Abstract
Recent outbreaks linked to Salmonella-contaminated produce heightened the need to develop simple, rapid, and accurate detection methods, particularly those capable of determining cell viability. In this study, we examined a novel strategy for the rapid detection and quantification of viable salmonellae in produce by coupling a simple propidium monoazide sample treatment with loop-mediated isothermal amplification (PMA-LAMP). We first designed and optimized a LAMP assay targeting Salmonella. Second, the performance of PMA-LAMP for detecting and quantifying viable salmonellae was determined. Finally, the assay was evaluated in experimentally contaminated produce items (cantaloupe, spinach, and tomato). Under the optimized condition, PMA-LAMP consistently gave negative results for heat-killed Salmonella cells with concentrations up to 108 CFU/ml (or CFU/g in produce). The detection limits of PMA-LAMP were 3.4 to 34 viable Salmonella cells in pure culture and 6.1 × 103 to 6.1 × 104 CFU/g in spiked produce samples. In comparison, PMA-PCR was up to 100-fold less sensitive. The correlation between LAMP time threshold (TT) values and viable Salmonella cell numbers was high (R2 = 0.949 to 0.993), with a quantification range (102 to 105 CFU/reaction in pure culture and 104 to 107 CFU/g in produce) comparable to that of PMA in combination with quantitative real-time PCR (PMA-qPCR). The complete PMA-LAMP assay took about 3 h to complete when testing produce samples. In conclusion, this rapid, accurate, and simple method to detect and quantify viable Salmonella cells in produce may present a useful tool for the produce industry to better control potential microbial hazards in produce.
INTRODUCTION
Nontyphoidal Salmonella is a leading cause of food-borne illness worldwide, with an estimated 1.03 million cases, 19,336 hospitalizations, and 378 deaths occurring in the United States each year (40). According to the U.S. Centers for Disease Control and Prevention (CDC), in 2009, Salmonella was responsible for over 40% of the total laboratory-confirmed infections from 10 bacterial/parasitic enteric agents under FoodNet surveillance (4). Furthermore, in recent years, an increasing number of Salmonella outbreaks linked to produce has been observed, implicating a wide variety of items such as melons, tomatoes, sprouts, mangoes, and peppers (3, 13). Therefore, multifaceted approaches are needed to better ensure produce safety, among which simple, rapid, and accurate detection methods that determine cell viability are especially needed in order to identify potential live Salmonella contamination problems throughout the production, processing, and distribution of produce.
Traditional culture-based methods for detecting Salmonella are reliable but labor-intensive and time-consuming, demanding several days for a definitive result (1). Immunoassays such as enzyme-linked immunosorbent assay (ELISA) have been developed for Salmonella detection (22). However, low specificity has limited their use (6). Recently, molecular biology-based methods such as PCR and real-time quantitative PCR (qPCR) have been used widely to detect Salmonella (21, 36). Although they are rapid and sensitive, a sophisticated thermal cycling instrument is an essential requirement for these techniques. More recently, loop-mediated isothermal amplification (LAMP) (32) has emerged as a promising alternative to detecting food-borne bacterial and viral agents. LAMP uses four specially designed primers and a strand-displacing Bst DNA polymerase to produce a target-specific stem-loop DNA structure during initial assay steps, followed by quasiexponential amplification of this structure under isothermal conditions (60 to 65°C), resulting in 109 copies of target DNA within an hour (32). The addition of one to two loop primers accelerates the LAMP reaction by hybridizing to stem-loop DNAs and facilitating strand displacement and amplification (25). Since it is isothermal, LAMP can be performed in much simpler instruments such as a heater or water bath. To date, LAMP assays have been developed for Campylobacter spp. (47), Shiga toxin-producing Escherichia coli (14), norovirus (50), Staphylococcus aureus (9), Vibrio parahaemolyticus (5, 26), and Vibrio vulnificus (10, 12), as well as Salmonella (15, 19, 20, 33, 34, 45, 48). Although LAMP was reported to be rapid, specific, and sensitive, several of the Salmonella LAMP studies (33, 34, 48) targeted only one specific Salmonella serovar or serogroup, and none of the studies evaluated the quantification of salmonellae in produce samples by LAMP. Additionally, a major drawback associated with DNA-based molecular detection assays, LAMP included, is the inability to distinguish viable from dead cells.
Several strategies have been used in molecular detection assays to differentiate viable/dead cells. First, as bacterial mRNA degrades more rapidly than DNA after cell death, assays targeting mRNA such as reverse transcriptase PCR, nucleic acid sequence-based amplification (NASBA), and reverse transcriptase LAMP have greater potential to detect only viable cells (8, 41, 42). However, working with RNA is technically demanding and some mRNA molecules can persist in dead cells for extended periods, leading to false-positive results (27). More recently, ethidium monoazide (EMA) or propidium monoazide (PMA) sample treatment has been combined with qPCR to distinguish viable from dead cells in Campylobacter, E. coli O157:H7, Listeria monocytogenes, Salmonella, and V. vulnificus (17, 28, 31, 35, 38, 39, 44, 46). These DNA binding dyes selectively penetrate compromised membranes of dead cells but not intact membranes of viable cells and intercalate into DNA once inside the cell membrane (28). Upon exposure to intense visible light, the photoreactive azide group on the dye is converted to a highly reactive nitrene radical that cross-links with dead cell DNAs, making them unavailable for subsequent qPCR amplifications (28, 31). Both EMA (31, 44) and PMA (29) sample treatments in combination with qPCR have been previously tested in Salmonella. Very recently, EMA coupled with LAMP was examined to detect viable Salmonella cells (20). However, EMA has been reported previously to compromise EMA/qPCR results due to insufficient differentiation of live and dead bacterial cells (7), and PMA was demonstrated to be advantageous over EMA in terms of dead cell exclusivity (28). Additionally, in that EMA-LAMP study (20), no loop primers were designed, and the quantitative capability of LAMP and the application of the assay in foods were not examined.
Subsequently, the objectives of this study were 2-fold. First, we aimed to develop and evaluate a LAMP assay targeting Salmonella invA. Second, we examined the novel PMA-LAMP combination for the rapid and specific detection and quantification of viable Salmonella in spiked produce samples.
MATERIALS AND METHODS
Bacterial strains and DNA template preparation.
Salmonella enterica serovar Typhimurium LT2 was used for assay optimization and sensitivity testing. An additional 27 Salmonella strains of 10 serovars and 25 non-Salmonella strains (Table 1) were used to evaluate assay specificity. Salmonella strains were cultured using Trypticase soy agar or broth (TSA or TSB, respectively; BD Diagnostic Systems, Sparks, MD) at 37°C overnight. Non-Salmonella strains were grown on TSA or blood agar (BD Diagnostic Systems), and Campylobacter strains were grown under microaerophilic conditions (85% N2, 10% CO2, and 5% O2).
Table 1.
Bacterial strains used in this study
| Group/genus and species | Strain ID and serotype | Origin and reference/source |
|---|---|---|
| Salmonella (n = 28) | H9812; Braenderup | Unknown |
| LT2, UMD 373; Typhimurium | Unknown | |
| S133, S134; Agona | Chicken, retail, Louisiana (18) | |
| S32, S33, S61, S62; Braenderup | ||
| S49, S50; Enteritidis | ||
| S37, S38, S98, S99; Hadar | ||
| S67, S68, S70, S71, S127, S128; Kentucky | ||
| S16, S46, S47; Mbandaka | ||
| S8, S9; Montevideo | ||
| S25, S26; Thompson | ||
| Non-Salmonella (n = 25) | ||
| Campylobacter jejuni | 81-176 | Human |
| Campylobacter jejuni | ATCC 33560 | Bovine feces |
| Citrobacter freundii | ATCC 8090 | Unknown |
| Enterobacter aerogenes | ATCC 13048 | Sputum, South Carolina |
| Enterococcus faecalis | ATCC 29212 | Urine |
| Escherichia coli | ATCC 25922 | Human |
| Listeria monocytogenes | ATCC 13932; 4b | Spinal fluid, Germany |
| Litonella anguillarum | ATCC 19264 | Ulcerous lesion in cod, United Kingdom |
| Pseudomonas aeruginosa | ATCC 27853 | Human blood |
| Shigella flexneri | ATCC 12022; 2b | Unknown |
| Shigella sonnei | ATCC 25931 | Human feces, Panama |
| Staphylococcus aureus | ATCC 29213 | Wound |
| Streptococcus pneumoniae | ATCC 49619; type 59 | Sputum, Arizona |
| Vibrio alginolyticus | ATCC 17749 | Spoiled horse mackerel, Japan |
| ATCC 33787 | Seawater, Hawaii | |
| Vibrio cholerae | ATCC 14035; O1 | NCTC, United Kingdom |
| Vibrio cincinnatiensis | ATCC 35912 | Blood/cerebrospinal fluid, Ohio |
| Vibrio fluvialis | ATCC 33809 | Human feces, Bangladesh |
| Vibrio harveyi | ATCC 14126 | Dead amphipod, Massachusetts |
| ATCC 35084 | Brown shark, Maryland | |
| Vibrio mimicus | ATCC 33653 | Human ear, North Carolina |
| ATCC 33655 | Feces, Tennessee | |
| Vibrio natriegens | ATCC 14048 | Salt marsh mud, Georgia |
| Vibrio parahaemolyticus | ATCC 17802; O1:K1 | Shirasu food poisoning, Japan |
| Vibrio vulnificus | ATCC 27562 | Blood, Florida |
To make DNA templates for specificity testing, several single colonies were suspended in 500 μl of TE buffer (10 mM Tris, pH 8.0; 1 mM EDTA; Sigma-Aldrich, St. Louis, MO) and heated at 95°C for 10 min in a dry heating block. After centrifugation at 12,000 × g for 2 min, the supernatants were stored at −20°C until use. For sensitivity testing, overnight S. Typhimurium LT2 culture was diluted 100-fold in fresh TSB and grown for 8 h to achieve mid-log phase (optical density at 600 nm [OD600] = 1; approximately 109 CFU/ml). The culture was 10-fold serially diluted in TSB, and the exact cell number was determined by standard plate counting. To test LAMP sensitivity, aliquots (500 μl) of each dilution were used to prepare DNA templates similarly by the heating method. To test PMA-LAMP sensitivity in pure culture and spiked produce samples, aliquots (500 μl) of each dilution (representing viable cells) were mixed with equal volumes of 3.8 × 105 CFU/ml of heat-killed salmonellae (incubated in boiling water bath for 10 min, representing dead cells), and the mixtures were treated with PMA as described below.
LAMP primer design and reaction conditions.
The Salmonella invasion gene (invA; GenBank accession number M90846) was used as the target for designing LAMP primers. A set of six primers (Table 2), two outer (F3 and B3), two inner (FIP and BIP), and two loop (Loop-F and Loop-B), which recognized eight distinct regions of the target sequence, were designed by the PrimerExplorer 4 software (Fujitsu Limited, Japan).
Table 2.
LAMP and PCR primers used in this study to detect Salmonella by targeting the invA gene
| Primer name | Sequence (5′–3′) | Positiona | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| LAMP primers | ||||
| F3 | CGGCCCGATTTTCTCTGG | 503–520 | Ladder-like bands with variable sizes | This study |
| B3 | CGGCAATAGCGTCACCTT | 665–682 | ||
| FIP | GCGCGGCATCCGCATCAATA-TGCCCGGTAAACAGATGAGT | 573–592 (F1c), 527–546 (F2) | ||
| BIP | GCGAACGGCGAAGCGTACTG-TCGCACCGTCAAAGGAAC | 593–612 (B1c), 635–652 (B2) | ||
| Loop-F | GGCCTTCAAATCGGCATCAAT | 547–567 | ||
| Loop-B | GAAAGGGAAAGCCAGCTTTACG | 613–634 | ||
| PCR and qPCR primers | ||||
| 139 | GTGAAATTATCGCCACGTTCGGGCAA | 371–396 | 285 | 36 |
| 141 | TCATCGCACCGTCAAAGGAACC | 634–655 |
The positions are numbered based on the coding sequence of Salmonella serovar Typhimurium invA gene (GenBank accession number M90846).
Based on the prototypic condition recommended by the manufacturer (Eiken Chemical Co., Ltd., Tokyo, Japan), the LAMP reagent mix and reaction condition were optimized by varying each parameter one at a time. Upon optimization, the final LAMP reagent mix in a total volume of 25 μl contained 1× ThermoPol reaction buffer (New England BioLabs, Ipswich, MA), 6 mM MgSO4, 1.2 mM each deoxynucleoside triphosphate (dNTP), 0.1 μM F3 and B3, 1.8 μM FIP and BIP, 1 μM Loop-F and Loop-B, 10 U of Bst DNA polymerase (New England BioLabs), and 2 μl of DNA template. The reaction was carried out at 63°C for 40 min and terminated at 80°C for 5 min in a real-time turbidimeter (LA-320C; Eiken Chemical Co., Ltd.), which acquired the turbidity readings at 650 nm every 6 s. The time threshold (TT; min) values were obtained when the turbidity increase measurements (the differential value of the moving average of turbidity) exceeded a threshold of 0.1, as shown in turbidity judgment graphs (Fig. 1). The net turbidity values during amplification were monitored in turbidity amplification graphs (Fig. 2A and 3A). Additionally, to facilitate future field applications, detection of LAMP products was also performed by adding 1 μl of 1:10-diluted original SYBR green I dye (Invitrogen, Carlsbad, CA) and observed immediately visually for color change (from orange to green or greenish yellow).
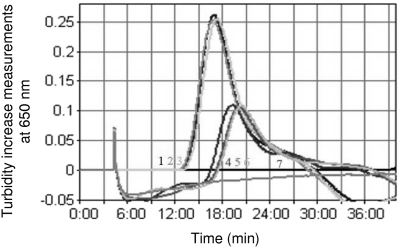
Fig. 1.
Comparison of LAMP amplification judgment graphs obtained when running the assay under optimized or prototypic conditions. Samples 1 to 3 and 4 to 6 are run under the optimized and prototypic conditions, respectively, and sample 7 is water.
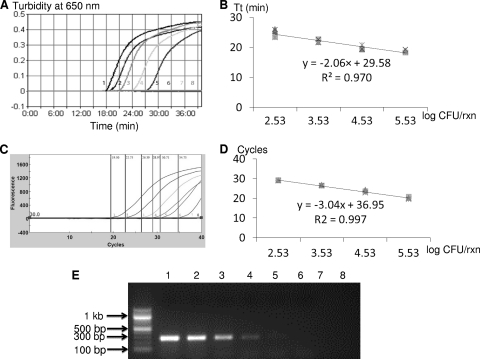
Fig. 2.
Comparison of sensitivities and quantitative capabilities of PMA-LAMP and PMA-qPCR assays when testing 10-fold serially diluted viable Salmonella enterica serovar Typhimurium LT2 in the background of 3.8 × 105 CFU/ml of dead Salmonella cells in pure culture. (A) Representative PMA-LAMP amplification graph. (B) Standard curve generated based on four independent repeats of PMA-LAMP. (C) Representative qPCR optical graph. (D) Standard curve generated based on four independent repeats of PMA-qPCR. (E) Representative gel image generated by PMA-PCR. Samples 1 to 7 correspond to 10-fold serial dilutions of viable S. Typhimurium LT2 cells ranging from 3.4 × 105 to 3.4 × 10−1 CFU/reaction; sample 8 is water.
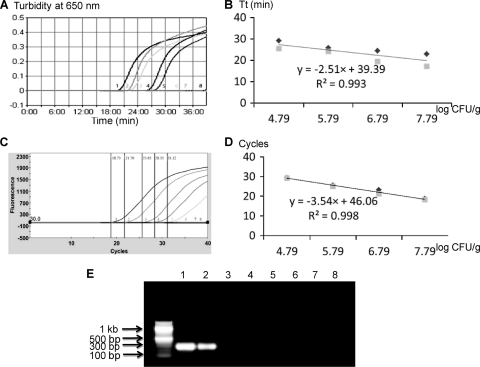
Fig. 3.
Quantitative detection of Salmonella enterica serovar Typhimurium LT2 in spiked cantaloupe samples by PMA-LAMP and PMA-qPCR. Three sets of independent spiking experiments were performed, and the LAMP reactions were repeated two times for each set of inoculations. (A) Representative PMA-LAMP amplification graph. (B) Standard curve generated for PMA-LAMP. (C) Representative PMA-qPCR optical graph. (D) Standard curve generated for PMA-qPCR. (E) Representative gel image generated by PMA-PCR. Samples 1 to 7 correspond to spiked cantaloupe samples containing 10-fold serially diluted viable Salmonella cells ranging from 6.1 × 107 to 61 CFU/g in the background of dead Salmonella at 4.2 × 106 CFU/g; sample 8 is water.
To confirm specific amplification of the Salmonella invA gene by LAMP, LAMP products were digested with 10 U of the restriction enzyme AluI (New England BioLabs). Digested and undigested LAMP products were analyzed side by side using electrophoresis on a 2% agarose gel containing ethidium bromide and visualized under UV light.
PCR and qPCR.
As comparison, a PCR assay targeting the Salmonella invA gene was performed side by side with LAMP using primers (Table 2) and conditions described previously (36). In addition, a SYBR green I-based qPCR assay was also carried out in parallel. The qPCR reagent mix (25 μl) consisted of 1× FastStart SYBR green Master Mix (Roche Applied Science, Indianapolis, IN), 0.3 μM (each) primer (36), and 2 μl of DNA template. The qPCRs were conducted using 40 cycles of denaturation at 95°C for 20 s, annealing at 64°C for 30 s, and extension at 72°C for 25 s in a SmartCycler II System (Cepheid, Sunnyvale, CA). Fluorescence readings were obtained using the 6-carboxyfluorescein (FAM) channel followed by melting curve analysis from 72°C to 94°C with increments of 0.2°C per second. The cycle threshold (CT) values were obtained when the fluorescence reading exceeded a threshold of 30 units.
LAMP specificity and sensitivity.
Fifty-three bacterial strains (Table 1) were used to determine LAMP specificity. Aliquots (2 μl) of each DNA template as prepared above were subjected to both LAMP and PCR/qPCR amplifications. Specificity tests were repeated twice.
To determine LAMP sensitivity (limit of detection), aliquots (2 μl) of the 10-fold serially diluted S. Typhimurium LT2 sensitivity templates prepared above were subjected to both LAMP and PCR/qPCR amplifications. Sensitivity tests were repeated four times.
PMA sample treatment and DNA extraction.
Freshly thawed PMA stock solution (20 mM in 20% dimethyl sulfoxide; Biotium Inc., Hayward, CA) was added to 1 ml Salmonella cell suspension (viable, dead, or viable/dead mix) to a final concentration of 100 μM. The mixture was incubated in a 2-ml light-transparent microcentrifuge tube in the dark for 5 min with periodic mixing. After the dark incubation, the tube was placed horizontally on ice and exposed for 2 min to a 650-W halogen lamp (FCW 120V; GE Lighting, Cleveland, OH) at a distance of 20 cm. During light exposure, the tube was gently shaken to ensure complete cross-linkage of dead cell DNA and photolysis of unbound PMA. Salmonella DNA was isolated from PMA-treated samples using a microbial DNA isolation kit (Mo Bio Laboratories Inc., Carlsbad, CA) according to the manufacturer's instructions. The extracted DNA was finally eluted with 100 μl elution buffer by centrifugation.
Specificity (for viable cells) and sensitivity of PMA-LAMP.
To determine whether PMA-LAMP could specifically detect only viable Salmonella cells but not dead ones, aliquots (2 μl) of the 10-fold serially diluted S. Typhimurium LT2 sensitivity DNA templates prepared above (dead cells) were subjected to PMA treatment and DNA extraction followed by both LAMP and PCR/qPCR amplifications. Specificity tests were repeated three times.
To determine the sensitivity of PMA-LAMP in detecting viable Salmonella in the background of dead Salmonella cells, mixtures of 10-fold serially diluted S. Typhimurium LT2 culture (viable cells) with 105 CFU/ml of heat-killed Salmonella (dead cells) were subjected to PMA treatment and DNA extraction followed by both LAMP and PCR/qPCR amplifications. Sensitivity tests were repeated four times.
Quantification of viable Salmonella in spiked produce.
Three replicate samples of each produce item (cantaloupe, spinach, and tomato) were obtained from local grocery stores and analyzed within 4 h of purchase. To facilitate homogenization, spinach leaves were cut into 4-cm2 squares using sterile scissors, and cantaloupe and tomato samples were sliced into fresh-cut-size pieces (2.5-cm3 cubes and 1/8 fruit wedge, respectively) using a sterile knife. Each sample (10 g) was mixed with 90 ml of buffered peptone water (BPW; BD Diagnostic Systems) and homogenized for 2 min in a food stomacher (Model 400; Tekmar Company, Cincinnati, OH) to produce 1:10 produce-BPW homogenate. The homogenate was analyzed for the presence/absence of endogenous Salmonella according to methods described previously (18).
Confirmed Salmonella-negative produce homogenates were spiked with mixtures of Salmonella viable/dead cells as described above and analyzed immediately. Briefly, 100 μl of Salmonella cell suspension (dead or viable/dead mix) was added to 900 μl of 1:10 produce-BPW homogenate, mixed thoroughly, and centrifuged at 900 × g for 3 min to remove large produce tissues. The supernatant was subjected to PMA treatment and DNA extraction as described above. Aliquots (2 μl) of the extracted DNA were used for both LAMP and PCR/qPCR amplifications. In addition to direct testing, enrichment was also performed by incubating the Salmonella-spiked produce homogenate at 37°C for 4 h. After enrichment, the homogenate was processed similarly as described above for direct testing. The produce tests were repeated three times.
Data analysis.
For specificity data, means and standard deviations of TT or CT values were calculated by using Microsoft Excel software (Microsoft, Seattle, WA). For sensitivity data, means and standard deviations of TT or CT values for detecting 10-fold serially diluted S. Typhimurium LT2 in pure culture and spiked produce samples were calculated similarly using Microsoft Excel. The detection limits (CFU/reaction in pure culture or CFU/g in spiked produce) were presented as the lowest number of cells that could be detected by the assays. In spiked produce samples, CFU/reaction was calculated by using CFU/g × 0.09 g/ml × 10 × 2 × 10−3, i.e., CFU/g × 1.8 × 10−3. Standard curves to quantify Salmonella in pure culture and spiked produce were generated by plotting TT or CT values against log CFU/reaction or log CFU/g, and linear regression was calculated using Microsoft Excel. Quantitative capabilities of the assays were derived based on the correlation coefficient (R2) values from the standard curves.
RESULTS
The optimized LAMP assay.
Besides completely eliminating betaine (0.8 M in the prototype), the optimized reagent mix contained slightly modified concentrations of dNTP, primers (inner, outer, and loop), and Bst DNA polymerase compared to those in the prototype. Additionally, a reaction temperature of 63°C was found to be optimal in the present study. Figure 1 shows the turbidity judgment graphs generated using the optimized condition in comparison with those using the prototypic one. Besides decreasing TT values (15 min using the optimized condition versus 19.3 min using the prototypic one), the turbidity increase measurements were also strikingly greater under the optimized condition, with maximum values of 0.26 and 0.11 under optimized and prototypic conditions, respectively.
LAMP specificity.
Among 53 bacterial strains (Table 1) used to evaluate the specificity of the invA-based LAMP assay, no false-positive or false-negative results were observed. For the 28 Salmonella strains of 10 serotypes, the TT values ranged from 15 to 17.8 min with an average of 16.3 ± 0.4 min. For the 25 non-Salmonella strains, no TT value was obtained, indicating negative results for LAMP. Similarly, PCR and qPCR assays included as comparisons successfully detected 28 Salmonella strains while showing negative results for 25 non-Salmonella strains (data not shown).
Additionally, digestion of the LAMP products with AluI yielded the expected fragment of 165 bp (data not shown), indicating the specific amplification of the target invA sequence by LAMP.
LAMP sensitivity and quantitative capability.
Table 3 summarizes the sensitivity and quantitative capability of the invA-based LAMP when testing 10-fold serially diluted S. Typhimurium LT2 DNA templates in comparison with those of PCR and qPCR. For pure culture templates ranging from 1.3 × 105 to 13 CFU/reaction, the average TT values for LAMP based on four repeats fell between 20.4 and 27.3 min (data not shown). In one out of four repeats, amplification of the 1.3-CFU template occurred, yielding a TT value of 37.7 min. Therefore, the detection limit for the invA-based LAMP assay was 1.3 to 13 CFU/reaction. Similarly, the invA-based qPCR had a detection limit of 1.3 CFU/reaction with CT values averaging between 23.3 and 35.3 cycles and melting temperatures consistently falling at around 81°C (data not shown). In contrast, the invA-based PCR had a detection limit of 130 CFU/reaction, up to 100-fold less sensitive than that of LAMP or qPCR. Additionally, the correlation coefficients (R2) of LAMP and qPCR were calculated to be 0.983 and 0.997, respectively, indicating excellent linear relationship between Salmonella cell numbers (log CFU/reaction) and the amplification signals (TT or CT). PCR, on the other hand, is not quantitative.
Table 3.
Comparison of sensitivities and quantitative capabilities of LAMP, PCR, and qPCR assays alone or in combination with PMA when testing serially diluted Salmonella enterica serovar Typhimurium LT2 viable cells in pure culture and spiked produce samples
| Sample typea | Methodb | Produce type | Detection limit (CFU/reaction or CFU/g) | Quantification equationg | Linear R2g |
|---|---|---|---|---|---|
| Pure culture | LAMP | NAh | 1.3–13c | y = −0.85x + 24.82 | 0.983 |
| PCR | NA | 130 | NA | NA | |
| qPCR | NA | 1.3 | y = −1.82x + 32.72 | 0.997 | |
| PMA-LAMP | NA | 3.4–34d | y = −2.06x + 29.58 | 0.970 | |
| PMA-PCR | NA | 340 | NA | NA | |
| PMA-qPCR | NA | 3.4–34e | y = −3.04x + 36.95 | 0.997 | |
| Spiked produce | PMA-LAMP | Cantaloupe | 6.1 × 103 | y = −2.51x + 39.39 | 0.993 |
| Spinach | 6.1 × 104 | y = −4.63x + 55.47 | 0.977 | ||
| Tomato | 6.1 × 104 | y = −1.78x + 36.57 | 0.949 | ||
| PMA-PCR | Cantaloupe | 6.1 × 105 | NA | NA | |
| Spinach | 6.1 × 105 | NA | NA | ||
| Tomato | 6.1 × 105 | NA | NA | ||
| PMA-qPCR | Cantaloupe | 6.1 × 103 | y = −3.54x + 46.06 | 0.998 | |
| Spinach | 6.1 × 102–6.1 × 103f | y = −3.50x + 46.08 | 0.993 | ||
| Tomato | 6.1 × 102 | y = −3.47x + 46.03 | 0.987 |
Four independent repeats were conducted for pure culture testing, and three repeats were conducted for spiked produce testing.
For testing involving PMA, dead Salmonella cells were present at a level of 3.8 × 103 CFU/reaction (or 2.1 × 105 CFU/g).
One out of four repeats was positive for the 1.3-CFU/reaction level.
One out of four repeats was positive for the 3.4-CFU/reaction level.
Three out of four repeats were positive for the 3.4-CFU/reaction level.
One out of two repeats was positive for the 6.1 × 102-CFU/g level.
Quantitative equation and R2 were calculated based on the linear relationship of average T T or C T values and log CFU/reaction between viable cell levels ranging from 102 to 105 CFU/reaction for pure culture and between 104 and 107 CFU/g for spiked produce samples.
NA, not applicable.
Specificity of PMA-LAMP for viable Salmonella cells.
The testing of 10-fold serially diluted dead Salmonella cells by PMA-LAMP, PMA-PCR, and PMA-qPCR indicated that after PMA treatment, LAMP consistently gave negative results for dead Salmonella cells ranging in concentration from 3.8 × 102 to 3.8 ×108 CFU/ml (i.e., 7.5 × 101 to 7.5 × 106 CFU/reaction). Amplification occurred at the 109-CFU/ml level (i.e., 7.5 × 107 CFU/reaction) with an average TT value of 29.6 min (data not shown). With qPCR, amplifications of dead Salmonella cells at both 108- and 109-CFU/ml levels occurred with average CT values of 33 and 33.4 cycles, respectively, and melting temperatures at around 81°C, suggesting false-positive (i.e., viable cells) results for dead Salmonella cells. On the other hand, PCR consistently gave negative results for dead Salmonella cells at up to 109 CFU/ml.
Sensitivity and quantitative capability of PMA-LAMP.
Table 3 shows the sensitivity and quantitative capability of PMA-LAMP when testing 10-fold serially diluted S. Typhimurium LT2 viable culture in the presence of 3.8 × 105 CFU/ml (i.e., 3.8 × 103 CFU/reaction) of dead Salmonella cells. For viable Salmonella cells between 3.4 × 105 and 34 CFU/reaction, after PMA treatment, consistent LAMP positive results were obtained with average TT values ranging from 19.3 to 29.6 min (data not shown). In one out of four repeats, amplification (TT = 29.7 min) also occurred for the sample with 3.4 CFU/reaction of viable Salmonella cells. No amplification was observed for the reaction tube containing 0.34 viable Salmonella cells and 3.8 × 103 dead ones. Therefore, the detection limit of PMA-LAMP was determined to be 3.4 to 34 CFU/reaction (Table 3). A similar sensitivity was observed for qPCR following PMA treatment (PMA-qPCR) with average CT values ranging from 20 to 33.4 cycles for samples containing 3.4 × 105 to 3.4 viable Salmonella cells/reaction (data not shown). In contrast, PMA-PCR had a detection limit of 340 CFU/reaction, up to 100-fold less sensitive than that of PMA-LAMP or PMA-qPCR (Table 3).
Figure 2 shows typical amplification graphs and standard curves generated when testing 10-fold serially diluted S. Typhimurium LT2 viable culture in the presence of 3.8 × 105 CFU/ml of dead Salmonella cells by PMA-LAMP (Fig. 2A and B) and PMA-qPCR (Fig. 2C and D), as well as a PCR gel (Fig. 2E). The correlation coefficients (R2) for PMA-LAMP and PMA-qPCR were calculated to be 0.970 and 0.997, respectively.
Rapid and specific quantification of viable Salmonella in produce by PMA-LAMP.
For produce samples spiked only with 10-fold serially diluted dead S. Typhimurium LT2 cells, PMA-LAMP consistently gave negative results for samples with dead cell concentrations ranging from 4.2 × 102 to 4.2 × 108 CFU/g (equivalent to 0.75 × 101 to 7.5 × 105 CFU/reaction). However, amplifications for produce samples containing 4.2 × 109 CFU/g (7.5 × 106 CFU/reaction) of dead Salmonella cells occurred with average TT values of 28.5, 38.8 and 25.2 min for cantaloupe, spinach, and tomato, respectively. In comparison, neither PMA-qPCR nor PMA-PCR showed amplifications for dead Salmonella cells up to 4.2 × 109 CFU/g, which was equivalent to 7.5 × 106 CFU in the reaction tube.
The sensitivities and quantitative capabilities of PMA-LAMP, PMA-PCR, and PMA-qPCR in detecting 10-fold serially diluted viable S. Typhimurium in the presence of 2.1 × 105 CFU/g (i.e., 3.8 × 102 CFU/reaction) of dead Salmonella cells are also summarized in Table 3. In three independent spiking experiments, PMA-LAMP consistently detected viable Salmonella cells down to 6.1 × 103 CFU/g (11 CFU/reaction) in cantaloupe samples with average TT values ranging from 20.1 to 30.9 min, whereas for spinach and tomato samples, the detection limit for both was at 6.1 × 104 CFU/g with average TT values of 19.9 to 34.2 min and 22.2 to 27.7 min, respectively. In comparison, PMA-qPCR could detect viable Salmonella cells down to 6.1 × 103 CFU/g in cantaloupe and 6.1 × 102 CFU/g (110 CFU/reaction) in spinach and tomato samples. The average CT values ranged between 18.5 and 31.8 cycles, 18.6 and 33.1 cycles, and 18.9 and 31.5 cycles for cantaloupe, spinach, and tomato samples, respectively. For PMA-PCR, the detection limits of viable Salmonella were 6.1 × 105 CFU/g in all three produce items, up to 100- and even 1,000-fold less sensitive than those of PMA-LAMP or PMA-qPCR. The R2 values ranged from 0.949 to 0.993 for PMA-LAMP and 0.987 to 0.998 for PMA-qPCR (Fig. 3).
After 4 h of enrichment, both PMA-LAMP and PMA-qPCR were able to detect an initial spiking of 40 viable Salmonella cells per gram of cantaloupe, spinach, or tomato, up to 1,000-fold more sensitive compared to direct testing without enrichment (data not shown).
DISCUSSION
The Salmonella invA-based LAMP assay developed in the present study was rapid (15 to 40 min), specific (no false-positive or false-negative results for 53 strains tested), sensitive (1.3 to 13 CFU/reaction), and quantitative (R2 = 0.983). When coupled with a simple PMA sample treatment, PMA-LAMP demonstrated good dead cell exclusivity (up to 108 CFU/ml in pure culture and 108 CFU/g in spiked produce), viable cell sensitivity (3.4 to 34 CFU/reaction in pure culture and 6.1 × 103 to 6.1 × 104 CFU/g in spiked produce), and quantitative capability (R2 = 0.949 to 0.993). To our knowledge, this is the first report examining the novel combination of PMA and LAMP in detecting and quantifying viable bacterial cells.
We chose the Salmonella invA gene as the target for designing LAMP primers. Previously, invA-based molecular detection assays using multiple platforms such as PCR, qPCR, and LAMP have been designed to accurately detect Salmonella with a broad specificity for more than 100 Salmonella serovars while demonstrating excellent exclusivity for non-Salmonella strains (8, 15, 36, 45). Findings of this study corroborated with these previous reports on the high specificity of invA-based molecular detection assays for Salmonella. A closer examination of primer sequences in this study and previously published invA-based LAMP studies (15, 20, 45) showed that the region (5′ end of F3 and 3′ end of B3) covered by our primers (503 to 682 bp) and those of Hara-Kudo et al. (225 to 468 bp) (15) overlapped with that (371 to 655 bp) targeted by the widely used Salmonella invA PCR primers (36). Primers reported in the other two studies (20, 45) were essentially the same with only one nucleotide deletion at the 3′ end of each FIP and BIP primer in the EMA-LAMP study (20), and the region (672 to 912 bp) covered was downstream of the invA PCR primers without any overlap. In addition, two loop primers were each incorporated in this study and the study by Hara-Kudo et al. (15), while the other two studies had no loop primers (20, 45).
It is noteworthy that the optimized LAMP reagent mix in the present study differed from that described in many previous LAMP publications (5, 10, 15, 19, 34), which essentially followed the formulation of the LoopAmp DNA amplification kit (Eiken Chemical Co., Ltd.). Comparing the turbidity judgment graphs (Fig. 1) clearly indicated that under the optimized condition, the LAMP reaction progressed faster and the turbidity signals increased markedly more speedily. A major deviation of the optimized conditions from the prototype was the elimination of betaine. Our recent study optimizing a LAMP assay for potentially virulent V. vulnificus also indicated that betaine had no beneficial effect on LAMP amplification (12). Several previous studies, however, suggested that higher betaine concentration resulted in elevated LAMP amplification efficiency and increased target selectivity (32, 49). Betaine was capable of isostabilizing DNA and preventing secondary structure formation in GC-rich region, thus reducing base stacking and promoting DNA amplification (16, 37). However, unlike LAMP, betaine has not been used routinely in PCRs. Therefore, our data indicate that when amplifying non-GC-rich target sequences, eliminating betaine may be preferable in order to increase LAMP amplification efficiency.
The LAMP assay developed in this study was capable of detecting 1.3 to 13 Salmonella cells per reaction in pure culture. This level of sensitivity was comparable to that of qPCR but up to 100-fold more sensitive than that of PCR run in parallel. The first published invA-based LAMP for Salmonella detection had a sensitivity of 2.2 CFU/test tube (15), whereas a more recent one reported a detection limit of 100 fg DNA/tube (45), approximately 20 CFU/tube (23). In both studies, LAMP was found to be 10-fold more sensitive than PCR for all serotypes tested (15, 45). Additionally, another LAMP assay for Salmonella detection that targeted the phoP gene was able to detect down to 35 CFU per reaction (19), and two LAMP assays specific for Salmonella O4 or O9 groups possessed a detection limit of 103 CFU/ml (equivalent to 100 CFU/tube), 100-fold more sensitive than PCR (33, 34). A very recent study compared the sensitivity of LAMP and TaqMan qPCR in detecting Salmonella enterica serovar Enteritidis and reported a detection limit of 4 copies per reaction by both assays (48). Therefore, the sensitivities of current LAMP assays for Salmonella, including the one developed in the present study, fell between 100 and 101 CFU/reaction, 10- to 100-fold more sensitive than PCR but similar to qPCR. This improved sensitivity (at least 10-fold) of LAMP compared to PCR has also been reported in previous LAMP studies on the detection of other food-borne pathogens (5, 11, 14). It is noteworthy that LAMP is markedly faster than qPCR by at least 10 min.
LAMP amplicons were commonly detected by gel electrophoresis, naked eye observation of turbidity or color change, and real-time turbidimeter monitoring (5, 10, 11, 15, 19, 24, 42, 45, 48). Since LAMP synthesizes a large amount of DNA (10 to 20 μg/25-μl reaction mixture), open-tube procedures after amplification such as gel electrophoresis potentially act as a significant source of cross-contamination, whereas a closed-tube procedure such as monitoring with a real-time turbidimeter is preferred (11). In addition, among these LAMP amplicon detection methods, real-time turbidimeter monitoring is the only one that is potentially quantitative. However, very few studies have examined the quantitative capability of LAMP. One study monitoring ammonia-oxidizing bacteria using LAMP reported that it possessed good quantitative capability between 104 and 1010 DNA copies (2). Two other studies demonstrated strong linear correlation coefficients (R2 = 0.94 to 0.99) of LAMP in detecting V. parahaemolyticus and V. vulnificus in spiked oysters (5, 11). In the present study, the R2 values were found to be 0.983 for cell concentrations ranging between 102 and 105 CFU/reaction in pure culture and 0.949 to 0.993 for viable Salmonella cells ranging from 104 to 107 CFU/g in spiked produce samples, suggesting an excellent quantitative capability.
From a public health perspective, determining cell viability is a critical requirement for pathogen testing methods in foods in order to accurately assess the potential risks (17). This is the first report examining the novel combination of PMA and LAMP in detecting viable bacterial cells. Dead Salmonella cells, up to 3.8 × 108 CFU/ml (4.2 × 108 CFU/g in spiked produce), were not detected by PMA-LAMP, illustrating excellent dead cell exclusivity. In comparison, PMA-PCR had 10-fold-better dead cell exclusivity, which was possibly due to the lower sensitivity associated with PCR compared to LAMP. On the other hand, PMA-qPCR gave positive signals for Salmonella dead cells at 3.8 × 108 CFU/ml, likely attributable to the high sensitivity of qPCR (Table 3). Previous studies of EMA or PMA in combination with qPCR found that qPCR consistently gave late signals for samples containing only dead bacterial cells due to its superior sensitivity, implying a great potential to generate false-positive results when detecting viable bacteria in the background of high concentrations of dead cells (29, 30, 43). Nonetheless, this level of dead cell concentration (107 to 109 CFU/g) is not commonly encountered in an agriculture field or in produce samples. Additionally, optimizing PMA treatment parameters, including final concentration, PMA incubation time, and light exposure time, may be able to further improve the dead cell exclusivity of PMA-LAMP.
In pure culture testing, PMA-LAMP possessed a similar sensitivity as PMA-qPCR, i.e., 3.4 to 34 viable Salmonella cells, which was comparable to the detection limit of 1.3 to 13 Salmonella cells obtained by LAMP alone in pure culture. Additionally, the R2 value of PMA-LAMP was 0.970 for viable Salmonella cell concentrations ranging from 102 to 105 CFU/reaction, indicating a comparable quantitative capability to that obtained using LAMP alone. This suggested that PMA did not have significant inhibitory effect on the overall assay sensitivity and quantitative ability, contrary to findings reported in a recent study (43). The recent EMA-LAMP study (20) reported the same level of sensitivity as that of LAMP (45), i.e., 100 fg DNA of Salmonella cells, suggesting EMA had no inhibitory effect on LAMP. In terms of sensitivity comparison, a recent study detecting viable Salmonella using a TaqMan-based reverse transcriptase qPCR assay targeting invA mRNA reported a detection limit of ca. 120 viable Salmonella cells at mid-exponential growth stage (8). Another study using reverse transcriptase LAMP had 105 CFU/ml by visual observation and 101 CFU/ml by gel electrophoresis for unenriched Salmonella overnight culture (42). However, a sensitivity of 101 CFU/ml would be equivalent to 0.05 CFU (0.05 DNA copy) per LAMP reaction tube, theoretically unattainable by molecular detection assays due to the absence of template DNA. The use of overnight culture where a high proportion of cells do not produce CFU was used to partly explain the detection limit of less than one cell (e.g., 0.1 cell) in a LAMP assay designed for virulent V. parahaemolyticus (26). In addition, in that reverse transcriptase LAMP study (42), the big discrepancy of sensitivity (4 logs) between visual observation and gel electrophoresis reported was rather uncommon. In the present study, we observed a general agreement between visual observation of color change after adding SYBR green I and real-time turbidity monitoring (data not shown).
Without enrichment, the detection limits of PMA-LAMP for viable Salmonella in spiked produce samples ranged from 6.1 × 103 CFU/g (11 CFU/reaction) in cantaloupe to 6.1 × 104 CFU/g in spinach and tomato, up to 100-fold more sensitive than those of PMA-PCR but less sensitive than PMA-qPCR. The differences observed in sensitivity for different produce items may be due to inherent factors such as pH and minerals and warrant further evaluations. Adding 4 h of enrichment, PMA-LAMP could detect an initial spiking of 40 CFU/g of viable Salmonella cells, comparable to that obtained by PMA-qPCR. This short-term enrichment procedure allowed for sample processing and LAMP confirmation within an 8-h working day. In comparison, Gonzalez-Escalona et al. (8) demonstrated a sensitivity of 2 viable Salmonella cells per 25 g of bagged spinach by reverse transcriptase qPCR after 24 h of preenrichment. Techathuvanan et al. (42) used reverse transcriptase LAMP to detect S. Typhimurium from pork and reported detection limits of 102 CFU/25 g with 10 h of enrichment and 106 CFU/25 g without enrichment. Neither study included dead Salmonella cells in the background to ascertain that the assay did not detect dead Salmonella DNAs. On the other hand, a recent study using PMA-qPCR to quantify viable Campylobacter cells on chicken carcasses reported a detection limit of 100 CFU/ml of chicken carcass rinse (17). Again, no dead Campylobacter cells were added in the background to ascertain that the assay did not detect dead Campylobacter cell DNAs.
In conclusion, the overall advantages of the PMA-LAMP assay were well demonstrated in terms of sensitivity, quantitative capability, rapidity, and simplicity. First, PMA-LAMP had comparable sensitivity to PMA-qPCR but up to 100-fold more sensitivity than PMA-PCR in both pure culture and produce samples. Second, PMA-LAMP showed excellent quantitative capabilities (R2 = 0.949 to 0.993) comparable to PMA-qPCR. Third, the total assay time for PMA-LAMP in produce without enrichment was 3 h, faster than either PMA-qPCR or PMA-PCR. Furthermore, PMA-LAMP is technically simpler than PMA-PCR as it eliminated gel electrophoresis. However, one limitation of this study was that log-phase cells were used to inoculate produce samples in order to obtain accurate counts of live Salmonella cells. In naturally contaminated produce samples, Salmonella cells are unlikely to be in this active physiological state; therefore, a longer enrichment step may be necessary. It is also helpful to apply this PMA-LAMP to examine the survival and persistence of Salmonella in produce samples with conditions mimicking industry practices. Therefore, upon further evaluation, this rapid, accurate, and simple method to detect and quantify viable Salmonella in produce may present a valuable tool for the produce industry to better control potential microbial hazards in produce.
ACKNOWLEDGMENTS
We thank Feifei Han for technical assistance and helpful discussion.
This study was supported in part by funding from the Center for Produce Safety (contract SA7498) at the University of California, Davis.
Footnotes
Published ahead of print on 15 April 2011.
REFERENCES
- 1.Andrews W. H., Hammack T. S.2007. [Last accessed 10 June 2010]. Bacteriological analytical manual online. Chapter 5: Salmonella. Food and Drug Administration, Silver Spring, MD. http://www.fda.gov/food/foodsafety/foodborneillness/foodborneillness foodbornepathogensnaturaltoxins/badbugbook/ucm069966.htm.
- 2. Aoi Y., Hosogai M., Tsuneda S. 2006. Real-time quantitative LAMP (loop-mediated isothermal amplification of DNA) as a simple method for monitoring ammonia-oxidizing bacteria. J. Biotechnol. 125:484–491 [DOI] [PubMed] [Google Scholar]
- 3. CDC 2008. Outbreak of Salmonella serotype Saintpaul infections associated with multiple raw produce items—United States, 2008. MMWR Morb. Mortal. Wkly. Rep. 57:929–934 [PubMed] [Google Scholar]
- 4. CDC 2010. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2009. MMWR Morb. Mortal. Wkly. Rep. 59:418–422 [PubMed] [Google Scholar]
- 5. Chen S., Ge B. 2010. Development of a toxR-based loop-mediated isothermal amplification assay for detecting Vibrio parahaemolyticus. BMC Microbiol. 10:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eriksson E., Aspan A. 2007. Comparison of culture, ELISA and PCR techniques for Salmonella detection in faecal samples for cattle, pig and poultry. BMC Vet. Res. 3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Flekna G., et al. 2007. Insufficient differentiation of live and dead Campylobacter jejuni and Listeria monocytogenes cells by ethidium monoazide (EMA) compromises EMA/real-time PCR. Res. Microbiol. 158:405–412 [DOI] [PubMed] [Google Scholar]
- 8. Gonzalez-Escalona N., et al. 2009. Detection of live Salmonella sp. cells in produce by a TaqMan-based quantitative reverse transcriptase real-time PCR targeting invA mRNA. Appl. Environ. Microbiol. 75:3714–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goto M., Hayashidani H., Takatori K., Hara-Kudo Y. 2007. Rapid detection of enterotoxigenic Staphylococcus aureus harbouring genes for four classical enterotoxins, SEA, SEB, SEC and SED, by loop-mediated isothermal amplification assay. Lett. Appl. Microbiol. 45:100–107 [DOI] [PubMed] [Google Scholar]
- 10. Han F., Ge B. 2008. Evaluation of a loop-mediated isothermal amplification assay for detecting Vibrio vulnificus in raw oysters. Foodborne Pathog. Dis. 5:311–320 [DOI] [PubMed] [Google Scholar]
- 11. Han F., Ge B. 2010. Quantitative detection of Vibrio vulnificus in raw oysters by real-time loop-mediated isothermal amplification. Int. J. Food Microbiol. 142:60–66 [DOI] [PubMed] [Google Scholar]
- 12. Han F., Wang F., Ge B. 2011. Detecting potentially virulent Vibrio vulnificus strains in raw oysters by quantitative loop-mediated isothermal amplification. Appl. Environ. Microbiol. 77:2589–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanning I. B., Nutt J. D., Ricke S. C. 2009. Salmonellosis outbreaks in the United States due to fresh produce: sources and potential intervention measures. Foodborne Pathog. Dis. 6:635–648 [DOI] [PubMed] [Google Scholar]
- 14. Hara-Kudo Y., et al. 2007. Sensitive and rapid detection of Vero toxin-producing Escherichia coli using loop-mediated isothermal amplification. J. Med. Microbiol. 56:398–406 [DOI] [PubMed] [Google Scholar]
- 15. Hara-Kudo Y., Yoshino M., Kojima T., Ikedo M. 2005. Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol. Lett. 253:155–161 [DOI] [PubMed] [Google Scholar]
- 16. Henke W., Herdel K., Jung K., Schnorr D., Loening S. A. 1997. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 25:3957–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Josefsen M. H., et al. 2010. Rapid quantification of viable Campylobacter bacteria on chicken carcasses, using real-time PCR and propidium monoazide treatment, as a tool for quantitative risk assessment. Appl. Environ. Microbiol. 76:5097–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lestari S. I., Han F., Wang F., Ge B. 2009. Prevalence and antimicrobial resistance of Salmonella serovars in conventional and organic chickens from Louisiana retail stores. J. Food Prot. 72:1165–1172 [DOI] [PubMed] [Google Scholar]
- 19. Li X., et al. 2009. A loop-mediated isothermal amplification method targets the phoP gene for the detection of Salmonella in food samples. Int. J. Food Microbiol. 133:252–258 [DOI] [PubMed] [Google Scholar]
- 20. Lu Y., et al. 2009. Specific detection of viable Salmonella cells by an ethidium monoazide-loop mediated isothermal amplificaiton (EMA-LAMP) method. J. Health Sci. 55:820–824 [Google Scholar]
- 21. Malorny B., Lofstrom C., Wagner M., Kramer N., Hoorfar J. 2008. Enumeration of Salmonella bacteria in food and feed samples by real-time PCR for quantitative microbial risk assessment. Appl. Environ. Microbiol. 74:1299–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mansfield L. P., Forsythe S. J. 2000. The detection of Salmonella using a combined immunomagnetic separation and ELISA end-detection procedure. Lett. Appl. Microbiol. 31:279–283 [DOI] [PubMed] [Google Scholar]
- 23. McClelland M., et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 24. Mori Y., Nagamine K., Tomita N., Notomi T. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150–154 [DOI] [PubMed] [Google Scholar]
- 25. Nagamine K., Hase T., Notomi T. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16:223–229 [DOI] [PubMed] [Google Scholar]
- 26. Nemoto J., et al. 2009. Rapid and specific detection of the thermostable direct hemolysin gene in Vibrio parahaemolyticus by loop-mediated isothermal amplification. J. Food Prot. 72:748–754 [DOI] [PubMed] [Google Scholar]
- 27. Nocker A., Camper A. K. 2009. Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiol. Lett. 291:137–142 [DOI] [PubMed] [Google Scholar]
- 28. Nocker A., Cheung C. Y., Camper A. K. 2006. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67:310–320 [DOI] [PubMed] [Google Scholar]
- 29. Nocker A., Mazza A., Masson L., Camper A. K., Brousseau R. 2009. Selective detection of live bacteria combining propidium monoazide sample treatment with microarray technology. J. Microbiol. Methods 76:253–261 [DOI] [PubMed] [Google Scholar]
- 30. Nocker A., Sossa K. E., Camper A. K. 2007. Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J. Microbiol. Methods 70:252–260 [DOI] [PubMed] [Google Scholar]
- 31. Nogva H. K., Dromtorp S. M., Nissen H., Rudi K. 2003. Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease PCR. Biotechniques 34:804–813 [DOI] [PubMed] [Google Scholar]
- 32. Notomi T., et al. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okamura M., et al. 2008. Loop-mediated isothermal amplification for the rapid, sensitive, and specific detection of the O9 group of Salmonella in chickens. Vet. Microbiol. 132:197–204 [DOI] [PubMed] [Google Scholar]
- 34. Okamura M., et al. 2009. Rapid, sensitive, and specific detection of the O4 group of Salmonella enterica by loop-mediated isothermal amplification. Avian Dis. 53:216–221 [DOI] [PubMed] [Google Scholar]
- 35. Pan Y., Breidt F., Jr. 2007. Enumeration of viable Listeria monocytogenes cells by real-time PCR with propidium monoazide and ethidium monoazide in the presence of dead cells. Appl. Environ. Microbiol. 73:8028–8031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rahn K., et al. 1992. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6:271–279 [DOI] [PubMed] [Google Scholar]
- 37. Rees W. A., Yager T. D., Korte J., von Hippel P. H. 1993. Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry 32:137–144 [DOI] [PubMed] [Google Scholar]
- 38. Rudi K., Moen B., Dromtorp S. M., Holck A. L. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71:1018–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rudi K., Naterstad K., Dromtorp S. M., Holo H. 2005. Detection of viable and dead Listeria monocytogenes on gouda-like cheeses by real-time PCR. Lett. Appl. Microbiol. 40:301–306 [DOI] [PubMed] [Google Scholar]
- 40. Scallan E., et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simpkins S. A., Chan A. B., Hays J., Popping B., Cook N. 2000. An RNA transcription-based amplification technique (NASBA) for the detection of viable Salmonella enterica. Lett. Appl. Microbiol. 30:75–79 [DOI] [PubMed] [Google Scholar]
- 42. Techathuvanan C., Draughon F. A., D'Souza D. H. 2010. Loop-mediated isothermal amplification (LAMP) for the rapid and sensitive detection of Salmonella Typhimurium from pork. J. Food Sci. 75:M165–M172 [DOI] [PubMed] [Google Scholar]
- 43. Varma M., et al. 2009. Quantitative real-time PCR analysis of total and propidium monoazide-resistant fecal indicator bacteria in wastewater. Water Res. 43:4790–4801 [DOI] [PubMed] [Google Scholar]
- 44. Wang L., Mustapha A. 2010. EMA-real-time PCR as a reliable method for detection of viable Salmonella in chicken and eggs. J. Food Sci. 75:M134–M139 [DOI] [PubMed] [Google Scholar]
- 45. Wang L., Shi L., Alam M. J., Geng Y., Li L. 2008. Specific and rapid detection of foodborne Salmonella by loop-mediated isothermal amplification method. Food Res. Int. 41:69–74 [Google Scholar]
- 46. Wang S., Levin R. E. 2006. Discrimination of viable Vibrio vulnificus cells from dead cells in real-time PCR. J. Microbiol. Methods 64:1–8 [DOI] [PubMed] [Google Scholar]
- 47. Yamazaki W., et al. 2008. Development and evaluation of a loop-mediated isothermal amplification assay for rapid and simple detection of Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 57:444–451 [DOI] [PubMed] [Google Scholar]
- 48. Yang J. L., et al. 2010. Simple and rapid detection of Salmonella serovar Enteritidis under field conditions by loop-mediated isothermal amplification. J. Appl. Microbiol. 109:1715–1723 [DOI] [PubMed] [Google Scholar]
- 49. Yeh H. Y., Shoemaker C. A., Klesius P. H. 2005. Evaluation of a loop-mediated isothermal amplification method for rapid detection of channel catfish Ictalurus punctatus important bacterial pathogen Edwardsiella ictaluri. J. Microbiol. Methods 63:36–44 [DOI] [PubMed] [Google Scholar]
- 50. Yoda T., et al. 2007. Evaluation and application of reverse transcription loop-mediated isothermal amplification for detection of noroviruses. J. Med. Virol. 79:326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]