Abstract
Ralstonia solanacearum is a Gram-negative bacterium and the causative agent of bacterial wilt in many important crops. We treated R. solanacearum with three lytic phages: φRSA1, φRSB1, and φRSL1. Infection with φRSA1 and φRSB1, either alone or in combination with the other phages, resulted in a rapid decrease in the host bacterial cell density. Cells that were resistant to infection by these phages became evident approximately 30 h after phage addition to the culture. On the other hand, cells infected solely with φRSL1 in a batch culture were maintained at a lower cell density (1/3 of control) over a long period. Pretreatment of tomato seedlings with φRSL1 drastically limited penetration, growth, and movement of root-inoculated bacterial cells. All φRSL1-treated tomato plants showed no symptoms of wilting during the experimental period, whereas all untreated plants had wilted by 18 days postinfection. φRSL1 was shown to be relatively stable in soil, especially at higher temperatures (37 to 50°C). Active φRSL1 particles were recovered from the roots of treated plants and from soil 4 months postinfection. Based on these observations, we propose an alternative biocontrol method using a unique phage, such as φRSL1, instead of a phage cocktail with highly virulent phages. Using this method, φRSL1 killed some but not all bacterial cells. The coexistence of bacterial cells and the phage resulted in effective prevention of wilting.
INTRODUCTION
Bacterial wilt is an important crop disease, caused by the soilborne Gram-negative bacterium Ralstonia solanacearum. This bacterium has an unusually wide host range, infecting more than 200 species belonging to more than 50 botanical families, including economically important crops (9, 10). R. solanacearum strains represent a heterogeneous group subdivided into five races based on host range, five biovars based on physiological and biochemical characteristics (8), and four phylotypes roughly corresponding to geographic origins. Phylotype I includes strains originating primarily from Asia, phylotype II from America, phylotype III from Africa and surrounding islands in the Indian Ocean, and phylotype IV from Indonesia (4). In the field, R. solanacearum is easily disseminated via soil, contaminated irrigation water, surface water, farm equipment, and infected biological material (14). Bacterial cells can survive for many years in association with alternate hosts, and soil fumigation with methyl bromide, vapam, or chloropicrin is of limited efficacy. Because methyl bromide depletes the stratospheric ozone layer, the production and use of this gas was phased out in 2005 under the Montreal Protocol and the Clean Air Act. Due to the limited effectiveness of the current integrated management strategies, bacterial wilt continues to be an economically serious problem for field-grown crops in many tropical, subtropical, and warmer areas of the world (9, 10).
Like other methods of biological control, one advantage of phage therapy (also called phage biocontrol) is the reduction in the use of chemical agents against pathogens. This avoids problems associated with environmental pollution, ecosystem disruption and residual chemicals on crops. Phage therapy in agricultural settings was extensively explored 40 to 50 years ago as a means of controlling plant pathogens (3, 18). Two major problems arose in those trials: (i) extracellular polysaccharides produced by pathogenic bacteria prevented phage adsorption, and (ii) there were various degrees of susceptibility among bacterial strains (8). Nevertheless, over recent decades, the use of phage therapy to control the growth of plant-based bacterial pathogens has been explored with increased enthusiasm. To control R. solanacearum, two bacteriophages have already been isolated, and their physical and physiological properties have been characterized: phage P4282 (19, 21) and phage PK101 (22). Both of these phages demonstrate very narrow host ranges and infect only a few strains of R. solanacearum. Phage P4282, which infects R. solanacearum strain M4S, was used to control bacterial wilt in tobacco plants under laboratory conditions, and possible phage-mediated protection was observed (21). However, for the practical use of phages as biocontrol agents against bacterial wilt, it is believed that multiple phages with wide host ranges and strong lytic activity are required (22). Recently, Yamada et al. (23) isolated and characterized several different kinds of phage that specifically infected R. solanacearum strains belonging to different races and/or biovars. Phage φRSA1 is a P2-like head-tail virus (Myoviridae) with a very wide host range; all 15 strains tested from race 1, 3, or 4 and biovar 1, N2, 3, or 4 were susceptible to this phage (6). Phage φRSL1 is another myovirus containing ∼231 kb in its genome; this phage was able to lyse 10 of 15 tested strains (24). The most recently isolated phage, φRSB1, displayed a T7-like morphology (Podoviridae) and also had the widest host range, with 14 of 15 strains from race 1, 3, or 4 being susceptible (17). φRSB1 lyses host cells and forms very large clear plaques that are 10 to 15 mm in diameter on assay plates.
The three phages—φRSA1, φRSB1, and φRSL1—appear to be useful in the eradication of the bacterial wilt pathogen. To increase the antibacterial efficacy of these phages in biocontrol, a thorough understanding of phage ecology and complex phage-host interactions in various environments is necessary. Genomic information on these phages and their host bacteria will be useful for understanding the phage characteristics and the history and molecular mechanisms involved in the phage-bacterium interactions.
MATERIALS AND METHODS
Bacterial strains and phages.
Strains of R. solanacearum were obtained from the culture collections as listed in Table 1. The avirulent strain M4S was used for routine purposes (21). For plant inoculation, a few virulent strains, such as Ps29 and MAFF 730138, were used. Strain MAFF 106611 was used mainly for in planta detection of bacterial cells. The bacterial cells were cultured in CPG medium containing 0.1% Casamino Acids, 1% peptone, and 0.5% glucose (11) at 28°C with shaking at 200 to 300 rpm. Bacteriophages φRSA1, φRSB1, and φRSL1 (Table 2) were routinely propagated using strain M4S as the host. Bacterial cells in the stationary phase (16 to 24 h postinoculation) grown in CPG medium were diluted 100-fold with 100 ml of fresh CPG medium in a 500-ml flask. To collect sufficient phage particles, a total of 1 liter of bacterial culture was grown. When the optical density at 600 nm (OD600) of the cultures reached 0.1 to 0.3, the phage was added at a dose of 0.5 PFU/host cell (0.5 × 108 PFU/ml) for φRSA1 and φRSB1 or 5.0 PFU/cell (5 × 108 PFU/ml) for φRSL1. After further growth for 6 to 24 h, the cells were removed by centrifugation in a Hitachi Himac CR21E centrifuge equipped with a R12A2 rotor (Hitachi Koki Co., Ltd., Tokyo, Japan) at 8,000 × g for 20 min at 4°C. To increase φRSA1 recovery, EGTA (final concentration, 1 mM) was added to the φRSA1-infected culture at 6 to 9 h postinfection (p.i.). The supernatant was passed through a 0.45-μm-pore-size membrane filter, and phage particles were precipitated by centrifugation in a Hitachi CP100β centrifuge with a P28S rotor at 40,000 × g for 1 h at 4°C and then suspended in SM buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM MgSO4, 0.01% gelatin). Purified phages were stored at 4°C until needed.
Table 1.
Bacterial strains used in this study
| R. solanacearum strain | Description |
Sourcea | ||
|---|---|---|---|---|
| Race | Biovar | Phylotype | ||
| M4S | 1 | 3 | I | LTRC |
| Ps29 | 1 | 3 | I | LTRC |
| MAFF 106611 | 1 | 4 | I | NIAS |
| MAFF 730138 | 1 | 3 | I | NIAS |
Table 2.
Bacteriophages used in this study
DNA manipulations.
Standard molecular biological techniques for DNA isolation, digestion with restriction enzymes and other nucleases, and construction of recombinant DNAs were as described by Sambrook and Russell (20). Phage DNA was isolated from the purified phage particles by phenol extraction (2, 23).
In planta detection of R. solanacearum cells.
Seeds of the tomato (Lycopersicon esculentum) cultivar Oogata-Fukuju were obtained from Takii Co., Ltd. (Kyoto, Japan). For aseptic cultures, seeds were surface sterilized with sodium hypochlorite and cultured in a square dish (sterile square Schale no. 2; Eiken Chemical Co., Ltd., Tokyo, Japan) containing solid medium (0.15% Hyponex powder; Hyponex Japan Corp., Ltd., Osaka, Japan), 0.5% sucrose, and 1.5% agar adjusted to pH 5.8. Plants were grown in an incubator (Sanyo growth cabinet; Sanyo, Osaka, Japan) at 28°C under a 16-h light (300 μmol photons/s/m2) and 8-h dark cycle. During the culture period, the dishes in the chamber were tilted to a 45° angle to encourage roots to grow on the surface of the medium. This made it easy to access the roots and monitor the inoculated bacterial cells under a stereomicroscope. To inoculate plants, bacterial cells (i.e., strain MAFF 106611 bearing pRSS12, a green fluorescent protein [GFP]-expressing plasmid with no effects on host virulence [5, 16]) were cultured in CPG medium and suspended in sterile distilled water at a density of 107 to 108 cells/ml. Tomato seedlings grown in culture dishes were cut at the tip of the taproot, 10 mm from the apex, with a razor blade and then pretreated with phages; 1.0 μl of phage preparation (109 PFU/ml) was added to the cut. For a mock control, 1.0 μl of distilled water was added. After 12 to 24 h from the phage treatment, 1.0 μl of bacterial suspension (107 to 108 cells/ml) was applied to the section. After inoculation, the plants in the dishes were cultured in the incubator until observation. Bacterial cells in the plants were observed by using an MZ16F fluorescence stereomicroscope (Leica Microsystems, Heidelberg, Germany) equipped with GFP2 and GFP3 filters and/or an Olympus BH2 fluorescence microscope (Olympus, Tokyo, Japan). Microscopic images were recorded with a charge-coupled device camera (VB-6010; Keyence, Osaka, Japan). For prolonged observation of plants, bacterium-treated plants were transferred from culture dishes to pots containing a mixture of peat moss and expanded vermiculite. Plants were grown under natural conditions.
Detection of φRSL1 remaining in plants and soil.
Two surviving plant cultures, 4 months after treatment, were subjected to assays to detect any remaining bacteriophages. Two parts of the stem (2.5 g at 10 cm above the soil surface and 0.7 to 0.8 g at 1 to 5 cm above the soil surface) were excised from the plant. For the root, 2 to 3 g (12 to 13 cm) was removed after washing the sample in running tap water. After the addition of 2 ml of CPG medium, the plant material was ground using a mortar and pestle at room temperature. After centrifugation at 4,200 × g for 5 min at 4°C, the supernatant was filtered through a 0.45-μm-pore-size membrane filter (Steradisc; Krabo Co., Okayama, Japan). The filtrate (100-μl aliquot) was subjected to plaque assays with strain Ps29 as the host on CPG plates containing 0.45% agar. A 2-g mass of pot soil was suspended in 5 ml of SM buffer, and then, after mixing and filtration through a membrane filter as described above, a 100-μl aliquot of the suspension was subjected to plaque assays.
Treatment of tomato plants in soil with phages and inoculation with R. solanacearum cells.
Tomato seeds (cv. Oogata-Fukuju) were planted in Jiffy 7 peat pellets (42 mm in diameter; Sakata Seed Co., Ltd., Kanagawa, Japan), which were soaked with a phage solution (1.3 × 1010 PFU/pot). For controls, pellets were soaked with tap water. After cultivation for 1 month, the plants (20 to 23 cm in height) were treated again with the phage solution (1.3 × 1010 PFU/pot); the pellet was soaked with the phage solution. Two days later, the plants were cut at the root tips with scissors and dipped in a bacterial suspension containing strain MAFF 106611 cells at 108 cells/ml for 30 s. The bacterium-treated plants were transferred to pots (9 cm in diameter) containing peat moss and expanded vermiculite and grown in an incubator (Sanyo) at 28°C under a 16-h light and 8-h dark cycle. Symptoms of wilting were graded from 0 to 5 as follows: 0, no symptoms; 1, only one petiole is wilting; 2, two to three petioles were wilting; 3, all but two to three petioles were wilting; 4, all petioles were wilting; and 5, the plant died.
Effect of temperature on the stability of φRSL1.
Phage preparations of φRSA1 (109 PFU/ml), φRSB1 (109 PFU/ml), and φRSL1 (109 PFU/ml) in SM buffer (0.5 ml) were incubated in sealed tubes at 4, 11, 28, 37, and 50°C for various periods before the plaque assay with strain Ps29 as the host. A 10-ml volume of each phage solution was added to 100 g of autoclaved field soil. The phage-soil mixture was divided into five equal amounts and separately filled into 15-ml tubes. After sealing, the tubes were incubated at 4, 11, 28, 37, and 50°C for various periods before the plaque assay was carried out. These experiments were performed twice.
RESULTS
Treatment of R. solanacearum cells with three different phages.
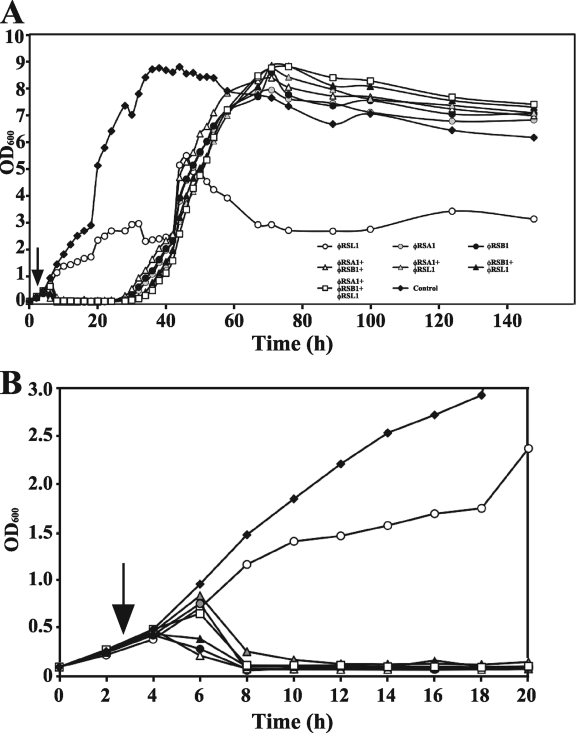
R. solanacearum M4S cells were treated with three lytic phages—φRSA1, φRSB1, and φRSL1—alone or in combination. The optimal phage doses per bacterial cell to give the highest phage yield were determined to be 0.5, 0.5, and 5.0 for φRSA1, φRSB1, and φRSL1, respectively. Figure 1 shows the effect of phage infection on bacterial growth. φRSA1 and φRSB1 infection, irrespective of whether they were used solely or mixed with other phages, were able to readily lyse growing cultures. However, at approximately 30 h p.i., resistant cells started to grow and reached the stationary state at 70 h p.i. The results were the same at different phage doses (PFU/bacterial cell) for each of the phages. Recovering cells demonstrated a general resistance to both φRSA1 and φRSB1, irrespective of the initial phage species they were infected with (Table 3). The growth recovery pattern at 30 h p.i. was not affected by the addition of other phage species at 40 h p.i. (data not shown). In contrast, cells infected solely with φRSL1 grew slowly until 40 to 50 h p.i., instead of quickly lysing, and were maintained at a low and steady level until 60 h p.i. (Fig. 1). This steady-state growth pattern may indicate an equilibrium between cell growth and lysis by the phage or resistant bacterial cells that are less robust in their growth (1). Infection of φRSL1 in combination with φRSA1 and/or φRSB1, however, resulted in cell lysis and recovering growth patterns similar to that seen in bacterial cells infected solely with φRSA1, φRSB1, or a mixture. Our previous results have shown that these three phages are virulent and form clear plaques on culture plates (23). Both φRSA1 and φRSB1 lyse the host cells at 2 to 3 h p.i. (6, 17), whereas φRSL1 takes longer for lysis to occur (3 to 4 h) (24). Therefore, one possible explanation for the observation in the mixed infection with φRSL1 is that φRSA1 and/or φRSB1 predominantly infected and lysed the host cells, and over time the titers of these phages were greatly increased compared to that of φRSL1, resulting in the lysis-recovery pattern of cell growth (Fig. 1). We confirmed this through plaque assays with the culture fluid after 100 h p.i. In the case of the φRSL1 and φRSA1 mixed infection, 74% of plaques (∼108) formed on the plates were due to φRSA1, easily distinguishable by plaque morphology. For the culture coinfected with φRSL1 and φRSB1, 92% of plaques (2 × 108) were due to φRSB1. φRSB1 was also predominant (>90% of 9 × 108 plaques) when combined with φRSA1. Phage-resistant cells from cultures infected with φRSA1 and φRSB1 were susceptible to φRSL1, as shown in Table 3. Therefore, φRSL1 presumably has a different host recognition way from the other phages. These results obtained with strain M4S as a host were reproducible with other R. solanacearum strains such as Ps29 and MAFF 730138.
Fig. 1.
Time course of bacterial growth after infection with bacteriophages. The first 20 h region is enlarged in panel B (the symbols are the same as shown in panel A). Cells of R. solanacearum strain M4S (OD600 = 0.3 corresponding to ∼108 cells/ml; vertical arrow) were infected solely or by mixing with three phages: φRSA1, dose = 0.5 × 108 PFU/ml; φRSB1, dose = 0.5 × 108 PFU/ml; and φRSL1, dose = 5.0 × 108 PFU/ml. At about 30 h p.i., resistant cells started to grow when treated with φRSA1 and/or φRSB1, either alone or as part of a phage mixture. Cells solely infected with φRSL1 were kept at a low cell density. Similar results were obtained with different R. solanacearum strains (data not shown).
Table 3.
Phage resistance of R. solanacearum cells in a culture treated with φRSA1 or φRSB1
| Single colonya | Phage resistanceb |
||
|---|---|---|---|
| φRSA1 | φRSB1 | φRSL1 | |
| Ar-1 | + | + | – |
| Ar-2 | + | + | – |
| Ar-3 | + | + | – |
| Br-1 | + | + | – |
| Br-2 | + | + | – |
| Br-3 | + | + | – |
Data for the single colonies isolated from φRSA1- or φRSB1-treated cultures at 100 h p.i. shown in Fig. 1 are presented. Ar-1, Ar-2, and Ar-3 and Br-1, Br-2, and Br-3 were obtained from cultures treated with φRSA1 and φRSB1, respectively.
Phage resistance is indicated as resistant (+) or sensitive (-).
From these results, we concluded that for control R. solanacearum cells, a cocktail of φRSA1, φRSB1, and φRSL1 may not be suitable due to the presence of phage-resistant cells. Instead, φRSL1 infection somehow maintains the cell population at lower levels and limits the growth of resistant cells recovering from φRSA1 and/or φRSB1 infection. Therefore, we attempted to use φRSL1 as an agent to control the growth of R. solanacearum cells and bacterial wilt disease.
In planta inhibition of R. solanacearum growth and movement by treatment with φRSL1.
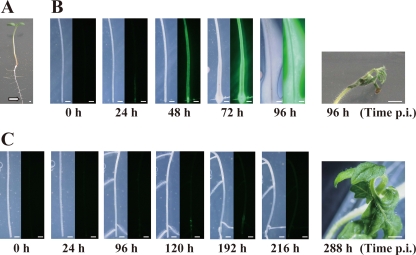
The effect of φRSL1 infection to stably limit the growth of R. solanacearum cells in vitro leads us to examine bacterial growth and movement in planta after treatment with φRSL1. We monitored real-time bacterial dynamics in inoculated tomato plants grown on solid agar medium using a previously described method (5, 16). Seven-day-old seedlings of the tomato cultivar Oogata-Fukuju were first treated with φRSL1 (1.0 μl containing 106 PFU) at a cut made in the tip of the taproot, 10 mm from the apex. At various times after treatment, bacterial cells were inoculated at the cut and observed (Fig. 2 A). Every experiment was performed in triplicate with 10 individual plants. Figure 2B shows the time course of bacterial growth and movement in a tomato taproot without φRSL1 treatment (control). After bacterial inoculation, GFP fluorescence intensity increased with time, and GFP-labeled bacterial cells moved upward through xylem vessels until 48 h p.i. At 72 h p.i., GFP fluorescence was apparent outside the taproot, suggesting cell movement, and growth occurred outside the taproot. Slimy colonies of cells covered the entire taproot by 96 h p.i. At this stage, the hypocotyls and young leaves were wilting (Fig. 2B).
Fig. 2.
In planta inhibition of R. solanacearum growth and movement by treatment with φRSL1. (A) Tomato seedlings grown in culture dishes (7 days old) were cut at the tip of the taproot (arrow) and treated with 1 μl of φRSL1 (106 PFU). After 12 to 24 h, bacterial cells (∼105 cells) of strain MAFF 106611 harboring pRSS12 were inoculated at the cut. (B) Without phage treatment (control), bacterial penetration into the taproot, successive upward movement, and growth in the tissues was evident at 24 h p.i. (left panel, bright-field image; right panel, dark-field image). GFP fluorescence was apparent outside the taproot at 72 h p.i. The hypocotyl and young leaves were wilted at 96 h p.i. No lateral shoot formation was observed throughout the period. (C) With phage treatment, no bacterial growth and movement in plant bodies was apparent until 96 h p.i. Faint GFP fluorescence was retained at the inoculation point. During this period, lateral shoots frequently formed; even though at 120 h p.i. some GFP-labeled cells were visible, their growth and movement were quite limited. The cotyledon, leaves, and the meristem appeared healthy with no symptoms of wilting at 288 h p.i. The 30 different plants tested displayed similar results. Scale bar, 0.5 mm.
Remarkably different phenomena were observed when tomato plants were treated with φRSL1 preceding bacterial inoculation. As shown in Fig. 2C, up until 96 h p.i. no bacterial growth and movement into plant bodies were apparent; faint GFP fluorescence was retained at the inoculation point. During this period, lateral shoots frequently formed as in healthy plants; this was never observed in control plants that were bacterially challenged but not phage treated. At 120 h p.i., GFP fluorescence intensity increased slightly, and GFP-labeled cells were observed to be moving upward along the taproot. However, the growth and movement was quite limited; at 192 h p.i., the GFP fluorescence pattern remained unchanged. The GFP fluorescence was unclear again at 216 h p.i. The cotyledons, leaves, and the meristem looked healthy with no symptoms of wilting (Fig. 2C). When tomato seedlings were treated in the same way with φRSA1 or φRSB1, bacterial penetration and growth in xylem vessels were obvious at 48 h p.i. (data not shown). These results indicated that bacterial growth and movement were drastically limited in tomato plants by pretreatment with φRSL1. Thus, pretreatment of tomato seedlings with φRSL1 may limit infection by R. solanacearum or prevent bacterial wilt disease.
Prolonged cultivation in soil of tomato plants treated with φRSL1 after inoculation with R. solanacearum.
Six tomato plants were treated with φRSL1 after inoculation with R. solanacearum (Fig. 2), transferred from culture dishes to pots, and then cultivated in a natural environment for prolonged observation. At 49 days posttransfer (1.5 months p.i.), four plants still grew well without any symptoms of wilting. Two plants died soon after transfer (2 to 3 days); we considered that this was caused by physical/physiological damage such as water shock or root damage and not by bacterial infection because few bacterial cells were detected in plant organs.
Persistence and stability of φRSL1 particles in plants and soil.
The stems, leaves, and roots of plants and culture soil from two pot cultures described above were subjected to phage titer assays 4 months later. One plant, 52 cm in height, was treated with φRSL1 only (control plant 1), and the other plant, 62 cm in height, was treated with φRSL1 followed by the pathogen (plant 2), and harvested 4 months p.i. The data shown in Table 4 indicated that no φRSL1 phage was detected from the stems or leaves of either plant. However, a considerably large number of phages were retained in the roots of both plants. The plants inoculated with bacterial cells gave titers 10 times higher than those without inoculation. Similar results were obtained in the in vitro experiments shown in Fig. 2). The soils from both cultures also yielded phage plaques. Plaques appearing on the assay plates always displayed homologous morphology, resembling φRSL1 plaques (23). The genomic DNA isolated from random plaques coincided with φRSL1 DNA by restriction digestion patterns (data not shown). These results indicated that φRSL1 phages were stably retained in plants roots, as well as in soils, with a lower concentration.
Table 4.
Persistence and stability of φRSL1 particles in plants and soil
| Plant or soila | Wt (g) | Mean phage titerb (PFU/g) ± SD |
|---|---|---|
| Plant 1 | ||
| Stems and leaves (10 cm) | 2.50 | ND |
| Stems (1 to 5 cm parts above ground) | 0.75 | ND |
| Roots (13 cm) | 2.50 | 1.8 × 103 ± 2.0 × 102 |
| Soil (rhizosphere) | 2.00 | 2.7 × 101 ± 8.5 × 100 |
| Plant 2 | ||
| Stems and leaves (10 cm) | 2.50 | ND |
| Stems (1 to 5 cm parts above ground) | 0.80 | ND |
| Roots (13 cm) | 3.10 | 1.9 × 104 ± 2.5 × 103 |
| Soil (rhizosphere) | 2.00 | 5.5 × 102 ± 1.5 × 102 |
| Plant 3 | ||
| Roots | 0.51 | 5.7 × 105 ± 3.0 × 104 |
| Soil (rhizosphere) | 2.00 | 7.1 × 105 ± 1.8 × 105 |
Plant 1, a control plant treated with φRSL1 only (in vitro treated and transferred to soil); plant 2, a plant treated with φRSL1, followed by the pathogen (in vitro treated and transferred to soil); plant 3, a φRSL1-treated plant with no wilting symptom 18 days after the pathogen challenge (in soil experiments).
Assays were repeated three times. ND, not detected.
Prevention of bacterial wilt by treatment with φRSL1.
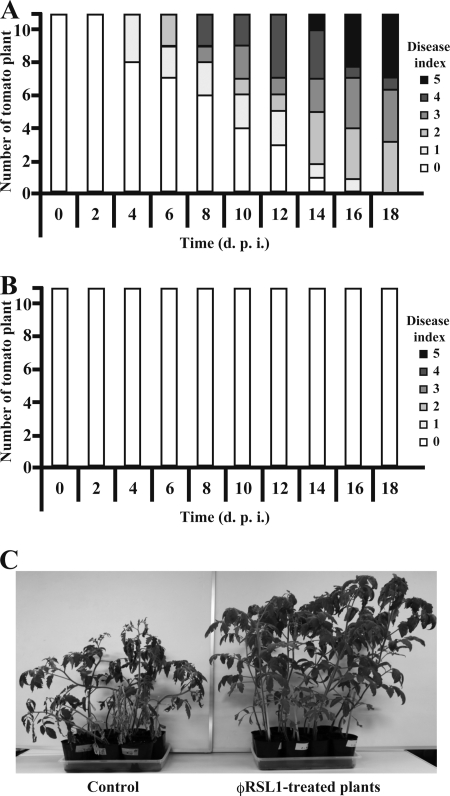
The effect of φRSL1 infection to stably limit the growth of R. solanacearum cells in vitro and in planta led us to examine whether φRSL1 treatment of tomato plants under soil conditions prevents wilting by inoculated R. solanacearum cells. One-month-old tomato plants (20 to 23 cm in height) pretreated with φRSL1 were inoculated with R. solanacearum cells as described in Materials and Methods. Wilting symptoms were recorded every 2 days. The results shown in Fig. 3 indicated an efficient prevention of wilting by treatment with φRSL1. Plants without φRSL1 started to show wilting symptoms 4 days p.i. and all 11 plants showed wilting symptoms 18 days p.i. (Fig. 3A and C). In contrast, all 11 φRSL1-treated plants showed no symptoms of wilting during the experimental period (Fig. 3B and C). The φRSL1 phages were stably retained in plant roots as well as in soils at this stage, as shown in Table 4. Three random tomato cultures treated with φRSL1 were subjected to phage titer assay as described above. Because all three cultures showed almost the same values, only one example (plant 3) is included in Table 4. These results suggest the effectiveness of φRSL1 in the practical application as a biocontrol agent against bacterial wilt. Plants can be treated with φRSL1 during the early stages of growth. When tomato plants were pretreated with a phage mixture containing two phages (φRSA1 and φRSB1) or three phages (φRSA1, φRSB1, and φRSL1) and challenged by R. solanacearum cells in the same way, as described above, wilting symptoms appeared by 4 days p.i., and all of them wilted after 14 days p.i. (see Fig. S1 in the supplemental material). The wilting patterns were generally similar to those observed in control plants without phage treatment. These results were consistent with the growth of resistant cells observed in the in vitro and in planta experiments in which a phage mixture was used.
Fig. 3.
Prevention of bacterial wilt by treatment with φRSL1. One-month-old tomato plants (20 to 23 cm in height) pretreated with tap water (A, control) or φRSL1 (B) were inoculated with R. solanacearum cells as described in Materials and Methods. Wilting symptoms were graded from 0 to 5 as follows: 0, no symptoms; 1, only one petiole was wilting; 2, two to three petioles were wilting; 3, all but two to three petioles were wilting; 4, all petioles were wilting; and 5, the plant died. (C) Tomato plants observed at 18 days p.i.
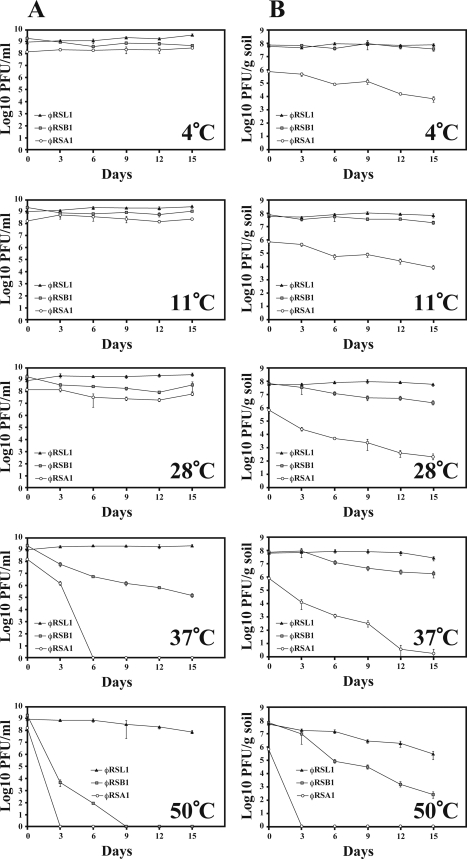
Effects of temperature on the stability of φRSL1 in soil.
We further studied the effects of temperature on the stability of φRSL1 compared to φRSA1 and φRSB1. Phage preparations were kept at different temperatures in the range 4 to 50°C, in the presence or absence of soil. As shown in Fig. 4 A, in the absence of soil, all three phages were essentially stable below 28°C, whereas φRSL1 exhibited greater stability compared to other phages at higher temperatures (37 and 50°C). After a 15-day incubation, ca. 10% φRSL1 survived at 50°C. No phages were detected after a 3-day incubation with φRSA1 and after a 9-day incubation with φRSB1. Similar stability patterns were observed under different soil temperatures. At low temperatures, the titers of some phages, especially φRSA1, were decreased probably due to nonspecific adsorption to soil particles. φRSL1 also exhibited the highest stability in soil (Fig. 4B).
Fig. 4.
Effect of temperature on the stability of φRSL1 compared to φRSA1 and φRSB1. Phage preparations were kept at a range of temperatures (4 to 50°C) for various periods without (A) or with (B) soil. φRSL1 displayed significant stability at higher temperatures (37 and 50°C). Error bars indicate the standard error (n = 3).
DISCUSSION
Alternative phage biocontrol using φRSL1.
For phage biocontrol, only virulent phages are used, thereby avoiding the problem of lysogeny. Phage cocktails are recommended to prevent the problem of resistance. These cocktails ideally contain several phages with different host specificities, replication mechanisms, and/or infection cycles (7, 15). In the present study, when host R. solanacearum cells were quickly lysed by treatment with φRSA1 or φRSB1, resistant cells presumably preexisting in the population at a very low frequency were increased at 30 h p.i. Given that the majority of the cell population contains susceptible cells, the elimination of these cells may allow minor cells to predominate to the next generation. Since such recovering cells were somehow resistant to both φRSA1 and φRSB1 (Table 3), treatment with a mixture of these phages resulted in the same death and recovery patterns as bacterial cells treated with a single phage (Fig. 1). The resistance mechanisms used by these cells are currently unknown, and the host specificity was different between φRSA1 (a P2-like myovirus) and φRSB1 (a T7-like podovirus) (23). In our preliminary observations, several phage-resistant mutants, induced by transposon mutagenesis, displayed differences in resistance to φRSA1 and φRSB1. Therefore, cells recovering from the phage treatment may include resistant cells caused by different mechanisms and with different characteristics. A cocktail containing three phages—φRSA1, φRSB1, and φRSL1—also failed to stably prevent bacterial growth (Fig. 1). Although φRSL1 could lyse some recovering cells after φRSA1 and/or φRSB1 treatment (Table 3), in mixed phage treatments, cells may have been quickly lysed by φRSA1 or φRSB1, thereby interrupting the replication of φRSL1 and reducing the φRSL1 titer. φRSL1 may not be able to effect the lysis of cultures if its density is too low. Compared to φRSA1 and φRSB1, RSL1 takes longer for lysis to occur (3 to 4 h) with a latent period of 2.5 h (24). This infection cycle is rather longer than the doubling time of host cells (3 h) under routine culture conditions. Therefore, a higher dose may be required to efficiently infect and lyse the host cells. This was supported by the observation that little φRSL1 was detected in the culture treated with three phages at 100 h p.i.
Consequently, the strategy of using a phage cocktail in the biocontrol of R. solanacearum cells did not work in the present study. Instead, treatment with φRSL1 alone did not rapidly kill cells but kept the cell density at a low level (Fig. 1). In planta monitoring of bacterial cells showed that treatment with φRSL1 resulted in a blockage of the growth and movement of bacterial cells in the tomato root. Treated plants survived for as long as 4 months. φRSA1 or φRSB1 were not able to induce similar plant-protecting effects. Based on these observations, we propose an alternative phage biocontrol method, using a unique phage such as φRSL1, instead of a phage cocktail containing highly lytic phages. With this method, bacterial cells are not killed altogether but a sustainable state of phage-bacterium coexistence is maintained, like a carrier state or pseudolysogeny (1). To apply this method practically, variants of φRSL1-type phages would be required that have different host ranges covering most strains of R. solanacearum.
Coexistence of R. solanacearum cells and φRSL1.
φRSL1 is a unique phage with a very large genome of 231 kb containing 343 open reading frames. A lysogenic cycle or episomal replication of φRSL1 has not been elucidated; genomic Southern blot analysis of many field-isolated strains has not identified any significant hybridizing signals with φRSL1 DNA as a probe (23; data not shown). After infection with φRSL1, cell density was maintained stably at low levels, as shown in Fig. 1, suggesting an equilibrium between cell growth and lysis. The establishment of equilibrium may be explained by a long latent period and small burst size of a phage. Our previous study revealed that φRSL1 takes longer for lysis to occur (3 to 4 h) with a latent period of 2.5 h and a burst size of 80 to 90 PFU per infected cell (24). Other possible explanations for the cell-phage balance may come from some specific regulation of phage infectivity or a low frequency of resistant cells and the production of growth-inhibitory factors due to phage infection. We are interested in the expression of many unique genes, including those for lysis genes encoded in the φRSL1 genomic DNA (24). Further characterization of the expression and function of these genes may reveal the mechanism for establishment of equilibrium. It is also interesting that in some cases, lysogenic filamentous phages were observed to be induced in the remaining host cells after infection with φRSL1, which may also contribute to the equilibrium.
Stability of φRSL1 in soil and its practical application.
Phages are utilized for controlling plant pathogens either in the rhizosphere or phyllosphere. The direct application of phages to the phyllosphere is subject to serious phage stability problems (15). Field and laboratory studies have demonstrated that phages are inactivated rapidly by exposure to sunlight, high temperatures, extremes in pH, oxidative conditions, and flowing water (12, 13). In the case of bacterial wilt, phages for biocontrol can be applied to the rhizosphere. Sunlight, the most destructive environmental factor, and oxidative inactivation are not as relevant in this case. φRSL1 was shown to be relatively stable in soil, especially at higher temperatures (Fig. 4). In fact, active φRSL1 particles were consistently recovered from the roots of treated plants and soils at 4 months p.i. (Table 4). This φRSL1 stability (Fig. 4) in soil appears to be another advantage of φRSL1 as a biocontrol agent. Moreover, bulk production at high concentrations of φRSL1 particles (∼1012 PFU/ml) is possible by centrifugation at 15,800 × g for 20 min. Prolonged disease control may be possible if φRSL1 is applied to plants at the seedling stage.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by the Industrial Technology Research Grant Program (04A09505) from the New Energy and Industrial Technology Development Organization of Japan and by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (21580095 to T.Y. and 20.3533 to A.F.).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Abedon S. T. 2009. Disambiguating bacteriophage pseudolysogeny: a historical analysis of lysogeny, pseudolysogeny, and the phage carrier state, p. 285–307 In Adams H. T. (ed.), Contemporary trends in bacteriophage research. Nova Science Publishers, Inc., Hauppauge, NY [Google Scholar]
- 2. Ausubel F., et al. 1995. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 3. Balogh B., Jones J. B., Iriarte F. B., Momol M. T. 2010. Phage therapy for plant disease control. Curr. Pharm. Biotechnol. 11:48–57 [DOI] [PubMed] [Google Scholar]
- 4. Fegan M., Prior P. 2005. How complex is the Ralstonia solanacearum species complex? p. 449–461 In Allen C., Prior P., Hayward A. C. (ed.), Bacterial wilt: the disease and the Ralstonia solanacearum species complex. American Phytopathology Society, St. Paul, MN [Google Scholar]
- 5. Fujie M., Takamoto H., Kawasaki T., Fujiwara A., Yamada T. 2010. Monitoring growth and movement of Ralstonia solanacearum cells harboring plasmid pRSS12 derived from bacteriophage φRSS1. J. Biosci. Bioeng. 109:153–158 [DOI] [PubMed] [Google Scholar]
- 6. Fujiwara A., Kawasaki T., Usami S., Fujie M., Yamada T. 2008. Genomic characterization of Ralstonia solanacearum phage φRSA1 and its related prophage (φRSX) in strain GMI1000. J. Bacteriol. 190:143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gill J., Abedon S. T. 2003. Bacteriophage ecology and plants. American Phytopathological Society, St. Paul, MN: http://apsnet.org/online/feature/phages/ [Google Scholar]
- 8. Goto N. 1992. Fundamentals of bacterial plant physiology. Academic Press, Inc., New York, NY [Google Scholar]
- 9. Hayward A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29:65–87 [DOI] [PubMed] [Google Scholar]
- 10. Hayward A. C. 2000. Ralstonia solanacearum, p. 32–42 In Lederberg J. (ed.), Encyclopedia of microbiology, vol. 4 Academic Press, Inc., San Diego, CA [Google Scholar]
- 11. Horita M., Tsuchiya K. 2002. Causal agent of bacterial wilt disease Ralstonia solanacearum, p. 5–8 In National Institute of Agricultural Sciences (ed.), MAFF microorganism genetic resources manual no. 12. National Institute of Agricultural Sciences, Tsukuba, Japan [Google Scholar]
- 12. Ignoffo C. M., Garcia C. 1992. Combination of environmental factors and simulated sunlight affecting activity of inclusion bodies of the heliothis (Lepidoprera: Noctuidae) nucleopolyhedrosis virus. Environ. Entomol. 21:210–213 [Google Scholar]
- 13. Iriarte F. B., et al. 2007. Factors affecting survival of bacteriophage on tomato leaf surfaces. Appl. Environ. Microbiol. 73:1704–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Janse J. 1996. Potato brown rot in western Europe-history, present occurrence and some remarks on possible origin, epidemiology, and control strategies. Bull. OEPP/EPPO 26:679–695 [Google Scholar]
- 15. Jones J. B., et al. 2007. Bacteriophages for plant disease control. Annu. Rev. Phytopathol. 45:245–262 [DOI] [PubMed] [Google Scholar]
- 16. Kawasaki T., Satsuma H., Fujie M., Usami S., Yamada T. 2007. Monitoring of phytopathogenic Ralstonia solanacearum cells using green fluorescent protein-expressing plasmid derived from bacteriophage φRSS1. J. Biosci. Bioeng. 104:451–456 [DOI] [PubMed] [Google Scholar]
- 17. Kawasaki T., et al. 2009. Genomic characterization of Ralstonia solanacearum phage φRSB1, a T7-like wide-host-range phage. J. Bacteriol. 191:422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okabe N., Goto M. 1963. Bacteriophages of plant pathogens. Annu. Rev. Phytopathol. 1:397–418 [Google Scholar]
- 19. Ozawa H., Tanaka H., Ichinose Y., Shiraishi H., Yamada T. 2001. Bacteriophage P4282, a parasite of Ralstonia solanacearum, encodes a bacteriolytic protein important for lytic infection of its host. Mo. Genet. Genom. 265:95–101 [DOI] [PubMed] [Google Scholar]
- 20. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21. Tanaka H., Negishi H., Maeda H. 1990. Control of tobacco bacterial wilt by an avirulent strain of Pseudomonas solanacearum M4S and its bacteriophage. Ann. Phytopathol. Soc. Japan 56:243–246 [Google Scholar]
- 22. Toyoda H., et al. 1991. Characterization of deoxyribonucleic acid of virulent bacteriophage and its infectivity to host bacterium, Pseudomonas solanacearum. J. Phytopathol. 131:11–21 [Google Scholar]
- 23. Yamada T., et al. 2007. Isolation and characterization of bacteriophages that infect the phytopathogen Ralstonia solanacearum. Microbiology 153:2630–2639 [DOI] [PubMed] [Google Scholar]
- 24. Yamada T., et al. 2010. A jumbo phage infecting the phytopathogen Ralstonia solanacearum defines a new lineage of the Myoviridae family. Virology 398:135–147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.