Abstract
The Online Chemical Modeling Environment is a web-based platform that aims to automate and simplify the typical steps required for QSAR modeling. The platform consists of two major subsystems: the database of experimental measurements and the modeling framework. A user-contributed database contains a set of tools for easy input, search and modification of thousands of records. The OCHEM database is based on the wiki principle and focuses primarily on the quality and verifiability of the data. The database is tightly integrated with the modeling framework, which supports all the steps required to create a predictive model: data search, calculation and selection of a vast variety of molecular descriptors, application of machine learning methods, validation, analysis of the model and assessment of the applicability domain. As compared to other similar systems, OCHEM is not intended to re-implement the existing tools or models but rather to invite the original authors to contribute their results, make them publicly available, share them with other users and to become members of the growing research community. Our intention is to make OCHEM a widely used platform to perform the QSPR/QSAR studies online and share it with other users on the Web. The ultimate goal of OCHEM is collecting all possible chemoinformatics tools within one simple, reliable and user-friendly resource. The OCHEM is free for web users and it is available online at http://www.ochem.eu.
Keywords: On-line web platform, Modeling workflow, Estimation of accuracy of predictions, Applicability domain, Data sharing, Open access
Introduction
Nowadays, the development of new drugs relies heavily on computational methods. The pioneering works on Quantitative Structure Activity/Property Relationships (QSAR/QSPR), started by Hansch and his colleagues more than 50 years ago, have reached maturity and represent an important part in modern drug discovery. The prediction of biological and physicochemical properties of chemical compounds with computational models reduces significantly the amount of experimental measurements. This is especially helpful for screening of a large number of compounds, some of which might have not been yet synthesized, in order to test whether they have a particular biological activity or physicochemical property. One of the prominent examples, where computational models can significantly reduce the amount of experimental measurements is ADME/Tox (Absorption, Distribution, Metabolism, Excretion and Toxicity), i.e., a prediction of pharmacokinetic properties that are important for all drug candidate compounds [1, 2].
Creation of a successful predictive model requires a number of time-consuming and tedious steps, including data acquisition and preparation, selection and calculation of appropriate molecular descriptors, application of a particular machine learning method, evaluation of results and assessment of the model applicability domain. A particularly difficult step is the collection of high quality experimental data. This involves time-consuming work with scientific literature, manual extraction of experimental data from the literature and preparation of the data for further steps of the modeling process. Thereafter, a researcher normally uses external tools to calculate molecular descriptors for the data and, finally, to train a model using a machine learning method of choice. Further follows the evaluation of the new models performance, assessment of the domain of applicability, investigation of outliers, modification of the initial dataset and repetition of the whole process. To sum up, the process of modeling is tedious and iterative.
It is thought that every year hundreds or possibly even thousands of models are published in the scientific literature (e.g., more than 50 models were estimated to be published only for lipophilicity, logP, and water solubility in 2005 alone) [3, 4]. However, for most models a publication marks the end of their life cycle. Very rarely models become available as software tools and perform predictions for new data, i.e., serve the purpose for which they were developed. Thus, after putting a substantial effort in data acquisition, model development and a preparation for publication, there is virtually no practical use of these models at the end of this endeavor. Attempts to reproduce published models are not always successful and could be considered as an art of their own. For example, twofold differences (1 and 2 log units) were observed using two different implementations of the MLOGP algorithm [5] for predictions of logP for 95,809 Pfizer compounds [6]. At the same time, an opposite performance of both implementations was observed for the prediction of 882 Nycomed compounds, thus leaving open the question which implementation is more accurate [6].
One of the major problems for model reconstruction is the unavailability of the initial data. Publications often contain only names of molecules or only a set of calculated descriptors. However, models built with so-called memory-based approaches, such as k-Nearest Neighbors (kNN), Support Vector Machines, Probabilistic Neural Networks, etc. require the initial dataset to be reproduced correctly. Nevertheless, many models still continue to be published without such data. Chemical names are sometimes ambiguous and it is not obligatory for authors to provide unified chemical identifiers such as CAS-RNs (Chemical Abstract Registration Numbers). Thus, it is not trivial to match the name with a chemical structure if the dataset of molecules is not provided in the publication. On the other hand, the accuracy of calculated descriptors is often a subject of the software implementation that is used for their generation. Different software programs could produce slightly different descriptors, which also affects the reproducibility of the published model. Thus, substantial efforts are required from a user to reproduce already published results.
There is a variety of online tools to store information on chemical compounds (i.e., online databases) and tools to create predictive models. However, most of them lack essential features required for modeling. For example, DrugBank [7], ChemSpider [8], Chempedia [9], ChemExper [10] and PubChem [11, 12] provide storage for chemical information but have no QSAR/QSPR modeling tools. Moreover, some databases do not store essential information required for data verification and modeling: the source of information as well as conditions under which experiments have been conducted. The quality of such data, which is very important for a predictive model, cannot be easily verified. Other online tools (e.g., VCCLAB [13], OpenTox [14], ChemBench [15], QSAR DataBank [16]) provide modeling facilities but lack an integration with a chemical database and, therefore, cannot support a typical workflow of QSAR/QSPR research. None of the aforementioned tools allow automated tracking and verification of the compounds used in the modeling process. Moreover, these tools do not have possibilities to be easily extendable by incorporating new descriptors and machine learning methods.
In this article, we present an online platform for QSAR/QSPR research, the Online Chemical Modeling Environment (OCHEM, http://www.ochem.eu), which allows to perform a full QSAR/QSPR cycle in a semiautomated manner. The platform includes two major subsystems: the database of experimentally measured properties and the modeling framework. The database subsystem includes the storage of experimental endpoints and the tools to efficiently introduce, search and manipulate the data. The modeling framework provides facilities to use these data in the modeling process and perform all the steps of a typical modeling workflow. Most important, the developed and published OCHEM models (together with the data used to develop them) are publicly available to the scientific community and can be freely used on the web site to predict new molecules. Moreover, the OCHEM is fully extendable for new descriptors, modeling tools and models.
The ultimate goal of this project is to provide the QSAR scientific community with the top-quality curated data combined with a comprehensive set of QSAR modeling tools.
The database of experimental measurements
This section describes the structure of the database of experimental measurements and provides an insight on its main features.
Structure overview
The database contains experimentally measured biological and physicochemical properties of small molecules together with the conditions under which the experiments have been conducted and references to the sources where the data were published. The structure of the database is shown schematically in Fig. 1.
Fig. 1.
A schematic overview of an individual record in the OCHEM database
The experimental measurements are the central entities of the database. They combine all the information related to the experiment, in particular the result of the measurement, which can be either numeric or qualitative depending on the measured property. The central system component, where the experimental measurements can be introduced, searched and manipulated, is the compound property browser.
An experimental measurement includes information about the property that was measured and the associated chemical compound. The compounds and the properties can be marked with particular keywords, also known as tags, that allow convenient filtering and grouping of the data.
For every measurement stored in the OCHEM, it is obligatory to specify the source of the data. The source is usually a publication in a scientific journal or a book. The strict policy of OCHEM is to accept only those experimental records, that have their source of information specified. This improves the quality of the data and allows it to be verified by checking the original publication. Although a user can also introduce an unpublished article and link the data to it, records from such sources should be treated with caution. The ways to browse, introduce and automatically fetch the publications from the PubMed database are described below in the “Sources of information” section.
Every numeric property has a corresponding category of units, for example, the category of units for Inhibition Concentration 50% (IC50) is “Concentration”. By default the OCHEM database keeps experimental endpoints in the original format (i.e., in units as reported in the publication). For this purpose all units are grouped into strictly defined unit categories, for example Kelvin, Celsius and Fahrenheit degrees belong to the “Temperature” category. For the purpose of compatibility and for modeling of the combined sets from different publications, the system provides on the fly conversion between different units.
An important feature of our database, which is also unique among other chemical databases, is the possibility to store the conditions of experiments. This information is crucial for modeling: in many cases, the result of an experimental measurement is senseless without knowing the conditions under which the experiment has been conducted. For example, it does not make sense to specify the boiling point for a compound without specifying the air pressure. Such conditions should be introduced as obligatory conditions, i.e., a new record will be rejected by the system if there is no information about these conditions provided. Conditional values stored in the database can be numerical (with units of measurement), qualitative or descriptive (textual). Moreover, in the “conditions” section it is possible to note additional information related to the experiment, even if it is not a “condition” in the classical sense. Examples of such additional information are assay descriptions, a target of the ligand (the receptor) or species on which the biological activity has been tested. For simplicity, we further universally refer to all this information as “conditions”.
Features overview
In brief, the distinguishing features of the OCHEM database are as follows.
The wiki principle: most of the data can be accessed, introduced and modified by users
Different access levels: guests, registered users, verified users, administrators
Tracking of all the changes
Obligatory indications of the source of the data
Possibility to indicate conditions of the experiment, which can be later used for QSAR modeling
Search by substructure, molecule names, by publication where the measurements were referenced, by conditions of experiments, etc.
Control of duplicated records
Batch upload and batch modification of large amounts of data
Different units of measurements and utilities to interconvert between units
Organizing the records in re-usable sets (“baskets”)
Introduction of data
Basics
There are two ways to introduce experimental data to the OCHEM database: the first way is the manual record-by-record input, where each experimental measurement is entered separately; the second way is the batch upload facility that allows upload of large amounts of data from Excel or SDF (Structure Data File). This functionality is described in more detail below in the “Batch upload” section. Other types of database entities (new physicochemical properties, units of measurements, publications, etc.) can be introduced via special interface windows called browsers. Basically, every entity in the OCHEM database has a corresponding browser: the compound properties browser for experimental measurements, the publications browser for articles and books, the properties browser, the units browser, etc. Additionally, for every entity there is a specific dialog window where a user can create a new entry or edit an existing one.
This also applies to experimental measurements: every measurement is created and modified in the record editor window, where a user specifies all relevant information: compound structure, corresponding publication, conditions of the experiment, units of measurement, etc.
Similarly, there is the molecule editor, that allows introduction of compound structures in several ways: a user can either explicitly draw the structure in the JME molecule editor [17], upload an SDF or MOL2 file, specify a SMILES string or paste a file in one of these formats. The JME editor used at the OCHEM is probably the most used structure input tool on the internet. The program allows users to draw, edit, and display molecules and reactions as described at http://www.molinspiration.com/jme/. Additionally, the structure of a molecule can be automatically retrieved from the PubChem database [11] by the name of molecule, though this leaves the potential for incorrect retrieval based on incorrect name-structure association and the user should validate the correctness of the structure that is downloaded.
Batch upload
Since OCHEM relies on user contributions, it is essential to provide the user with simple and efficient tools to introduce data into the system. The record editor mentioned above is useful for data correction or single record introduction, but is not suitable for the introduction of hundreds or thousands of records. For efficient and fast introduction of large amounts of data, the OCHEM offers the “batch upload” tool. This section provides an insight into the main features of this tool.
Input data. The input data for the batch upload tool is a specially prepared Excel workbook, CSV or SDF file. The file is processed by the tool to create records according to the provided data. To unify the upload process, SDF and CSV files are internally converted to the Excel file format with tags represented as columns and molecular structures and names put into additional columns. An example Excel file with all possible columns and explanations can be downloaded directly at the first page of the batch upload tool.
As described in the “Structure overview” section, the essential information contained in the record is the value of a biological or chemical property for a specific molecule published in a specific article. Although the Excel file format allows a user to provide all the detailed information about a record (number of a page in an article, where a particular value was published, accuracy of measurements, textual comments, record evidence, measurement units, etc.), a minimal valid file must contain only the information on property value, molecular structure (or name) and article details for every uploaded record. In case some information is not provided (i.e., unit of measurement), the default values specified for the uploaded property (or condition) are used.
Information about a molecular structure can be provided in the form of SMILES, SDF or MOL2. If the structure of the molecule is not available, it is possible to provide a molecule name or identifier, e.g., Chemical Abstract Registration Number (CAS-RN)—if possible, the structure will be retrieved automatically from the PubChem database. The publication can be specified either in the form of an internal OCHEM article identifier or a PubMed identifier. The sheet can also contain information about the measurement conditions. The information about the property itself and all the required conditions and units should already be present in the database. For numerical properties, users can provide predicates, such as >, <, ≥, ≤, ~, ≫, ≪, ≈ as well as the errors of the measurements.
After the file has been created, the user can use the batch upload tool to introduce data to OCHEM. The tool is created in the form of a “wizard” with a step-by-step approach for the upload process.
In the first step, the user must choose the file to upload. The file should be a valid Excel, CSV or SDF file not larger than 25 MB.
Sheet preview. The second step of the batch upload is the data sheet preview (see Fig. 2). At this stage the user can choose a particular sheet from the uploaded workbook (only one sheet at a time can be uploaded), deselect unnecessary columns (especially useful in the case of multiple property columns in a sheet since only one property can be uploaded at a time) and override default units for properties and conditions.
Fig. 2.
A screenshot of the second step of the batch upload process
At this stage, it is also possible to provide details of the associated article for the records that have no article explicitly specified in the uploaded file. This is often the case for SDF files. In case of mistakes in the property and column names, the user can rename (or “remap”) columns.
Once the required columns are selected, all property units are set to the desired values and column names are remapped, the user can proceed with the upload.
Data preview. After an intermediate page that shows the upload progress, the user proceeds to next step, as displayed in Fig. 3.
Fig. 3.
A screenshot of data preview, the third step of the batch upload
At this stage, the user is presented a summary of the batch upload. The summary indicates the recognized or non-recognized columns, missing required columns, as well as some basic statistics regarding the molecular structures—the number of structures retrieved from OCHEM or from PubChem. It also contains the preview browser, which allows the user to preview the data almost exactly in the same way as it would be stored in the OCHEM database.
At the data preview step, the records are generally divided into four groups—valid records, internal duplicates, external duplicates and unrecoverable errors.
A valid record is a record that fulfills all the requirements and is ready to be uploaded and saved. This record has a white or yellow background. It can be marked for upload or skipped.
An internal duplicate is a record that from the OCHEM point of view is an exact duplicate of some other record in the uploaded sheet. These records can not be uploaded.
An external duplicate is a record that from the OCHEM point of view is a duplicate of some other record in the database, e.g., a duplicate of a record uploaded before. The rules for duplicate detection are described in more detail below in the “Data quality and consistency” section. The external duplicate can either be skipped, saved as a duplicate, or overwritten. Saving as a duplicate saves the record, but marks it as an error. External duplicates should be reviewed and corrected after uploading. When an overwrite option is selected, the system attempts to replace the existing record with a new one only if the user has sufficient access privileges. Otherwise, any attempt to overwrite the existing record will be rejected.
An unrecoverable error is any record that according to the system rules is invalid, e.g., wrong format or invalid value in the property column, a missing article or obligatory condition. A user can use a drop-down button to review the cause of the error. Such records cannot be saved to the database in their current form.
The preview browser has special filters and navigation buttons to help a user to review specific subsets of the uploaded dataset. At this stage it is also possible to download the Excel file generated with all the changes made in the previous step.
Data upload. After revision and verification of the data in the preview mode, the uploading process can be finalized by saving the records into the OCHEM database. A final upload report is generated and displayed. It contains error messages for every skipped record with a brief explanation of the reason why the record has not been uploaded. It is possible to export the report as an Excel file for further revision.
After the upload process is complete, the user can proceed to the compound property browser to inspect the freshly uploaded records and to work with them.
Sources of information
One of the basic principles of the OCHEM database policy is a strict requirement to provide the source of information for every experimental measurement introduced to the database. Most chemical databases [7–9] do not store this information, which makes it difficult to verify the data and to correct errors.
OCHEM supports two types of sources: articles (publications in scientific journals) and books (or chapters of books). There are a number of supplementary fields for every type of source: the title, the abstract, the journal, PubMed identifier, DOI identifier, ISBN number, web link, etc. For every source it is possible to store a PDF file which makes it easier to verify the data later on. For legal reasons a PDF file uploaded by a particular user is accessible by this user only.
There are several ways to add a new article to the database:
automatically retrieve the article from the PubMed database [18]. This requires a PubMed identifier.
upload from an external citation file. Currently the database supports the RIS, EndNote, BibTex and ISI formats. Such files are frequently provided by publishers.
input all the fields manually. This is the most tedious and error prone way and should be used only if the PubMed identifier is not known and no article citation file is available
Similarly to the addition of a new article, there are several ways to introduce a new book:
automatic retrieval by the ISBN number
manual input of all fields
Manually introduced articles and books can be edited later. If a publication has been retrieved automatically (via PubMed or ISBN identifier), further modifications are forbidden to ensure the consistency of information.
All publications stored in the OCHEM database can be searched for and accessed from the article browser. Similar to the other browsers in our system, the article browser has a set of filters, which allow search by author, PubMed ID, ISBN, title, journal, etc. From this browser, the user can easily navigate to the experimental measurements associated with a particular publication.
Data access and manipulation
All data records stored in the OCHEM database can be easily modified, filtered, searched and eventually grouped and organized for further convenient work.
Batch editing
Data modification can be done in two ways: by either editing a single record separately or by using the batch editing tool to work with multiple records simultaneously. In the editor window, the user can change all fields of a record: measured property, article information, structure and conditions. Batch editing is useful to correct systematic errors that might have occurred during batch upload, e.g., wrong units or missing conditions of experiment.
Filtering and search
Every browser of the OCHEM, e.g., molecules, articles, properties, etc., has a panel for data filtering. Filters are used to focus on a certain subset of the data, e.g., a set of certain properties, a set of specific organisms or conditions, etc. Records can be filtered by literature source (article or book where the data has been published), physicochemical property or experimental condition and structural information, e.g., molecule name or InChI key as well as by molecular sub-fragments. Comprehensive filter options to find duplicates, errors or non-validated entries are available.
The OCHEM supports relational filtering: for a given record, the user can find other records with the same structure (preserving or ignoring stereochemistry) or the records that have been modified at approximately the same time as this record.
Data can also be filtered by tags, i.e., labels assigned to molecules or properties. A desired set of tags, referred to as an “area of interest”, restricts all the displayed information (experimental measurements, publications, properties and compounds) to the selected tags.
Data organization
By applying various filters, a user can specify and select records of interest that can be stored separately in sets called baskets. A typical use of a basket is to assign both training and validation sets for modeling. The content of a basket can be browsed and modified from the compound properties browser or from the basket browser.
Users and access policies
The OCHEM database supports access to the data for guest and registered users. Registration is free and open for everyone. We strongly encourage registration since only registered users have access to the full set of features. The OCHEM database is based on the Wiki principle: all users can introduce new experimental data and modify the data introduced by others, with the particular access restrictions discussed below.
Access levels
There are different access levels for users, summarized in Fig. 4: guest (anonymous) users, normal users, privileged users and administrators. By default, a new registered user has a “normal” access level and can be elevated to “privileged” by the system administrator. Users can modify data introduced by other users from the same or lower access level group. For example, privileged users can modify data introduced by other privileged users, as well as by normal and guest users. Guest users have the lowest privileges and a limited functionality, e.g., they cannot store and modify models. For every entry OCHEM stores and displays the original depositor and the last modifier of the record, with timestamps of record creation and last modification. Additionally, the OCHEM stores all record changes with a reference to the date of change and the user who modified the record. Thus, all the changes are traceable and, if necessary, can be manually reverted to the previous state.
Fig. 4.
An overview of different user levels in the OCHEM. User’s rights decrease with level. Users can edit/delete entities of the same or lower user levels
Public and private records
By default, all records introduced to the database are publicly available unless the user explicitly makes the records private. All the access rules discussed above apply to the public data. Private records can be viewed and modified only by the introducer or by members of the same group.
Groups of users
The administrator can combine users in groups. The members of the same group can view and edit private records as well as access models and baskets of each other. This allows a group to work on the same project without making their data and models public.
User registration
The registration is open for everybody and requires a user to fill out a standard registration form with obligatory specification of login, password and email address. Immediately after registration, the user can access his account and use the OCHEM.
Data download
The users who contributed any public data to OCHEM will be allowed to download the data of other users (e.g., in Excel or SDF format). The number of records that can be downloaded by a user will be proportional to the number of new data records that (s)he uploaded and were validated by the administrator. We expect that this feature will motivate users to upload new data and to make these data public to the community. This feature is currently under development.
Data quality and consistency
Control of errors, data origin and quality
An experimental measurement can be marked as an “error”. Such records are highlighted with a red background and indicate a possible problem. The system allows users to manually mark a record as an error if they believe there is a mistake. In this case, the user should provide an explanation of the problem in the comment or discussion field related to this record. The OCHEM system can also automatically mark records as erroneous if they do not comply with the system rules. Namely, a record is automatically marked as an error if:
an obligatory condition of the experiment has not been specified (for example, a boiling point measurement without specifying the pressure is ambiguous and would be marked as an error automatically)
a duplicate of the record exists in the database (see the next section for the definition of “duplicate”)
Another quality indicator is the “to be verified” flag. This flag signals that the record has been introduced from a referencing article, e.g., benchmarking/methodological article and should be verified against the original publication. This flag can be set either manually or automatically by the system (e.g., in case of batch data upload, see the “Batch upload” section for details).
Duplicates management
To ensure data consistency, it is essential to avoid redundancy in the database. Thus, there is a need for strict rules for the definition of duplicates. In OCHEM two experimental records of a physicochemical or biological property are considered to be duplicates if they are obtained for the same compound under the same conditions, had the same measured value (with a precision up to 3 significant digits) and are published in the same article. We refer to these records as strong duplicates, as opposed to weak duplicates, for which only part of the information is the same. The OCHEM database does not forbid strong duplicates completely, but forces all the duplicates (except for the record introduced first) to be explicitly marked as errors. This ensures that there are no strong duplicates among the valid (i.e., non-error) records.
The uniqueness of chemical compounds is controlled by special molecular hashes, referred to as InChI-Keys [19]. Namely, for the determination of duplicated experimental measurements, two chemical structures are considered the same if they have identical Inchi-keys.
OCHEM allows weak duplicates (for example, completely identical experimental values, published in different articles) and provides facilities to find them. Moreover, in the modeling process, it is always automatically ensured that the same compounds in the training set appear only in onefold of the N-fold cross-validation process.
Experimental data origin
Each record has a colored dot indicating the origin of the data. Green dots indicate “original records” from publications with a description of experimental protocols; these are usually the publications where the property was originally measured (original data). The users can verify experimental conditions and experiments by reading these articles. These are the most reliable records in the database. The weak duplicates of original records have magenta dots. The other records have red dots and originate from articles that re-use the original data but for which the original records are not stored. These are frequently methodological QSAR/QSPR studies. The original records can be easily filtered out by checking a corresponding box in the compound property browser. Another filter, “primary records”, eliminates all weak duplicates except the record with the most early publication date.
Modeling framework
An essential part of the OCHEM platform is the modeling framework. Its main purpose is to provide facilities for the development of predictive computational models for physicochemical and biological properties of compounds. The framework is integrated with the database of experimental data and includes all the necessary steps required to build a computational model: data preparation, calculation and filtering of molecular descriptors, application of machine learning methods and analysis of a models’ performance. This section gives an overview of these features and of the steps required to build a computational model in the OCHEM.
Features overview
Concisely, the main features of the modeling framework within the OCHEM include:
Support of regression and classification models
Calculation of various molecular descriptors ranging from molecular fragments to quantum chemical descriptors. Both whole-molecule and per-atom descriptors are supported.
Tracking of each compound from the training and validation sets
Basic and detailed model statistics and evaluation of model performance on training and validation sets
Assessment of applicability domain of the models and their prediction accuracy
Pre-filtering of descriptors: manual selection, decorrelation filter, principal component analysis (PCA) based selection
Various machine learning methods including both linear and non-linear approaches
N-fold cross-validation and bagging validation of models
Multi-learning: models can predict several properties simultaneously
Combining data with different conditions of measurements and the data in different measurement units
Distribution of calculations to an internal cluster of Linux and Mac computers
Scalability and expendability for new descriptors and machine learning methods
The steps of a typical QSAR research in the OCHEM system and the corresponding features are summarized in a diagram in Fig. 5.
Fig. 5.
The workflow of a typical QSAR research in the OCHEM system
Model development
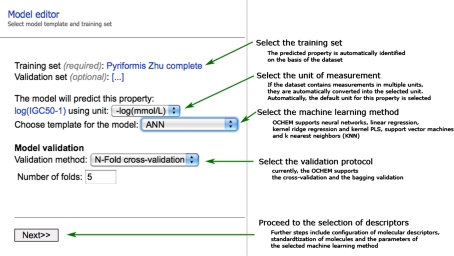
To create a new QSAR model in OCHEM the user must prepare the training and (optional) validation sets, configure the preprocessing of molecules (standardization and 3D optimization), choose and configure the molecular descriptors and the machine learning method, select the validation protocol (N-fold cross-validation or bagging) and, when the model has been calculated, review the predictive statistics and save or discard the model.
The following sections describe each of the aforementioned steps in detail.
Training and validation sets, machine learning method and validation
Training and validation datasets. One of the most important steps in model development is the preparation of input data, i.e., a training set that contains experimentally measured values of the predicted property.
The property that will be predicted by the model is identified automatically based on the contents of the training set. If the training set contains multiple properties, they will be predicted by the model simultaneously. This allows knowledge about different (but related) properties to be combined into a single model, so called multi-learning [20]. Multi-learning was shown to significantly increase the overall performance in comparison to models developed for each property separately [20].
The OCHEM system allows a user to combine heterogeneous data reported in different units of measurements into a single unit set. For every property, the user must select a unit; all the input data will be automatically converted to this unit and, therefore, the final model will give predictions in this unit.
Machine learning method. After assigning the training and validation sets (see Fig. 6), the user selects a machine learning method and a validation protocol. Currently OCHEM supports linear and Kernel Ridge Regression (KRR) [21], ASsociative Neural Networks (ASNN) [22], Kernel Partial Least Squares (KPLS) [23], a correction-based LogP-LIBRARY model [24], Support Vector Machines (SVM) [25], Fast Stagewise Multivariate Linear Regression (FSMLR) and k Nearest Neighbors (kNN) [26].
Fig. 6.
The first step of model creation: selection of a training and validation set, a machine learning method and a validation protocol
Validation of models. OCHEM offers two possibilities to validate a model: N-fold cross-validation and bagging. We recommend always using one of the two validation options to avoid a common pitfall of model over-fitting, which results to misleading predictions [27, 28]. The validation procedure is also used to calibrate the estimation of prediction accuracy [29].
If cross-validation is chosen, the whole process of model development, including the filtering of descriptors, is repeated N (by default 5) times with a different split of the initial set into training and validation sets. Only the respective training set is used in each step for model development.
In the case of bagging validation, the system generates N (by default 100) training sets and builds N models, based on these sets. The N sets are generated from the initial training set by random sampling with replacement. The compounds not included in the training set are used to validate the performance of this model; the final prediction for each compound is the mean prediction over all the models where this compound was in the validation set [30].
No matter what validation method is chosen, duplicated molecules (regardless of stereochemistry) are used either in training or validation sets but never in both simultaneously, which ensures the proper assessment of the model predictive ability.
Data preprocessing
Before chemical compounds are passed to the further steps of the modeling, they undergo a user-specified preprocessing procedure. Currently OCHEM standardizes different forms of the same molecule, i.e., mesomers and tautomers, by replacing them with a single unique representation. Since most descriptor generating software cannot work with salts (when two or more disconnected parts are present) all salts are automatically replaced with the largest component of the compound. For the same reason charged compounds are neutralized by adding/deleting hydrogens.
Furthermore, preprocessing steps are performed only during the modeling stage. In the database, all compounds are stored “as is”, i.e., original representations are kept as they were uploaded or provided by users.
Descriptors
The descriptors available in OCHEM are grouped by the software name that contributes them: ADRIANA.Code [31], CDK descriptors [32], Chemaxon descriptors [33], Chirality codes [34–38], Dragon descriptors [39], E-State indices [40], ETM descriptors [41, 42], GSFrag molecular fragments [43], inductive descriptors [44], ISIDA molecular fragments [45], quantum chemical MOPAC 7.1 descriptors [46], MERA and MerSy descriptors [47–50], MolPrint 2D descriptors [51], ShapeSignatures [52] and logP and aqueous solubility calculated with ALOGPS program [24]. The descriptor selection screen is shown in Fig. 7.
Fig. 7.
A screenshot of the descriptor selection and configuration panel
The following section briefly describes available descriptors. If at least one descriptor in the block requires 3D structures, the block is marked as 3D and as 2D otherwise.
ADRIANA.Code (3D) comprises a unique combination of methods for calculating molecular descriptors on a sound geometric and physicochemical basis [31, 53]. Thus, they are all prone to an interpretation and allow the understanding of the influence of various structural and physicochemical effects on the property under investigation. ADRIANA.Code performs calculations with user-supplied 3D structures or applies built-in methods to generate 3D structures based on rapid empirical models. In addition, it contains a hierarchy of increasing levels of sophistication in representing chemical compounds from the constitution through the 3D structure to the surface of a molecule. At each level, a wide range of physicochemical effects can be included in the molecular descriptors.
ALOGPS descriptors (2D) predict logP [54] and the aqueous solubility [55] of chemicals. This program was recently top-ranked amid 18 competitors for logP prediction using >96,000 in house molecules from Pfizer and Nycomed [24]. It was also reported to be “the best available ‘off-the-shelf package’ for intrinsic aqueous solubility prediction” at F. Hoffmann-La Roche [56]. ALOGPS does not have additional configuration options.
CDK descriptors (3D) are calculated by the DescriptorsEngine, which is a part of the Chemistry Development Kit (CDK) [32]. The CDK descriptors include 204 molecular descriptors of 5 types: topological, geometrical, constitutional, electronic and hybrid descriptors. The CDK also provides local atomic and bond-based descriptors, which will be included in OCHEM in future.
Chemaxon descriptors (also known as calculator plugins) calculate a range of physico-chemical and life-science related properties from chemical structures and are developed by ChemAxon. These calculators are usually part of the Marvin and JChem cheminformatics platforms. The descriptors are divided into 7 different groups: elemental analysis, charge, geometry, partitioning, protonation, isomers and “other” descriptors, which is a collective group for all heterogeneous descriptors that do not directly fall under any of the previous categories. For some descriptors, such as distribution coefficient (logD), the pH value is essential for calculation. By default, the descriptor value is calculated over the spectrum of pH from 0 to 14 with 1 pH unit intervals. However, it is possible to explicitly designate a specific pH value or range of pH values.
Chirality codes (3D) are molecular descriptors that represent chirality using a spectrum-like, fixed-length code, and include information on geometry and atomic properties. Conformation-independent chirality codes (CICC) [34] are derived from the configuration of chiral centers, properties of the atoms in their neighborhoods, and bond lengths. Conformation-dependent chirality codes (CDCC) [35] characterize the chirality of a 3D structure considered as a rigid set of points (atoms) with properties (atomic properties), connected by bonds. Physicochemical atomic stereo-descriptors (PAS) [36] were implemented to represent the chirality of an atomic chiral center on the basis of empirical physicochemical properties of its ligands—the ligands are ranked according to a specific property, and the chiral center takes an “S/R-like” descriptor relative to that property. The procedure is performed for a series of properties, yielding a chirality profile. All three types of chirality descriptors can distinguish between enantiomers. Examples of applications include the prediction of chromatographic elution order [35], the prediction of enantioselectivity in chemical reactions [34, 37], and the representation of metabolic reactions catalyzed by racemates and epimerases of E.C. subclass 5.1 [57].
DRAGON (v. 5.4) descriptors (3D) include more than 1,600 descriptors organized into 20 different sub-types that can be selected separately. DRAGON is an application for the calculation of molecular descriptors developed by the Milano Chemometrics and QSAR Research Group. The DRAGON descriptors include not only the simplest atom type, functional group and fragment counts, but also several topological and geometrical descriptors; molecular properties such as logP, molar refractivity, number of rotatable bonds, H-donors, H-acceptors, and topological surface area (TPSA) [58] are also calculated by using well-known published models.
E-State indices (2D) are separated on atom/bond type. In addition to indices it is also possible to select E-state counts, which correspond to counts of atom or bond types according to the respective indices. In some studies E-state counts were reported to produce better models than E-state indices [26].
ETM (Electronic-Topological Method [41, 42]) descriptors (3D) are based on the comparison of 3D structures of molecules. The molecules are represented as matrices where diagonal elements are atom charges and non-diagonal elements are distances between them. The molecules are compared with a template molecule and common fragments become ETM descriptors (i.e., 3D pharmacophores). There are usually two templates representing the most active and inactive molecules.
GSFRAG descriptors (2D) are the occurrence numbers of certain special fragments containing 2–10 non-hydrogen atoms; GSFRAG-L is an extension of GSFRAG that considers fragments that contain a labeled vertex, allowing one to capture the effect of heteroatoms. It was shown that the occurrence numbers of these fragments produce a unique code of a chemical structure for wide sets of compounds [43].
Inductive descriptors (3D) have been derived from the LFER (Linear Free Energy Relationships)-based equations for inductive and steric substituent constants and can be computed for bound atoms, groups and molecules using intra-molecular distances, atomic electronegativities and covalent radii [44].
ISIDA descriptors (2D) include two types of fragments: sequences and atom centered fragments, each of which includes explicitly atoms, bonds or both [45]. In the current version of OCHEM only sequences of atoms and bonds are used. The user can specify their minimum and maximum length.
MERA descriptors (3D) are calculated using the non-parametrical 3D MERA algorithm and include (a) geometrical MERA descriptors (linear and quadratic geometry descriptors, descriptors related to molecular volume, proportions of a molecule, ratios of molecular sizes, quantitative characteristics of molecular symmetry, dissymmetry, chirality), (b) energy characteristics (inter- and intra-molecular Van der Waals and Coulomb energies; decomposition of intermolecular energies) and (c) physicochemical characteristics (probabilities of association, heat capacity, entropy, pKa) [47–50, 59].
MerSy (MERA Symmetry, 3D) descriptors are calculated using 3D representation of molecules in the framework of the MERA algorithm (see above) and include the quantitative estimations of molecular symmetry with respect to symmetry axes from C2 to C6 and the inversion-rotational axis from S1 to S6 in the space of principal rotational invariants about each orthogonal component. Additionally, the molecular chirality is quantitatively evaluated in agreement with the negative criterion of chirality (the absence of inversion-rotational axes in the molecular point group) [47–50, 59].
MolPrint descriptors [51] (2D) are circular fingerprints [60] based on Sybyl mol2 atom types. They are very efficient and can be easily calculated even for libraries comprising millions of molecules. Circular fingerprints capture a lot of information that relates molecular structure to its bioactivity. It has been shown in large-scale comparative virtual screening studies that MolPrint descriptors often outperform other fingerprinting algorithms in enrichment [61, 62]. Given the binary nature of MolPrint 2D fingerprints, they are ideally suited for virtual screening and clustering of molecules, as well as to the generation of numerical bioactivity models, which are able to accommodate the presence/absence nature of the descriptor.
MOPAC descriptors (3D) include whole-molecule and atom-type descriptors. The latter can be used to model local (atom-dependent) properties of molecules, such as pKa or the site of metabolism [63].
Shape Signatures (3D) can be viewed as a very compact descriptor that encodes molecular shape and electrostatics in a single entity. It reduces the dimensionality of 3D molecular shape and surface charge by representing complex 3D molecules as simple histograms. These signatures lend themselves to rapid comparison with each other for virtual screening of large chemical databases. Shape Signatures can be used by itself or in conjunction with currently available computational modeling approaches commonly employed in drug discovery and predictive toxicology, such as traditional virtual screening, descriptor-based (e.g., QSAR) models, ligand-receptor docking, and structure-based drug design [52, 64–67].
The set of descriptors can be easily extended by incorporating new modules that could also be provided by external contributors. It is possible to use the output of previously created models as input for a new model: this option is sometimes referred to as a feature net [20].
For the descriptors that require 3D structures of molecules, users can either rely on 3D structures generated by CORINA [68] or retrieve molecules optimized by MOPAC and the AM1 Hamiltonian [46] calculated by the web services implemented within the CADASTER project (http://mopac.cadaster.eu). If additional parameters are required for the calculation of descriptors, e.g., pKa value for ChemAxon descriptors, they are specified explicitly in the interface of the corresponding descriptor block. In this case, the parameters are saved with descriptors and are then used exactly in the same form for new molecular sets during the model application.
ATOMIC descriptors (3D) are defined for a particular atom (active center) of a molecule. Atomic descriptors can be used to describe reactive centers (e.g., for pKa calculation, prediction of reactivity). These descriptors are applicable for the prediction of particular “local” properties of molecules that depend on the specified active center (currently, only macro pKa constants are supported). The currently available atomic descriptors are based on MOPAC descriptors and E-State indices.
New development includes descriptors that characterize ligand–protein interactions. These descriptors will allow using the information about 3D structure of proteins for modeling. For example, a number of docking derived descriptors based on Vina software [69] were added recently and are currently in use for an ongoing study for prediction of CYP450 inhibitors [70]. There is also a plan to extend OCHEM with other types of descriptors, e.g., those used in the COMBINE method [71].
Users can export most of the descriptor values (with an exception of commercial descriptors) for offline model development. Descriptor values can be exported as an Excel file or as a simple text file in a comma-separated values (CSV) format.
Before descriptors are passed to the machine-learning method, it is possible to filter part of them out by several criteria to avoid redundancy. Currently, OCHEM supports the following filtering options for descriptors: the number of unique descriptor values, the pairwise correlation of descriptors and the variance of principal components, obtained from the principal component analysis (PCA). Thus, it is possible to exclude highly correlated descriptors or descriptors that do not pass user specified thresholds.
Conditions of experiments
A unique feature of OCHEM is the possibility to use the conditions of experiments in modeling. Usually, chemical properties and biological activities depend on a number of conditions under which the experiment was carried out. Exemplary conditions are temperature, pressure, pH, measurement method, etc. OCHEM allows using these conditions in the modeling process as descriptors and as such permits combining data measured under different conditions into one modeling set. For example, boiling point data measured under different pressures can be combined into a single training set and used to develop a computational model. Another example is a combination of logP values measured in different buffers, e.g., pure water and 30% methanol.
The obligatory conditions of the experiments are selected in the same dialog as the molecular descriptors. For every selected condition, the user must provide (a) the default value that will be used for the records where the condition has not been specified, and (b) the unit to convert all the values to.
Configuration of the machine learning method
There are a number of configuration options that are specific for every particular machine learning method. These options are configured in separate dialog windows. Here, we briefly provide an overview of the methods and their parameters.
k Nearest Neighbors (kNN) predicts the property using the average property value of those k compounds from the training set that are nearest (in the descriptor space) to the target compound. The configurable options are: metrics type (Euclidean distance or the Pearson correlation coefficient) and the number of neighbors. By default the number of neighbors is determined automatically by the method itself.
ASsociative Neural Network (ASNN). This method uses the correlation between ensemble responses (each molecule is represented in the space of neural network models as a vector of predictions of neural network models) as a measure of distance amid the analyzed cases for the nearest neighbor technique [22, 72]. Thus ASNN performs kNN in the space of ensemble predictions. This provides an improved prediction by the bias correction of the neural network ensemble. The configurable options are: the number of neurons in the hidden layer, the number of iterations, the size of the model ensemble and the method of neural network training.
Fast Stagewise Multivariate Linear Regression (FSMLR) is a procedure for stagewise building of linear regression models by means of greedy descriptor selection. It can be viewed as a special case of the additive regression procedure (regression boosting) specially designed to be compatible with the three-set approach based on the use of three different sets for learning: training set, internal validation set and external test set [73]. The main configurable parameters are: (1) shrinkage—its lower values result in the large number of required iterations but may provide higher generalization performance, and (2) the relative size of the internal validation set used for stopping descriptor selection procedure.
Kernel Partial Least Squares (KPLS) and Kernel Ridge regression (KRR) are modifications of partial least squares (PLS) and ridge regression (RR) that use a non-linear kernel (for an introduction to kernel methods see book by Schölkopf and Smola [74]). The most important parameter for kernel-based methods is the type of kernel, as that determines non-linear relations. Available kernels are: linear, polynomial, and radial basis functions, as well as the iterative similarity optimal assignment kernel (ISOAK) [21]. The first three kernels are used with molecular descriptors. The ISOAK kernel is defined directly on the molecular structure graph. The individual parameters for every kernel can be either specified manually or configured to be selected automatically by the method itself in an inner loop of cross-validation.
The LOGP-LIBRARY model does not require any additional configuration options. This model is based on ASNN and corrects the ALOGPS logP model [75] using so called LIBRARY correction [76] with the training set. The idea of this method is to adjust the LogP model to predict other properties. The success of this methodology was shown for prediction of logD of chemical compounds at pH 7.4 [76, 77] and it was extended to prediction of arbitrary properties in the OCHEM database.
Multiple Linear Regression Analysis (MLRA) uses step-wise variable selection. The method eliminates at each step one variable that has regression coefficients that are not significantly different from zero (according to the t test). Thus, MLRA has only one parameter which corresponds to the p value of variables to be kept for the regression.
Support Vector Machine (SVM) uses the LibSVM program [78]. The SVM method has two important configurable options: the SVM type (ε-SVR and μ-SVR) and the kernel type (linear, polynomial, radial basis function and sigmoid). The other options can be optimized by the method automatically using grid search.
Monitoring of the model calculation
After assigning the training and validation sets, specifying descriptors and configuration parameters, the user is forwarded to a screen that displays the current status of the model calculation. As it can be quite a long process—up to several days or even weeks, if large datasets and/or large number of descriptors are used, it is possible to fetch results afterwards, which would allow working with OCHEM while the model is being calculated. The calculated results will be stored until they are retrieved by the user for further inspection.
All completed models are stored until the user checks them and decides to save or discard them. However, all completed models are deleted automatically after 1 month. It is possible to check the status of the pending models and to continue working with them (see Fig. 8).
Fig. 8.
The list of pending models. The models being calculated and the completed models waiting for an inspection by the user are listed here
The “pending tasks” dialog shows:
models being calculated at the moment and the current status of the calculation
models that successfully finished the training process and are waiting for the user’s decision to save or to discard them. These models are denoted as “ready” in the model “status” column.
failed models (e.g., terminated by user or failed because of errors during calculation process). The corresponding error message is displayed in the “Details” column.
From the pending tasks dialog, it is possible to terminate, delete or recalculate the models.
Analysis of models
OCHEM provides a variety of statistical instruments to analyze the performance of models, to find outliers in the training and validation sets, to discover the reasons for the outliers and to assess the applicability domain of the model. In this section, we briefly overview these instruments.
Basic statistics
Regression models. Commonly used measures of a regression model performance are the root mean square error (RMSE), the mean absolute error (MAE) and the squared correlation coefficient (r 2). The OCHEM system calculates these statistical parameters for both the training and the validation sets.
For a convenient visual inspection of the results OCHEM is equipped with a graphical tool that allows the user to create an observed-versus-predicted chart. This type of chart is traditionally used to visualize the model performance and to discover outliers. Each compound from the input dataset is represented as a dot on this chart, where the x-coordinate of the dot corresponds to the value of the experimentally observed property and the y-coordinate is the value predicted by the model. Each dot on the chart is interactive; a click on the dot opens a window with the detailed information about the compound: name, measured and predicted property values, publication, conditions of experiment, etc. The possibility to track each compound to the reference source is a very important step for understanding the reasons why the compound is considered to be an outlier. A user can quickly check why a bad prediction happened. Is it due to an error in the dataset, differences in the experimental conditions or due to the failure of the model to predict the compound properly? (see Fig. 9). This seemingly simple feature is a good example of the advantage of integrating the database with the modeling framework.
Fig. 9.
Basic statistics for a predictive model. The training set has a link that opens a browser of experimental records where a user can examine properties of all compounds used in the model. A click on a dot in the observed versus predicted chart opens a similar browser information window for the corresponding compound
Classification models. The OCHEM system uses the average correct classification rate (as a percentage) as a measure of the performance of the classification models. The correct classification rate is complemented with a confusion matrix that shows a number of compounds classified correctly for every class as well as details of the misclassified compounds, e.g., how many compounds from a class A are classified to belong to a class B (see Fig. 10).
Fig. 10.
Statistics of a classification model. Summarized are the prediction accuracies for the training and test sets as well as confusion matrices
Applicability domain assessment
A unique feature of OCHEM is the automatic assessment of the prediction accuracy. The estimation of the accuracy is based on the concept of “distance to a model” (DM) [29], i.e., some numerical value estimated solely from molecular structures and experimental conditions, which correlates with the average model performance. Currently several DMs are supported: the standard deviation of an ensemble of models (STDEV), the correlation in the space of models (CORREL) [79] and Mahalanobis distance (LEVERAGE). The DMs are calibrated against the accuracy of models for the training set using N-fold cross-validation as described elsewhere [29]. The estimated accuracy of predictions as a function of the respective DM is visualized on the accuracy averaging plot (see Fig. 11), which shows the absolute values of the prediction residuals versus a respective DM. The DMs are used to estimate the prediction accuracies for new molecules. The same methodology has been recently extended for classification models [80]. Currently, estimation of the prediction accuracy is readily available for the ASNN (CORREL, STDEV) and linear regression models (LEVERAGE). For other methods, e.g., kNN, KRR and KPLS, the estimation of the prediction accuracy can be performed using the bagging approach which generates an ensemble of models and uses the standard deviation of the ensemble predictions (referred to as BAGGING-STDEV).
Fig. 11.
The plot shows residuals of the predictions for the training set, mapped against the selected “distance to model”, in this case ASNN-STDEV, the standard deviation of ensemble prediction vector. This information is used to estimate accuracy of predictions, when the model is applied to new compounds
Comparison of models
Often it is useful to compare different models that are built using the same data but with different descriptors and machine learning methods. OCHEM supports a collective view of the models with the same training set. This screen is available from the basket browser as the model overview link.
Sustainability of models
Since OCHEM is a public database that is populated by users, the data may contain errors. Therefore, data may be changed during verification and correction by other users over time. It may lead to a significant alteration of the training sets and to invalidation of the previously developed models. To address this problem, OCHEM provides the possibility to recalculate the existing model preserving the previous workflow parameters (e.g., by applying the same machine learning method with the same parameters and descriptors) and to compare new results with the original model. This option is available solely for the user who has published the original model and for the OCHEM system administrators.
Application of models
List of available models
After the model has been successfully trained and saved, it can be applied to the prediction of new compounds (see a screen on Fig. 12).
Fig. 12.
Selection of models to predict properties for new compounds
To predict new compounds, the models are selected (by checking the rightmost checkbox) from previously trained and saved models. The model browser displays a brief summary of the model: the name, the predicted property, the training and validation sets and their sizes, the date of creation of the model. The following additional options are available from the model browser:
Download of model summary in Excel or CSV formats. Summary includes descriptors (where available), observed and predicted values and applicability domain values for training and validation sets.
Access to the model profile screen that displays complete information on the model and its statistics. In the model profile, it is possible to publish the model and recalculate it
Inspection of the training and validation sets by clicking on their names in the list
Deletion of models (only for non-published models)
Unpublished models are accessible only by the owner of a model and users of the same working group. Published models are available for revision for everyone. For security reasons, the OCHEM system assigns a random public identifier for each model. Any user who knows the identifier can access the model in a read-only mode. This is a convenient way to share a model with other users without publicly revealing it. It may be very helpful, for example, to provide an internal link for reviewers while submitting a paper for publication. A model becomes visible and accessible to all users once it is published on the OCHEM website. Published models cannot be deleted.
Predictions for new compounds
Any model in OCHEM can be applied to predict the target property for new compounds. A set of new compounds can either be provided in an SDF file, drawn manually in a molecule editor or selected as a basket, if the analyzed molecules are already present in the OCHEM database. After the molecules have been selected, the user is forwarded to the waiting screen. When the model calculation is completed the user is provided with the predictions by the selected models for all the target compounds. The predictions can be exported into an Excel file for further offline analysis.
For every prediction OCHEM estimates its accuracy (see the “Applicability domain assessment” section), which is very helpful for the users to decide whether the results of the given model are adequate for the purposes of their study.
Additionally, the predictions of a model are accessible via web services technology, which could be seamlessly integrated with other developing approaches in this area. The user can submit a molecule or a set of molecules using the web services and retrieve the predicted values. A tutorial and several examples on how to access predictions via the web services technology are provided on the OCHEM web site.
Interaction with users
The OCHEM system is under continuous development and, therefore, not all the features are covered in this article. The current status of the development and extended help is available at the project’s wiki (http://wiki.ochem.eu). Most of the interface windows of the OCHEM contain links to corresponding articles on the project’s wiki. We strongly encourage any feedback from the users. Registered users can submit bug reports (http://bugzilla.ochem.eu) or participate in a forum (http://forum.ochem.eu) to discuss limitations, advantages and requirements important to advance the OCHEM. Registered users can also use the internal mail to exchange messages or to get notifications about changes in the database, e.g., if a user deletes records of another user, the latter will receive an automatic e-mail about this change. Users may suggest their descriptors or new machine learning methods, that will then be considered for integration into OCHEM. The OCHEM developers are open for collaborations and joint studies that may result in joint publications and/or development of a new model on the OCHEM web site.
The detailed step-by-step tutorials on how to use the basic elements of the OCHEM are provided at http://www.ochem.eu/tutorials.
Implementation aspects
The OCHEM is mainly based on the Java platform. Resource-intensive calculation methods (ASNN, kNN, MLRA) were developed using C++ code. The data is stored in a MySQL database. All queries are executed using the Java Hibernate technology that provides an intermediate abstract layer between Java code and the database.
The JAXB library is used to create XML files and XSLT transformations to convert XML files to HTML web-pages. To connect design and functionality we used MVC methodology with the Java Spring framework. To make the user interface dynamic and user friendly, we used Java-script and AJAX, which makes the system look more like a dynamic online application rather than a static Web site.
For chemistry-related features we used the JME molecule editor [17], the CDK toolkits [81] and the ChemAxon (http://www.chemaxon.com) Standardizer. The CDK is used for various chemoinformatics tasks such as preprocessing and fragmentation of molecules and the calculation of descriptors. The visualization of molecules as well as interconversion of molecules between SDF, SMILES and MOL2 molecular formats is done using the ChemAxon toolkit.
The calculations are performed on a self-developed system for distributed calculations on a cluster with more than 300 CPUs. When possible, the tasks are calculated in parallel, e.g., different descriptor types, individual models from a model ensemble and models within a cross-validated model are calculated in parallel.
OCHEM comprises about 100,000 lines of Java, C++, and shell script code. Several of its critical components, e.g., the task management system, were inspired by the Virtual Computational Chemistry Laboratory (VCCLAB, http://www.vcclab.org) [13].
Summary and outlook
The Online Chemical Modeling Environment (OCHEM) contains a set of tools for easy creation, publication and use of predictive models for physicochemical and biological properties. The user-contributed database allows the uploading of large amounts of experimental data and supplementary information, like conditions of experiments, units of measurements with automatic interconversions, sources of the data (scientific publications, books), etc.
The database is tightly integrated with the modeling framework; the data can be flexibly filtered and used for the training of predictive computational models. The OCHEM modeling framework supports all the typical steps of QSAR/QSPR modeling: data preparation, calculation and filtering of molecular descriptors, application of machine learning methods (both classification and regression), analysis of the model, assessment of the modeling domain of applicability and, finally, using the model to predict target properties for new molecules. Importantly, OCHEM allows for the combining of data with different units of measurements, different conditions of experiments and even different properties and activities. The complexity of the modeling process is hidden behind a convenient and well documented user interface. Models can be published on the Web and publicly used by others.
OCHEM is available at http://www.ochem.eu and comes in two versions: as the main database and as the “sandbox”. The latter is intended for testing and to allow a user to become acquainted with the system. Currently, the main database contains about 120,000 publicly available experimental measurements for about 300 properties. Moreover, we developed tools that facilitate the migration of data from other databases and used them to introduce about 1,700,000 experimental measurements from the ChEMBL database (http://www.ebi.ac.uk/chembldb). These data are readily available for verification and work in the “sandbox”. Recently, we have also uploaded more than 23,000 records for physical properties such as boiling point, melting point and density from ChemExper (http://www.chemexper.com). To keep the data up-to-date, the update server periodically uploads new records from the ChemExper database. A similar server is currently being implemented for automatic data retrieval from the ChemSpider (http://www.chemspider.com). The developed methodology can be easily adapted for a quick integration of any other database.
At the moment OCHEM has been used to collect data and to develop QSARs in several published studies. For example, the Ames mutagenicity studies [80] included the collection of 6,542 experimental measurements. The model developed in this study is available at OCHEM at http://www.ochem.eu/models/1. The study for prediction of toxicity against T. Pyriformis bacteria [29] included a collection of 1,093 experimental measurements used for the model, which is available at http://www.ochem.eu/models/3. A number of QSAR studies for pKa calculation, calculation of lipophilicity of platinum compounds, inhibition of CYP450 1A2B, prediction of blood–brain barrier coefficients, protein-plasma binding as well as prediction of boiling and melting points are currently in preparation for a publication and is in peer-review. Furthermore, the platform is rapidly growing and is being used for a number of ongoing studies, e.g., within the FP7 CADASTER (http://www.cadaster.eu) and FP7 MC ITN “Environmental Chemoinformatics” (http://www.eco-itn.eu) projects.
Large collaborative projects have become a pervasive hallmark of research in physics and biology and, with the advent of large free databases of small molecules, to a lesser extent also in chemistry. The OCHEM project aims to contribute in this direction by providing a platform that integrates a large and richly annotated compound repository with modeling tools provided by a large number of collaborators that are recognized experts in the field.
OCHEM was initially developed with the help of a GO-Bio award (http://www.go-bio.de/projekte) from the German Federal Ministry of Education and Research (BMBF). The future support of the public version of OCHEM will be provided by the company eADMET GmbH (http://www.eadmet.com), which will also distribute a commercial version to the interested partners. This will guarantee that the project will be further developed and extended in the future.
Our vision of the OCHEM is to make it the platform of choice to contribute new data, descriptors, modeling tools, to perform on-line QSPR/QSAR studies and to share them with other users on the Web. “Computing chemistry on the web” [82] is becoming a reality.
Acknowledgments
This project was partially supported by GO-Bio BMBF project “Development of ADME/T methods using Associative Neural Networks: A novel self-learning software for confident ADME/T predictions”, project number 0313883, DLR German-Ukraine collaboration project UKR 08/006 and MC ITN “Environmental Chemoinformatics” ECO, project number 238701. We thank ChemAxon (http://www.chemaxon.com) for providing the Standardizer, calculator plugins and the molecule depiction tool. We also thank CDK for their chemoinformatics tools as well as the Java and MySQL communities for development of toolkits used in this project. Dr. Vladyslav Kholodovych is a recipient of Alexander von Humboldt Research Fellowship wants to express his gratitude to AvH Foundation for its support in this research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Iurii Sushko, Sergii Novotarskyi, Robert Körner and Anil Kumar Pandey made equal contributions to the article.
References
- 1.Patani GA, LaVoie EJ. Bioisosterism: a rational approach in drug design. Chem Rev. 1996;96:3147–3176. doi: 10.1021/cr950066q. [DOI] [PubMed] [Google Scholar]
- 2.Cronin MTD, Schultz TW. Pitfalls in QSAR. J Mol Struct Theochem. 2003;622:39–51. doi: 10.1016/S0166-1280(02)00616-4. [DOI] [Google Scholar]
- 3.Balakin KV, Savchuk NP, Tetko IV. In silico approaches to prediction of aqueous and DMSO solubility of drug-like compounds: trends, problems and solutions. Curr Med Chem. 2006;13:223–241. doi: 10.2174/092986706775197917. [DOI] [PubMed] [Google Scholar]
- 4.Tetko IV, Livingstone DJ. Rule-based systems to predict lipophilicity. Oxford: comprehensive medicinal chemistry II. Elsevier; 2007. pp. 649–668. [Google Scholar]
- 5.Moriguchi I, Hironon S, Liu Q, Nakagome I, Matsushita Y. Simple method of calculating octanol/water partition coefficient. Chem Pharm Bull. 1992;40:127–130. [Google Scholar]
- 6.Mannhold R, Poda GI, Ostermann C, Tetko IV. Calculation of molecular lipophilicity: state-of-the-art and comparison of log P methods on more than 96, 000 compounds. J Pharm Sci. 2009;98:861–893. doi: 10.1002/jps.21494. [DOI] [PubMed] [Google Scholar]
- 7.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucl Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams AJ. Internet-based tools for communication and collaboration in chemistry. Drug Discov Today. 2008;13:502–506. doi: 10.1016/j.drudis.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 9.The Chempedia Project (2011) http://www.chempedia.com/. Accessed 24 May 2011
- 10.Patiny L. Sharing product physical characteristics over the internet. Internet J Chem. 2000;3:1–6. [Google Scholar]
- 11.Bolton E, Wang Y, Thiessen PA, Bryant SH. PubChem: Integrated platform of small molecules and biological activities. Annu Rep Comput Chem. 2008;4:217–241. doi: 10.1016/S1574-1400(08)00012-1. [DOI] [Google Scholar]
- 12.Kaiser J. Science resources: chemists want NIH to curtail database. Science. 2005;308:774a. doi: 10.1126/science.308.5723.774a. [DOI] [PubMed] [Google Scholar]
- 13.Tetko IV, Gasteiger J, Todeschini R, Mauri A, Livingstone D, Ertl P, Palyulin VA, Radchenko EV, Zefirov NS, Makarenko AS, Tanchuk VY, Prokopenko VV. Virtual computational chemistry laboratory—design and description. J Comput Aided Mol Des. 2005;19:453–463. doi: 10.1007/s10822-005-8694-y. [DOI] [PubMed] [Google Scholar]
- 14.The OpenTox Project (2011) http://www.opentox.org/. Accessed 24 May 2011
- 15.Walker T, Grulke CM, Pozefsky D, Tropsha A. Chembench: A cheminformatics workbench. Bioinformatics. 2010;26:3000–3001. doi: 10.1093/bioinformatics/btq556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.QSAR DataBank (2011) An emerging proposal for the electronic organization and archiving of QSAR/QSPR model information. http://www.qsardb.org/. Accessed 24 May 2011
- 17.Ertl P. Molecular structure input on the web. J Cheminf. 2010;2:1. doi: 10.1186/1758-2946-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The PubMed Database (2011) http://www.ncbi.nlm.nih.gov/pubmed/. Accessed 24 May 2011
- 19.McNaught A. The IUPAC international chemical identifier: InChl-A new standard for molecular informatics. Chem Int. 2006;28:12–15. [Google Scholar]
- 20.Varnek A, Gaudin C, Marcou G, Baskin I, Pandey AK, Tetko IV. Inductive transfer of knowledge: application of multi-task learning and feature net approaches to model tissue-air partition coefficients. J Chem Inf Model. 2009;49:133–144. doi: 10.1021/ci8002914. [DOI] [PubMed] [Google Scholar]
- 21.Rupp M, Proschak E, Schneider G. Kernel approach to molecular similarity based on iterative graph similarity. J Chem Inf Model. 2007;47:2280–2286. doi: 10.1021/ci700274r. [DOI] [PubMed] [Google Scholar]
- 22.Tetko IV. Associative neural network. Methods Mol Biol. 2008;458:185–202. doi: 10.1007/978-1-60327-101-1_10. [DOI] [PubMed] [Google Scholar]
- 23.Rosipal R, Trejo LJ. Kernel partial least squares regression in reproducing kernel hilbert space. J Mach Learn Res. 2002;2:97–123. doi: 10.1162/15324430260185556. [DOI] [Google Scholar]
- 24.Tetko IV, Poda GI, Ostermann C, Mannhold R. Large-scale evaluation of log P predictors: local corrections may compensate insufficient accuracy and need of experimentally testing every other compound. Chem Biodivers. 2009;6:1837–1844. doi: 10.1002/cbdv.200900075. [DOI] [PubMed] [Google Scholar]
- 25.Cristianini N, Shawe-Taylor J. An introduction to support vector machines and other kernel-based learning methods. Cambridge: Cambridge University Press; 2000. [Google Scholar]
- 26.Tetko IV, Solov’ev VP, Antonov AV, Yao X, Doucet JP, Fan B, Hoonakker F, Fourches D, Jost P, Lachiche N, Varnek A. Benchmarking of linear and nonlinear approaches for quantitative structure-property relationship studies of metal complexation with ionophores. J Chem Inf Model. 2006;46:808–819. doi: 10.1021/ci0504216. [DOI] [PubMed] [Google Scholar]
- 27.Livingstone DJ, Manallack DT, Tetko IV. Data modelling with neural networks: advantages and limitations. J Comput Aided Mol Des. 1997;11:135–142. doi: 10.1023/A:1008074223811. [DOI] [PubMed] [Google Scholar]
- 28.Tropsha A, Gramatica P, Gombar VK. The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. QSAR Comb Sci. 2003;22:69–77. doi: 10.1002/qsar.200390007. [DOI] [Google Scholar]
- 29.Tetko IV, Sushko I, Pandey AK, Zhu H, Tropsha A, Papa E, Oberg T, Todeschini R, Fourches D, Varnek A. Critical assessment of QSAR models of environmental toxicity against Tetrahymena pyriformis: focusing on applicability domain and overfitting by variable selection. J Chem Inf Model. 2008;48:1733–1746. doi: 10.1021/ci800151m. [DOI] [PubMed] [Google Scholar]
- 30.Breiman L. Bagging predictors. Mach Learn. 1996;24:123–140. [Google Scholar]
- 31.Adriana (2011) Code web-page. http://www.molecular-networks.com/products/adrianacode/. Accessed 24 May 2011
- 32.Steinbeck C, Hoppe C, Kuhn S, Floris M, Guha R, Willighagen EL. Recent Developments of the Chemistry Development Kit (CDK)—An Open-Source Java Library for Chemo- and Bioinformatics. Curr Pharm Des. 2006;12:2111–2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- 33.Chemaxon (2010) Chemaxon—toolkits and desktop applications for chemoinformatics: calculator Plugins. http://www.chemaxon.com/library/scientific-presentations/calculator-plugins/. Accessed 24 May 2011
- 34.Aires-de-Sousa J, Gasteiger J. New description of molecular chirality and its application to the prediction of the preferred enantiomer in stereoselective reactions. J Chem Inf Comput Sci. 2001;41:369–375. doi: 10.1021/ci000125n. [DOI] [PubMed] [Google Scholar]
- 35.Aires-de-Sousa J, Gasteiger J. Prediction of enantiomeric selectivity in chromatography. Application of conformation-dependent and conformation-independent descriptors of molecular chirality. J Mol Graph Model. 2002;20:373–388. doi: 10.1016/S1093-3263(01)00136-X. [DOI] [PubMed] [Google Scholar]
- 36.Zhang QY, Aires-de-Sousa J. Physicochemical stereodescriptors of atomic chiral centers. J Chem Inf Model. 2006;46:2278–2287. doi: 10.1021/ci600235w. [DOI] [PubMed] [Google Scholar]
- 37.Aires-de-Sousa J, Gasteiger J. Prediction of enantiomeric excess in a combinatorial library of catalytic enantioselective reactions. J Comb Chem. 2005;7:298–301. doi: 10.1021/cc049961q. [DOI] [PubMed] [Google Scholar]
- 38.Aires F, Prigent C, Rossow WB. Neural network uncertainty assessment using bayesian statistics: a remote sensing application. Neural Comput. 2004;16:2415–2458. doi: 10.1162/0899766041941925. [DOI] [PubMed] [Google Scholar]
- 39.Todeschini R, Consonni V. Molecular descriptors for chemoinformatics. New York: Wiley-VCH; 2009. [Google Scholar]
- 40.Hall LH, Kier LB, Brown BB. Molecular similarity based on novel atom-type electrotopological state indices. J Chem Inf Comput Sci. 1995;35:1074–1080. [Google Scholar]
- 41.Dimoglo AS, Shvets NM, Tetko IV, Livingstone DJ. Electronic-topological investigation of the structure—acetylcholinesterase inhibitor activity relationship in the series of N-benzylpiperidine derivatives. Q Struct Act Relat. 2001;20:31–45. doi: 10.1002/1521-3838(200105)20:1<31::AID-QSAR31>3.0.CO;2-S. [DOI] [Google Scholar]
- 42.Dimoglo AS. Compositional approach to electronic structure description of chemical compounds, oriented computer analysis of structure-activity relation. Khim Pharm Zh. 1985;4:438–444. [Google Scholar]
- 43.Skvortsova MI, Baskin II, Skvortsov LA, Palyulin VA, Zefirov NS, Stankevich IV. Chemical graphs and their basis invariants. J Mol Struct Theochem. 1999;466:211–217. doi: 10.1016/S0166-1280(98)00467-9. [DOI] [Google Scholar]
- 44.Cherkasov A, Ban F, Santos-Filho O, Thorsteinson N, Fallahi M, Hammond GL. An updated steroid benchmark set and its application in the discovery of novel nanomolar ligands of sex hormone-binding globulin. J Med Chem. 2008;51:2047–2056. doi: 10.1021/jm7011485. [DOI] [PubMed] [Google Scholar]
- 45.Varnek A, Fourches D, Horvath D, Klimchuk O, Gaudin C, Vayer P, Solov’ev V, Hoonakker F, Tetko IV, Marcou G. ISIDA—Platform for Virtual Screening Based on Fragment and Pharmacophoric Descriptors. Curr Comp Aid Drug Des. 2008;4:191–198. doi: 10.2174/157340908785747465. [DOI] [Google Scholar]
- 46.Stewart JJP. Optimization of parameters for semiempirical methods I. Method. J Comput Chem. 1989;10:209–220. doi: 10.1002/jcc.540100208. [DOI] [Google Scholar]
- 47.Potemkin VA, Grishina MA. A new paradigm for pattern recognition of drugs. J Comput Aided Mol Des. 2008;22:489–505. doi: 10.1007/s10822-008-9203-x. [DOI] [PubMed] [Google Scholar]
- 48.Grishina MA, Bartashevich EV, Potemkin VA, Belik AV. Genetic Algorithm for Predicting Structures and Properties of Molecular Aggregates in Organic Substances. J Struct Chem. 2002;43:1040–1044. doi: 10.1023/A:1023663115138. [DOI] [Google Scholar]
- 49.Potemkin VA, Pogrebnoy AA, Grishina MA. Technique for energy decomposition in the study of “receptor–ligand” complexes. J Chem Inf Model. 2009;49:1389–1406. doi: 10.1021/ci800405n. [DOI] [PubMed] [Google Scholar]
- 50.Potemkin VA, Bartashevich EV, Belik AV. New approaches to prediction of thermodynamic parameters of substances using molecular data. Russ J Phys Chem. 1996;70:411–416. [Google Scholar]
- 51.Bender A, Mussa HY, Glen RC, Reiling S. Molecular similarity searching using atom environments, information-based feature selection, and a naive Bayesian classifier. J Chem Inf Comput Sci. 2004;44:170–178. doi: 10.1021/ci034207y. [DOI] [PubMed] [Google Scholar]
- 52.Zauhar RJ, Moyna G, Tian L, Li Z, Welsh WJ. Shape signatures: a new approach to computer-aided ligand- and receptor-based drug design. J Med Chem. 2003;46:5674–5690. doi: 10.1021/jm030242k. [DOI] [PubMed] [Google Scholar]
- 53.Gasteiger J. Of molecules and humans. J Med Chem. 2006;49:6429–6434. doi: 10.1021/jm0608964. [DOI] [PubMed] [Google Scholar]
- 54.Tetko IV, Tanchuk VY, Villa AE. Prediction of n-octanol/water partition coefficients from PHYSPROP database using artificial neural networks and E-state indices. J Chem Inf Comput Sci. 2001;41:1407–1421. doi: 10.1021/ci010368v. [DOI] [PubMed] [Google Scholar]
- 55.Tetko IV, Tanchuk VY, Kasheva TN, Villa AE. Estimation of aqueous solubility of chemical compounds using E-state indices. J Chem Inf Comput Sci. 2001;41:1488–1493. doi: 10.1021/ci000392t. [DOI] [PubMed] [Google Scholar]
- 56.Du-Cuny L (2006) Aqueous solubility of drug-like compounds, PhD diss., Rheinische Friedrich-Wilhelms-University Bonn. http://hss.ulb.uni-bonn.de/2006/0744/0744.htm. Accessed 3 June 2011
- 57.Latino DARS, Zhang Q-Y, Aires-de-Sousa J. Genome-scale classification of metabolic reactions and assignment of EC numbers with self-organizing maps. Bioinformatics. 2008;24:2236–2244. doi: 10.1093/bioinformatics/btn405. [DOI] [PubMed] [Google Scholar]
- 58.Ertl P, Rohde B, Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem. 2000;43:3714–3717. doi: 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
- 59.Bartashevich EV, Potemkin VA, Grishina MA, Belik AV. A method for multiconformational modeling of the three-dimensional shape of a molecule. J Struct Chem. 2002;43:1033–1039. doi: 10.1023/A:1023611131068. [DOI] [Google Scholar]
- 60.Glem RC, Bender A, Arnby CH, Carlsson L, Boyer S, Smith J. Circular fingerprints: flexible molecular descriptors with applications from physical chemistry to ADME. IDrugs. 2006;9:199–204. [PubMed] [Google Scholar]
- 61.Sastry M, Lowrie JF, Dixon SL, Sherman W. Large-scale systematic analysis of 2d fingerprint methods and parameters to improve virtual screening enrichments. J Chem Inf Model. 2010;50:771–784. doi: 10.1021/ci100062n. [DOI] [PubMed] [Google Scholar]
- 62.Bender A, Jenkins JL, Scheiber J, Sukuru SCK, Glick M, Davies JW. How similar are similarity searching methods? a principal component analysis of molecular descriptor space. J Chem Inf Model. 2009;49:108–119. doi: 10.1021/ci800249s. [DOI] [PubMed] [Google Scholar]
- 63.Rupp M, Körner R, Tetko IV. Estimation of acid dissociation constants using graph kernels. Mol Inf. 2010;29:731–740. doi: 10.1002/minf.201000072. [DOI] [PubMed] [Google Scholar]
- 64.Hartman I, Gillies AR, Arora S, Andaya C, Royapet N, Welsh WJ, Wood DW, Zauhar RJ. Application of screening methods, shape signatures and engineered biosensors in early drug discovery process. Pharm Res. 2009;26:2247–2258. doi: 10.1007/s11095-009-9941-z. [DOI] [PubMed] [Google Scholar]
- 65.Chekmarev D, Kholodovych V, Kortagere S, Welsh W, Ekins S. Predicting inhibitors of acetylcholinesterase by regression and classification machine learning approaches with combinations of molecular descriptors. Pharm Res. 2009;26:2216–2224. doi: 10.1007/s11095-009-9937-8. [DOI] [PubMed] [Google Scholar]
- 66.Chekmarev DS, Kholodovych V, Balakin KV, Ivanenkov Y, Ekins S, Welsh WJ. Shape signatures: new descriptors for predicting cardiotoxicity in silico. Chem Res Toxicol. 2008;21:1304–1314. doi: 10.1021/tx800063r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meek PJ, Liu Z, Tian L, Wang CY, Welsh WJ, Zauhar RJ. Shape signatures: speeding up computer aided drug discovery. Drug Discov Today. 2006;11:895–904. doi: 10.1016/j.drudis.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 68.Sadowski J, Gasteiger J, Klebe G. Comparison of automatic three-dimensional model builders using 639 X-ray structures. J Chem Inf Comput Sci. 1994;34:1000–1008. [Google Scholar]
- 69.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Novotarskyi S, Sushko I, Körner R, Pandey AK, Tetko IV (2011) A comparison of different QSAR approaches to modeling CYP450 1A2 inhibition. J Chem Inf Model. doi:10.1021/ci200091h [DOI] [PubMed]
- 71.Ortiz AR, Pisabarro MT, Gago F, Wade RC. Prediction of drug binding affinities by comparative binding energy analysis. J Med Chem. 1995;38:2681–2691. doi: 10.1021/jm00014a020. [DOI] [PubMed] [Google Scholar]
- 72.Tetko IV. Neural network studies. 4. Introduction to associative neural networks. J Chem Inf Comput Sci. 2002;42:717–728. doi: 10.1021/ci010379o. [DOI] [PubMed] [Google Scholar]
- 73.Zhokhova N, Baskin I, Palyulin V, Zefirov A, Zefirov N. Fragmental descriptors with labeled atoms and their application in QSAR/QSPR studies. Dokl Chem. 2007;417:282–284. doi: 10.1134/S0012500807120026. [DOI] [Google Scholar]
- 74.Schölkopf B, Smola AJ. Learning with kernels. Cambridge: MIT Press; 2002. [Google Scholar]
- 75.Tetko IV, Tanchuk VY. Application of associative neural networks for prediction of lipophilicity in ALOGPS 2.1 program. J Chem Inf Comput Sci. 2002;42:1136–1145. doi: 10.1021/ci025515j. [DOI] [PubMed] [Google Scholar]
- 76.Tetko IV, Poda GI. Application of ALOGPS 2.1 to predict log D distribution coefficient for Pfizer proprietary compounds. J Med Chem. 2004;47:5601–5604. doi: 10.1021/jm049509l. [DOI] [PubMed] [Google Scholar]
- 77.Tetko IV, Bruneau P. Application of ALOGPS to predict 1-octanol/water distribution coefficients, logP, and logD, of AstraZeneca in-house database. J Pharm Sci. 2004;93:3103–3110. doi: 10.1002/jps.20217. [DOI] [PubMed] [Google Scholar]
- 78.Fan R-E, Chen P-H, Lin C-J. Working Set Selection Using Second Order Information for Training Support Vector Machines. J Mach Learn Res. 2005;6:1889–1918. [Google Scholar]
- 79.Tetko IV, Bruneau P, Mewes H-W, Rohrer DC, Poda GI. Can we estimate the accuracy of ADME-Tox predictions? Drug Discov Today. 2006;11:700–707. doi: 10.1016/j.drudis.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 80.Sushko I, Novotarskyi S, Körner R, Pandey AK, Kovalishyn VV, Prokopenko VV, Tetko IV. Applicability domain for in silico models to achieve accuracy of experimental measurements. J Chemom. 2010;24:202–208. doi: 10.1002/cem.1296. [DOI] [Google Scholar]
- 81.Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner J, Willighagen EL. The Blue Obelisk-interoperability in chemical informatics. J Chem Inf Model. 2006;46:991–998. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tetko IV. Computing chemistry on the web. Drug Discov Today. 2005;10:1497–1500. doi: 10.1016/S1359-6446(05)03584-1. [DOI] [PubMed] [Google Scholar]