Abstract
Aims/hypothesis
The adiponectin signalling pathway is largely unknown, but recently the adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain and leucine zipper motif (APPL1), has been shown to interact directly with adiponectin receptor (ADIPOR)1. APPL1 is present in C2C12 myoblasts and mouse skeletal muscle, but its presence in human skeletal muscle has not been investigated.
Methods
Samples from type 2 diabetic, and lean and non-diabetic obese participants were analysed by: immunoprecipitation and western blot; HPLC-electrospray ionisation (ESI)-mass spectrometry (MS) analysis; peak area analysis by MS; HPLC-ESI-MS/MS/MS analysis; and RT-PCR analysis of APPL1 mRNA.
Results
Immunoprecipitation and western blot indicated a band specific to APPL1. Tryptic digestion and HPLC-ESI-MS analysis of whole-muscle homogenate APPL1 unambiguously identified APPL1 with 56% sequence coverage. Peak area analysis by MS validated western blot results, showing APPL1 levels to be significantly increased in type 2 diabetic and obese as compared with lean participants. Targeted phosphopeptide analysis by HPLC-ESI-MS/MS/MS showed that APPL1 was phosphorylated specifically on Ser401. APPL1 mRNA expression was significantly increased in obese and type 2 diabetic participants as compared with lean participants. After bariatric surgery in morbidly obese participants with subsequent weight loss, skeletal muscle APPL1 abundance was significantly reduced (p < 0.05) in association with an increase in plasma adiponectin (p < 0.01), increased levels of ADIPOR1 (p < 0.05) and increased muscle AMP-activated protein kinase (AMPK) phosphorylation (p < 0.05).
Conclusions/interpretation
APPL1 abundance is significantly higher in type 2 diabetic muscle; APPL1 is phosphorylated in vivo on Ser401. Improvements in hyperglycaemia and hypoadiponectinaemia following weight loss are associated with reduced skeletal muscle APPL1, and increased plasma adiponectin levels and muscle AMPK phosphorylation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-011-2173-x) contains supplementary material, which is available to authorised users.
Keywords: Adiponectin, APPL1, Obesity, Type 2 diabetes
Introduction
The adipocyte hormone, adiponectin, has recently received much attention due to its insulin-sensitising effects on lipid and glucose metabolism [1–3]. The molecular causes of insulin resistance are currently unknown, but one possible mechanism is altered or impaired adiponectin signalling. Defects in adiponectin signalling can arise from alterations in total adiponectin concentrations, defects in adiponectin receptor (ADIPOR) levels or defects in intracellular signalling molecules. Low serum concentrations of adiponectin accompany insulin resistance [4] and have been associated with disorders such as obesity [5–7], hyperlipidaemia [8, 9], cardiovascular disease [10] and type 2 diabetes [11]. mRNA of both ADIPORs (ADIPOR1, ADIPOR2) is highly expressed in human skeletal muscle [12], as opposed to mouse, where Adipor2 is most abundantly expressed in liver and Adipor1 is predominantly expressed in myotubes [13].
The intracellular adiponectin signalling pathway is still largely unknown, despite the identification of two ADIPORs, ADIPOR1 and ADIPOR2 [13]. ADIPORs are seven transmembrane domain receptors, but are unique from G-protein coupled receptors due to their opposite location of the intracellular N-terminal and the extracellular C-terminal [13]. Insulin-resistant ob/ob mice have decreased abundance of ADIPOR1 and ADIPOR2 [14]; but mRNA expression of ADIPOR1 and ADIPOR2 showed no difference between healthy, lean control and obese type 2 diabetic individuals [15]. However, Jang et al. reported that ADIPOR2 levels are significantly lower in type 2 diabetic participants than in lean controls, with ADIPOR1 following the same trend, albeit without statistical significance [16]. ADIPOR1 and ADIPOR2 mRNA expression is significantly decreased in normal glucose-tolerant individuals with a strong family history of diabetes as compared with those with no history of diabetes [12]. In addition, insulin-stimulated glucose disposal is positively associated with ADIPOR1 and ADIPOR2 expression [12]. Finally, cultured myotubes from obese controls and obese diabetic participants showed increased levels of ADIPOR1 relative to lean controls under basal conditions [17]. These findings suggest that circulating levels of adiponectin and expression of ADIPOR genes play an important role in the regulation of skeletal muscle insulin action.
It is unclear whether intracellular adiponectin signalling might also be abnormal in muscle from insulin-resistant individuals. Previous work indicates that AMP-activated protein kinase (AMPK) and p38 mitogen-activated protein kinase (p38MAPK) are downstream of the ADIPOR, but do not interact directly with ADIPORs. This work also identified the adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain and leucine zipper motif (APPL1) as a protein that interacts with ADIPOR1, using the intracellular N-terminal portion of ADIPOR1 as bait in a yeast two-hybrid screen [18]. APPL1 is a 79 kDa protein, which contains several distinct domains, including a pleckstrin homology domain, a phosphotyrosine-binding domain and a leucine zipper motif embedded in the coiled-coil region [19]. APPL1 interacts with numerous other receptors, including follicle-stimulating hormone receptor, EGF receptor, androgen receptor and the deleted in colorectal cancer receptor [20–23]. In addition, APPL1 interacts with components of the insulin signalling pathway such as v-akt murine thymoma viral oncogene homologue 2, phosphoinositide 3-kinase, regulatory subunit 1 and the p110 subunit of phosphatidylinositol 3-kinase [19, 22]. This indicates that APPL1 may act as a scaffold protein. Overabundance of APPL1 increases fatty acid oxidation and glucose metabolism upon adiponectin stimulation by increasing the activity of AMPK and p38MAPK [18]. Although APPL1 does not directly interact with AMPK, adiponectin-stimulated activity of AMPK seems to be regulated in part by APPL1 interaction with the ADIPOR [18].
Some [24], but not all [25], studies suggest that AMPK protein levels and mRNA expression are not altered in skeletal muscle of obese or type 2 diabetic patients. In light of the evidence connecting adiponectin signalling and AMPK activity, and given both the decreased fat oxidation in insulin-resistant muscle and the role of APPL1 in adiponectin signalling, we sought to determine whether APPL1 abundance or expression is altered in insulin resistance. To test our hypothesis, we first sought to detect the presence of APPL1 in human skeletal muscle, using western blot techniques and mass spectrometry (MS). We then established a novel approach for quantifying protein abundance using MS to detect changes between lean control, obese control and type 2 diabetic participants. We also performed mRNA analysis to determine whether changes in APPL1 and ADIPOR1 were present between these groups. Finally, we examined the effect of weight loss following bariatric surgery on skeletal muscle ADIPOR1/2, APPL1 and AMPK phosphorylation in obese insulin-resistant individuals.
Methods
Participants for protein analysis
We recruited 29 participants for protein abundance and quantification studies. These included lean control participants, obese control participants and type 2 diabetic participants (Table 1). All studies were approved by the Institutional Review Board of the University of Texas Health Science Center at San Antonio or Arizona State University. Results for some participants used in this study have been published previously [26].
Table 1.
Characteristics of participants used for protein analysis
| Characteristic | Lean control | Obese control | Type 2 diabetes |
|---|---|---|---|
| Age (years) | 36 ± 3 | 41 ± 3 | 48 ± 2* |
| Women (n) | 4 | 4 | 5 |
| Men (n) | 6 | 5 | 5 |
| BMI (kg/m2) | 25.8 ± 1.0 | 31.6 ± 0.9 | 31.8 ± 2.4 |
| Fasting glucose (mmol/l) | 5.5 ± 0.2 | 5.4 ± 0.1 | 8.9 ± 1.3† |
| Lean participants (%) | 76.2 ± 2.3 | 67.3 ± 2.8 | 64.2 ± 3.3* |
| HbA1c (%) | 4.8 ± 0.1 | 5.0 ± 0.1 | 7.6 ± 0.8‡ |
| M value (mg kg−1 min−1) | 6.7 ± 0.6 | 4.1 ± 0.4 | 1.9 ± 0.7*** |
Values are given as mean±SEM, unless indicated otherwise
*p < 0.05 and ***p < 0.0001 vs lean control; † p < 0.005 vs lean and obese control; ‡ p < 0.0005 vs lean and obese control
Participants for mRNA analysis
Muscle (vastus lateralis) biopsies from 21 participants were used for real-time quantitative PCR analysis. Participant categories were lean control, obese control and type 2 diabetic (Table 2). The study protocol was approved by the Institutional Review Board of University of Texas Health Science Center at San Antonio. All participants gave written informed consent.
Table 2.
Characteristics of participants used for mRNA analysis
| Characteristic | Lean control | Obese control | Type 2 diabetes |
|---|---|---|---|
| Age (years) | 32 ± 3 | 34 ± 3 | 50 ± 4† |
| Women (n) | 3 | 3 | 3 |
| Men (n) | 4 | 4 | 4 |
| BMI (kg/m2) | 24.2 ± 1.0 | 29.3 ± 1.0 | 34.1 ± 3.3* |
| Fasting glucose (mmol/l) | 5.1 ± 0.2 | 5.2 ± 0.1 | 7.2 ± 0.7† |
| Lean participants (%) | 81.6 ± 6.9 | 69.2 ± 2.6 | 70.5 ± 3.1 |
| HbA1c (%) | 4.7 ± 0.1 | 5.1 ± 0.1 | 6.9 ± 0.4† |
| M value (mg kg−1 min−1) | 6.3 ± 0.5 | 4.1 ± 0.6* | 2.7 ± 0.5*** |
Values are given as mean±SEM, unless indicated otherwise
*p < 0.05 and ***p < 0.001 vs lean control; † p < 0.005 vs lean and obese control
Effects of weight loss on adiponectin signalling
We also studied six (age 38 ± 4 years, BMI 48.5 ± 4.8 kg/m2, HbA1c 6.0 ± 0.1% [mean±SEM]) non-diabetic morbidly obese participants with the metabolic syndrome, who underwent bariatric surgery (Roux-en-Y gastric bypass) (Table 3). Vastus lateralis muscle biopsies were obtained during surgery and 6 months following surgery, and were used for real-time quantitative PCR analysis and determination of AMPK phosphorylation. These studies were approved by the Baylor College of Medicine Institutional Review Board.
Table 3.
Metabolic and anthropometric variables before and after weight loss subsequent to bariatric surgery
| Variable | Before weight loss | After weight lost |
|---|---|---|
| Body weight (kg) | 135.2 ± 13.8 | 100.3 ± 10.1** |
| BMI (kg/m2) | 48.5 ± 4.8 | 35.1 ± 2.9** |
| Fasting insulin (pmol/l) | 161.8 ± 10.4 | 84.7 ± 6.3** |
| Fasting glucose (mmol/l) | 5.8 ± 0.3 | 4.7 ± 0.1** |
| HOMA-IR | 6.0 ± 0.5 | 2.6 ± 0.2** |
| Adiponectin (μg/ml) | 6.5 ± 0.6 | 11.6 ± 0.9** |
Values are given as mean±SEM
**p < 0.01 for difference between before vs after weight loss
HOMA-IR, HOMA of insulin resistance
Oral glucose tolerance test
Baseline blood samples for determining plasma glucose, NEFA and insulin concentrations were drawn at −30, −15 and 0 min. At time 0, participants ingested 75 g glucose in 300 ml water. Plasma glucose, NEFA and insulin concentrations were measured at 15 min intervals for 2 h.
Euglycaemic–hyperinsulinaemic clamp with muscle biopsies
A euglycaemic–hyperinsulinaemic clamp was performed to determine insulin sensitivity and to obtain human skeletal muscle biopsy as previously described [27]. Basal biopsies were used for all protein abundance and MS analyses. A clamp was completed to determine participants’ insulin sensitivity.
Analytical determinations
Plasma glucose was measured using the glucose oxidase method (Beckman Instruments, Fullerton, CA, USA). Plasma insulin concentrations were measured by radioimmunoassay (Diagnostic Products, Los Angeles, CA, USA). Plasma adiponectin concentration was measured by ELISA (Linco Research, St Charles, MO, USA)
Muscle biopsy processing
Frozen muscle biopsies were homogenised in detergent containing lysis buffer as described [28]. Briefly, muscle biopsies were homogenised while still frozen in buffer (10 μl/mg tissue) consisting of (final concentrations): 50 mmol/l HEPES, pH 7.6, 150 mmol/l NaCl, 20 mmol/l sodium pyrophosphate, 20 mmol/l β-glycerophosphate, 10 mmol/l NaF, 2 mmol/l sodium orthovanadate, 2 mmol/l EDTA, pH 8.0, 1% (vol./vol.) IGEPAL, 10% (vol./vol.) glycerol, 2 mmol/l phenylmethylsulfonyl fluoride, 1 mmol/l MgCl2, 1 mmol/l CaCl, 10 μg/ml leupeptin and 10 μg/ml aprotinin. Biopsies were homogenised with a polytron homogeniser for 20 s, cooled on ice for 30 min and then centrifuged for 20 min at 10,000 g and 4°C. Biopsy supernatant fractions were frozen until use. Protein concentrations were determined using the Lowry method [29].
Immunoprecipitation, gel electrophoresis and western blot analysis
APPL1 was immunoprecipitated using an anti-APPL1 antibody (gift from L. Q. Dong). Protein A sepharose beads (Sigma, St Louis, MO, USA) were rehydrated for 2 h at 4°C in 1× PBS (Gibco, Grand Island, NY, USA). Beads were distributed equally between samples, washed three times in PBS and then incubated for 1 h at room temperature in 300 μl PBS with anti-APPL1 antibody (2 μl). Beads were again washed three times with PBS and aspirated till dry. Muscle lysates (2.5 mg) were added to beads and incubated overnight at 4°C. After incubation, lysates were removed and the beads washed three times with PBS, after which 35 μl 2× SDS was added to beads and the beads boiled at 95°C for 4 min. Protein (immunoprecipitated or whole muscle lysate) was separated on a 10% SDS-polyacrylamide gel, transferred onto a nitrocellulose membrane and blocked for 1 h in 1% (wt/vol.) BSA at room temperature. Membranes were incubated for 1 h at room temperature with anti-APPL1 or anti-β-actin (Cell Signaling Technology, Danvers, MA, USA) primary antibodies. Membranes were washed three times and incubated for 1 h at room temperature with secondary antibodies (anti-rabbit, 1:1500; GE Healthcare, Piscataway, NJ, USA). Membranes were then washed and proteins visualised using an electrochemoluminscence reaction (Perkin Elmer, Boston, MA, USA). The densities of the bands corresponding to the proteins of interest were measured using a software package (Quantity One, Versadoc; Bio-Rad, Hercules, CA, USA). The ratio of the densities of APPL1 to β-actin was analysed by one-way ANOVA. Significance was defined as p < 0.05.
Antibodies
The APPL1 antibody was raised in rabbits as previously described [18]. Briefly, antiserum to APPL1 was raised in rabbits using a glutathione-S-tranferase-APPL1c. An antibody to β-actin was purchased from Cell Signaling.
Electrophoresis, staining and in-gel digestion
Proteins were separated on a 10% SDS-polyacrylamide gel containing 100 μg of muscle lysate per participant. Proteins were visualised with Coomassie blue staining. The gel portions containing APPL1 (80–90 kDa) and actin (37–50 kDa) were excised and prepared as previously described [28, 30], and as outlined in the electronic supplementary material (ESM, Preparation of samples).
Mass spectrometry
HPLC-electrospray ionisation (ESI)-MS was performed on a hybrid linear ion trap Fourier transform ion cyclotron resonance mass spectrometer (LTQ FT; Thermo Fisher, San Jose, CA, USA), which was fitted with a PicoView nanospray source (New Objective, Woburn, MA, USA). The mass spectrometer was calibrated weekly according to manufacturer’s instructions, achieving mass accuracies of the calibrants within 2 ppm. Online capillary HPLC was performed using a device (Paradigm MS4 micro HPLC; Michrom BioResources, Auburn, CA, USA) with a PicoFrit column (New Objective) (75 μmol/l i.d.; packed with ProteoPep II C18 material, 30 nm), mobile phase linear gradient of 2 to 27% (vol./vol.) acetonitrile in 0.1% (vol./vol.) formic acid in 45 min, a hold of 5 min at 27% acetonitrile, followed by a step to 50% acetonitrile, a hold of 5 min and then a step to 80% (flow rate 250 nl/min).
A ‘top ten’ approach was used to identify APPL1 in muscle homogenate, whereas a ‘targeted’ approach was used for peak area analysis; both of these methods have been described and validated previously by our lab [28, 31]. The ‘top ten’ approach was used for immunoprecipitated APPL1 from human skeletal muscle and to verify the accuracy of the antibody. All further experiments for MS analysis were performed on whole-muscle homogenates.
Peak area analysis was used to determine differences in protein levels between the three participant groups studied. To validate western blot data and mRNA data, β-actin was used as a normalisation protein for APPL1 levels; peptides targeted are listed in ESM Table 1. We reconstructed ion chromatograms of the fragment ions of seven peptides for APPL1 and β-actin quantification. The mass:charge ratio (m/z) values of the seven representative peptides were placed on the target list, based on ion intensity. The fragment ion m/z values had to be higher than the precursor ion in order to reduce interference from the background or other co-eluting peptide ions. We assessed the linearity of the β-actin to APPL1 normalisation quantification; complete methods for this can be found in the ESM (Validation of the use of the APPL1). This method demonstrates that our choice of β-actin as an internal normalisation peptide is valid for protein quantification using the peak area technique. Once these data were generated, 100 μg of whole-muscle homogenate was run on a gel, and bands for APPL1 and β-actin were excised, trypsinised, desalted and analysed by MS as described above. The ratios of APPL1:β-actin were used for our quantification analysis.
Real-time quantitative
RT-PCR Total RNA was isolated using STAT-60 (Tel-Test, Friendswood, TX, USA) and purified with an mRNA kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Concentration was determined by spectrophotometer at 260/280 nm. Quantitative real-time PCR was used to quantify mRNA. The specific primers used for quantification are listed in ESM Table 2. mRNA fold changes were calculated relative to the average lean control value quantified relative to β-actin. A device (ICycler; Bio-Rad) was used to quantify mRNA levels and fold changes were expressed using the 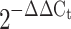 method [32].
method [32].
AMPK phosphorylation
For western blotting, muscle lysates were prepared as previously described. Proteins (50 μg) from muscle lysates were separated by 10% SDS-PAGE and transferred to nitrocellulose membranes. After blocking the membranes with 5% (wt/vol.) BSA/TBST, membranes were probed overnight at 4°C with antibodies against anti-phospho-AMPK (Thr172) (1:1,000; Cell Signaling). AMPK phosphorylation was normalised for total AMPK α protein content using a pan-α antibody (1:1000; Upstate Biotechnology, Lake Placid, NY, USA). Bound antibodies were detected using anti-rabbit immunoglobulin horseradish-peroxidase-linked antibody and electrochemoluminscence reagents (Perkin Elmer), and the bands were quantified using ImageQuant (GE Healthcare, Piscataway, NJ, USA) software.
Statistical analysis
Changes in APPL1 using western blot and MS were analysed by ANOVA. Changes in mRNA expression are expressed as fold change relative to lean control average and statistical significance compared with lean control assessed by ANOVA. Changes after weight loss in obese participants were compared using paired t tests. All data are presented as means±SEM. A value of p < 0.05 was considered statistically significant.
Results
APPL1 protein abundance determined by western blotting
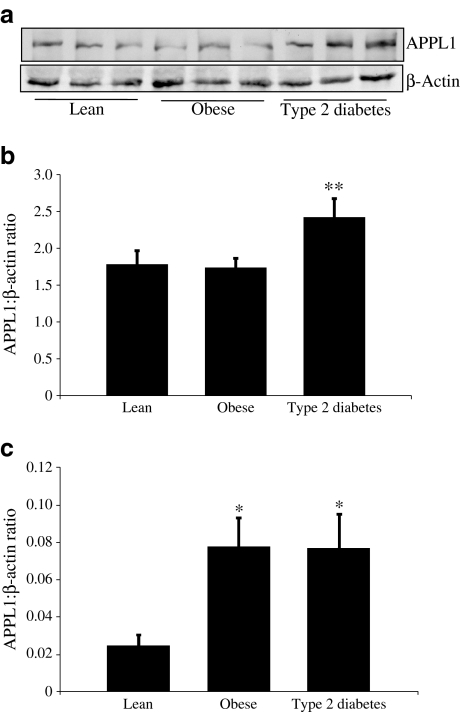
The detection of APPL1 in human skeletal muscle by immunoprecipitation indicates that measurable quantities of the protein were available for analysis (ESM Fig. 1). Whole-muscle homogenates (not immunoprecipitated) were separated by SDS-PAGE and western blots analysed for densitometry. APPL1 and actin antibodies showed specific bands for the corresponding molecular weights of APPL1 and β-actin. APPL1 protein levels were significantly increased in type 2 diabetic participants (2.42 ± 0.25 arbitrary units [means±SEM]) as compared with lean control (1.78 ± 0.18) and obese control participants (1.74 ± 0.12; p < 0.05 for all) (Fig. 1a, b). Our attempts to co-immunoprecipate endogenous ADIPORs from human skeletal muscle were unsuccessful. Undetectability of co-immunoprecipitation may only mean that the interaction was too weak to be maintained during the immunoprecipitation procedure. Although previous investigators have reported co-immunoprecipation of APPL1 and ADIPORs, this was done in a cell-based system [18]. This is not uncommon, as strong co-immunoprecipitation of the insulin receptor and IRS1 in a yeast two-hybrid system has also been reported, but this interaction is very difficult to observe in human skeletal muscle (L. J. Mandarino, unpublished results) [33].
Fig. 1.
Human skeletal muscle APPL1 protein level changes. a Vastus lateralis muscle biopsies were homogenised, separated by SDS-PAGE and proteins transferred electrophorectically to nitrocellulose membrane. Membranes were exposed to anti-APPL1 or β-actin antibody and visualised by chemiluminescence. Density of each band was determined using Versadoc software. b Protein quantification, expressed as ratio of APPL1:β-actin, showing that APPL1 levels increased significantly in type 2 diabetic participants as compared with lean control and obese control participants. Values mean±SEM (error bars). c Whole skeletal muscle homogenates were used for peak area analysis. Biopsies were homogenised, separated by SDS-PAGE and stained with Coomassie blue. Bands were cut for APPL1 and β-actin, subjected to trypsin digestion and quantified by MS as described. Protein levels are expressed as ratio of APPL1: β-actin (×102). Values are mean±SEM (error bars); n = 8 or 9 per group. *p < 0.05
APPL1 quantification using peak area analysis
Validation of APPL1 quantification using MS is shown in ESM Fig. 2, with complete methods for APPL1 identification described in the ESM (APPL1 identification). To confirm changes of APPL1 levels as visualised by western blots, HPLC-ESI-MS analysis of muscle homogenates was performed. A ‘targeted’ approach using peptide m/z values for each APPL1 and β-actin peptide was performed. Seven unique peptides for APPL1 or β-actin were targeted. Those identified were quantified ±95% CI. ESM Fig. 3 shows one of seven peptides used for APPL1 quantification as an example. The peak area is calculated for the unique spectrum identified as the particular peptide targeted. Peak area was obtained for β-actin and APPL1 peptides, and the ratio of APPL1:β-actin was used to numerically evaluate protein amount in the sample. Peak area analysis of APPL1 protein showed significantly higher levels in obese control and type 2 diabetic participants than in lean control participants (Fig. 1c).
Identification of phosphorylation of APPL1 at Ser401 in human muscle
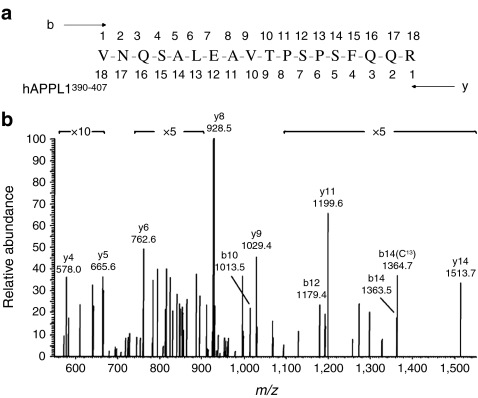
We had previously identified Ser430 and Ser401 as APPL1 phosphorylation sites in cultured myocytes (L. Q. Dong, unpublished data). In addition, previous reports have identified Ser401 as a phosphorylation site on APPL1 in HeLa nuclear extracts [34]. Since phosphorylation was undetectable using the phospho-specific antibodies developed for that study, we used MS to assess phosphorylation of APPL1. We targeted these and other potential phosphorylation sites as predicted by Phosphosite (www.phosphosite.org; accessed 13 April 2011), calculated as a loss of 80 Da or 98 Da on the basis of a 2+ or 3+ charge state, respectively. Due to the low levels of APPL1 phosphorylated peptides in whole-muscle homogenates, immunoprecipitation of APPL1 was performed. Muscle homogenate (2.5 mg) was immunoprecipitated with the APPL1 antibody, and phospho-Ser401 targeted and identified on the basis of the 3+ charge state. The tandem mass spectrum of pSer401 peptide is shown in Fig. 2.
Fig. 2.
Identification of Ser401 as a phosphorylation site. APPL1 phosphorylation at Ser401 was detected in tryptic digests of APPL1 isolated from a human muscle biopsy. a APPL1390–407 VNQSALEAVTPpSPSFQQR (pSer401) peptide was analysed (b) by tandem mass spectrum using HPLC-ESI-MS/MS/MS. The presence of y-series ions containing phosphate starting with y5 support phosphorylation of Ser401
Changes in APPL1 and ADIPOR1 mRNA expression between participant groups
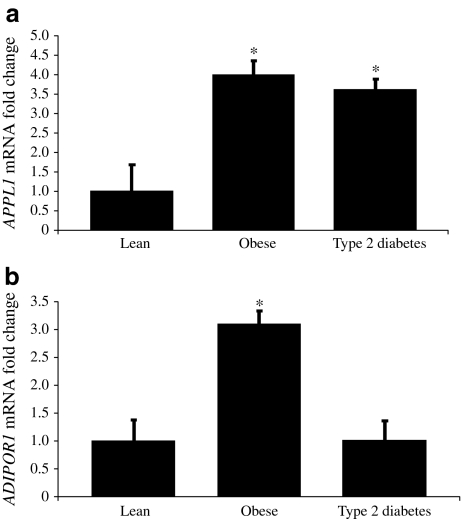
To determine whether changes in mRNA expression of APPL1 and ADIPOR1 were present in lean, obese and diabetic participants, real-time PCR was used. Results indicate a similar trend to that seen in protein levels of APPL1. Obese controls and type 2 diabetic patients had significantly increased mRNA expression of APPL1 compared with lean controls (Fig. 3a). Skeletal muscle ADIPOR1 expression increased approximately threefold (p < 0.05) in obese participants compared with lean controls, although type 2 diabetic patients did not differ from lean control individuals (Fig. 3b).
Fig. 3.
Human skeletal muscle expression of (a) APPL1 and (b) ADIPOR1. Quantitative real-time PCR was used to quantify mRNA. mRNA fold changes were calculated relative to average lean control value quantified relative to β-actin. mRNA expression and fold changes are expressed using the 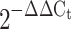 method [32]. *p < 0.05 vs lean
method [32]. *p < 0.05 vs lean
Changes in skeletal muscle adiponectin signalling after weight loss
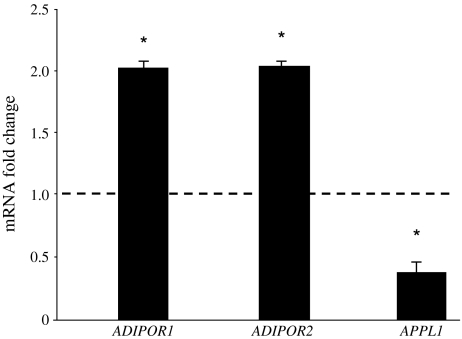
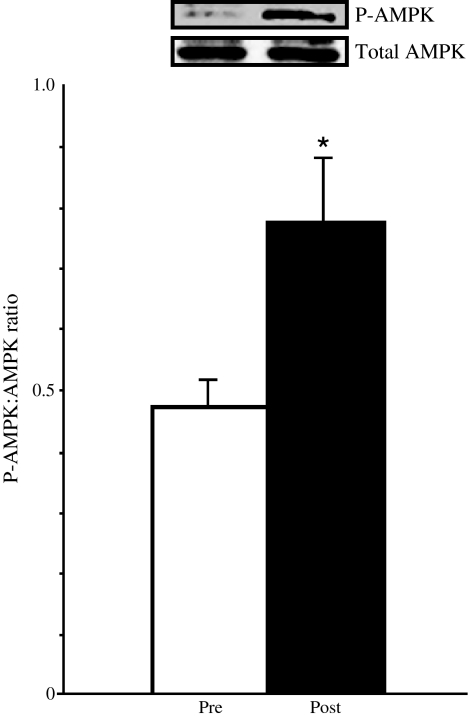
At 6 months following Roux-en-Y gastric bypass surgery [35], body weight (p < 0.01), mean fasting plasma glucose (p < 0.01), plasma insulin (p < 0.01) and insulin resistance (HOMA of insulin resistance) (p < 0.01) decreased significantly (Table 3). The improvement in insulin sensitivity was associated with an increase in plasma adiponectin levels (6.5 ± 0.6 to 11.6 ± 0.9 μg/ml, p < 0.01). Skeletal muscle ADIPOR1 and ADIPOR2 expression increased approximately twofold (p < 0.05), while APPL1 expression decreased by approximately 50% (p < 0.05) following weight loss (Fig. 4). Skeletal muscle AMPK phosphorylation (P-AMPK: AMPK ratio) increased significantly following weight loss (0.47 ± 0.05 to 0.78 ± 0.13, p < 0.05) (Fig. 5).
Fig. 4.
Effect of weight loss following bariatric surgery on ADIPOR1, ADIPOR2 and APPL1 expression. Quantitative real-time PCR was used to quantify mRNA. mRNA fold changes were calculated relative to average pre-surgery value quantified relative to 18S values. mRNA expression and fold changes are expressed using the 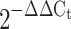 method [32]. *p < 0.05
method [32]. *p < 0.05
Fig. 5.
Skeletal muscle AMPK expressed as phosphorylated AMPK (P-AMPK):total AMPK ratio in six morbidly obese patients before (pre) and after (post) weight loss following bariatric surgery. *p < 0.05
Discussion
Adiponectin signalling is an intriguing area of research due to its anti-inflammatory, glucose-lowering and insulin-sensitising properties [1–3, 36–38]. Although some components of the pathway have been elucidated, many questions regarding the regulation and interaction of proteins involved remain unanswered. Previous findings have shown that adiponectin interacts with the C-terminal extracellular portion of the ADIPOR [13] and, upon binding, recruits APPL1 to the N-terminal intracellular portion [18]. This binding may facilitate the downstream effects of adiponectin, which include AMPK and p38MAPK activation, thus leading to increased fatty acid oxidation and glucose uptake [18, 37].
Previous reports have indicated that APPL1 plays an important role in the adiponectin signalling pathway by transmitting signals from the receptor to downstream targets [18, 19]. Although APPL1 has been identified and characterised in cells, to date no studies have identified its presence in human skeletal muscle. Here we have identified APPL1 in biopsies taken from human vastus lateralis muscle and quantified APPL1 protein and mRNA in samples from lean, obese and type 2 diabetic participants. To determine whether APPL1 was present in human skeletal muscle, immunoprecipitated and supernatant fractions of muscle homogenates were run on western blots, revealing a band specific for APPL1. To verify that this band was indeed APPL1, we ran identical gels, using one for western blot and the other for Coomassie staining. The Coomassie stain showed a very light band at ~80 kDa, similar to the size of APPL1 (82 kDa). We cut this band and performed a ‘top ten’ approach using the MASCOT (version 2.2; Matrix Science, London UK) search engine, which detects the top ten most abundant proteins in a sample. MASCOT detected APPL1 in the muscle homogenate sample with sequence coverage of 56%. We also tested whether APPL2 was present in the sample, but did not find this protein (data not shown). Upon verification that the western blot band corresponded to APPL1, we ran whole-muscle homogenates to quantify western blots using the APPL1 antibody. Western blot quantification showed APPL1 levels to be significantly higher in type 2 diabetic patients than in lean and obese controls.
Although western blot can be useful for quantifying protein levels, we validated our APPL1 quantification results using MS. In fact, in the present study, we implemented a unique MS quantification technique using β-actin peptides as normalisation factor. To verify the validity of this method for quantification purposes, increasing amounts of muscle homogenate samples were analysed (50 μg, 75 μg and 100 μg). Peptides for β-actin and APPL1 were targeted and Scaffold (version 02_00_06; Proteome Software, Portland, OR, USA) was used to quantify peak areas. The peak areas of the peptides for each protein were averaged and plotted for regression analysis. The averages of the peptides were quantified so that the ratio of APPL1:β-actin levels represented protein present in the sample. The CV from these ratios was found to be 6.59%, indicating that increasing amounts of sample were consistent based on the APPL1:β-actin ratio. For this study, we quantified proteins based on peak area as opposed to ion intensity, because peak area is the average peak of many scans, as opposed to using just one scan for ion intensity, and therefore reduces variability.
After determining that β-actin could be used as a normalisation factor, we then quantified APPL1 levels in human skeletal muscle samples using peak area analysis. The peak area ratio showed significantly higher APPL1 levels in obese control and type 2 diabetic participants compared with lean control. The difference in data between western blot and MS could be explained by the variability in immunoblot analysis. Due to the mRNA and MS data, and also based on linearity validation, we believe that MS analysis of protein abundance is more likely to be correct. This indicates that APPL1 levels are higher in an insulin-resistant state (obese controls and type 2 diabetic patients) and that the body may be compensating for decreased levels of adiponectin, and therefore trying to increase signalling downstream of the ADIPOR through increased APPL1 production.
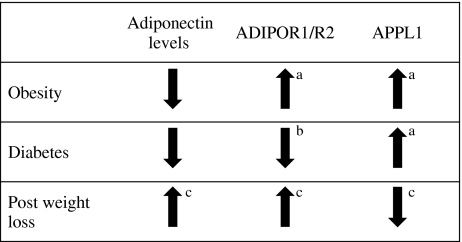
We also used quantitative real-time PCR to determine changes in APPL1 mRNA among the three experimental groups. APPL1 mRNA increased significantly in obese control and type 2 diabetic participants, as compared with lean control. This verifies our protein data, showing that in a state of insulin resistance, APPL1 mRNA expressison is increased, possibly representing a compensatory mechanism in skeletal muscle to enhance adiponectin signalling. Additionally, ADIPOR1 mRNA expression was significantly increased in obese compared with lean controls, but was not altered in type 2 diabetic patients. The increase in ADIPOR1 mRNA in obese controls may be an additional compensatory mechanism triggered by the decreased adiponectin levels in obese and type 2 diabetic patients. ADIPOR1 mRNA expression in type 2 diabetic patients may become exhausted, thus leading to levels similar to those in lean controls. The combination of compensatory mechanisms for APPL1 and ADIPOR1 due to decreased adiponectin levels in the obese state illustrates how this signalling pathway is altered in insulin resistance (Fig. 6).
Fig. 6.
Schematic representation of protein or mRNA changes in adiponectin, ADIPOR1/2 and APPL1 associated with obesity, diabetes and post weight loss. aCompensatory mechanism compared with lean; bexhausted system compared with lean; cnormalised system compared with pre-weight loss
To further understand how APPL1 and ADIPORs change in relation to insulin resistance, skeletal muscle ADIPOR1/2 and APPL1 expression was determined following weight loss after bariatric surgery in morbidly obese, non-diabetic participants with the metabolic syndrome. Following weight loss, plasma adiponectin increased significantly and was associated with a twofold increase in both ADIPOR1 and ADIPOR2 expression. APPL1 expression was significantly reduced and skeletal muscle AMPK phosphorylation increased significantly, suggesting that the correction of hyperglycaemia and hypoadiponectinaemia following weight loss is associated with a reduction in APPL1 expression. This normalisation of APPL1 and ADIPOR1/R2 levels after weight loss illustrates the compensatory mechanism to changes in adiponectin levels pre and post-surgery (Fig. 6).
In addition to mRNA and protein level changes, we also have identified Ser401 as a phosphorylation site in APPL1 in human skeletal muscle. This phosphorylation site has previously been identified in HeLa cell nuclear extract, although this is the first time it has been reported in human skeletal muscle [34]. Currently, we are testing the kinase responsible for this phosphorylation site, and whether this site plays a functional role in regulating APPL1 function in adiponectin signalling.
In conclusion, APPL1 levels are associated with insulin resistance, during which the body may be attempting to compensate for the decreased adiponectin levels found in this state. In addition, APPL1 is phosphorylated in human skeletal muscle at Ser401, a mechanism of functional regulation that is now under investigation and may shed new light on the adiponectin signalling pathway. Improvements in hyperglycaemia and hypoadiponectinaemia following weight loss are associated with enhanced skeletal muscle adiponectin signalling, as well as AMPK phosphorylation and a reduction in APPL1 levels.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 110 kb)
Targeted peptides for β-actin and APPL1 (PDF 57 kb)
RT-PCR primers (PDF 49 kb)
Identification of APPL1 using both western blots and MS techniques. a Human skeletal muscle homogenates were immunoprecipitated with anti-APPL1 antibody and then blotted for APPL1 protein. b Coomassie staining of the SDS-PAGE gel indicates a band corresponding to the size of APPL1. Gel slices were cut as indicated and subjected to trypsin digest and preparation for HPLC-ESI linear ion trap Fourier transform ion cyclotron resonance analysis. c Sequence coverage (black italics) of APPL1, using gel slices as indicated (b) and showing 56% sequence coverage (PDF 20 kb)
Regression analysis of β-actin and APPL1 peptides based on peak area analysis. Vastus lateralis muscle biopsies were homogenised, separated by SDS-PAGE and stained with Coomassie blue. Bands were cut at 80 to 90 kDa (for APPL1) and 37 to 50 kDa (for β-actin). Peak area analysis was used to determine increasing amounts of protein, as described in Methods. Peak areas shown are 1 × 108 and 1 × 105 for β-actin and APPL1, respectively. Values represent mean±SEM (error bars) (PDF 34 kb)
Example of peak area for APPL1 peptide VNQSALEAVTPSPSFQQR. Retention time (RT) is the time of elution for this particular peptide, which elutes at 37.41 min. The peptide was verified by retention time and spectrum matching. Peak Area (AA), or AUC, was used for quantification by MS analysis. In this example the peak area is 111,243 arbitrary units. The peak area is then used for quantification purposes, and seven different peptides for each APPL1 and β-actin were averaged. NL, normalised level; F, filter; ITMS, ion trap mass spectrometry; cNSI, centroid nano spray inlet; SM, smoothing points (PDF 17 kb)
Acknowledgements
This work was supported in part by grants from the American Diabetes Association 1-06-CR-03 (to M. Bajaj) and the Ron MacDonald Foundation at St Luke’s Episcopal Hospital 08RDM008 (to M. Bajaj), as well as by a Marilyn Fishman Grant for Diabetes Research from the Endocrine Fellows Foundation (to S. Shahani) and grants NIH R01DK081750-01A1 (to Z. Yi) and NIH R01DK066483 (to L. J. Mandarino).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- ADIPOR
Adiponectin receptor
- AMPK
AMP-activated protein kinase
- APPL1
Adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain and leucine zipper motif
- ESI
Electrospray ionisation-mass
- MS
Mass spectrometry
- m/z
Mass:charge ratio
- p38MAPK
p38 Mitogen-activated protein kinase
Contributor Information
L. J. Mandarino, Email: Lawrence.mandarino@asu.edu
M. Bajaj, Email: mandeepbajaj@hotmail.com
References
- 1.Fruebis J, Tsao TS, Javorschi S, et al. Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc Natl Acad Sci USA. 2001;98:2005–2010. doi: 10.1073/pnas.041591798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamauchi T, Kamon J, Waki H, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 3.Berg AH, Combs TP, Du X, et al. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat Med. 2001;7:947–953. doi: 10.1038/90992. [DOI] [PubMed] [Google Scholar]
- 4.Matsubara M, Maruoka S, Katayose S. Inverse relationship between plasma adiponectin and leptin concentrations in normal-weight and obese women. Eur J Endocrinol. 2002;147:173–180. doi: 10.1530/eje.0.1470173. [DOI] [PubMed] [Google Scholar]
- 5.Statnick MA, Beavers LS, Conner LJ, et al. Decreased expression of apM1 in omental and subcutaneous adipose tissue of humans with type 2 diabetes. Int J Exp Diabetes Res. 2000;1:81–88. doi: 10.1155/EDR.2000.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arita Y, Kihara S, Ouchi N, et al. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun. 1999;257:79–83. doi: 10.1006/bbrc.1999.0255. [DOI] [PubMed] [Google Scholar]
- 7.Weyer C, Funahashi T, Tanaka S. Hypoadiponectinemia in obesity and type 2 diabetes: close association with insulin resistance and hyperinsulinemia. J Clin Endocrinol Metab. 2001;86:1930–1935. doi: 10.1210/jc.86.5.1930. [DOI] [PubMed] [Google Scholar]
- 8.Matsubara M, Maruoka S, Katayose S. Decreased plasma adiponectin concentrations in women with dyslipidemia. J Clin Endocrinol Metab. 2002;87:2764–2769. doi: 10.1210/jc.87.6.2764. [DOI] [PubMed] [Google Scholar]
- 9.Cnop M, Havel PJ, Utzschneider KM, et al. Relationship of adiponectin to body fat distribution, insulin sensitivity and plasma lipoproteins: evidence for independent roles of age and sex. Diabetologia. 2003;46:459–469. doi: 10.1007/s00125-003-1074-z. [DOI] [PubMed] [Google Scholar]
- 10.Ouchi N, Kihara S, Arita Y, et al. Adiponectin, an adipocyte-derived plasma protein, inhibits endothelial NF-kappaB signaling through a cAMP-dependent pathway. Circulation. 2000;102:1296–1301. doi: 10.1161/01.cir.102.11.1296. [DOI] [PubMed] [Google Scholar]
- 11.Hotta K, Funahashi T, Arita Y. Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler Thromb Vasc Biol. 2000;20:1595–1599. doi: 10.1161/01.ATV.20.6.1595. [DOI] [PubMed] [Google Scholar]
- 12.Civitarese AE, Jenkinson CP, Richardson D, et al. Adiponectin receptors gene expression and insulin sensitivity in non-diabetic Mexican Americans with or without a family history of type 2 diabetes. Diabetologia. 2004;47:816–820. doi: 10.1007/s00125-004-1359-x. [DOI] [PubMed] [Google Scholar]
- 13.Yamauchi T, Kamon J, Ito Y, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423:762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- 14.Tsuchida A, Yamauchi T, Ito Y, et al. Insulin/Foxo1 pathway regulates expression levels of adiponectin receptors and adiponectin sensitivity. J Biol Chem. 2004;279:30817–30822. doi: 10.1074/jbc.M402367200. [DOI] [PubMed] [Google Scholar]
- 15.Debard C, Laville M, Berbe V, et al. Expression of key genes of fatty acid oxidation, including adiponectin receptors, in skeletal muscle of type 2 diabetic patients. Diabetologia. 2004;47:917–925. doi: 10.1007/s00125-004-1394-7. [DOI] [PubMed] [Google Scholar]
- 16.Jang C, Inder WJ, Obeyesekere VR, et al. Adiponectin, skeletal muscle adiponectin receptor expression and insulin resistance following dexamethasone. Clin Endocrinol Oxf. 2008;69:745–750. doi: 10.1111/j.1365-2265.2008.03242.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen MB, McAinch AJ, Macaulay SL, et al. Impaired activation of AMP-kinase and fatty acid oxidation by globular adiponectin in cultured human skeletal muscle of obese type 2 diabetics. J Clin Endocrinol Metab. 2005;90:3665–3672. doi: 10.1210/jc.2004-1980. [DOI] [PubMed] [Google Scholar]
- 18.Mao X, Kikani CK, Riojas RA, et al. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat Cell Biol. 2006;8:516–523. doi: 10.1038/ncb1404. [DOI] [PubMed] [Google Scholar]
- 19.Mitsuuchi Y, Johnson SW, Sonoda G, et al. Identification of a chromosome 3p14.3-21.1 gene, APPL, encoding an adaptor molecule that interacts with the oncoprotein-serine/threonine kinase AKT2. Oncogene. 1999;18:4891–4898. doi: 10.1038/sj.onc.1203080. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Yao F, Wu R, et al. Mediation of the DCC apoptotic signal by DIP13 alpha. J Biol Chem. 2002;277:26281–26285. doi: 10.1074/jbc.M204679200. [DOI] [PubMed] [Google Scholar]
- 21.Nechamen CA, Thomas RM, Cohen BD, et al. Human follicle-stimulating hormone (FSH) receptor interacts with the adaptor protein APPL1 in HEK 293 cells: potential involvement of the PI3K pathway in FSH signaling. Biol Reprod. 2004;71:629–636. doi: 10.1095/biolreprod.103.025833. [DOI] [PubMed] [Google Scholar]
- 22.Yang L, Lin HK, Altuwaijri S, et al. APPL suppresses androgen receptor transactivation via potentiating Akt activity. J Biol Chem. 2003;278:16820–16827. doi: 10.1074/jbc.M213163200. [DOI] [PubMed] [Google Scholar]
- 23.Lin DC, Quevedo C, Brewer NE, et al. APPL1 associates with TrkA and GIPC1 and is required for nerve growth factor-mediated signal transduction. Mol Cell Biol. 2006;26:8928–8941. doi: 10.1128/MCB.00228-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinberg GR, Smith AC, van Denderen BJ, et al. AMP-activated protein kinase is not down-regulated in human skeletal muscle of obese females. J Clin Endocrinol Metab. 2004;89:4575–4580. doi: 10.1210/jc.2004-0308. [DOI] [PubMed] [Google Scholar]
- 25.Koistinen HA, Galuska D, Chibalin AV, et al. 5-amino-imidazole carboxamide riboside increases glucose transport and cell-surface GLUT4 content in skeletal muscle from subjects with type 2 diabetes. Diabetes. 2003;52:1066–1072. doi: 10.2337/diabetes.52.5.1066. [DOI] [PubMed] [Google Scholar]
- 26.Berria R, Wang L, Richardson DK, et al. Increased collagen content in insulin-resistant skeletal muscle. Am J Physiol Endocrinol Metab. 2006;290:E560–E565. doi: 10.1152/ajpendo.00202.2005. [DOI] [PubMed] [Google Scholar]
- 27.DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Physiol. 1979;237:E214–E223. doi: 10.1152/ajpendo.1979.237.3.E214. [DOI] [PubMed] [Google Scholar]
- 28.Yi Z, Langlais P, de Filippis EA, et al. Global assessment of regulation of phosphorylation of insulin receptor substrate-1 by insulin in vivo in human muscle. Diabetes. 2007;56:1508–1516. doi: 10.2337/db06-1355. [DOI] [PubMed] [Google Scholar]
- 29.Lowry OH, Rosebrough NJ, Farr AL, et al. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 30.Hojlund K, Yi Z, Hwang H, et al. Characterization of the human skeletal muscle proteome by one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. Mol Cell Proteomics. 2008;7:257–267. doi: 10.1074/mcp.M700304-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langlais P, Mandarino LJ, Yi Z. Label-free relative quantification of co-eluting isobaric phosphopeptides of insulin receptor substrate-1 by HPLC-ESI-MS/MS. J Am Soc Mass Spectrom. 2010;21:1490–1499. doi: 10.1016/j.jasms.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.O’Neill TJ, Craparo A, Gustafson TA. Characterization of an interaction between insulin receptor substrate 1 and the insulin receptor by using the two-hybrid system. Mol Cell Biol. 1994;14:6433–6442. doi: 10.1128/mcb.14.10.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beausoleil SA, Jedrychowski M, Schwartz D, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Folli F, Pontiroli AE, Schwesinger WH. Metabolic aspects of bariatric surgery. Med Clin North Am. 2007;91:393–414. doi: 10.1016/j.mcna.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Yokota T, Oritani K, Takahashi I, et al. Adiponectin, a new member of the family of soluble defense collagens, negatively regulates the growth of myelomonocytic progenitors and the functions of macrophages. Blood. 2000;96:1723–1732. [PubMed] [Google Scholar]
- 37.Yamauchi T, Kamon J, Minokoshi Y, et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat Med. 2002;8:1288–1295. doi: 10.1038/nm788. [DOI] [PubMed] [Google Scholar]
- 38.Tomas E, Tsao TS, Saha AK, et al. Enhanced muscle fat oxidation and glucose transport by ACRP30 globular domain: acetyl-CoA carboxylase inhibition and AMP-activated protein kinase activation. Proc Natl Acad Sci USA. 2002;99:16309–16313. doi: 10.1073/pnas.222657499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 110 kb)
Targeted peptides for β-actin and APPL1 (PDF 57 kb)
RT-PCR primers (PDF 49 kb)
Identification of APPL1 using both western blots and MS techniques. a Human skeletal muscle homogenates were immunoprecipitated with anti-APPL1 antibody and then blotted for APPL1 protein. b Coomassie staining of the SDS-PAGE gel indicates a band corresponding to the size of APPL1. Gel slices were cut as indicated and subjected to trypsin digest and preparation for HPLC-ESI linear ion trap Fourier transform ion cyclotron resonance analysis. c Sequence coverage (black italics) of APPL1, using gel slices as indicated (b) and showing 56% sequence coverage (PDF 20 kb)
Regression analysis of β-actin and APPL1 peptides based on peak area analysis. Vastus lateralis muscle biopsies were homogenised, separated by SDS-PAGE and stained with Coomassie blue. Bands were cut at 80 to 90 kDa (for APPL1) and 37 to 50 kDa (for β-actin). Peak area analysis was used to determine increasing amounts of protein, as described in Methods. Peak areas shown are 1 × 108 and 1 × 105 for β-actin and APPL1, respectively. Values represent mean±SEM (error bars) (PDF 34 kb)
Example of peak area for APPL1 peptide VNQSALEAVTPSPSFQQR. Retention time (RT) is the time of elution for this particular peptide, which elutes at 37.41 min. The peptide was verified by retention time and spectrum matching. Peak Area (AA), or AUC, was used for quantification by MS analysis. In this example the peak area is 111,243 arbitrary units. The peak area is then used for quantification purposes, and seven different peptides for each APPL1 and β-actin were averaged. NL, normalised level; F, filter; ITMS, ion trap mass spectrometry; cNSI, centroid nano spray inlet; SM, smoothing points (PDF 17 kb)