toward 10% FBS for 24 h. Statistical significance was determined by Student’s t-test (****, p<0.0001). Results are representative of two independent experiments.
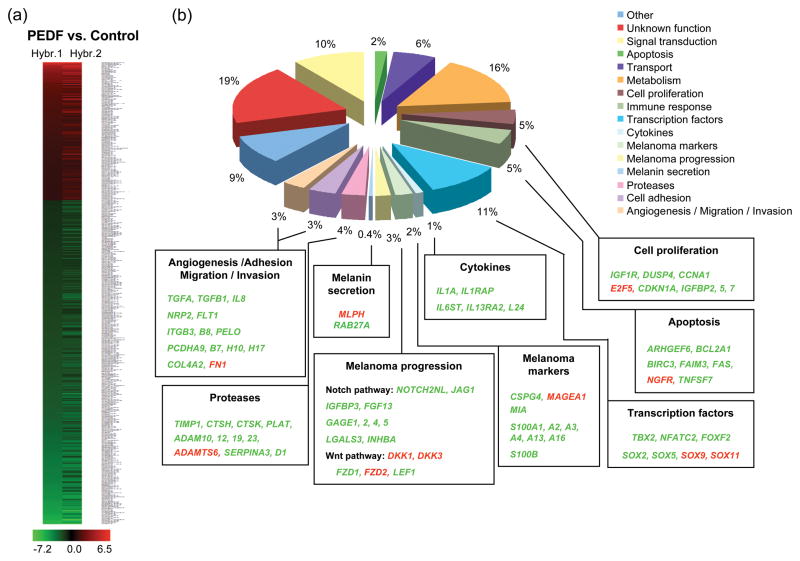
We hybridized RNAs isolated from A375-pCEP4-PEDF Pool and A375-pCEP4 Pool cells to oligonucleotide Affymetrix microarrays. The results from two independent experiments were analyzed. Considering a two-fold cut-off, 757 probe sets showed differential expression (Supplementary Table 2). Among these, 470 corresponded to characterized genes annotated in the National Center for Biotechnology Information (NCBI). The majority of genes were downregulated by PEDF over-expression (340 genes); while 140 genes were upregulated (heat map is shown in Fig. 2a). The differentially regulated genes were classified according to their function into biological process categories from the Gene Ontology database. We expanded this analysis to include additional categories specifically related to melanoma biology, which are not included at Gene Ontology database, using literature searches for the data supporting the functional role of genes regulated by PEDF (Fig. 2b and Table 1). Significant number of genes (highlighted in the lower boxes in Fig. 2b) involved in angiogenesis, adhesion, migration, invasion and matrix degradation were regulated by PEDF. The trend of the regulation was in agreement with its previously described inhibitory effects on melanoma [20]. Interestingly, several genes shown in other studies as relevant for melanoma progression or lineage-specific functions were also modulated by PEDF.
Fig. 2.
Distribution in functional categories of genes regulated by PEDF in A375 melanoma cells. (a) Heat map of expression of genes regulated by PEDF in two independent hybridizations (Hybr. 1 and Hybr. 2) of A375-pCEP4-PEDF Pool cells compared to control A375-pCEP4 Pool cells. Coloring represents normalized signal intensity of the ratio log2 (A375-pCEP4-PEDF Pool/A375-pCEP4 Pool): red, upregulation; black, no change; green, downregulation. (b) Diagram showing genes regulated by PEDF grouped into biological processes and/or functional categories as described in the main text. Percentage of regulated genes in each category relative to total number of regulated genes is shown. For some categories the most relevant genes are listed in the lower boxes, being shown in red when upregulated and in green when downregulated in A375-pCEP4-PEDF Pool compared to A375-pCEP4 Pool.
Table 1. Distribution in functional categories of genes regulated by PEDF in A375 human melanoma cell line.
Selected relevant genes regulated by PEDFe are shown classified into functional categories described in the main text. Upregulated genes are included at the top of the list followed by downregulated genes. Shown are gene symbol, description and the linear ratio of GCRMA-normalized expression PEDF/control in the two hybridizations from independently isolated RNAs from A375-pCEP4-PEDF Pool y A375-pCEP4 Pool cells (Hybr. 1 and Hybr. 2).
| ANGIOGENESIS/ADHESION/MIGRATION/INVASION | |||
|---|---|---|---|
| Gene symbol | Description | PEDF/Control | |
| Hybr. 1 | Hybr. 2 | ||
| FN1 | Fibronectin 1 | 14.22 | 16.52 |
| VEGFC | vascular endothelial growth factor C | 3.13 | 4.64 |
| FLT1 | Fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | 0.05 | 0.07 |
| IL8 | Interleukin 8 | 0.36 | 0.36 |
| ITGB3 | Integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | 0.30 | 0.26 |
| ITGB8 | Integrin, beta 8 | 0.11 | 0.19 |
| NRP2 | Neuropilin 2 | 0.40 | 0.41 |
| PCDH10 | Protocadherin 10 | 0.10 | 0.12 |
| PCDH17 | Protocadherin 17 | 0.34 | 0.27 |
| PCDHA9 | Protocadherin alpha 9 | 0.46 | 0.48 |
| PCDHB7 | Protocadherin beta 7 | 0.34 | 0.34 |
| PELO | Integrin, alpha 1 | 0.12 | 0.15 |
| TGFA | Transforming growth factor, alpha | 0.03 | 0.04 |
| TGFBI | Transforming growth factor, beta-induced, 68kDa | 0.41 | 0.43 |
| PROTEASES | |||
|---|---|---|---|
| Gene symbol | Description | PEDF/Control | |
| Hybr. 1 | Hybr. 2 | ||
| ADAMTS6 | A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 6 | 3.39 | 8.66 |
| ADAM10 | A disintegrin and metalloproteinase domain 10 | 0.46 | 0.46 |
| ADAM12 | A disintegrin and metalloproteinase domain 12 (meltrin alpha) | 0.27 | 0.47 |
| ADAM19 | A disintegrin and metalloproteinase domain 19 (meltrin beta) | 0.06 | 0.09 |
| ADAM23 | A disintegrin and metalloproteinase domain 23 | 0.08 | 0.10 |
| CTSH | Cathepsin H | 0.17 | 0.30 |
| CTSK | Cathepsin K (pycnodysostosis) | 0.08 | 0.10 |
| PLAT | Plasminogen activator, tissue | 0.39 | 0.40 |
| PRSS11 | protease, serine, 11 (IGF binding) | 0.07 | 0.04 |
| SERPINA3 | Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | 0.05 | 0.04 |
| SERPIND1 | Serine (or cysteine) proteinase inhibitor, clade D (heparin cofactor), member 1 | 0.07 | 0.07 |
| TIMP1 | Tissue inhibitor of metalloproteinase 1 (erythroid potentiating activity, collagenase inhibitor) | 0.33 | 0.32 |
| MELANIN SECRETION | |||
|---|---|---|---|
| Gene symbol | Description | PEDF/Control | |
| Hybr. 1 | Hybr. 2 | ||
| MLPH | Melanophilin | 15.70 | 28.90 |
| RAB27A | RAB27A, member RAS oncogene family | 0.31 | 0.22 |
| MELANOMA PROGRESSION | |||
|---|---|---|---|
| Gene symbol | Description | PEDF/Control | |
| Hybr. 1 | Hybr. 2 | ||
| DKK1 | Dickkopf homolog 1 (Xenopus laevis) | 2.86 | 2.87 |
| DKK3 | Dickkopf homolog 3 (Xenopus laevis) | 2.28 | 2.85 |
| FZD2 | Frizzled homolog 2 (Drosophila) | 2.60 | 2.38 |
| FGF13 | Fibroblast growth factor 13 | 0.14 | 0.08 |
| FZD1 | Frizzled homolog 1 (Drosophila) | 0.37 | 0.42 |
| GAGE1 | G antigen 1 | 0.35 | 0.46 |
| GAGE2 | G antigen 2 | 0.11 | 0.10 |
| GAGE4 | G antigen 4 | 0.45 | 0.38 |
| GAGE5 | G antigen 5 | 0.24 | 0.31 |
| IGFBP3 | Insulin-like growth factor binding protein 3 | 0.11 | 0.22 |
| INHBA | Inhibin, beta A (activin A, activin AB alpha polypeptide) | 0.18 | 0.24 |
| JAG1 | Jagged 1 (Alagille syndrome) | 0.14 | 0.13 |
| LEF1 | Lymphoid enhancer-binding factor 1 | 0.48 | 0.37 |
| LGALS3 | Lectin, galactoside-binding, soluble, 3 (galectin 3) /// galectin-3 internal gene | 0.23 | 0.21 |
| NOTCH2NL | Notch homolog 2 (Drosophila) N-terminal like | 0.29 | 0.26 |
| MELANOMA MARKERS | |||
|---|---|---|---|
| Gene symbol | Description | PEDF/Control | |
| Hybr. 1 | Hybr. 2 | ||
| MAGEA1 | Melanoma antigen family A, 1 (directs expression of antigen MZ2-E) | 2.78 | 2.78 |
| CSPG4 | Chondroitin sulfate proteoglycan 4 (melanoma-associated) | 0.19 | 0.24 |
| MIA | Melanoma inhibitory activity | 0.48 | 0.44 |
| S100A1 | S100 calcium binding protein A1 | 0.33 | 0.13 |
| S100A13 | S100 calcium binding protein A13 | 0.38 | 0.33 |
| S100A16 | S100 calcium binding protein A16 | 0.29 | 0.36 |
| S100A2 | S100 calcium binding protein A2 | 0.30 | 0.27 |
| S100A3 | S100 calcium binding protein A3 | 0.15 | 0.12 |
| S100A4 | S100 calcium binding protein A4 (calcium protein, calvasculin, metastasin, murine placental homolog) | 0.10 | 0.09 |
| S100B | S100 calcium binding protein, beta (neural) | 0.13 | 0.10 |
| TRANSCRIPTION FACTORS | |||
|---|---|---|---|
| Gene symbol | Description | PEDF/Control | |
| Hybr. 1 | Hybr. 2 | ||
| FOXF2 | Forkhead box F2 | 2.16 | 2.32 |
| SOX11 | SRY (sex determining region Y)-box 11 | 3.85 | 4.10 |
| SOX9 | SRY (sex determining region Y)-box 9 (campomelic dysplasia, autosomal sex-reversal) | 2.27 | 12.34 |
| NFATC2 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | 0.29 | 0.22 |
| SOX2 | SRY (sex determining region Y)-box 2 | 0.41 | 0.46 |
| SOX5 | SRY (sex determining region Y)-box 5 | 0.20 | 0.08 |
| TBX2 | T-box 2 | 0.24 | 0.19 |
| APOPTOSIS | |||
|---|---|---|---|
| Gene symbol | Description | PEDF/Control | |
| Hybr. 1 | Hybr. 2 | ||
| BID | BH3 interacting domain death agonist | 2.17 | 2.28 |
| NGFR | Nerve growth factor receptor (TNFR superfamily, member 16) | 6.67 | 4.56 |
| ARHGEF6 | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 | 0.46 | 0.23 |
| BCL2A1 | BCL2-related protein A1 | 0.04 | 0.05 |
| BIRC3 | Baculoviral IAP repeat-containing 3 | 0.36 | 0.18 |
| FAIM3 | Fas apoptotic inhibitory molecule | 0.32 | 0.30 |
| FAS | Fas (TNF receptor superfamily, member 6) | 0.19 | 0.19 |
| TNFSF7 | Tumor necrosis factor (ligand) superfamily, member 7 | 0.33 | 0.35 |
| CELL PROLIFERATION | |||
|---|---|---|---|
| Gene symbol | Description | PEDF/Control | |
| Hybr. 1 | Hybr. 2 | ||
| CCNJ | Cyclin J | 2.04 | 2.91 |
| CLEC11A | C-type lectin domain family 11, member A | 3.94 | 3.52 |
| CYR61 | Cysteine-rich, angiogenic inducer, 61 | 2.06 | 2.55 |
| E2F5 | E2F transcription factor 5, p130-binding | 3.72 | 3.91 |
| HDAC9 | Histone deacetylase 9 | 2.51 | 3.35 |
| MDK | Midkine (neurite growth-promoting factor 2) | 7.29 | 9.42 |
| CCNA1 | Cyclin A1 | 0.10 | 0.08 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 0.21 | 0.37 |
| DUSP4 | Dual specificity phosphatase 4 | 0.39 | 0.32 |
| HIPK2 | Homeodomain interacting protein kinase 2 | 0.41 | 0.41 |
| IGF1R | Insulin-like growth factor 1 receptor | 0.25 | 0.25 |
| IGFBP2 | Insulin-like growth factor binding protein 2, 36kDa | 0.24 | 0.14 |
| IGFBP5 | Insulin-like growth factor binding protein 5 | 0.27 | 0.27 |
| IGFBP7 | Insulin-like growth factor binding protein 7 | 0.32 | 0.45 |
| NDN | Necdin homolog (mouse) | 0.07 | 0.09 |
| CYTOKINES | |||
|---|---|---|---|
| Gene symbol | Description | PEDF/Control | |
| Hybr. 1 | Hybr. 2 | ||
| IL13RA2 | Interleukin 13 receptor, alpha 2 | 0.41 | 0.44 |
| IL1A | Interleukin 1, alpha | 0.16 | 0.11 |
| IL1RAP | Interleukin 1 receptor accessory protein | 0.17 | 0.14 |
| IL24 | Interleukin 24 | 0.06 | 0.07 |
| IL6ST | Interleukin 6 signal transducer (gp130, oncostatin M receptor) | 0.38 | 0.38 |
| METABOLISM | |||
|---|---|---|---|
| Gene symbol | Description | PEDF/Control | |
| Hybr. 1 | Hybr. 2 | ||
| PLTP | phospholipid transfer protein | 2.73 | 2.71 |
| ACOX2 | acyl-Coenzyme A oxidase 2, branched chain | 0.49 | 0.30 |
| ACSL1 | acyl-CoA synthetase long-chain family member 1 | 0.34 | 0.39 |
| ADPGK | ADP-dependent glucokinase | 0.39 | 0.38 |
| B3GAT1 | beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) | 0.35 | 0.29 |
| CHST11 | carbohydrate (chondroitin 4) sulfotransferase 11 | 0.36 | 0.18 |
| IRS2 | insulin receptor substrate 2 | 0.44 | 0.40 |
| MGLL | monoglyceride lipase /// monoglyceride lipase | 0.24 | 0.44 |
| PLA2G4A | phospholipase A2, group IVA (cytosolic, calcium-dependent) | 0.24 | 0.38 |
Genes involved in the control of angiogenesis and invasion modulated by PEDF in A375 human melanoma cells
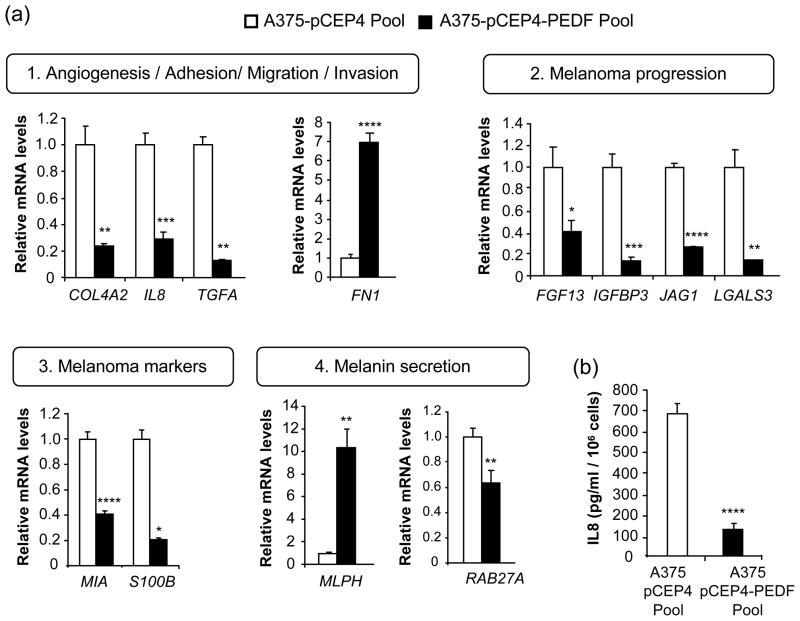
Genes that determine the angiogenic phenotype and the migratory and invasive ability of melanoma cells were modulated by PEDF over-expression in A375 cells (Fig. 2b and Table 1). A number of soluble factors with pro-angiogenic or chemotactic properties were downregulated upon PEDF over-expression, including interleukin 8 (IL8), transforming growth factor alpha (TGFA) and transforming growth factor beta-induced (TGFBI) [3]. The downregulation was confirmed by quantitative RT-PCR for IL8 and TGFA (Fig. 3a) and by ELISA for IL8 (Fig. 3b).
Fig. 3.
Validation of candidate target genes regulated by PEDF in A375 melanoma cells. (a) Quantitative RT-PCR analysis of mRNA levels of genes related to: 1) angiogenesis/adhesion/migration/invasion: COL4A2, FN1, IL8 and TGFA; 2) melanoma progression: FGF13, IGFBP3, JAG1 and LGALS3; 3) melanoma markers: MIA and S100B; 4) melanin secretion: MLPH and RAB27A. Empty bars correspond to A375-pCEP4 Pool cells and filled bars to A375-pCEP4-PEDF Pool cells. For each gene mRNA levels are shown relative to A375-pCEP4 Pool after normalization to GAPDH. Bars represent average ± SD. Statistical significance was determined by Student’s t-test or Welch-corrected t-test (the latter for COL4A2, TGFA, LGALS3, MLPH and S100B) (*, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001). (b) ELISA analysis of secreted IL8 protein levels in 48 h CM from A375-pCEP4 Pool and A375-pCEP4-PEDF Pool cells. Bars represent average ± SD. Statistical significance was determined by Student’s t-test (****, p<0.0001). Results are representative of two independent experiments.
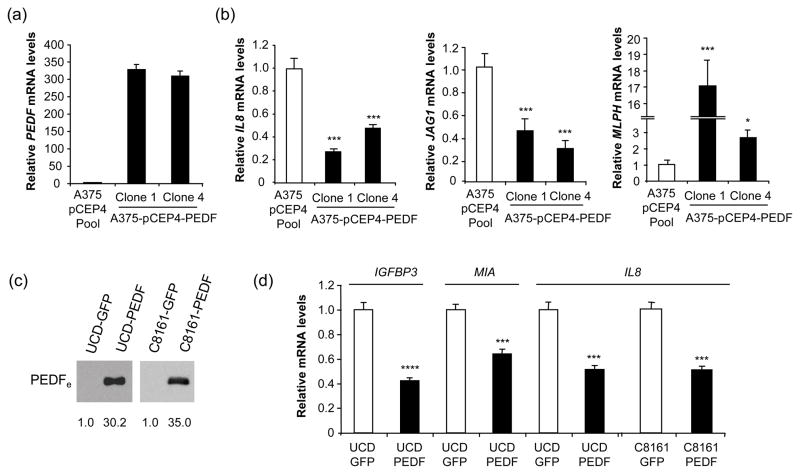
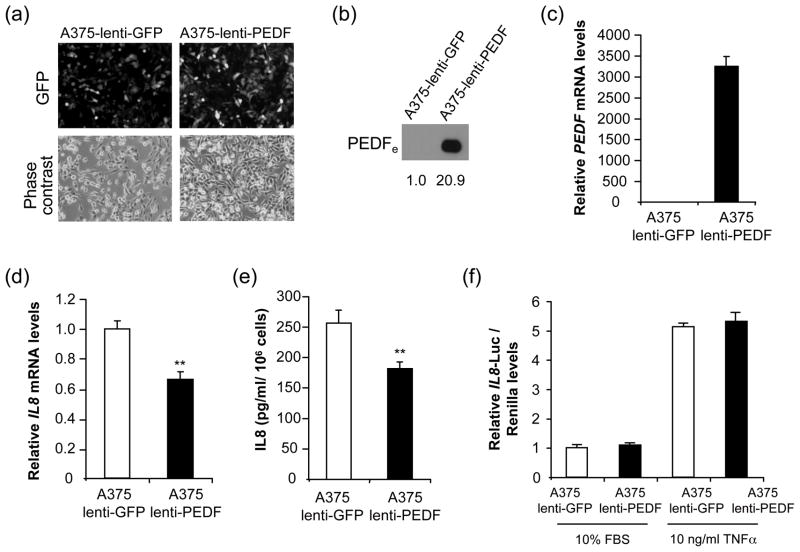
IL8 augments the angiogenic potential of melanoma cells and activates their proliferation and migration [36], and its levels directly correlate with melanoma metastatic potential [37]. Considering IL8 key role in melanoma progression [36], we sought to further analyze its regulation by PEDF. We confirmed IL8 downregulation in PEDF-over-expressing A375 clones and UCD-Mel-N and C8161 melanoma cell lines (Fig. 4). We also analyzed the effect of transient PEDF over-expression on IL8 levels. A375 cells were efficiently transduced (>95% infected cells) with a lentivirus encoding PEDF (A375-lenti-PEDF) or control lentivirus (A375-lenti-GFP) (Fig. 5a). PEDF expression was measured 72 h after transduction by Western blot of CM (Fig. 5b) and quantitative RT-PCR (Fig. 5c). IL8 mRNA (Fig. 5d) and protein (Fig. 5e) levels were significantly decreased by transient PEDF over-expression. Low levels of IL8 expression were maintained at least for two weeks after lentiviral transduction (data not shown). We also assessed if the decreased IL8 expression levels by PEDF was mediated by transcriptional regulation. We analyzed IL8 promoter activity using a reporter construct encompassing 2000 bp upstream of the IL8 transcription start site. In melanoma cells transiently or stably over-expressing PEDF and cultured in 10% serum or treated with TNFα to stimulate IL8 production [33], we found no change in the IL8 promoter activity (Fig. 5f and data not shown).
Fig. 4.
Confirmation of regulation of several target genes in other PEDF-over-expressing melanoma cell lines. (a) Quantitative RT-PCR analysis of PEDF mRNA levels in A375-pCEP4 Pool, A375-pCEP4-PEDF Clone 1 and A375-pCEP4-PEDF Clone 4 cells. PEDF mRNA levels are shown relative to A375-pCEP4 Pool after GADPH normalization. Bars represent average ± SD. (b) Quantitative RT-PCR analysis of IL8 (left panel), JAG1 (middle panel) and MLPH (right panel) mRNA levels in A375-pCEP4 Pool, A375-pCEP4-PEDF Clone 1 and A375-pCEP4-PEDF Clone 4 cells. For each gene mRNA levels are shown relative to A375-pCEP4 Pool after GAPDH normalization. Bars represent average ± SD. Statistical significance was determined by one-way ANOVA test using Tukey-Kramer post-test (*, p<0.05; ***, p<0.001). Results are representative of two independent experiments. (c) Western blot analysis of PEDFe protein in 48 h CM from control (UCD-GFP or C8161-GFP) and PEDF-over-expressing (UCD-PEDF or C8161-PEDF) cells. Numbers below blots show densitometry values normalized to each control cells expression. (d) Quantitative RT-PCR analysis of IGFBP3, MIA and IL8 mRNA levels in UCD-GFP, UCD-PEDF, C8161-GFP and C8161-PEDF cells. For each gene mRNA levels are shown relative to control cells after GAPDH normalization. Bars represent average ± SD. Statistical significance was determined by Student’s t-test (***, p<0.001; ****, p<0.0001). Results are representative of two independent experiments.
Fig. 5.
Validation of regulation of IL8 by transient transduction of A375 melanoma cells using lentivirus-PEDF. (a) Transduction efficiency of A375 melanoma cell line after infection with control (A375-lenti-GFP) or PEDF (A375-lenti-PEDF) lentivirus. Fluorescence images show more than 95% GFP-positive cells after 72 h of infection. Corresponding phase-contrast images are shown. (b) Western blot analysis of secreted extracellular PEDF (PEDFe) protein in 48 h CM from A375 melanoma cell line after 72 h of transduction with control lentivirus-GFP (A375-lenti-GFP) or lentivirus-PEDF (A375-lenti-PEDF). Numbers below blot show densitometry values relative to A375-lenti-GFP. (c) Quantitative RT-PCR analysis of PEDF mRNA levels in A375 melanoma cell line after 72 h of transduction with control lentivirus-GFP (A375-lenti-GFP) or lentivirus-PEDF (A375-lenti-PEDF). PEDF mRNA levels are shown relative to A375-lenti-GFP after normalization to GAPDH. Bars represent average ± SD. (d) Quantitative RT-PCR analysis of IL8 mRNA levels in A375 melanoma cell line after 72 h of transduction with control lentivirus-GFP (A375-lenti-GFP) or lentivirus-PEDF (A375-lenti-PEDF). IL8 mRNA levels are shown relative to A375-lenti-GFP after normalization to GAPDH. Bars represent average ± SD. Statistical significance was determined by Student’s t-test (*, p<0.01). (e) ELISA analysis of secreted IL8 protein levels in 48 h CM from A375 melanoma cell line after 72 h of transduction with control lentivirus-GFP (A375-lenti-GFP) or lentivirus-PEDF (A375-lenti- PEDF). Bars represent average ± SD. Statistical significance was determined by Student’s t-test (**, p<0.01). (f) IL8 transcriptional activity reporter assay in the A375-lenti-GFP and A375-lenti-PEDF melanoma cells mentioned above after 24 h treatment with 10% FBS or 10 ng/ml TNFα. Luciferase levels relative to A375-lenti-GFP in 10% FBS after normalization to renilla levels are shown. Bars represent average ± SD. Results are representative of two independent experiments.
Proteases or protease inhibitors from diverse families were predominantly downregulated by PEDF over-expression in A375 cells. This group included TIMPs (tissue inhibitors of metalloproteinases), ADAMs (a disintegrin and metalloproteinase domain), ADAMTSs (a disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif) or SERPINs (serine (or cysteine) proteinase inhibitor) (Fig. 2b and Table 1). Moreover, PEDF modified the expression of extracellular matrix proteins involved in migration as well as angiogenesis, such as collagen IV (COL4A2) [38] or fibronectin (FN1) [39], whose regulation was validated by quantitative RT-PCR (Fig. 3a). Even though the downregulation of COL4A2 by PEDF was less than two-fold in the microarray, a more pronounced decrease was found by quantitative RT-PCR. Other factors involved in adhesion and migration, including integrins, cadherins and protocadherins, were also downregulated in the presence of PEDF (Fig. 2b and Table 1). Furthermore, several molecules of the VEGF pathway, which could potentially contribute to control of melanoma proliferation and/or migration [22], such as fms-related tyrosine kinase 1 (FLT1) and neuropilin 2 (NRP2) (Fig. 2b and Table 1), were reduced by PEDF.
Genes implicated in melanoma progression modulated by PEDF in A375 human melanoma cells
Multiple genes previously involved in melanoma progression [40–42] were modulated by PEDF over-expression in A375 cells with the trend consistent with decreased aggressiveness (Fig. 2b and Table 1). This group included genes of the Notch pathway [43], such as notch homolog 2 (NOTCH2NL) and jagged 1 (JAG1); and of the Wnt pathway [44], including Dickkopf 1 and 3 (DKK1 and DKK3), frizzled 1 and 2 (FZD1, FZD2) and lymphoid enhancer-binding factor 1 (LEF1). The decreased expression of JAG1 in the presence of PEDF was confirmed by quantitative RT-PCR in transfection pools (Fig. 3a) and two independent clones (Fig. 4). Likewise, PEDF downregulated other genes previously described to participate in melanoma progression by enhancing proliferation or invasion, such as fibroblast growth factor 13 (FGF13) [41], insulin-like growth factor binding protein 3 (IGFBP3) [45], inhibin, beta A (INHBA) [42] and galectin 3 (LGALS3) [46]. We validated the regulation of FGF13, IGFBP3 and LGALS3 by quantitative RT-PCR (Figs. 3a and 4).
PEDF also altered the expression of transcription factors potentially relevant for melanocyte transformation and/or melanoma progression including T-box 2 (TBX2) [47], SOX2 and SOX5 [48], which were downregulated; and SOX9 and SOX11 [49], whose expression was increased. This regulation trend was consistent with reduced tumorigenicity and delayed progression. Moreover, nuclear factor of activated T-cells (NFATC2), involved in mediating PEDF’s intracellular signaling [50], also had its expression decreased by PEDF (Fig. 2b and Table 1).
Interestingly, a number of melanoma markers whose expression increases in the course of malignant progression were predominantly downregulated in PEDF over-expressing A375 cells, such as melanoma inhibitory activity (MIA) and several S100 calcium binding protein family members (Fig. 2b and Table 1); the decreased MIA and S100B expression was confirmed by quantitative RT-PCR (Figs. 3a and 4).
Finally, PEDF also diminished the expression of factors involved in the inhibition of apoptosis, such as Bcl2-related protein A1 (BCL2A1); or in cell proliferation, like insulin-like growth factor 1 receptor (IGF1R) and IGFBP 2, 5 and 7, among others; as well as a number of cytokines and genes involved in metabolism (Fig. 2b and Table 1).
Genes specific for melanocyte lineage functions modulated by PEDF in A375 human melanoma cells
Genes involved in melanocytic lineage-specific functions, such as melanogenesis, were modulated by PEDF over-expression in A375 cells [51]. Melanosomes are specialized membrane vesicles where melanin is synthesized, stored and eventually delivered to the cell membrane, to be transferred to adjacent keratinocytes [52]. Two regulators of melanosome trafficking, the small GTP-binding protein RAB27A and its effector melanophilin (MLPH) [53], were regulated by PEDF over-expression (Fig. 2b and Table 1) and this regulation was confirmed by quantitative RT-PCR in transfection pools (Fig. 3a) and two independent clones (Fig. 4).
Discussion
PEDF is an emerging agent for the targeted anti-cancer therapies. The combination of the potent and diverse effects on tumor cells, as well as its anti-angiogenic action; make it especially compelling, particularly for advanced melanoma, a highly aggressive cancer which responds poorly to the currently available treatments. Despite the growing interest in the therapeutic applications of PEDF, very few studies used global expression analysis to seek molecular mediators underlying its multiple biological actions [54–56]. In this study we describe genome-wide changes in the expression profile of A375 aggressive human melanoma cells caused by PEDF.
In general, PEDF decreased the expression of numerous genes involved in key tumor-specific functions critical for melanoma progression and whose products are likely to contribute to PEDF known anti-cancer properties.
Firstly, several factors that contribute to angiogenesis and chemotaxis were diminished due to PEDF expression. This is consistent with previous observations by us and others describing PEDF-dependent inhibition of angiogenic potential and motility of the melanoma cells [20, 35, 57]. Decreased expression of these genes weakened the ability of melanoma cells to induce neovascularization. The decrease of VEGF mRNA in PEDF-expressing cells was less than two-fold; hence, it was not included in our master list. However, VEGF protein levels were decreased to a larger extent than mRNA [20, 58, 59], suggesting additional post-transcriptional events. A recent expression profiling study in prostate adenocarcinoma has also shown downregulation of angiogenesis-related genes upon treatment with recombinant PEDF, which included fibroblast growth factor 3, neuropilin 1, brain-specific angiogenesis inhibitor 2 and endothelial PAS domain protein 1 [55]. In our study, neuropilin and fibroblast growth factor family members were also regulated by PEDF in A375 melanoma cells. IL8, whose expression is also diminished in the presence of PEDF, falls in the same category. IL8 is a multifunctional cytokine, which stimulates angiogenesis, proliferation and migration [36]. IL8 levels directly correlate with the metastatic potential and aggressiveness of melanoma [37, 60]. IL8 protein and mRNA were significantly decreased due to the transient or stable PEDF over-expression in A375 melanoma cells. However, IL8 regulation by PEDF in A375 melanoma cells seemed to be mainly post-transcriptional as we were unable to register a decrease in the activity of IL8 reporter constructs upon PEDF over-expression, despite the presence of multiple binding sites for the transcription factors potentially regulated by PEDF in A375 melanoma cells and other cell types, and involved in IL8 regulation, like NFATC2 [50, 61]. The promoter region used in our study has been previously shown to respond to a number of IL8 stimulating factors including hypoxia [62], acidosis [63], TNFα and IL1 [64].
Altered expression of adhesion and extracellular matrix–related molecules by PEDF may, in addition to the chemokine expression, also contribute to PEDF-dependent inhibition of melanoma migration and invasion. In our study, PEDF decreased collagen IV (COL4A2) expression, an essential component of the basement membrane, which is important for angiogenesis, adhesion and migration [38, 65]. PEDF also enhanced expression of fibronectin, an extracellular matrix protein involved in adhesion, proliferation and migration through its binding to integrins [39]. Increased fibronectin could enhance cell adhesion which may impair invasiveness. A recent report shows that silencing of fibronectin in thyroid carcinoma cells enhances tumor growth and metastasis [66].
PEDF over-expression in A375 cells modified the expression of a number of genes attributed in other studies to the malignant progression of human melanoma, including activin A, insulin-like growth factor binding protein 3 and galectin 3, several members of the SOX transcription factor family, and the genes that belong to the Notch (NOTCH2NL, JAG1) [43] or Wnt (DKK1, FZD1, LEF1) [44, 67] pathways. Importantly, PEDF downregulated several melanoma markers currently utilized in clinico-pathological diagnosis of melanoma, such as S100β [68]; or used as prognostic factors for relapse and metastasis, like S100β or MIA [69]. Collectively, these changes in the expression pattern of the genes related to melanoma progression may reflect the switch to a less aggressive phenotype by PEDF-expressing melanoma cells and supports the therapeutic potential of PEDF in this type of cancer.
Interestingly, PEDF over-expression in A375 cells altered the expression of genes involved in functions specific for melanocyte lineage, upregulating MLPH and downregulating RAB27A. These genes are involved in melanosome trafficking and therefore play a key role in melanin transfer [52]. PEDF has been detected in immature melanosomes of melanoma cells [70], and prior reports have proposed its participation in pigment production through induction of tyrosinase [71] and promotion of melanosome maturation in retinal pigment epithelium cells [72]. We have previously described that PEDF is expressed at high levels in melanocytes, and restricts their proliferative and migratory potential [26]. Modulation of MLPH and RAB27A expression by PEDF in A375 cells suggests that it could also mediate melanocyte-specific processes, such melanin distribution in the skin. In keeping with this new role, we found that PEDF increases the expression of SOX9, which was recently implicated in control of melanocyte differentiation and pigmentation, acting upstream of microphthalmia-associated transcription factor (MITF) [73].
In summary, our study points to novel molecular targets and signaling pathways that may potentially contribute to determine PEDF’s ability to restrict the aggressiveness of A375 and other human melanoma cells. We also stumbled upon unanticipated PEDF targets involved in melanocytic lineage-specific functions. Our results presented set the stage for further in-depth studies of the molecular mechanisms underlying PEDF effects in melanoma, which would be essential for the clinical translation of this factor.
Supplementary Material
Acknowledgments
We acknowledge with gratitude all researchers that contributed with cell lines and/or reagents: F Vidal-Vanaclocha, A Bernad and J Raingeaud. We also thank P Fernández for her technical assistance in lentivirus production. Supported by grants Ministerio de Educacion y Ciencia grant SAF2007-62292 (BJ), Comunidad de Madrid SAL-0311-2006 (BJ). JLO has been supported by a Ministerio de Educacion y Ciencia fellowship and a SAF2007-62292 contract.
References
- 1.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Bouck N, Stellmach V, Hsu SC. How tumors become angiogenic. Adv Cancer Res. 1996;69:135–174. doi: 10.1016/s0065-230x(08)60862-3. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 5.Jimenez B, Volpert OV. Mechanistic insights on the inhibition of tumor angiogenesis. Journal of molecular medicine (Berlin, Germany) 2001;78:663–672. doi: 10.1007/s001090000178. [DOI] [PubMed] [Google Scholar]
- 6.Brekken RA, Overholser JP, Stastny VA, Waltenberger J, Minna JD, Thorpe PE. Selective inhibition of vascular endothelial growth factor (VEGF) receptor 2 (KDR/Flk-1) activity by a monoclonal anti-VEGF antibody blocks tumor growth in mice. Cancer research. 2000;60:5117–5124. [PubMed] [Google Scholar]
- 7.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Y. Angiogenesis in malignancy. Semin Cancer Biol. 2009;19:277–278. doi: 10.1016/j.semcancer.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Eikesdal HP, Kalluri R. Drug resistance associated with antiangiogenesis therapy. Semin Cancer Biol. 2009;19:310–317. doi: 10.1016/j.semcancer.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrara N, Kerbel RS. Angiogenesis as a therapeutic target. Nature. 2005;438:967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 11.Folkman J. Endogenous angiogenesis inhibitors. APMIS. 2004;112:496–507. doi: 10.1111/j.1600-0463.2004.apm11207-0809.x. [DOI] [PubMed] [Google Scholar]
- 12.Nyberg P, Xie L, Kalluri R. Endogenous inhibitors of angiogenesis. Cancer research. 2005;65:3967–3979. doi: 10.1158/0008-5472.CAN-04-2427. [DOI] [PubMed] [Google Scholar]
- 13.Reiher FK, Volpert OV, Jimenez B, Crawford SE, Dinney CP, Henkin J, et al. Inhibition of tumor growth by systemic treatment with thrombospondin-1 peptide mimetics. International journal of cancer. 2002;98:682–689. doi: 10.1002/ijc.10247. [DOI] [PubMed] [Google Scholar]
- 14.Ribatti D. Endogenous inhibitors of angiogenesis: a historical review. Leuk Res. 2009;33:638–644. doi: 10.1016/j.leukres.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Barnstable CJ, Tombran-Tink J. Neuroprotective and antiangiogenic actions of PEDF in the eye: molecular targets and therapeutic potential. Prog Retin Eye Res. 2004;23:561–577. doi: 10.1016/j.preteyeres.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Becerra SP, Sagasti A, Spinella P, Notario V. Pigment epithelium-derived factor behaves like a noninhibitory serpin. Neurotrophic activity does not require the serpin reactive loop. The Journal of biological chemistry. 1995;270:25992–25999. doi: 10.1074/jbc.270.43.25992. [DOI] [PubMed] [Google Scholar]
- 17.Bouck N. PEDF: anti-angiogenic guardian of ocular function. Trends in molecular medicine. 2002;8:330–334. doi: 10.1016/s1471-4914(02)02362-6. [DOI] [PubMed] [Google Scholar]
- 18.Doll JA, Stellmach VM, Bouck NP, Bergh AR, Lee C, Abramson LP, et al. Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nature medicine. 2003;9:774–780. doi: 10.1038/nm870. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Garcia NI, Volpert OV, Jimenez B. Pigment epithelium-derived factor as a multifunctional antitumor factor. Journal of molecular medicine (Berlin, Germany) 2007;85:15–22. doi: 10.1007/s00109-006-0111-z. [DOI] [PubMed] [Google Scholar]
- 20.Garcia M, Fernandez-Garcia NI, Rivas V, Carretero M, Escamez MJ, Gonzalez-Martin A, et al. Inhibition of xenografted human melanoma growth and prevention of metastasis development by dual antiangiogenic/antitumor activities of pigment epithelium-derived factor. Cancer research. 2004;64:5632–5642. doi: 10.1158/0008-5472.CAN-04-0230. [DOI] [PubMed] [Google Scholar]
- 21.Mahabeleshwar GH, Byzova TV. Angiogenesis in melanoma. Semin Oncol. 2007;34:555–565. doi: 10.1053/j.seminoncol.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Streit M, Detmar M. Angiogenesis, lymphangiogenesis, and melanoma metastasis. Oncogene. 2003;22:3172–3179. doi: 10.1038/sj.onc.1206457. [DOI] [PubMed] [Google Scholar]
- 23.Filleur S, Nelius T, de Riese W, Kennedy RC. Characterization of PEDF: a multi-functional serpin family protein. J Cell Biochem. 2009;106:769–775. doi: 10.1002/jcb.22072. [DOI] [PubMed] [Google Scholar]
- 24.Filleur S, Volz K, Nelius T, Mirochnik Y, Huang H, Zaichuk TA, et al. Two functional epitopes of pigment epithelial-derived factor block angiogenesis and induce differentiation in prostate cancer. Cancer research. 2005;65:5144–5152. doi: 10.1158/0008-5472.CAN-04-3744. [DOI] [PubMed] [Google Scholar]
- 25.Mirochnik Y, Aurora A, Schulze-Hoepfner FT, Deabes A, Shifrin V, Beckmann R, et al. Short pigment epithelial-derived factor-derived peptide inhibits angiogenesis and tumor growth. Clin Cancer Res. 2009;15:1655–1663. doi: 10.1158/1078-0432.CCR-08-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orgaz JL, Ladhani O, Hoek KS, Fernandez-Barral A, Mihic D, Aguilera O, et al. Loss of pigment epithelium-derived factor enables migration, invasion and metastatic spread of human melanoma. Oncogene. 2009;28:4147–4161. doi: 10.1038/onc.2009.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 28.Giard DJ, Aaronson SA, Todaro GJ, Arnstein P, Kersey JH, Dosik H, et al. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. Journal of the National Cancer Institute. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 29.Kozlowski JM, Hart IR, Fidler IJ, Hanna N. A human melanoma line heterogeneous with respect to metastatic capacityin athymic nude mice. Journal of the National Cancer Institute. 1984;72:913–917. [PubMed] [Google Scholar]
- 30.Reed JA, Bales E, Xu W, Okan NA, Bandyopadhyay D, Medrano EE. Cytoplasmic localization of the oncogenic protein Ski in human cutaneous melanomas in vivo: functional implications for transforming growth factor beta signaling. Cancer research. 2001;61:8074–8078. [PubMed] [Google Scholar]
- 31.Welch DR, Bisi JE, Miller BE, Conaway D, Seftor EA, Yohem KH, et al. Characterization of a highly invasive and spontaneously metastatic human malignant melanoma cell line. International journal of cancer. 1991;47:227–237. doi: 10.1002/ijc.2910470211. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez-Garcia NI, Palmer HG, Garcia M, Gonzalez-Martin A, del Rio M, Barettino D, et al. 1alpha,25-Dihydroxyvitamin D3 regulates the expression of Id1 and Id2 genes and the angiogenic phenotype of human colon carcinoma cells. Oncogene. 2005;24:6533–6544. doi: 10.1038/sj.onc.1208801. [DOI] [PubMed] [Google Scholar]
- 33.Raingeaud J, Pierre J. Interleukin-4 downregulates TNFalpha-induced IL-8 production in keratinocytes. FEBS Lett. 2005;579:3953–3959. doi: 10.1016/j.febslet.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 34.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. Journal of the American Statistical Association. 2004;99:909–917. [Google Scholar]
- 35.Abe R, Shimizu T, Yamagishi S, Shibaki A, Amano S, Inagaki Y, et al. Overexpression of pigment epithelium-derived factor decreases angiogenesis and inhibits the growth of human malignant melanoma cells in vivo. The American journal of pathology. 2004;164:1225–1232. doi: 10.1016/s0002-9440(10)63210-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melnikova VO, Bar-Eli M. Bioimmunotherapy for melanoma using fully human antibodies targeting MCAM/MUC18 and IL-8. Pigment cell research/sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2006;19:395–405. doi: 10.1111/j.1600-0749.2006.00331.x. [DOI] [PubMed] [Google Scholar]
- 37.Singh RK, Gutman M, Radinsky R, Bucana CD, Fidler IJ. Expression of interleukin 8 correlates with the metastatic potential of human melanoma cells in nude mice. Cancer research. 1994;54:3242–3247. [PubMed] [Google Scholar]
- 38.Pasco S, Brassart B, Ramont L, Maquart FX, Monboisse JC. Control of melanoma cell invasion by type IV collagen. Cancer Detect Prev. 2005;29:260–266. doi: 10.1016/j.cdp.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 39.Danen EH, Yamada KM. Fibronectin, integrins, and growth control. J Cell Physiol. 2001;189:1–13. doi: 10.1002/jcp.1137. [DOI] [PubMed] [Google Scholar]
- 40.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer research. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 42.Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment cell research/sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 43.Pinnix CC, Herlyn M. The many faces of Notch signaling in skin-derived cells. Pigment cell research/sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2007;20:458–465. doi: 10.1111/j.1600-0749.2007.00410.x. [DOI] [PubMed] [Google Scholar]
- 44.Weeraratna AT. A Wnt-er wonderland--the complexity of Wnt signaling in melanoma. Cancer Metastasis Rev. 2005;24:237–250. doi: 10.1007/s10555-005-1574-z. [DOI] [PubMed] [Google Scholar]
- 45.Xi Y, Nakajima G, Hamil T, Fodstad O, Riker A, Ju J. Association of insulin-like growth factor binding protein-3 expression with melanoma progression. Molecular cancer therapeutics. 2006;5:3078–3084. doi: 10.1158/1535-7163.MCT-06-0424. [DOI] [PubMed] [Google Scholar]
- 46.Mourad-Zeidan AA, Melnikova VO, Wang H, Raz A, Bar-Eli M. Expression profiling of Galectin-3-depleted melanoma cells reveals its major role in melanoma cell plasticity and vasculogenic mimicry. The American journal of pathology. 2008;173:1839–1852. doi: 10.2353/ajpath.2008.080380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vance KW, Carreira S, Brosch G, Goding CR. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer research. 2005;65:2260–2268. doi: 10.1158/0008-5472.CAN-04-3045. [DOI] [PubMed] [Google Scholar]
- 48.Laga AC, Lai C, Zhan Q, Huang SJ, Velazquez EF, Yang Q, et al. Expression of The Embryonic Stem Cell Transcription Factor SOX2 in Human Skin: Relevance to Melanocyte and Merkel Cell Biology. The American journal of pathology. 2010;176:903–913. doi: 10.2353/ajpath.2010.090495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Passeron T, Valencia JC, Namiki T, Vieira WD, Passeron H, Miyamura Y, et al. Upregulation of SOX9 inhibits the growth of human and mouse melanomas and restores their sensitivity to retinoic acid. J Clin Invest. 2009;119:954–963. doi: 10.1172/JCI34015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaichuk TA, Shroff EH, Emmanuel R, Filleur S, Nelius T, Volpert OV. Nuclear factor of activated T cells balances angiogenesis activation and inhibition. The Journal of experimental medicine. 2004;199:1513–1522. doi: 10.1084/jem.20040474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 52.Barral DC, Seabra MC. The melanosome as a model to study organelle motility in mammals. Pigment cell research/sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2004;17:111–118. doi: 10.1111/j.1600-0749.2004.00138.x. [DOI] [PubMed] [Google Scholar]
- 53.Fukuda M. Versatile role of Rab27 in membrane trafficking: focus on the Rab27 effector families. J Biochem. 2005;137:9–16. doi: 10.1093/jb/mvi002. [DOI] [PubMed] [Google Scholar]
- 54.Kozulin P, Natoli R, O’Brien KM, Madigan MC, Provis JM. Differential expression of anti-angiogenic factors and guidance genes in the developing macula. Mol Vis. 2009;15:45–59. [PMC free article] [PubMed] [Google Scholar]
- 55.Liu W, Wu Z, Guan M, Lu Y. cDNA microarray analysis of pigment epithelium-derived factor-regulated gene expression profile in prostate carcinoma cells. Int J Urol. 2009;16:323–328. doi: 10.1111/j.1442-2042.2008.02199.x. [DOI] [PubMed] [Google Scholar]
- 56.Yabe T, Herbert JT, Takanohashi A, Schwartz JP. Treatment of cerebellar granule cell neurons with the neurotrophic factor pigment epithelium-derived factor in vitro enhances expression of other neurotrophic factors as well as cytokines and chemokines. J Neurosci Res. 2004;77:642–652. doi: 10.1002/jnr.20196. [DOI] [PubMed] [Google Scholar]
- 57.Yang LP, Cheng P, Peng XC, Shi HS, He WH, Cui FY, et al. Anti-tumor effect of adenovirus-mediated gene transfer of pigment epithelium-derived factor on mouse B16-F10 melanoma. J Exp Clin Cancer Res. 2009;28:75. doi: 10.1186/1756-9966-28-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guan M, Pang CP, Yam HF, Cheung KF, Liu WW, Lu Y. Inhibition of glioma invasion by overexpression of pigment epithelium-derived factor. Cancer Gene Ther. 2004;11:325–332. doi: 10.1038/sj.cgt.7700675. [DOI] [PubMed] [Google Scholar]
- 59.Takenaka K, Yamagishi S, Jinnouchi Y, Nakamura K, Matsui T, Imaizumi T. Pigment epithelium-derived factor (PEDF)-induced apoptosis and inhibition of vascular endothelial growth factor (VEGF) expression in MG63 human osteosarcoma cells. Life Sci. 2005;77:3231–3241. doi: 10.1016/j.lfs.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 60.Nurnberg W, Tobias D, Otto F, Henz BM, Schadendorf D. Expression of interleukin-8 detected by in situ hybridization correlates with worse prognosis in primary cutaneous melanoma. J Pathol. 1999;189:546–551. doi: 10.1002/(SICI)1096-9896(199912)189:4<546::AID-PATH487>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 61.Maldonado-Perez D, Brown P, Morgan K, Millar RP, Thompson EA, Jabbour HN. Prokineticin 1 modulates IL-8 expression via the calcineurin/NFAT signaling pathway. Biochim Biophys Acta. 2009;1793:1315–1324. doi: 10.1016/j.bbamcr.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu L, Xie K, Mukaida N, Matsushima K, Fidler IJ. Hypoxia-induced elevation in interleukin-8 expression by human ovarian carcinoma cells. Cancer research. 1999;59:5822–5829. [PubMed] [Google Scholar]
- 63.Xu L, Fidler IJ. Acidic pH-induced elevation in interleukin 8 expression by human ovarian carcinoma cells. Cancer research. 2000;60:4610–4616. [PubMed] [Google Scholar]
- 64.Mukaida N, Mahe Y, Matsushima K. Cooperative interaction of nuclear factor-kappa B- and cis-regulatory enhancer binding protein-like factor binding elements in activating the interleukin-8 gene by pro-inflammatory cytokines. The Journal of biological chemistry. 1990;265:21128–21133. [PubMed] [Google Scholar]
- 65.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 66.Liu W, Cheng S, Asa SL, Ezzat S. The melanoma-associated antigen A3 mediates fibronectin-controlled cancer progression and metastasis. Cancer research. 2008;68:8104–8112. doi: 10.1158/0008-5472.CAN-08-2132. [DOI] [PubMed] [Google Scholar]
- 67.O’Connell MP, Weeraratna AT. Hear the Wnt Ror: how melanoma cells adjust to changes in Wnt. Pigment Cell Melanoma Res. 2009 doi: 10.1111/j.1755-148X.2009.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Utikal J, Schadendorf D, Ugurel S. Serologic and immunohistochemical prognostic biomarkers of cutaneous malignancies. Arch Dermatol Res. 2007;298:469–477. doi: 10.1007/s00403-006-0726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deichmann M, Benner A, Bock M, Jackel A, Uhl K, Waldmann V, et al. S100-Beta, melanoma-inhibiting activity, and lactate dehydrogenase discriminate progressive from nonprogressive American Joint Committee on Cancer stage IV melanoma. J Clin Oncol. 1999;17:1891–1896. doi: 10.1200/JCO.1999.17.6.1891. [DOI] [PubMed] [Google Scholar]
- 70.Chi A, Valencia JC, Hu ZZ, Watabe H, Yamaguchi H, Mangini NJ, et al. Proteomic and bioinformatic characterization of the biogenesis and function of melanosomes. J Proteome Res. 2006;5:3135–3144. doi: 10.1021/pr060363j. [DOI] [PubMed] [Google Scholar]
- 71.Abul-Hassan K, Walmsley R, Tombran-Tink J, Boulton M. Regulation of tyrosinase expression and activity in cultured human retinal pigment epithelial cells. Pigment cell research/sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2000;13:436–441. doi: 10.1034/j.1600-0749.2000.130605.x. [DOI] [PubMed] [Google Scholar]
- 72.Malchiodi-Albedi F, Feher J, Caiazza S, Formisano G, Perilli R, Falchi M, et al. PEDF (pigment epithelium-derived factor) promotes increase and maturation of pigment granules in pigment epithelial cells in neonatal albino rat retinal cultures. Int J Dev Neurosci. 1998;16:423–432. doi: 10.1016/s0736-5748(98)00014-8. [DOI] [PubMed] [Google Scholar]
- 73.Passeron T, Valencia JC, Bertolotto C, Hoashi T, Le Pape E, Takahashi K, et al. SOX9 is a key player in ultraviolet B-induced melanocyte differentiation and pigmentation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13984–13989. doi: 10.1073/pnas.0705117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.