Abstract
Purpose
Identify novel tumor suppressor genes in melanoma utilizing an integrative genomic approach
Methods
Data from: 1) prior reports of DNA loss and gain in malignant melanoma accompanied by CGH high-definition array data of the entire human genome, 2) Microarray expression data from melanoma derived cell lines identifying genes with significantly increased expression due to methylation using a pharmacologic demethylating strategy, and 3) publicly available RNA expression microarray data of primary tumors and benign nevi was integrated utilizing statistical tools in order to define a population of candidate tumor suppressor genes.
Results
27 genes were identified in areas of deletion that demonstrated diminished expression in primary melanomas relative to benign nevi and were significantly increased in expression by ‘5-Aza treatment. Seven genes of these genes demonstrated methylation and deletion in a validation cohort of 14 separate primary tumors. These were: CHRDL1, SFRP1, TMEM47, LPL, RARRES1, PLCXD1, and KOX15. All of these genes demonstrated growth suppressive properties with transfection into melanoma derived cell lines.
Conclusions
7 putative tumor suppressor genes in malignant melanoma were identified utilizing a novel integrative technique.
Keywords: DNA hypermethylation, epigenetics, malignant melanoma, tumor suppressor
Introduction
Cytogenetic and epigenetic changes are the primary mechanisms by which tumor suppressor genes are inactivated. DNA methylation is one form of epigenetic change and involves the covalent addition of a methyl group to cytosine residues in CpG dinucleotides by DNA methyltransferases. CpG-rich sequences (CpG islands) are infrequent in the genome. Examination of familial cancer genes has revealed that tumor-specific hypermethylation may act as an inactivating event by preferentially targeting the wild-type allele of tumor suppressors, whereas the promoter region of the mutant allele is not affected by methylation. Inhibition of de novo methylation enzymes has been shown to reduce tumor formation in several settings. Epigenetic inactivation of individual tumor suppressors by DNA methylation has also been shown in malignant melanoma. A high incidence of methylation in melanoma tissue samples has been reported for RARB (70%), RASSF1A (55%), PYCARD (50%), MGMT (34%), DAPK (19%), and APC (19%), which may play tumor suppressor roles in several tumor settings[1].
Exposure to ultraviolet radiation is the environmental factor most strongly associated with development of melanoma. Many genetic and epigenetic changes associated with melanomas have been described [2-4]. Yet, the molecular mechanisms of melanoma carcinogenesis are still undergoing intensive investigation. Neoplastic cells undergo a variety of genetic alterations, from point mutations to chromosomal aberrations, affecting the function or expression of both oncogenes and tumor-suppressor genes. Deletion can result in silencing via uniallelic or biallelic loss. Advances in techniques over the past few years, including: array-based comparative genomic hybridization (aCGH), and array-based single nucleotide polymorphism (aSNP) have allowed high-throughput, highly detailed studies of chromosomal loss or gain.
New high-throughput screens of cancer genes are being developed at a rapid pace making the need for efficient approaches for integration of large datasets that use diverse technologies to describe genetic alterations in human cancers. [5]. Examples of such genome-scale datasets include: array CGH (DNA loss or gain), RNA expression microarray (tissues or cell lines), small molecule cell line screens, and various proteomic approaches. Tumors may be susceptible to targeted therapies once their essential molecular alterations have been found. Integrative approaches to these genome-scale data sets allow multiple pieces of salient information to be combined in a manner that may yield novel and powerful new insights into cancer biology [6]. One prominent example of the utility of these integrative genetic approaches led to finding the oncogene MITF in melanoma via application of two genome-scale datasets (array SNP and expression microaray). This gene may represent a new class of “lineage addiction oncogenes”-- a fundamental tumor survival mechanism with important therapeutic implications[7] . Other examples of integrative genomic approaches have also improved the genetic understanding of other cancers [8-11]
Tumor suppressor inactivation is associated with loss or inactivation of both genomic copies of DNA. This has been associated with epigenetic[1, 2] and cytogenetic mechanisms in malignant melanoma. Identifying genes that demonstrate inactivation by multiple mechanisms may serve to more intelligently enrich a search for putative tumor suppressors in melanoma. We hypothesize that genes whose expression is decreased in melanoma relative to benign nevi and have evidence of methylation and deletion are potential tumor suppressor genes in this disease.
We report a study utilizing high throughput genomic and informatic techniques to identify a set of putative tumor suppressor genes whose expression is regulated by methylation and whose expression is decreased in primary melanomas utilizing a set of previously published microarray data[12]. This information was combined with aCGH data identifying areas of known deletion in melanoma. Genes found to be regulated by methylation and found in areas of known deletion were analyzed in melanoma samples collected from an independent cohort to determine the incidence and coincidence of hypermethylation and deletion. Genes validated in this manner were transfected into melanoma cell lines to determine if growth suppression was induced.
Methods
Public datasets
The public databases used in this study were the University of California Santa Cruz (UCSC) Human Genome reference sequence and the annotation database from the May 2004 freeze (hg17). Array Comparative Genome Hybridization (aCGH) data for 178 malignant melanomas and tumor derived cell lines were obtained from progenetix.com[13]. Progenetix collects information about the genomic copy number profiles of cancer cases. It consists mainly of a compilation of published data from chromosomal and array/matrix Comparative Genomic Hybridization (aCGH) experiments. Gene Expression Omnibus (GEO) repository was utilized to locate microarray expression data for 45 melanoma and 18 benign melanocytic nevi [12]
5Aza-dC Treatment of Cells
These in vitro techniques employ treatment of cultured cells with 5-aza-deoxycytidine (a cytosine analog which cannot be methylated) and subsequent expression array analysis with validation of tumor suppressor gene targets. We treated melanoma cell lines with 5Aza-dC as described previously[14]. Briefly, cells were split to low density (1 × 106 cells/T-75 flask) 24 hours before treatment. Stock solutions of 5Aza-dC (Sigma, St. Louis, MO) were dissolved in DMSO (Sigma). Cells were treated with 5 μM 5-Aza-deoxycytidine for 2 days. Baseline expression was established by mock-treated cells with the same volume of DMSO.
Integrative analysis
For target discovery we employed three data sets. aCGH dataset from progenetix.com was utilized to identify detailed loci of DNA/chromosomal loss in melanoma (>10% incidence). Gene RNA expression microarray analysis of RNA extracted from 4 5′-Aza treated melanoma derived cell lines was conducted on the Affymetrix U133A and U133B platforms (33,000 genes). Genes, which were significantly upregulated by Aza treatment utilizing the techniques described previously, were identified. Publicly available data sets, published through GEO, which contained U133A data regarding gene expression in 45 melanoma tumors relative to 18 benign nevi [12]. Microarrays were studied with dChip and invariant-set normalized. Median tumor expression and median normal expression were calculated. P values were determined by STATA 9.0 utilizing Mann-Whitney U test (StataCorp LP, College Station, Texas). Genes which demonstrated significantly decreased median expression in melanomas as compared to melanocytic nevi were identified (p <.05). The intersection of the sets of genes found in areas of deletions, those upregulated by Aza treatment, and those with diminished expression in melanoma relative to nevi was identified. The list was ranked by equal weight of the ranking of the relative median expression difference and fold upregulation by Aza treatment. These sources of information were combined by using a rank product. These rankings were combined to rank all targets with a significance threshold of α = 0.005. Subsequent random permutation of our rank list resulted in 47 genes deemed significant. Validation of targets was based upon identification of targets with known CpG Island, and subsequent bisulfite sequencing, followed by qPCR for precise validation of gene amplification or deletion, and/or sequencing. (Supplemental Figure 1)
Oligonucleotide microarray analysis
Total cellular RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instruction. We carried out oligonucleotide microarray analysis using the GeneChip U133 A and U133B Affymetrix expression microarray that assays 33,000 genes (Affymetrix, Santa Clara, CA). Samples were converted to labeled, fragmented, cRNA per the Affymetrix protocol for use on the expression microarray. Signal intensity and statistical significance was established for each transcript using dChip version 2006. Default settings for dChip were used including the PM/MM difference model; invariant set normalization, check single/probe/array outlier algorithm.
DNA sample source and extraction
Tumor samples and lymphocyte DNA were obtained from 14 melanoma patients who had undergone enrolment in institutional review board approved protocols after obtaining informed consent. This population consisted of 13 men and one woman. (Table 1) Resected primary or metastatic melanoma lesions were collected in the operating room and immediately snap frozen with liquid nitrogen. Tumor specimens were microdissected on a cryostat so that the samples used for mutational analysis contained greater than 70% neoplastic cells. Samples were centrifuged and digested in a solution of detergent (sodium dodecylsulfate) and proteinase K, for removal of proteins bound to the DNA. Samples were first purified and desalted with phenol/chloroform extraction. Digested sample was subjected twice to ethanol precipitation, and subsequently resuspended in 500 μL of LoTE (EDTA 2.5 mmol/L and Tris-HCl 10 mmol/L) and stored at −80°C.
Table 1.
Tumor and patient information for samples utilized.
| Sample | Age | Sex | Stage at Presentation |
T | N | M | Disease State | Disease Free Interval (Months) |
Primary | Sample Derivation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 50 | M | 4 | 3b | 2b | 1 | NED | - | Scalp | Primary |
| 2 | 82 | M | 3c | 4a | 2b | 0 | DOD | 1 | Shoulder | Lymph Node |
| 3 | 81 | M | 2a | 3a | 0 | 0 | NED | - | Scalp | Primary |
| 4 | 77 | M | 3a | X | 2b | 0 | DOD | 6 | Unknown | Lymph Node |
| 5 | 87 | M | 2b | 4 | 0 | 0 | NED | - | Cheek | Recurrence |
| 6 | 39 | M | 1b | 2a | 0 | 0 | AWD | 10 | Neck | Primary |
| 7 | 84 | M | 3c | 4 | 3 | 0 | DOC | - | Shoulder | Primary |
| 8 | 83 | M | 3b | 2a | 2b | 0 | DOD | 22 | Ear | Recurrence |
| 9 | 61 | F | 1a | 1 | 0 | 0 | NED | 45 | Mucosal | Primary |
| 10 | 42 | M | 3a | X | 2a | 0 | DOD | 18 | Unknown | Recurrence |
| 11 | 73 | M | 4 | X | X | 1 | AWD | 10 | Unknown | Lymph Node |
| 12 | 57 | M | 3a | 1a | 1a | 0 | NED | - | Cheek | Primary |
| 13 | 76 | M | 4 | X | 1a | 1 | NED | - | Unknown | Lymph Node |
| 14 | 92 | M | 4 | X | X | 1 | NED | - | Unknown | Lymph Node |
NED=No Evidence of Disease AWD=Alive with Disease DOC=Dead of other causes DOD=Dead of Disease
Bisulfite Treatment
DNA derived from tumor specimen was subjected to bisulfite treatment, as described previously[14]. In short, 2 μg of genomic DNA was denatured in 0.2 M NaOH for 30 minutes at 50°C. This denatured DNA was then diluted into 500 μL of a solution of 10 mmol/L hydroquinone and 3 M sodium bisulfite. This was incubated for 3 hours at 70°C. After the DNA sample was purified with a sepharose column (Wizard DNA Clean-Up System; Promega, Madison, WI). Eluted DNA was treated with 0.3 M of NaOH for 10 minutes at room temperature, and precipitated with ethanol. This bisulfite-modified DNA was subsequently resuspended in 120 μL of LoTE (EDTA 2.5 mmol/L and Tris-HCl 10 mmol/L) and stored at −80°C.
Bisulfite Sequencing
Bisulfite sequence analysis was performed to check the methylation status in primary tumors for putative tumor suppressors identified by integrative methods. Bisulfite-treated DNA was amplified using primers designed by MethPrimer to span areas of CpG islands in the promoter or first intron [15].Primer sequences were designed to not have CG dinucleotides. Detailed primer sequences and PCR conditions are available upon request. The PCR products were gel-purified using the QIAquick Gel Extraction Kit (Qiagen), according to the manufacturer’s instructions. Each amplified DNA sample was applied with nested primers to the Applied Biosystems 3700 DNA analyzer using BD terminator dye (Applied Biosystems, Foster City, CA).
qPCR
Total DNA was measured and adjusted to the same amount for each tissue sample. The DNA was used as the templates for quantitative real-time PCR with primers designed to specifically measure the DNA copy number of each candidate gene. Three genes (LRTM4, Pelo, Bid) were identified which were not located in areas of loss or gain. These were quantitated and relative amounts averaged to ensure accurate relative quantitation of copy number in qPCR. Detailed PCR conditions and primer sequences are available upon request.
Transfection
Full-length ORF cDNAs in expression vectors of zinc finger protein 22 (KOX15), lipoprotein lipase (LPL), chordin-like 1 (CHRDL1), secreted frizzled related protein 1 (SFRP1), transmembrane protein 47 (TMEM47), Phosphatidylinositol-specific phospholipase C, X domain containing1 (PLCXD1), and Retinoic acid receptor responder 1 (RARRES1) were obtained for transient transfections from Invitrogen (Carlsbad, CA). Cell lines were plated at 1 × 105/well using 96-well plates and simultaneously transfected with either empty vector (created by restriction digest removal of ORF and blunt end ligation) or gene of interest using the FuGene 6 Transfection Reagent (Roche, Basel, Switzerland) according to the manufacturer’s protocol.
Cell Count
Calcein florescence was measured by the Spectramax M2e 96-well fluorescence plate reader Molecular Devices (Sunnyvale, California). Live cells are distinguished by the presence of ubiquitous intracellular esterase activity, determined by the enzymatic conversion of the virtually nonfluorescent cell-permeable calcein AM to the intensely fluorescent calcein. The polyanionic calcein dye is well retained within live cells, producing an intense uniform green fluorescence (excitation/emission ~495 nm/515 nm).
Statistical analysis
All statistical computations were made with STATA SE version 9.0 (STATACORP, College Station, TX).
Results
Integrative approaches identifying potential tumor suppressors in melanoma
In order to identify novel tumor suppressor genes in malignant melanoma, we hypothesized that those genes that were differentially expressed in melanoma relative to benign melanocytic nevi were candidate tumor suppressor genes. Furthermore, we postulated genes that had a higher propensity for inactivation were more likely to be tumor suppressors. While there are a variety of mechanisms of gene inactivation, we focused on deletion and methylation as a basis for our investigation.
We chose to adapt prior methods of epigenetic screening using 5-aza/TSA treatment that have been found to be successful in defining candidate tumor suppressor genes. Four malignant melanoma derived cell lines were treated with 5μM 5-aza deoxycytidine for three days prior to harvesting total RNA for expression array analysis using Affymetrix U133 AB array and dChip analysis software.
Concurrently, we performed a comparative approach utilizing microarray data utilizing 46 primary melanoma and 18 benign nevi assayed for mRNA expression on the Affymetrix U133A mRNA expression microarray platform (16,383 probe sets) compiled from prior work[12].
Additionally, we identified known loci of deletions in melanomas compiled from previous study of cytogenetic alterations in melanoma[16] and a public repository of CGH data from 176 melanoma samples (progenetix.com).
We determined gene ranks in two ways: 1) Based upon degree of disparity of median expression between melanomas and benign nevi, ranking higher those genes which demonstrated decreased expression in tumors, and 2) Upfold regulation after pharmacologic demethylation in cell lines. Only genes located in known loci of deletion with an incidence of greater than 10% in primary melanoma were included in this analysis. An integrative rank product was calculated. Using a significance threshold (α= 0.005) and subsequent random permutation of our rank-lists, we identified 47 genes that were significantly differentially upregulated (Supplemental Table 1). Twenty-nine of 47 genes contained promoter-associated CpG islands for which primers could be designed utilizing the MethPrimer software [15]. These were selected for further studies.
Tumor specific promoter methylation of target genes
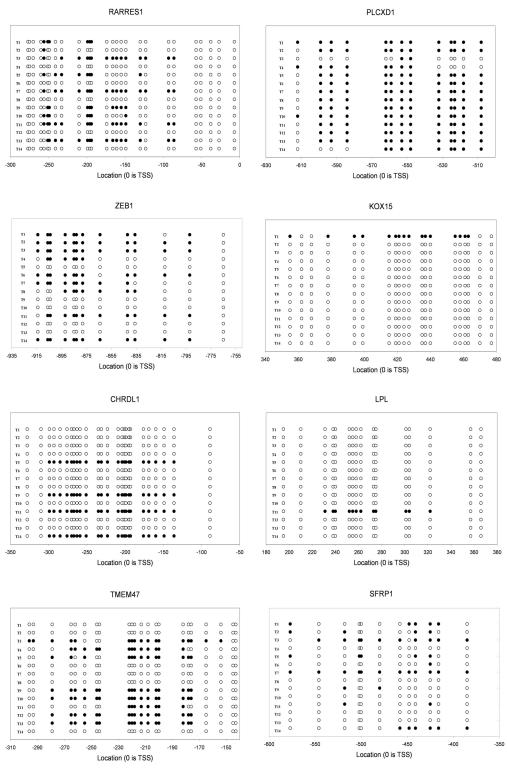
CpG islands in the promoter region of the 29 selected gene targets with CpG islands were bisulfite sequenced in fourteen melanoma tumor samples (Figure 1). Eight genes were identified containing some degree of tumor DNA promoter methylation. These were: SFRP1 (4/14 methylated, 28.6%), CHRDL1 (4/14 methylated, 28.6%), TMEM47 (8/14 methylated, 57.1%), KOX15 (1/14 methylated, 7.1%), LPL (1/14 methylated, 7.1%), ZEB1 (8/14 methylated, 57.1%), PLCXD1 (13/14 methylated, 92.9%), RARRES1 (5/14 methylated, 35.7%). (Table 2)
Figure 1.
Bisulfite sequencing of methylated candidate genes identified by integrative analysis. TSS= Transcriptional start site. ○ Unmethylated CpG ● Methylated CpG.
Table 2.
Targets identified by integrative approach and validation
| Gene | Abbreviation | Location | CGH Loss % | Median Tumor Expression vs Nevi |
Aza Fold Upregulation |
|---|---|---|---|---|---|
| Chordin-like 1 | CHRDL1 | Xq23 | −13.2 | 0.15 | 1.70 |
| Secreted frizzled-related protein 1 | SFRP1 | 8p12 | −12.3 | 0.17 | 1.72 |
| Transmembrane protein 47 | TMEM47 | Xp11 | −13.5 | 0.08 | 1.29 |
| Transcription factor 8 | ZEB1 | 10p11 | −20.8 | 0.33 | 1.85 |
| Lipoprotein lipase | LPL | 8p22 | −13.2 | 0.32 | 1.28 |
| Retinoic acid receptor responder 1 | RARRES1 | 3q25 | −19.8 | 0.46 | 1.74 |
| Phosphatidylinositol-specific phospholipase C, X domain containing 1 |
PLCXD1 | Xp22 | −16 | 0.41 | 1.32 |
| Zinc finger protein 22 | KOX 15 | 10q11 | −24.5 | 0.43 | 1.29 |
Tumor specific deletion of target genes
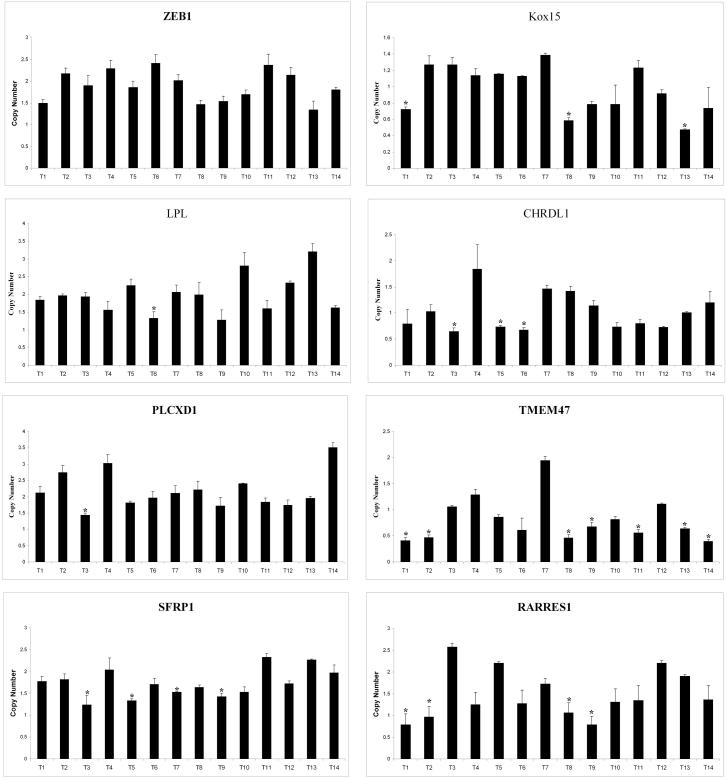
Quantitative PCR (qPCR) was performed on the 14 melanoma samples tested for methylation for the eight genes previously investigated. Copy number was calculated by comparison to the average quantitation of three housekeeping genes that are not found in areas of deletion or amplification in melanoma. This value was taken as the baseline amount corresponding to 2 alleles and utilized as a denominator to divide the quantitation of the genes of interest. A threshold of 1.5 alleles for somatic genes and 0.75 alleles for X linked genes in males was utilized to determine presence of a deletion. 7/8 (87.5%) genes studied demonstrated some evidence of deletion (Table 3). Incidences were: SFRP1 (3/14 deleted, 21.4%), CHRDL1 (3/14 deleted, 21.4%), TMEM47 (7/14 deleted, 50.0%), KOX15 (3/14 deleted, 21.4%), LPL (1/14 deleted, 7.1%), ZEB1 (0/14 deleted, 0.0%), PLCXD1 (1/14 deleted, 7.1%), RARRES1 (4/14 deleted, 28.6%). (Figure 2). ZEB1 demonstrated no evidence of deletion in the tumor samples assessed and was not analyzed further.
Table 3.
Incidence of deletion and methylation
| Gene | Deleted | Methylated | Deleted or Methylated | |
|---|---|---|---|---|
| CHRDL1 | 3 | 4 | 6 | 42.9% |
| SFRP1 | 3 | 4 | 6 | 42.9% |
| TMEM47 | 7 | 8 | 12 | 85.7% |
| ZEB1 | 0 | 8 | 8 | 57.1% |
| LPL | 1 | 1 | 2 | 14.3% |
| RARRES1 | 4 | 5 | 8 | 57.1% |
| PLCXD1 | 1 | 13 | 14 | 100.0% |
| KOX15 | 3 | 1 | 3 | 21.4% |
| Total Samples | 14 | |||
Figure 2.
Gene copy number analysis. Asterisk (*) indicates statistically significant decrease in copy number (relative to 2 for autosomal genes and 1 for X-linked genes) suggestive of deletion (p<.05). Error bars represent standard deviation.
Incidence of tumor specific gene methylation and deletion
Seven genes identified by our analysis demonstrated evidence of methylation and deletion. When combined, deletion or methylation in the fourteen primary melanoma assayed is present with high incidence for the genes identified by our analysis (Table 3), implying a significant role for these mechanisms in silencing of expression. Combined incidences were: SFRP1 (6/14, 42.9%), CHRDL1 (6/14, 42.9%), TMEM47 (12/14, 85.7%), KOX15 (3/14, 21.4%), LPL (2/14, 14.2%), PLCXD1 (14/14, 100%), RARRES1 (8/14, 57.1%). Co-incidence of methylation and deletion are shown in Supplemental Figure 2.
Candidate genes demonstrate growth suppression of melanoma derived cell lines
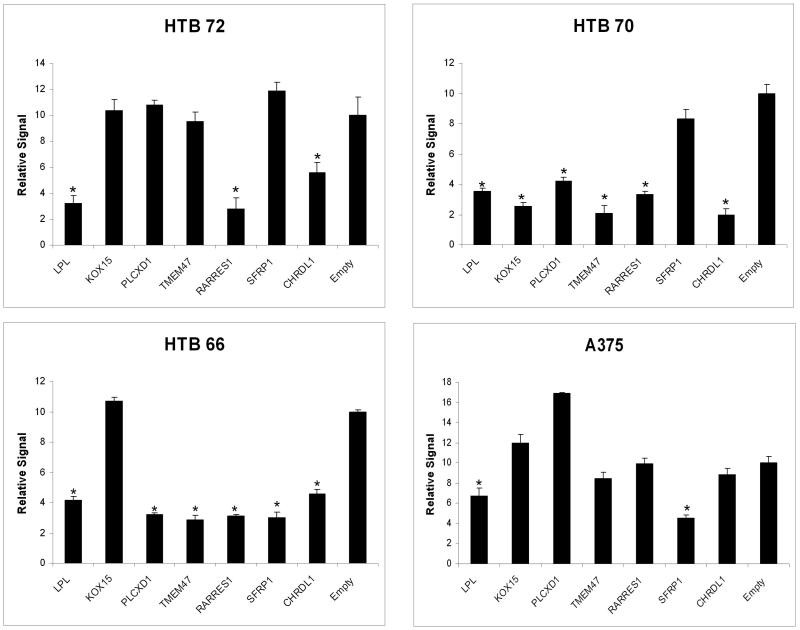
Transient transfections were performed to evaluate growth-suppressive effects in seven targets that show tumor-specific promoter hypermethylation and allele deletion (Figure 2). Four melanoma derived cell lines (HTB-66, HTB-70. HTB-72, and A375) were transfected with full-length cDNA ORFs for the seven genes of interest and empty vector controls to demonstrate the effects of constituitive overexpression by the putative tumor suppressors. Cell lines were selected based upon disparate derivation techniques and tumor stages in order to test tumor suppressor ability of genes of interest. Cell counts were performed on post transfection day 3 utilizing calcein fluorescence. Experiments were performed in multiples of 8. Statistically significant decreases in cell count were seen in 1 cell line with KOX15, in 2 cell lines with SFRP1, TMEM47 and PLCXD1, in 3 cell lines with RARRES1 and CHRDL1and in all 4 cell lines transfected with LPL. All genes showed growth inhibition in at least one cell line, confirming the validity of our approach.
Discussion
Using an integrative epigenetic/cytogenetic screening analysis, we identified 29 genes in malignant melanoma that are significantly decreased in expression relative to benign melanocytic nevi, are expression-responsive to demethylation, and found in areas of known deletion in melanoma. Of these genes, eight were validated by bisulfite sequencing of primary melanomas to demonstrate promoter hypermethylation. Seven of these genes were found to have some evidence of allelic deletion in primary melanoma. The expression of the validated targets in benign nevi relative to melanoma is shown in Supplemental Figure 3. Methylation and deletion appear to happen in an independent fashion that is not coordinated. All of these genes CHRDL1, SFRP1, TMEM47, LPL, RARRES1, PLCXD1, and KOX15 demonstrated some evidence of in vitro growth suppressive effects in melanoma derived cell lines. The heterogeneity and varying degree of growth suppression seen by transient transfection is likely a reflection of the genetic variability in the melanoma derived cell lines and the relative strength of the identified tumor suppressor genes.
The technique employed in this study to screen for potential tumor suppressor genes is a variation of techniques previously employed by our group to perform high throughput screening analysis[17, 18]. This approach is, in our estimation, a very stringent one. Its major drawbacks include the likelihood of exclusion of potential targets and the lack of multiple testing controls which limits precision of target identification. These limitations are minimized through the exhaustive validation employed in this study which eliminates false positive targets.
These seven genes are candidate tumor suppressor genes in malignant melanoma and are potential targets for further functional analysis to determine their role in tumorigenesis. Three of these 7: SFRP1, LPL, and RARRES1 have been identified as potential tumor suppressors in previous studies of different cancer types. The additional finding that RARRES1 has been recently reported to have significant methylation in melanoma by a group using disparate genome wide screening approach serves to validate further our approach[19].
CHRDL1 is an X-linked gene whose function is the source of ongoing investigation. Originally described as a BMP antagonist in retinal development[20], it has recently been found to be induced by hypoxia in a HIF 1α dependent fashion in retinal pericytes[21], as well as differentially expressed in colonic crypts[22]. This report is first describing a potential role CHRDL1 in any tumor type.
TMEM47 encodes a member of the PMP22/EMP/claudin protein family. The encoded protein is localized to the ER and the plasma membrane. Its expression has been noted in Ewing family tumor[23], but no functional studies have addressed its potential role in carcinogenesis. A microarray analysis melanoma cell lines and tumors has identified TMEM47 as one of a cluster of genes associated with aggressiveness in melanoma metastases[24]. This finding in association with its high rate of allelic loss in our validation studies (12/14 tumors with deletion or methylation, 85.7%), make it a compelling target for further investigation.
PLCXD1 is a pseudoautosomal gene located on both the X and Y chromosomes. It contains a PI-PLC X-box domain and its protein sequence and projected folding modeling suggest phospholipase C activity and possible role in intracellular transduction. No previous studies have assessed or analyzed its role in human cancer.
KOX15 is a zinc finger protein which has been implicated as a regulator of early tooth formation and amelogenesis[25]. It has been mapped to a locus of familial deficiency of permanent tooth development [26]. No previous studies have assessed or analyzed its role in human cancer.
SFRP1 is a member of the SFRP family that contains a cysteine-rich domain homologous to the putative Wnt-binding site of Frizzled proteins. SFRPs act as soluble modulators of Wnt signaling. Functionally, it may play a role in promotion of apoptosis[27-29]. Its loci is associated with LOH and deletion in a variety of human cancers [30]. Decreased expression of SFRP1 is demonstrated in breast cancer progression[31-33], but does not always correlate with deletion[34]. Promoter hypermethylation of SFRP1 has been demonstrated in colon[35, 36], prostate[37], ovarian[38], bladder[39], and gastric cancer[40]. Studies in non small cell lung cancer indicate that concomitant genetic and epigenetic events lead to expression silencing of SFRP1[41]. The role of SFRP1 as a possible tumor suppressor in melanoma has not previously been reported. Compelling evidence is present in other organ systems for a synergistic role of cytogenetic and epigenetic mechanisms in SFRP1 inactivation that makes it an appealing candidate for a tumor suppressor gene in melanoma.
LPL encodes lipoprotein lipase, which is expressed in heart, muscle, and adipose tissue. LPL functions as a homodimer, and has the dual functions of triglyceride hydrolase and ligand/bridging factor for receptor-mediated lipoprotein uptake. Severe mutations causing deficiency result in type I hyperlipoproteinemia. LPL has been implicated as a tumor suppressor. LPL expression may be regulated in an APC directed manner with upregulation of LPL mRNA decreasing the tumorigenic phenotype in APC −/− mice [42, 43]. Multiple mechanisms may be involved in the role of LPL in human carcinogenesis. LPL treatment has been shown to inactivate nuclear factor kappa B [NF-κB) [44], and trigger apoptosis[45]. LPL, was reported as one of the most frequently deleted loci in prostate cancer with LOH and deletion in up to 68% of prostate cancers[46] LPL deletion was reported in many other types of human cancers as well, including gastric, colon and oral cancer [47-49]. A recent study has also identified a role for synergistic hypermethylation and deletion of LPL in prostate cancer [50].
RARRES1 was identified as a retinoid acid (RA) receptor-responsive gene[51].. It encodes a type 1 membrane protein. The expression of this gene is upregulated by tazarotene as well as by retinoic acid receptors. The expression of this gene is found to be downregulated in prostate cancer, which is caused by the methylation of its promoter and CpG island. It has been shown to be methylated in human melanoma with concomitant decrease in mRNA expression [19]. Its expression has been associated with spontaneous regression of melanoma in an animal model[52]. RARRES1 is a putative tumor suppressor in prostate cancer [53]and demonstrates promoter hypermethylation in gastric carcinoma [54]. Down-regulation of expression in different cancer cell lines is related to hypermethylation of the promoter [55] It is posited that silencing of RARRES1 by promoter hypermethylation is common in human cancers and may contribute to the loss of retinoic acid responsiveness in some neoplastic cells[55]. RARRES1 demonstrates promise as a potential tumor suppressor in melanoma. Our findings of significant allelic loss due to deletion (4/14, 28.6%) and an overall 57.1% (8/14) incidence of allelic loss by either methylation or deletion underscore the potential importance of this gene as a tumor suppressor.
Several studies have been published in the interim since the initiation of our study which have utilized microarray analysis of primary melanoma and nevi to elucidate genetic differences between these lesions[56, 57]. These have revealed a spectrum of genes associated with different histopathologic characteristics. Other studies have similarly shown links between tumor invasiveness[24]or clinical outcomes[58] and a variety of gene expression profiles. As well, other studies have assessed epigenetic changes associated with melanoma[59]. The raw data from these studies would undoubtedly increase the power of a gene screening algorithm and should be employed in future iterations.
Our study attempts to include these disparate analyses in a single integrated approach, designed to identify genes whose downregulation may be multifactorial. As such, the discovery of a mostly novel set of putative tumor suppressor genes not previously identified in melanoma is an unsurprising outcome of our investigation.
Although much work remains to elucidate possible contributors to that malignant phenotype of melanoma, we did find 7 genes that are solid candidates for tumor suppressor genes that were associated with silencing by cytogenetic and epigenetic mechanisms. One pitfall of integrative genomic approaches is the problem of identifying true signal from the substantial baseline noise generated by such large amounts of data. In addition to utilizing stringent statistical criteria in our discovery phase, we utilized separate discovery and validation cohorts and complementary experimental techniques, in order to bolster the validity of our findings. This approach represents the attempted discovery and validation of genes in melanoma that are potential tumor suppressors. Future work to explore the implications of this study would include functional studies of CHRDL1, SFRP1, TMEM47, LPL, RARRES1, PLCXD1, and KOX15 in melanoma derived cell lines.
Supplementary Material
Supplemental Figure 1: Integrative genetic approach. We employed a three-pronged integrative approach to identify novel tumor suppressors in malignant melanoma. Changes in gene expression after chemical demethylation in 4 melanoma derived cell lines was assessed by expression microarray analysis, Genes were ranked by median upfold regulation after demethylation. Utilizing previously published expression microarray data, we ranked genes by median expression decrease in tumors relative to benign nevi. Sum ranks of expression data for each probe set were made. Pearson rank correlation was utilized to identify significant expression changes in genes identified in areas of known deletion in malignant melanoma (Threshold <. 005). Review of publicly available CGH data from 176 melanomas identified areas of chromosomal loss in malignant melanoma. 47 genes were identified by rank correlations that were in areas of loss. Of these, 29 were found to have potential CpG islands by sequence analysis and were selected for further evaluation by bisulfite sequencing (see Supplemental Table 1).
Supplemental Figure 2: Incidence of deletion and methylation in primary tumors. These events appear to be independent.
Supplemental Figure 3: Microarray gene expression of validated putative tumor suppressor genes in benign melanocytic nevi relative to malignant melanoma. Y-axes represent relative expression of each gene in each particular sample. Data was median-normalized by expression array and each gene was median normalized for this figure. Median difference in expression is listed in Table 2.
Supplementary Table 1: Results of integrative screening approach
Figure 3.
Transient transfection of melanoma derived cell lines with genes of interest. Cells harvested 72 hours after transfection. Cellular signal was quantitated with calcein fluorescence. Y axes represent the relative ratio of the detector signal absorption. Experiments performed in octuplicate. * indicates statistically significant difference from empty vector transfection (p<.05). Error bars represent standard deviation. A Transfection of LPL, RARRES1, and CHRDL1 results in growth inhibition of the HTB-72 cell line. B Transfection of LPL, KOX15, PLCXD1, TMEM47, RARRES1, and CHRDL1 results in growth inhibition of the HTB-70 cell line. C Transfection of LPL, PLCXD1, TMEM47, RARRES1, SFRP1 and CHRDL1 results in growth inhibition of the HTB-66 cell line. D Transfection of LPL, and SFRP1 results in growth inhibition of the A375 cell line.
Acknowledgments
Dr. Califano is supported by the National Cancer Institute SPORE (5P50CA096784-05).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Muthusamy V, Duraisamy S, Bradbury CM, et al. Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res. 2006;66:11187–93. doi: 10.1158/0008-5472.CAN-06-1274. [DOI] [PubMed] [Google Scholar]
- 2.Liu S, Ren S, Howell P, Fodstad O, Riker AI. Identification of novel epigenetically modified genes in human melanoma via promoter methylation gene profiling. Pigment Cell Melanoma Res. 2008;21:545–58. doi: 10.1111/j.1755-148X.2008.00484.x. [DOI] [PubMed] [Google Scholar]
- 3.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–42. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- 4.Schwabe M, Lubbert M. Epigenetic lesions in malignant melanoma. Curr Pharm Biotechnol. 2007;8:382–7. doi: 10.2174/138920107783018372. [DOI] [PubMed] [Google Scholar]
- 5.Strauss BS. Limits to the Human Cancer Genome Project? Science. 2007;315:762–4. author reply 4-5. [PubMed] [Google Scholar]
- 6.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 7.Garraway LA, Weir BA, Zhao X, et al. “Lineage addiction” in human cancer: lessons from integrated genomics. Cold Spring Harb Symp Quant Biol. 2005;70:25–34. doi: 10.1101/sqb.2005.70.016. [DOI] [PubMed] [Google Scholar]
- 8.Shen L, Kondo Y, Ahmed S, et al. Drug sensitivity prediction by CpG island methylation profile in the NCI-60 cancer cell line panel. Cancer Res. 2007;67:11335–43. doi: 10.1158/0008-5472.CAN-07-1502. [DOI] [PubMed] [Google Scholar]
- 9.Pujana MA, Ruiz A, Badenas C, et al. Molecular characterization of a t(9;12)(p21;q13) balanced chromosome translocation in combination with integrative genomics analysis identifies C9orf14 as a candidate tumor-suppressor. Genes Chromosomes Cancer. 2007;46:155–62. doi: 10.1002/gcc.20396. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q, Diskin S, Rappaport E, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–62. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- 11.Dahia PL, Hao K, Rogus J, et al. Novel pheochromocytoma susceptibility loci identified by integrative genomics. Cancer Res. 2005;65:9651–8. doi: 10.1158/0008-5472.CAN-05-1427. [DOI] [PubMed] [Google Scholar]
- 12.Talantov D, Mazumder A, Yu JX, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–42. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 13.Baudis M, Cleary ML. Progenetix.net: an online repository for molecular cytogenetic aberration data. Bioinformatics. 2001;17:1228–9. doi: 10.1093/bioinformatics/17.12.1228. [DOI] [PubMed] [Google Scholar]
- 14.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 16.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 17.Glazer CA, Smith IM, Ochs MF, et al. Integrative discovery of epigenetically derepressed cancer testis antigens in NSCLC. PLoS One. 2009;4:e8189. doi: 10.1371/journal.pone.0008189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith IM, Glazer CA, Mithani SK, et al. Coordinated activation of candidate proto-oncogenes and cancer testes antigens via promoter demethylation in head and neck cancer and lung cancer. PLoS One. 2009;4:e4961. doi: 10.1371/journal.pone.0004961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonazzi VF, Irwin D, Hayward NK. Identification of candidate tumor suppressor genes inactivated by promoter methylation in melanoma. Genes Chromosomes Cancer. 2009;48:10–21. doi: 10.1002/gcc.20615. [DOI] [PubMed] [Google Scholar]
- 20.Sakuta H, Suzuki R, Takahashi H, et al. Ventroptin: a BMP-4 antagonist expressed in a double-gradient pattern in the retina. Science. 2001;293:111–5. doi: 10.1126/science.1058379. [DOI] [PubMed] [Google Scholar]
- 21.Kane R, Godson C, O’Brien C. Chordin-like 1, a bone morphogenetic protein-4 antagonist, is upregulated by hypoxia in human retinal pericytes and plays a role in regulating angiogenesis. Mol Vis. 2008;14:1138–48. [PMC free article] [PubMed] [Google Scholar]
- 22.Kosinski C, Li VS, Chan AS, et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci U S A. 2007;104:15418–23. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung IY, Feng Y, Danis K, et al. Novel markers of subclinical disease for Ewing family tumors from gene expression profiling. Clin Cancer Res. 2007;13:6978–83. doi: 10.1158/1078-0432.CCR-07-1417. [DOI] [PubMed] [Google Scholar]
- 24.Xu L, Shen SS, Hoshida Y, et al. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res. 2008;6:760–9. doi: 10.1158/1541-7786.MCR-07-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao Y, Kobayashi H, Ganss B. The human KROX-26/ZNF22 gene is expressed at sites of tooth formation and maps to the locus for permanent tooth agenesis (He-Zhao deficiency) J Dent Res. 2003;82:1002–7. doi: 10.1177/154405910308201213. [DOI] [PubMed] [Google Scholar]
- 26.Liu W, Wang H, Zhao S, et al. The novel gene locus for agenesis of permanent teeth (He-Zhao deficiency) maps to chromosome 10q11.2. J Dent Res. 2001;80:1716–20. doi: 10.1177/00220345010800080701. [DOI] [PubMed] [Google Scholar]
- 27.Ko J, Ryu KS, Lee YH, et al. Human secreted frizzled-related protein is down-regulated and induces apoptosis in human cervical cancer. Exp Cell Res. 2002;280:280–7. doi: 10.1006/excr.2002.5649. [DOI] [PubMed] [Google Scholar]
- 28.Melkonyan HS, Chang WC, Shapiro JP, et al. SARPs: a family of secreted apoptosis-related proteins. Proc Natl Acad Sci U S A. 1997;94:13636–41. doi: 10.1073/pnas.94.25.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki H, Watkins DN, Jair KW, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–22. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 30.Leach RJ, Banga SS, Ben-Othame K, et al. Report of the Third International Workshop on Human Chromosome 8 Mapping. San Antonio, Texas, October 25-27, 1996. Cytogenet Cell Genet. 1996;75:71–84. doi: 10.1159/000134460. [DOI] [PubMed] [Google Scholar]
- 31.Klopocki E, Kristiansen G, Wild PJ, et al. Loss of SFRP1 is associated with breast cancer progression and poor prognosis in early stage tumors. Int J Oncol. 2004;25:641–9. [PubMed] [Google Scholar]
- 32.Ugolini F, Adelaide J, Charafe-Jauffret E, et al. Differential expression assay of chromosome arm 8p genes identifies Frizzled-related (FRP1/FRZB) and Fibroblast Growth Factor Receptor 1 (FGFR1) as candidate breast cancer genes. Oncogene. 1999;18:1903–10. doi: 10.1038/sj.onc.1202739. [DOI] [PubMed] [Google Scholar]
- 33.Ugolini F, Charafe-Jauffret E, Bardou VJ, et al. WNT pathway and mammary carcinogenesis: loss of expression of candidate tumor suppressor gene SFRP1 in most invasive carcinomas except of the medullary type. Oncogene. 2001;20:5810–7. doi: 10.1038/sj.onc.1204706. [DOI] [PubMed] [Google Scholar]
- 34.Armes JE, Hammet F, de Silva M, et al. Candidate tumor-suppressor genes on chromosome arm 8p in early-onset and high-grade breast cancers. Oncogene. 2004;23:5697–702. doi: 10.1038/sj.onc.1207740. [DOI] [PubMed] [Google Scholar]
- 35.Caldwell GM, Jones C, Gensberg K, et al. The Wnt antagonist sFRP1 in colorectal tumorigenesis. Cancer Res. 2004;64:883–8. doi: 10.1158/0008-5472.can-03-1346. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–9. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 37.Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res. 2005;65:4218–27. doi: 10.1158/0008-5472.CAN-04-4407. [DOI] [PubMed] [Google Scholar]
- 38.Takada T, Yagi Y, Maekita T, et al. Methylation-associated silencing of the Wnt antagonist SFRP1 gene in human ovarian cancers. Cancer Sci. 2004;95:741–4. doi: 10.1111/j.1349-7006.2004.tb03255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoehr R, Wissmann C, Suzuki H, et al. Deletions of chromosome 8p and loss of sFRP1 expression are progression markers of papillary bladder cancer. Lab Invest. 2004;84:465–78. doi: 10.1038/labinvest.3700068. [DOI] [PubMed] [Google Scholar]
- 40.To KF, Chan MW, Leung WK, et al. Alterations of frizzled (FzE3) and secreted frizzled related protein (hsFRP) expression in gastric cancer. Life Sci. 2001;70:483–9. doi: 10.1016/s0024-3205(01)01422-9. [DOI] [PubMed] [Google Scholar]
- 41.Fukui T, Kondo M, Ito G, et al. Transcriptional silencing of secreted frizzled related protein 1 (SFRP 1) by promoter hypermethylation in non-small-cell lung cancer. Oncogene. 2005;24:6323–7. doi: 10.1038/sj.onc.1208777. [DOI] [PubMed] [Google Scholar]
- 42.Mutoh M, Niho N, Wakabayashi K. Concomitant suppression of hyperlipidemia and intestinal polyp formation by increasing lipoprotein lipase activity in Apc-deficient mice. Biol Chem. 2006;387:381–5. doi: 10.1515/BC.2006.051. [DOI] [PubMed] [Google Scholar]
- 43.Niho N, Mutoh M, Takahashi M, Tsutsumi K, Sugimura T, Wakabayashi K. Concurrent suppression of hyperlipidemia and intestinal polyp formation by NO-1886, increasing lipoprotein lipase activity in Min mice. Proc Natl Acad Sci U S A. 2005;102:2970–4. doi: 10.1073/pnas.0500153102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kota RS, Ramana CV, Tenorio FA, Enelow RI, Rutledge JC. Differential effects of lipoprotein lipase on tumor necrosis factor-alpha and interferon-gamma-mediated gene expression in human endothelial cells. J Biol Chem. 2005;280:31076–84. doi: 10.1074/jbc.M412189200. [DOI] [PubMed] [Google Scholar]
- 45.Schwarz S, Hufnagel B, Dworak M, Klumpp S, Krieglstein J. Protein phosphatase type 2Calpha and 2Cbeta are involved in fatty acid-induced apoptosis of neuronal and endothelial cells. Apoptosis. 2006;11:1111–9. doi: 10.1007/s10495-006-6982-1. [DOI] [PubMed] [Google Scholar]
- 46.Gallucci M, Merola R, Farsetti A, et al. Cytogenetic profiles as additional markers to pathological features in clinically localized prostate carcinoma. Cancer Lett. 2006;237:76–82. doi: 10.1016/j.canlet.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 47.Liu CJ, Lin SC, Chen YJ, Chang KM, Chang KW. Array-comparative genomic hybridization to detect genomewide changes in microdissected primary and metastatic oral squamous cell carcinomas. Mol Carcinog. 2006;45:721–31. doi: 10.1002/mc.20213. [DOI] [PubMed] [Google Scholar]
- 48.Baffa R, Santoro R, Bullrich F, Mandes B, Ishii H, Croce CM. Definition and refinement of chromosome 8p regions of loss of heterozygosity in gastric cancer. Clin Cancer Res. 2000;6:1372–7. [PubMed] [Google Scholar]
- 49.Cunningham C, Dunlop MG, Wyllie AH, Bird CC. Deletion mapping in colorectal cancer of a putative tumour suppressor gene in 8p22-p21.3. Oncogene. 1993;8:1391–6. [PubMed] [Google Scholar]
- 50.Kim JW, Cheng Y, Liu W, et al. Genetic and epigenetic inactivation of LPL gene in human prostate cancer. Int J Cancer. 2009;124:734–8. doi: 10.1002/ijc.23972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagpal S, Patel S, Asano AT, Johnson AT, Duvic M, Chandraratna RA. Tazarotene-induced gene 1 (TIG1), a novel retinoic acid receptor-responsive gene in skin. J Invest Dermatol. 1996;106:269–74. doi: 10.1111/1523-1747.ep12340668. [DOI] [PubMed] [Google Scholar]
- 52.Rambow F, Malek O, Geffrotin C, et al. Identification of differentially expressed genes in spontaneously regressing melanoma using the MeLiM swine model. Pigment Cell Melanoma Res. 2008;21:147–61. doi: 10.1111/j.1755-148X.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- 53.Jing C, El-Ghany MA, Beesley C, et al. Tazarotene-induced gene 1 (TIG1) expression in prostate carcinomas and its relationship to tumorigenicity. J Natl Cancer Inst. 2002;94:482–90. doi: 10.1093/jnci/94.7.482. [DOI] [PubMed] [Google Scholar]
- 54.Shutoh M, Oue N, Aung PP, et al. DNA methylation of genes linked with retinoid signaling in gastric carcinoma: expression of the retinoid acid receptor beta, cellular retinol-binding protein 1, and tazarotene-induced gene 1 genes is associated with DNA methylation. Cancer. 2005;104:1609–19. doi: 10.1002/cncr.21392. [DOI] [PubMed] [Google Scholar]
- 55.Youssef EM, Chen XQ, Higuchi E, et al. Hypermethylation and silencing of the putative tumor suppressor Tazarotene-induced gene 1 in human cancers. Cancer Res. 2004;64:2411–7. doi: 10.1158/0008-5472.can-03-0164. [DOI] [PubMed] [Google Scholar]
- 56.Scatolini M, Grand MM, Grosso E, et al. Altered molecular pathways in melanocytic lesions. Int J Cancer. 2010;126:1869–81. doi: 10.1002/ijc.24899. [DOI] [PubMed] [Google Scholar]
- 57.Smith AP, Hoek K, Becker D. Whole-genome expression profiling of the melanoma progression pathway reveals marked molecular differences between nevi/melanoma in situ and advanced-stage melanomas. Cancer Biol Ther. 2005;4:1018–29. doi: 10.4161/cbt.4.9.2165. [DOI] [PubMed] [Google Scholar]
- 58.Winnepenninckx V, Lazar V, Michiels S, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–82. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 59.Schinke C, Mo Y, Yu Y, et al. Aberrant DNA methylation in malignant melanoma. Melanoma Res. 2010;20:253–65. doi: 10.1097/CMR.0b013e328338a35a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Integrative genetic approach. We employed a three-pronged integrative approach to identify novel tumor suppressors in malignant melanoma. Changes in gene expression after chemical demethylation in 4 melanoma derived cell lines was assessed by expression microarray analysis, Genes were ranked by median upfold regulation after demethylation. Utilizing previously published expression microarray data, we ranked genes by median expression decrease in tumors relative to benign nevi. Sum ranks of expression data for each probe set were made. Pearson rank correlation was utilized to identify significant expression changes in genes identified in areas of known deletion in malignant melanoma (Threshold <. 005). Review of publicly available CGH data from 176 melanomas identified areas of chromosomal loss in malignant melanoma. 47 genes were identified by rank correlations that were in areas of loss. Of these, 29 were found to have potential CpG islands by sequence analysis and were selected for further evaluation by bisulfite sequencing (see Supplemental Table 1).
Supplemental Figure 2: Incidence of deletion and methylation in primary tumors. These events appear to be independent.
Supplemental Figure 3: Microarray gene expression of validated putative tumor suppressor genes in benign melanocytic nevi relative to malignant melanoma. Y-axes represent relative expression of each gene in each particular sample. Data was median-normalized by expression array and each gene was median normalized for this figure. Median difference in expression is listed in Table 2.
Supplementary Table 1: Results of integrative screening approach