Abstract
Molecular methods incorporating nested PCR-restriction fragment length polymorphism (RFLP) analysis of the 18S rRNA gene of Cryptosporidium species were validated to assess performance based on limit of detection (LoD) and for detecting and resolving mixtures of species and genotypes within a single sample. The 95% LoD was determined for seven species (Cryptosporidium hominis, C. parvum, C. felis, C. meleagridis, C. ubiquitum, C. muris, and C. andersoni) and ranged from 7 to 11 plasmid template copies with overlapping 95% confidence limits. The LoD values for genomic DNA from oocysts on microscope slides were 7 and 10 template copies for C. andersoni and C. parvum, respectively. The repetitive nested PCR-RFLP slide protocol had an LoD of 4 oocysts per slide. When templates of two species were mixed in equal ratios in the nested PCR-RFLP reaction mixture, there was no amplification bias toward one species over another. At high ratios of template mixtures (>1:10), there was a reduction or loss of detection of the less abundant species by RFLP analysis, most likely due to heteroduplex formation in the later cycles of the PCR. Replicate nested PCR was successful at resolving many mixtures of Cryptosporidium at template concentrations near or below the LoD. The cloning of nested PCR products resulted in 17% of the cloned sequences being recombinants of the two original templates. Limiting-dilution nested PCR followed by the sequencing of PCR products resulted in no sequence anomalies, suggesting that this method is an effective and accurate way to study the species diversity of Cryptosporidium, particularly for environmental water samples, in which mixtures of parasites are common.
INTRODUCTION
DNA sequencing of rRNA genes has long been used to study microbially diverse populations in the environment. Underrepresentations of the genetic diversity in the microbial community have been attributed to differential PCR amplification, while overestimations of genetic diversity have been attributed to artifact PCR products (38). PCR biases can result from differential priming efficiencies of degenerate primers, differential denaturation conditions, GC contents in the primer and/or template, and amplicon length (22, 27, 34, 39). Artifact PCR products include heteroduplex formation between different PCR products having a high degree of sequence homology and PCR-mediated recombination resulting in chimera formation (16, 36).
The molecular characterization of Cryptosporidium species and genotypes in environmental water samples using the 18S rRNA gene has emerged as a powerful source-tracking tool for public health risk assessment purposes. Many PCR methods have been evaluated for their effectivenesses on environmental samples (11, 25, 42, 46). Regulatory and environmental monitoring of Cryptosporidium is typically carried out by using methods based on U.S. Environmental Protection Agency (EPA) Method 1623 (8, 37), and some applications have been developed for the molecular characterization of samples processed via these methods. The molecular characterization of environmental Cryptosporidium isolates by identifying species and genotypes of oocysts from positive microscope slides allows the collection of occurrence data and species and genotype identification from a single sample (6, 25, 26, 29, 30). None of these methods have been thoroughly validated to determine what biases or artifacts may occur as a result of the different molecular approaches used.
An accurate identification of Cryptosporidium species and genotypes using the 18S rRNA gene requires confirmation by DNA sequence analysis. Environmental samples have been shown to often contain more than one species or genotype in a sample (13, 14, 26, 29, 47). More than one species or genotype can be present in a single PCR mixture, reducing the ability to obtain sequence information from a sample containing multiple species. Various approaches have been used by research groups to resolve individual species and genotypes from mixed DNA templates: (i) using repetitive nested PCR-restriction fragment length polymorphism (RFLP) analysis (29, 30), (ii) varying the concentration of the template used for the nested PCR-RFLP analysis followed by sequencing (15, 41, 45, 47), (iii) cloning the nested PCR products and sequencing the cloned inserts (13, 14, 48), and (iv) using multiple target loci (26).
In this study, the performance of a nested PCR-RFLP approach was evaluated by examining the method sensitivity and accuracy with a range of Cryptosporidium species. This included determining the limit of detection (LoD) of our basic platform (repetitive nested PCR-RFLP) for the molecular characterization of Cryptosporidium oocysts from microscope slides processed according to Method 1623. The ability of replicate nested PCR-RFLP analysis to resolve multiple Cryptosporidium spp. from an individual microscope slide was examined, followed by an assessment of the suitability of cloning and limiting template dilution techniques for resolving mixtures of Cryptosporidium species from slides when required.
MATERIALS AND METHODS
Preparation of DNA templates from Cryptosporidium species.
To compare the limits of detection, DNA templates of a known concentration were required. In order to accurately determine gene copy numbers, full-length 18S rRNA plasmid templates were constructed for the following species: C. parvum, C. hominis, C. andersoni, C. muris, C. meleagridis, C. felis, and C. ubiquitum. DNA extracts from isolates previously identified as C. hominis, C. meleagridis, C. felis, and C. ubiquitum isolates were obtained from the UK Cryptosporidium Reference Unit (Swansea, United Kingdom) (4). Commercially available preparations of C. parvum (Hyperion Research Ltd., Medicine Hat, AB, Canada) and C. muris (Waterborne Inc., New Orleans, LA) containing approximately 106 oocysts were obtained. The oocyst stocks were mixed by manual shaking for 2 min, and 100 μl of the oocyst suspension was removed and combined with 100 μl double-strength Sarkosyl lysis buffer followed by DNA extraction (18, 29). Feces from an asymptomatic adult steer known to shed high numbers of C. andersoni oocysts were obtained from the Agriculture and Agri-Food Canada Research Center (Lethbridge, AB, Canada). A 1-ml volume of feces was diluted in 9 ml distilled water and purified by immunomagnetic separation (Dynabeads; Invitrogen Canada Inc., Burlington, ON, Canada). Oocyst-bead complexes were suspended in 200 μl of Sarkosyl lysis buffer, which was followed by DNA extraction as described above.
Full-length 18S rRNA gene amplicons for each species were created by using previously described primers and conditions (45). The product was then cloned into Escherichia coli cells by using the TOPO TA vector (Invitrogen Canada Inc.), and the plasmids were purified by using a Qiagen plasmid minikit (Qiagen, Mississauga, ON, Canada) and sequenced bidirectionally (Macrogen, Seoul, South Korea) to confirm the species identification. Plasmid DNA was diluted in Tris-EDTA (TE) buffer to approximately 50 ng/ml and quantified by using a Quant-iT PicoGreen double-stranded DNA (dsDNA) assay kit (Invitrogen Canada Inc.), with fluorescence determined by use of a LightCycler II instrument (Roche Applied Science, Laval, PQ, Canada). Template copy numbers for each plasmid were estimated based on the quantity of DNA, the size of the plasmid insert, the molecular weight constant for double-stranded DNA, and Avogadro's number. A working stock containing 106 template copies of the 18S rRNA gene per 5 μl was prepared, and serial dilutions (1-ml volumes) were prepared to 1 template copy per 5 μl. Additional dilutions resulting in 8 concentrations between 50 and 0.25 template copies per 5 μl (equal to the template copy number per PCR) were assayed by the genus-specific nested PCR described below.
Sources of Cryptosporidium oocysts for method validation.
Oocysts of C. andersoni were purified from the feces of the asymptomatic adult steer by first preparing a slurry of feces in 0.01% Tween 20, which was sequentially passed through sieves (3), followed by concentrating the particulates at 1,100 × g for 20 min and aspirating off the supernatant. The oocysts were purified from the particulates by flotation on a continuous cesium chloride gradient (1.4, 1.1, and 1.05 g/ml) using a Beckman L7-55 ultracentrifuge (Beckman Instruments, Palo Alto, CA) at 16,000 × g for 60 min using a swing-bucket rotor (3). The interface between the 1.4- and 1.1-g/ml cesium chloride concentrations was washed 3 times in 0.01% Tween 20 and further purified by using a Sorvall RC5-B fixed-angle centrifuge at 14,500 × g for 10 min with semicontinuous concentrations (0.075 and 0.06 g/ml) of cesium chloride (with washing 3 times in 0.01% Tween 20 between purifications). Oocysts were suspended in 0.01% Tween 20 containing 100 units/ml penicillin, 100 μg/ml streptomycin, and 100 μg/ml gentamicin and shipped on ice to the Wisconsin State Laboratory of Hygiene (WSLH). Purified C. parvum oocysts were obtained by the WSLH from Chuck Sterling at the University of Arizona (Sterling Parasitology Laboratories, Tucson, AZ). Parasite preparations were examined for general intactness by differential interference contrast (DIC) microscopy upon receipt at the WSLH.
Flow cytometry sorting of Cryptosporidium oocysts on microscope slides.
Single-well microscope slides (Spot-On; Invitrogen Canada Inc.) were prepared by using flow cytometry with cell sorting (FCCS) according to standard WSLH protocols. Briefly, oocyst suspensions were stained with Crypt-a-Glo (Waterborne Inc.) and analyzed by flow cytometry using a Becton Dickinson Aria II instrument (Becton Dickinson, San Jose, CA). Fluorescently labeled oocysts were sorted into the wells based on forward angle, log right angle, and fluorescent (fluorescein isothiocyanate [FITC]) light scatter properties. Slides were allowed to dry at room temperature and fixed with methanol, and oocyst numbers were confirmed by epifluorescence microscopy. Ten replicates of each oocyst concentration were prepared for single-species slides (1, 2, 3, 5, 6, 10, and 20 oocysts) and five replicates for the slides with both C. parvum and C. andersoni oocysts (3 and 3, 3 and 6, 3 and 30, 3 and 60, 6 and 6, 6 and 30, 6 and 60, 30 and 30, 30 and 60, 60 and 60, 6 and 3, 30 and 3, 60 and 3, 30 and 6, 60 and 6, and 60 and 30 oocysts, respectively). Slides were shipped to the Alberta Provincial Public Health Laboratory (ProvLab, Calgary, AB, Canada) and processed for molecular analysis. Oocysts were removed from the slides, and the DNA was extracted as previously described (29). Template copy numbers for PCR mixtures containing whole oocysts were based on 100-μl-volume DNA extracts and the assumption that each oocyst contained 20 copies of the 18S rRNA gene (20). DNA extracts from all slides were analyzed by using the repetitive nested PCR platform with five replicates of 5 μl of DNA extract in the primary reaction mixture for the nested PCR.
Genus-specific nested PCR-RFLP analysis.
Nested PCR mixtures contained 1× PCR buffer, 3 mM MgCl2, 200 μM deoxynucleoside triphosphate (dNTP) (Invitrogen Canada Inc.), 200 μg/ml nonacetylated bovine serum albumin (BSA) (primary PCR only) (Sigma-Aldrich, St. Louis, MO), 200 nM primer, and 1 U Platinum Taq polymerase (Invitrogen Canada Inc.). The volume of all reaction mixtures was 50 μl, with 5 μl of template added to the primary PCR mixture and 1 μl of the primary product transferred into the secondary PCR mixture. Primers 5′-TTCTAGAGCTAATACATGCG and 5′-CCCATTTCCTTCGAAACAGGA for the primary PCR and primers 5′-GGAAGGGTTGTATTTATTAGATAAAG and 5′-AAGGAGTAAGGAACAACCTCCA for the secondary PCR were described previously by Xiao et al. (45). Cycling conditions consisting of a hot start at 94°C for 3 min followed by 94°C for 45 s, 55°C for 45 s, 72°C for 1 min, and a final extension step of 72°C for 7 min were also previously described (45). PCR was carried out with GeneAmp 2700 thermal cyclers (Applied Biosystems, Carlsbad, CA) for 30 cycles of primary PCR and 25 cycles of secondary PCR. Electrophoresis of the products was carried out with 2% agarose gels and visualized with ethidium bromide staining. RFLP analysis using SspI (Promega Corporation, Madison, WI) was used to differentiate the PCR products containing multiple species (C. hominis or C. parvum from C. andersoni or C. ubiquitum) used in this study (29).
Resolution of mixtures of Cryptosporidium species.
Limiting the concentration of the template used for the nested PCR-RFLP analysis and sequencing was one approach examined for its utility in resolving samples containing multiple species. Rather than the 5 μl of template typically used for the first round of repetitive nested PCR, 5 replicates of 2.0, 1.0, and 0.5 μl were used. All positive nested PCR products were exposed to restriction enzyme digests (SspI) and visualized on 2% agarose gels. In some cases, nested PCR amplicons from slides seeded with C. parvum and C. andersoni and demonstrating both RFLP patterns were cloned as another tool for resolving mixed-template reactions. The cloning of secondary PCR products was carried out by using a TOPO TA cloning kit.
Purified plasmids (Miniprep; Qiagen) and gel-purified nested PCR products (QiaQuick; Qiagen) were sent to Macrogen Inc. (Seoul, South Korea) for bidirectional sequencing. Base calling was performed on the raw sequence data with Seqscape, version 2.6 (Applied Biosystems), using the KB base caller, and a consensus sequence was constructed. The sequences were aligned with ClustalW (35) at the Sun Center of Excellence for Visual Genomics at the University of Calgary.
RESULTS
Sensitivity of the genus-specific nested PCR to various Cryptosporidium species.
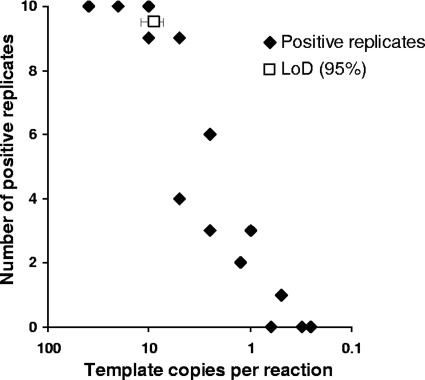
The sensitivity of the 18S rRNA gene nested PCR was assessed by defining the template copy number at which various species were detected 95% of the time. Nested PCR products were determined to be positive by the presence of a band of approximately 840 bp visualized by ethidium bromide staining after gel electrophoresis. Plasmid DNA was used to compare the limits of detection (LoDs) of the nested PCR for C. parvum, C. hominis, C. meleagridis, C. felis, C. muris, C. andersoni, and C. ubiquitum. Eight dilutions consisting of 10 replicates each and yielding between 40 and 0.25 template copies per reaction were prepared. Data were combined from two separate experimental setups to determine the LoD by Probit analysis for each species (SPSS Inc., Chicago, IL) along with 95% confidence limits (CLs). Figure 1 shows a typical dose-response plot of the relationship between rates of detection and template copy numbers.
Fig. 1.
Dose-response plot of template copy numbers (plasmid) versus numbers of replicates positive (out of 10 replicates) for two replicate experiments with Cryptosporidium meleagridis. The limit of detection (95% detection) with confidence intervals was determined by Probit analysis.
The LoD (Probit = 95) for the various species of Cryptosporidium ranged from 7.5 to 11.3 template copies with overlapping CLs, while 50% detection ranged from 2.5 to 4.0 template copy numbers (Table 1). The Pearson goodness-of-fit chi-square statistic was assessed, and the model fit was determined to be adequate (P > 0.05). The overlapping confidence limits for both the LoD and 50% detection suggest that there were no significant differences in the sensitivities of the nested PCR for all species of Cryptosporidium tested under ideal conditions. The limits of detection of C. parvum and C. andersoni from whole-oocyst DNA extracts (genomic DNA) were also determined. The LoD for C. parvum was 10.4 template copies and that for C. andersoni was 7.3 template copies, and they were determined not to be significantly different. Based on the overlapping LoD values for whole-oocyst DNA extracts and plasmid DNA, it was concluded that the Cryptosporidium species tested amplify equally well using the nested PCR protocols described.
Table 1.
Results of Probit analysis of the dose-response data (dose versus rate of detection) for Cryptosporidium nested PCR using plasmid templates of different species or genomic DNA templates of C. parvum and C. andersoni
| Species or genotype | Probit = 95a | 95% CL | Probit = 50 | 95% CL | χ2 (df, P)b |
|---|---|---|---|---|---|
| Plasmid DNA | |||||
| C. andersoni | 10.4 | 8.2–15.1 | 3.9 | 2.8–5.2 | 8.1 (10, 0.62) |
| C. parvum | 11.0 | 8.6–16.2 | 3.5 | 2.4–4.8 | 16.4 (14, 0.289) |
| C. muris | 11.3 | 8.2–20.4 | 4.3 | 2.7–6.7 | 20.8 (12, 0.054) |
| C. felis | 7.5 | 5.7–12.3 | 3.5 | 2.6–5.1 | 20.3 (14, 0.122) |
| C. ubiquitum | 8.0 | 6.0–13.2 | 3.0 | 1.9–4.4 | 20.2 (14, 0.124) |
| C. hominis | 8.5 | 6.6–12.6 | 2.6 | 1.6–3.6 | 18.1 (14, 0.201) |
| C. meleagridis | 8.8 | 7.2–12.0 | 4.0 | 3.2–5.1 | 18.2 (16, 0.310) |
| Genomic DNA | |||||
| C. andersoni | 7.3 | 4.1–9.1 | 2.5 | −8.3–4.8 | 81.8 (67, 0.106) |
| C. parvum | 10.4 | 8.6–13.0 | 3.7 | 0.2–5.7 | 95.1 (76, 0.068) |
Probit = 95 equals the LoD.
Model fit was assessed as a part of the Probit analysis and was determined by the Pearson goodness-of-fit chi-square statistic (a P value of >0.05 indicates an adequate model fit).
Probit analysis was also used to determine the detection limit of the basic testing platform of repetitive nested PCR for amplifying DNA from oocysts on slides after microscopic enumeration. The dose-response data for the C. parvum and C. andersoni slides were reclassified based on whether any of the 5 replicates from a slide were positive (i.e., the number of slides where Cryptosporidium was detected at each oocyst concentration). The dose-response data for C. andersoni and C. parvum were as follows: 6/10 and 6/10 for one oocyst, 9/10 and 8/10 for two oocysts, 9/10 and 9/10 for three oocysts, and 10/10 and 9/10 for five oocysts, with 10/10 slides being positive for all other concentrations tested (6, 10, and 20 oocysts), respectively. The C. andersoni and C. parvum results were combined to have sufficient data for determining a confidence interval. Probit analysis determined that the LoD for the slide protocol was 4 oocysts (95% CL, 3.1 to 7.3), and the model fit was determined to be adequate (Pearson goodness-of-fit chi-square = 4.24; df = 12; P = 0.979). It is worth noting that a single oocyst was detected on 60% of the slides (n = 20).
Detection of mixed populations of Cryptosporidium species by nested PCR-RFLP analysis.
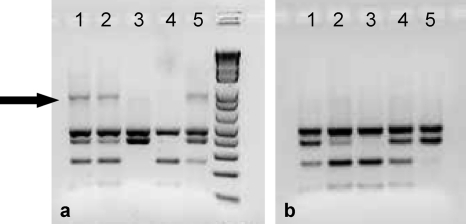
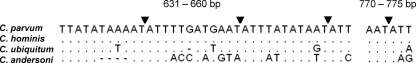
The ability to detect C. parvum and C. hominis in the presence of other Cryptosporidium species was assessed. Plasmid DNAs of more than one species (C. andersoni, C. ubiquitum, C. hominis, or C. parvum) were combined in the nested PCR, and detection was determined by RFLP analysis. An example of a nested PCR-RFLP analysis with controlled mixtures containing 10 plasmid copies of 18S rRNA genes from C. hominis and C. andersoni or C. hominis and C. ubiquitum is shown in Fig. 2. When multitemplate reaction mixtures containing C. hominis and C. andersoni were amplified by nested PCR, undigested secondary PCR products were observed (Fig. 2a, arrow), suggesting that the formation of heteroduplex DNA between amplicons of these species may have eliminated the restriction enzyme site. At plasmid template concentrations greater than the detection limit of the nested PCR (50, 100, and 1,000 template copies), undigested products were observed for every nested PCR, even if only one RFLP pattern could be detected. Undigested products were not observed when multitemplate reaction mixtures contained mixtures of C. hominis and C. ubiquitum (Fig. 2b), suggesting that any heteroduplex formation between these two species did not affect the fidelity of the restriction enzyme. The positions of SspI restriction enzyme recognition sites (5′-AAT▾ATT) for C. parvum and C. hominis are shown in Fig. 3, along with the mismatches that are likely to occur in the restriction enzyme recognition sites if heteroduplexes were formed with either C. andersoni or C. ubiquitum.
Fig. 2.
Detection and differentiation of C. hominis from C. andersoni (a) and C. hominis from C. ubiquitum (b) by RFLP analysis (SspI restriction enzyme digestion) of nested PCR products. Nested PCR-RFLP analysis was carried out in 5 replicates for plasmid templates mixed at 1:1 ratios (10 copies each). (a) Nested PCR-RFLP products in lanes 1, 2, and 5 contain a mixture of amplified products from C. hominis and C. andersoni, whereas only C. andersoni and C. hominis were amplified in lanes 3 and 4, respectively. (b) Nested PCR-RFLP products in lanes 1, 2, and 4 contain a mixture of amplified products from C. hominis and C. ubiquitum, whereas only C. hominis and C. ubiquitum were amplified in lanes 3 and 5, respectively. The arrow indicates undigested PCR products.
Fig. 3.
SspI restriction enzyme recognition sequences (cleavage site indicated by ▾) in the nested PCR products of C. parvum. Dots indicate base pair matches. C. hominis, C. ubiquitum, and C. andersoni were aligned to C. parvum by using a ClustalW alignment. Base pair mismatches in sequences when C. ubiquitum or C. andersoni was aligned to C. parvum or C. hominis are indicated by letters or gaps (–). The locations of the restriction sites correspond to the base pair locations in C. parvum reported under GenBank accession number AF164102.
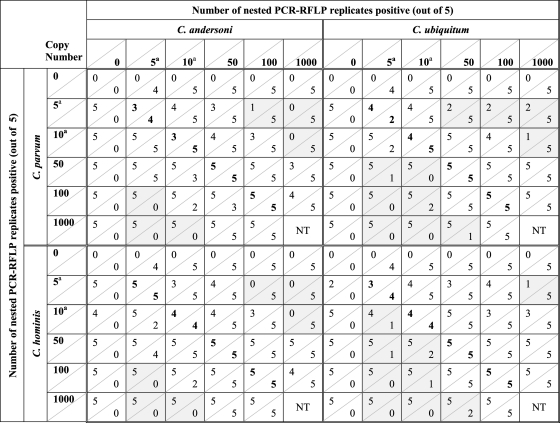
The 18S rRNA plasmids from C. parvum or C. hominis were mixed in the presence of C. andersoni at various template copy numbers as shown by the checkerboard pattern in Table 2. When template concentrations near or below the individual LoD were mixed at equal concentrations (5 and 10 template copies), both species were frequently detected, although the number of detections was not always equal to that of the single-template control. This is due to the variability in the numbers of detections observed at low template copy numbers (as shown in Fig. 1). At plasmid concentrations near or below the detection limit, undigested RFLP products were observed mostly for reaction mixtures that contained RFLP bands of both C. parvum and C. andersoni or C. hominis and C. andersoni (Fig. 2a). In general, the results showed that when template numbers of both species in the mixture were at equal template concentrations (5 to 100 template copy numbers), both species could be detected almost equally well (detection values in boldface type in Table 2).
Table 2.
Detection of C. parvum or C. hominis in the presence of C. andersoni or C. ubiquitum as determined by nested PCR-RFLP for reaction mixtures containing a mixture of plasmid templatesb
Template copy numbers that are near or below the detection limit of the assay, indicating there is a high degree of variability in the number of positive replicates obtained at these copy numbers even in single-template reactions.
Shading indicates plasmid template concentrations where a reduction or total loss of detection by nested PCR-RFLP occurred. Results in boldface type indicate results where plasmid templates were mixed at equal concentrations. The value below a diagonal line represents the number of nested PCR-RFLP mixtures positive for those species listed on the horizontal row; the value above the diagonal line is the number of nested PCR-RFLP mixtures positive for the species listed on the vertical column. NT, these ratios were not tested.
When 5 plasmid template copies of either C. parvum or C. hominis were mixed with 100 or 1,000 copies of C. andersoni (1:20 and 1:200 ratios), there appeared to be a reduction or loss of detection of the former species based on RFLP analysis (Table 2, shaded). When 10 plasmid template copies of C. parvum or C. hominis were mixed with 1,000 copies of C. andersoni (1:100 ratio), there was a loss in detection based on RFLP patterns. Although it would appear that the nested PCR-RFLP analysis may be biased toward C. andersoni in excess, when the template copy numbers were reversed (C. andersoni near the detection limit and C. parvum or C. hominis in excess), a similar result was observed, indicating a detection bias toward any species in excess of a 1:20 ratio. To further examine the effects of templates in excess, the plasmid experiments were repeated by using combinations of C. parvum or C. hominis with C. ubiquitum.
Once again, plasmid copy numbers near or below the detection limit showed variability in the numbers of detections at each template concentration, and when template numbers of both species in the mixture were at equal template concentrations, both species could be detected equally well (detection values in boldface type in Table 2). There were differences observed for the C. ubiquitum combinations (as opposed to the C. andersoni combinations) when one template was present in excess. At 5 and 10 template copies of C. parvum or C. hominis, detection was never completely eliminated when C. ubiquitum was in excess. When C. parvum or C. hominis was in excess, the number of detections of C. ubiquitum was lost or reduced at 1:5 ratios (10:50 template copies) or greater (5:50, 5:100, 5:1,000, 10:100, 10:1,000, and 50:1,000). No undigested RFLP bands were observed for any combination of C. parvum or C. hominis with C. ubiquitum.
The increases in the detection of C. parvum or C. hominis when mixed with C. ubiquitum as opposed to C. andersoni may be explained by heteroduplex formation, as mentioned above. Heteroduplex formation between PCR amplicons of C. andersoni and C. hominis or C. andersoni and C. parvum would remain undigested, resulting in a general loss of C. parvum or C. hominis detection sensitivity by nested PCR-RFLP analysis. Conversely, the heteroduplex formation of C. parvum or C. hominis with C. ubiquitum results in a greater integrity of the first restriction site (Fig. 3), resulting in the full digestion of amplicon products, therefore increasing the number of detections of C. parvum and C. hominis. The combined results of two experiments using plasmid DNA templates indicate that there is some bias when a template is in excess and that this bias is most likely to begin at approximately 1:10 ratios.
The ability to detect mixed Cryptosporidium species at different concentrations was also assessed by using whole-oocyst extracts. Slides with mixtures of oocysts of Cryptosporidium parvum and C. andersoni were analyzed by repetitive nested PCR-RFLP analysis. Although each oocyst theoretically contains 5 copies of the 18S rRNA gene, when the input DNA was normalized to extraction and PCR template volumes, slides containing 3, 6, 30, and 60 oocysts concomitantly had 3, 6, 30, and 60 template copies per PCR, respectively.
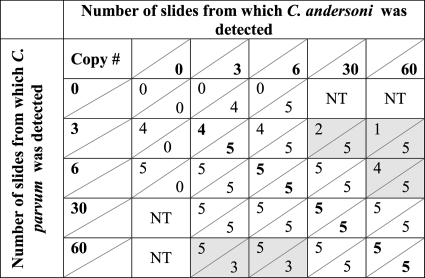
The results of mixed-template slides are shown in Table 3. When slides were seeded with 3 oocysts of C. parvum containing 30 or 60 oocysts of C. andersoni, C. parvum was detected in at least one replicate on 2 slides and 1 slide, respectively. Six C. parvum oocysts were not detected on one of the slides in the presence of 60 C. andersoni oocysts. Likewise, when C. andersoni was seeded at 3 or 6 oocysts and C. parvum was seeded at 60 oocysts, C. andersoni was not detected on 4 of 10 slides. Thus, when the DNA concentration from one species of Cryptosporidium exceeds that from another species by a ratio of 10:1, the ability of the repetitive nested PCR-RFLP assay to detect the less dominant species decreases, resulting in potential false-negative results for a portion of the samples.
Table 3.
Detection of C. parvum in the presence of C. andersoni by repetitive nested PCR-RFLP from 5 slides seeded with whole-oocyst combinationsa
Results in boldface type indicate results where oocysts were mixed at equal concentrations. Shading indicates plasmid template concentrations where a significant reduction or total loss of detection by repetitive nested PCR-RFLP occurred. The value below a diagonal line represents the number of nested PCR-RFLP mixtures positive for those species listed on the horizontal row; the value above the diagonal line is the number of nested PCR-RFLP mixtures positive for the species listed on the vertical column. NT, these ratios were not tested.
Resolution of mixtures of Cryptosporidium species.
The basic testing platform (5 replicates of a 5-μl template) was applied to the mixed-species slides to determine the likelihood of obtaining nested PCRs where a single species had been isolated (single-species RFLP analysis). For the 20 slides where 12 oocysts or fewer were seeded onto the slides (3 C. andersoni and 3 C. parvum, 3 C. andersoni and 6 C. parvum, 6 C. andersoni and 3 C. parvum, and 6 C. andersoni and 6 C. parvum oocysts), each species was isolated in at least one replicate from seven slides. A single species was isolated from 12 slides (single-species RFLP pattern for only one of the species in at least 1 of the 5 replicates), while all 5 replicates were mixed from 1 slide. When one species was present at 30 or 60 oocysts and the other species was present at 3 or 6 oocysts, only the species present at the high concentration was isolated in at least 1 reaction. When both species were present at a high concentration (30 or 60 oocysts), neither species was isolated in any of the replicate PCR reactions.
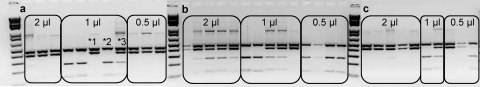
Based on results for the detection of mixtures described above, whole-oocyst DNA extracts under the experimental conditions where 6 oocysts of one species were mixed with various numbers of oocysts from the other species were selected for analysis. Template copy numbers for C. parvum and C. andersoni were 6 and 6, 6 and 30, and 30 and 6, respectively. Amplified PCR products displayed RFLP patterns indicating mixtures in all replicates tested. In the initial experiment, 5 μl of template was used per nested PCR; however, to resolve single species into individual tubes, replicate limiting template dilution was carried out (5 replicates of 2.0, 1.0, and 0.5 μl), reducing the initial template per reaction by 2.5, 5, and 10 times. This limiting template dilution scheme was selected based on knowing the LoD in template copy numbers and varies by one 2-fold dilution from that used previously by Xiao et al. (3 replicates of 1, 0.5, and 0.25 μl) (41). The limiting-template approach resulted in the resolution of species from each other where there was at least one reaction for each species which was not mixed, as indicated by the RFLP patterns (Fig. 4). The limiting-template approach was successful even when one species was present at 5 times the concentration of the other. Sequence analysis was attempted on all the PCR products shown in Fig. 4. High-quality bidirectional sequences were obtained for all samples that showed a single-species RFLP pattern. Sequence data could be grouped into 4 categories: (i) excellent quality, (ii) underlying sequence resulting in a sequence with reduced but acceptable quality, (iii) random mixed bases resulting in ambiguous base calls (i.e., R, Y, or M), or (iv) no full-length sequence. Often, a reasonable-quality sequence could be obtained from samples showing multiple-species RFLP patterns if the banding pattern of one species was significantly more intense than that of the other. Excellent- or acceptable-quality sequences had 100% DNA sequence identity to original isolates of C. parvum or C. andersoni used in this study. Those samples with ambiguous mixed bases or partial sequences of poor quality were relegated to the “no-usable-sequence” category. The distribution of sequences obtained from the direct sequencing of the nested PCR products is shown in Table 4. All C. parvum or C. andersoni sequences matched known reference sequences for C. parvum and C. andersoni (GenBank accession numbers AF164102 and AB089285, respectively), with no random single-nucleotide polymorphisms (SNPs) detected in any of these sequences. Thus, when limiting-template nested PCR-RFLP was used and DNA sequence analysis was performed on single-species nested PCR products, 100% identity was obtained by the DNA sequence analysis for both species present in the original sample.
Fig. 4.
Limiting-template nested PCR-RFLP results from slides where the initial 5-μl nested PCR products contained mixtures of the two species and template copy numbers were as follows: 6 C. parvum and 6 C. andersoni copies (a), 30 C. parvum and 6 C. andersoni copies (b), and 6 C. parvum and 30 C. andersoni copies (c). Although 15 reactions were carried out by limiting dilution on each sample, only the RFLP results for reactions that were positive by nested PCR are shown. *1 indicates an RFLP pattern for C. andersoni, *2 indicates an RFLP pattern for C. parvum, and *3 indicates an RFLP pattern where a reaction mixture contained both species.
Table 4.
Distribution of species obtained from direct sequencing of limiting-template nested PCRs of whole-oocyst DNA extracts
| No. of copies | No. of sequences detecteda |
||
|---|---|---|---|
| C. parvum | C. andersoni | No sequence | |
| 6 of C. parvum and 6 of C. andersoni | 3 | 14 | 2 |
| 6 of C. parvum and 30 of C. andersoni | 2 | 9 | 4 |
| 30 of C. parvum and 6 of C. andersoni | 13 | 5 | 6 |
Number of sequences detected from individual species by sequencing the cloned nested-PCR products from mixed, whole-oocyte DNA extracts.
Three nested PCR-RFLP products in which more than a single species were detected were selected for cloning. To be comparable to limiting dilution, template copy numbers for C. parvum and C. andersoni were also 6 and 6, 6 and 30, and 30 and 6, respectively. Plasmids from 20 clones from each of the template combinations were purified and sequenced bidirectionally. Sequence analysis of the cloned nested PCR products resulted in an excellent-quality sequence (with the exception of 1 plasmid, which did not provide a usable sequence); however, sequence analysis revealed that only 4 of the 59 clone sequences matched the original reference sequences determined by direct sequencing of PCR products (GenBank accession numbers AF164102 and AB089285 for C. parvum and C. andersoni, respectively). A high number of SNPs in conserved regions of the gene was observed for the sequences of the cloned material (approximately 1 SNP in every 600 bases). A total of 21 clones were identified as C. parvum, and 25 clones were identified as C. andersoni. The distribution of clones obtained from the nested PCRs are shown in Table 5. Not all the cloned products were identical or variations of either C. parvum or C. andersoni (Fig. 5), suggesting that the cloning of mixed PCR products may result in some form of genetic recombination. Sequence data demonstrated that 10 of the 59 sequences (17%) were recombinants of C. parvum and C. andersoni (Fig. 5). The distribution of recombinant clones among the 3 oocyst combinations is shown in Table 5. A higher proportion of recombinant clones was obtained when nested PCR products were cloned from mixtures containing 30 C. parvum and 6 C. andersoni template copies. Single-nucleotide polymorphisms were also obtained in the recombinant clones at a rate of approximately 1 in every 700 bp, occurring randomly and mostly in conserved areas of the gene (Fig. 5).
Table 5.
Distribution of species obtained from cloned nested PCRs of whole-oocyst DNA extracts
| No. of copies | No. of clones detecteda |
||
|---|---|---|---|
| C. parvum | C. andersoni | Recombinant | |
| 6 of C. parvum and 6 of C. andersoni | 10 | 8 | 2 |
| 6 of C. parvum and 30 of C. andersoni | 3 | 15 | 1 |
| 30 of C. parvum and 6 of C. andersoni | 9 | 4 | 7 |
Number of sequences detected from individual species by sequencing the cloned nested-PCR products from mixed, whole-oocyte DNA extracts.
Fig. 5.
Recombinant sequences resulting from cloning of PCR products containing mixtures of C. parvum (GenBank accession number AF164102) and C. andersoni (GenBank accession number AB089285). Shaded areas indicate regions homologous to C. andersoni, while unshaded areas indicate homology to C. parvum. Circled bases indicate sites of random single-nucleotide polymorphisms. (a) Nucleotide positions 324 to 523 of C. parvum (accession number AF164102). (b) Nucleotide positions 624 to 860 of C. parvum (accession number AF164102). Positions 524 to 623 show no sequence variation between C. parvum and C. andersoni.
DISCUSSION
The identification of more than one Cryptosporidium species or genotype in a single water sample has been reported in the literature (6, 13, 14, 25, 26, 30, 47). Previous studies of the Potomac River watershed and the South Nation watershed reported that 24% and 27% of the samples tested, respectively, had more than a single Cryptosporidium species and/or genotype identified. A study from Scotland reported that 16.9% of raw and finished drinking waters tested over a 1-year monitoring period contained multiple species or genotypes of Cryptosporidium (26). Several studies have demonstrated that C. andersoni is commonly detected in source waters (25, 26, 30, 47), raising questions about the sensitivity of and possible biases associated with PCR-based molecular methods used to detect and differentiate Cryptosporidium species and genotypes in environmental samples. The Scottish study attempted to address the potential for resolution and possible bias using the data from environmental monitoring (26), and several studies have been conducted to compare the sensitivities and specificities of primer sets and gene targets (5, 21, 28, 46). However, this is the first study to statistically compare the sensitivities of a genus-specific nested PCR-RFLP assay for a range of species under controlled conditions and to examine the performance of these molecular-based assays for samples containing template copies from more than one species of Cryptosporidium.
Ample quantities of DNA from multiple species and genotypes of Cryptosporidium are not readily available for research purposes. To overcome these issues, the full-length 18S rRNA gene was cloned from DNA extracts from multiple Cryptosporidium species and used to statistically determine the LoD against a broad range of parasites. Many of the Cryptosporidium species used in this study were of public health significance (C. parvum, C. hominis, C. meleagridis, and C. felis), have been found in humans (C. muris and C. ubiquitum), or are commonly identified in the environment (C. andersoni and C. ubiquitum) (26, 30, 44, 47). These species also reflect a broad range of genetic diversity based on phylogenetic analyses of the 18S rRNA gene (6, 10, 30).
Based on the detection of 18S rRNA plasmid constructs, the LoDs for all Cryptosporidium species tested were not significantly different (overlapping 95% CLs). Similarly, LoD determinations carried out with DNA extracts from genomic DNAs of C. parvum and C. andersoni resulted in overlapping confidence limits, indicating that the LoDs were not statistically different between the two species tested. Furthermore, when LoDs for cloned plasmid DNA and genomic DNA extracts were compared, the results were not significantly different, indicating comparable extraction and amplification efficiencies of the template during the DNA isolation and PCR processes, respectively. For both cloned plasmid constructs and genomic DNA, detection rates were variable when template levels for both species were near or below the LoD. A random distribution of templates in solution at low concentrations, fluctuations in the interaction of templates at low concentrations with PCR reagents in early cycles, and the reproducibility of pipetting small volumes contribute to the variability observed for the data. These parameters are known to be associated with molecular sampling errors, stochastic fluctuations, and PCR drift (24, 27, 39). Based on the range of LoDs and overlapping confidence limits of the species tested, it was concluded the genus-specific nested PCR described previously by Xiao et al. (45) is capable of detecting all Cryptosporidium species at the same level of sensitivity.
When the initial template concentration of one species was in excess of another (i.e., a 1:20 ratio), the ability to detect DNA from the less prevalent species was reduced or completely lost, suggesting a certain level of bias when DNAs from different species are present in a sample. The potential for biased results in multitemplate PCRs of this nature have been well documented in the scientific literature. PCR-associated biases of bacterial communities when 16S rRNA gene strategies are used have been extensively reviewed (7, 17, 38). Bias with PCR can occur as a result of degenerate primers (22, 27, 36), stringency (31), GC content (39), and amplicon length (39). Based on the observation that similar LoDs existed among the diverse Cryptosporidium species and genotypes with conserved primers, the uniform GC content of the 18S rRNA gene target region across diverse Cryptosporidium species and genotypes, and the relatively consistent amplicon length, these known causes of PCR bias do not provide rational explanations for the observed loss of detection when one Cryptosporidium species was present at a low template concentration compared to that of another species (1:20 template ratios). However, a possible explanation for these observations is heteroduplex formation in PCR products, resulting from a high level of similarity of DNA contents in multitemplate PCRs. Kanagawa previously outlined the three kinds of duplexes that will be formed during the annealing phase of a PCR: (i) a duplex between primers and templates, (ii) a homoduplex between complementary strands, and (iii) a heteroduplex caused by the cross-hybridizations of heterologous sequences (17). Heteroduplex formation has been shown to increase during later cycles of multitemplate PCRs when the amplified products reach concentrations high enough to compete with primers for binding sites. Research has demonstrated that when 16S rRNA gene fragments showing 99.3% sequence homology are mixed at 1:10 ratios, nearly all of the DNA from the less abundant species forms heteroduplexes during PCR, due to the fact that the DNA from the less abundant species had only a 10% chance to reanneal with its own species (40). This appears to explain the loss of detection of one species when mixed with another at 1:20 ratios or higher. Thus, it is not that the species at the higher ratio is preferentially amplified; rather, it is plausible that the product from the less abundant species exists predominantly in the heteroduplex form and that the homoduplex ratio is not high enough for RFLP detection using visualization by ethidium bromide staining.
Judo et al. showed previously that heteroduplex products may contain a restriction site on only one strand, preventing the cleavage of the duplex (16). We observed a similar phenomenon when C. parvum or C. hominis was used in multitemplate reactions with C. andersoni. The same undigested products (i.e., heteroduplex) were not observed when C. parvum or C. hominis was used in multitemplate reactions with C. ubiquitum. This can be attributed to the specificity of the SspI restriction enzyme. Although it is assumed that these enzymes have a high degree of sequence recognition, the specificity is not absolute. It was found previously that many single-nucleotide-mismatch substrates remain intact for double-strand DNA cleavage with many commonly used restriction enzymes (19). An examination of the SspI restriction sites of C. parvum and C. hominis compared to those of C. andersoni and C. ubiquitum shows a 4-bp deletion in C. andersoni immediately prior to the first recognition site. It is probable that heteroduplex formation between C. hominis or C. parvum and C. andersoni would result in a misalignment at this key recognition site (the only SspI site in C. andersoni), therefore preventing cleavage. Along with the mismatches found in the C. andersoni sequence occurring at the other restriction sites of C. parvum and C. hominis, it is expected that any heteroduplex formation between these species will remain undigested by the SspI restriction enzyme.
A duplex formed with C. ubiquitum retains the first key restriction site, which is why no undigested secondary PCR products were observed when C. ubiquitum was mixed with C. parvum and C. hominis. The second restriction site is also conserved among C. parvum, C. hominis, and C. ubiquitum. The lack of specificity for a 1-bp mismatch in the third and fourth cleavage sites may also explain why there was a slightly better sensitivity for C. hominis or C. parvum mixed with C. ubiquitum at a 1:10, 1:20, or 1:100 ratio. If the cleavage did not occur at the third and fourth restriction sites in the heteroduplex with C. parvum or C. hominis, the result would be an increased detection of C. ubiquitum. As the detection of C. ubiquitum is decreased, it is expected that all heteroduplex strands were cleaved, resulting in the same RFLP pattern as those of C. parvum and C. hominis, thereby increasing their detection.
Interestingly, heteroduplex formation during PCR amplification is known to result in erroneous cloning results, leading to artificially high sequence diversity (12, 33, 36). When heteroduplexes are cloned, the E. coli host-nick-directed mismatch repair system can convert a heteroduplex into a single hybrid sequence by excision repair (23). Artificial diversity was previously reported when mixtures of Cryptosporidium species were cloned, but the cause was attributed to PCR-mediated recombination rather than heteroduplex formation (48). PCR-mediated recombination was reported previously by other multitemplate studies to be a result of incomplete primer extension (16, 17, 32, 40).
When cloning was used in an attempt to further resolve mixtures of Cryptosporidium species in this study, increased genetic diversity was found. This was not only in the form of recombinant sequences possibly caused by both heteroduplex formation and chimeric PCR products but also from a high rate of SNPs. These SNPs could be attributed to either an increased detection of heterogeneous copies of the 18S rRNA gene (20, 43) or the misincorporation of bases during PCR due to errors caused by Taq polymerase (9). When the sequence data from the direct sequencing of PCR products are compared to those from cloned PCR products, it would appear that the frequencies of polymorphisms (caused by either sequence heterogeneity or Taq polymerase errors) are too low to be detected by direct sequencing, as any low-frequency sequence variation is obscured by the sequence present in great abundance in the sample. However, cloning has the ability to detect variations in a single rRNA gene molecule and therefore is more likely to detect low-level polymorphisms. The rate of detection and number of polymorphisms would be dependent upon how many clones are sequenced from each nested PCR product.
Cloning approaches have been used on environmental samples, which may contain multiple species of Cryptosporidium (13, 14). Although cloning may be more sensitive for detecting a greater number of polymorphisms, it is more likely to contribute to an artificially large amount of sequence diversity. Ashelford et al. previously reported high numbers of anomalies (1 in 20 records) in 16S rRNA gene clone libraries from public databases (1). It was determined that over 90% of the anomalies found in clone libraries could be attributed to recombinant or chimeric patterns (2). Therefore, cloning results are reliable only for those species and genotypes that are already well described.
The rate of occurrence of parasites in surface water is typically low, with approximately 40% (our unpublished data) of samples tested in our laboratory being positive for Cryptosporidium oocysts by U.S. EPA Method 1623. Of the samples that tested positive for the parasite, 83% of the slides contained fewer than 10 oocysts (our unpublished data), which is similar to the 88% previously reported (26). The basic testing platform of replicate nested PCR-RFLP analysis with 5 μl of template in the nested PCR mixture has been successful for detecting and resolving species and genotypes for many water samples (29, 30). Nichols et al. (26) previously examined the use of replicate nested PCR analysis of environmental samples and found that (i) the predominant species were more likely to be detected, (ii) replicate nested PCR detected all the species and genotypes present in the majority of samples, and (iii) detection is not ensured at low concentrations of DNA. Similar trends were observed in this study, including data which confirmed that there was no preferential bias (tendency toward the amplification of one species over another when template concentrations are similar) in the nested PCR. Rather, the data demonstrated the variability in detection due to the stochastic sampling of extracts near or below the LoD for the template concentration.
Based on the replicate nested PCR-RFLP results, there was a need for a further resolution of species for some samples. PCR amplification of limiting-dilution templates, followed by direct sequencing, proved to be an effective method for accurately determining species diversity and resolving mixed-template DNA extracts. This approach was successful when the species were mixed at up to 1:20 ratios of template. This finding has changed our basic testing platform to include an additional nested PCR-RFLP assay using 5 replicates each of 1 and 0.5 μl of template in an attempt to resolve samples suspected to contain mixtures based on the RFLP results of the initial nested PCR-RFLP assay.
Our data suggest that the use of cloning to identify and characterize Cryptosporidium species and genotypes in water may overestimate the true genetic diversity of these parasites in the sample and lead to erroneous conclusions about sources of contamination. Cloned Cryptosporidium DNA sequences from environmental samples that do not provide a 100% match to reference GenBank sequences may represent artifact chimeric or heteroduplexes from multitemplate PCRs. For this reason, we recommend that direct sequencing of PCR products be used to confirm the identity of Cryptosporidium species in water samples.
ACKNOWLEDGMENTS
Financial support for this study was provided by the Natural Sciences and Engineering Research Council (NSERC), Alberta Water Research Institute, and Alberta Provincial Laboratory for Public Health (ProvLab).
We give many thanks to Ricci Schafer (ProvLab) for her expertise in cloning the 18S rRNA plasmids, Tim McAllister (Agriculture and Agri-Food Canada) for the C. andersoni-positive cattle feces, Mike Belosevic (University of Alberta) for access to the ultracentrifuge, Kristen Elwin (UK Cryptosporidium Reference Unit) for providing the Cryptosporidium DNA extracts, and Marty Collins (Wisconsin State Laboratory of Hygiene) for the preparation of microscope slides by flow cytometry. We greatly appreciate these contributions.
Footnotes
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Ashelford K. E., Chuzhanova N. A., Fry J. C., Jones A. J., Weightman A. J. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ashelford K. E., Chuzhanova N. A., Fry J. C., Jones A. J., Weightman A. J. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belosevic M. and the AWWA Research Foundation 1997. Vital dye staining of Giardia and Cryptosporidium. AWWA Research Foundation, Denver, CO.
- 4. Chalmers R. M., Elwin K., Thomas A. L., Guy E. C., Mason B. 2009. Long-term Cryptosporidium typing reveals the aetiology and species-specific epidemiology of human cryptosporidiosis in England and Wales, 2000 to 2003. Euro Surveill. 14 (2):pii=19086http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19086 [DOI] [PubMed] [Google Scholar]
- 5. Chalmers R. M., et al. 2005. Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. Int. J. Parasitol. 35:397–410 [DOI] [PubMed] [Google Scholar]
- 6. Chalmers R. M., et al. 2010. Detection of Cryptosporidium species and sources of contamination with Cryptosporidium hominis during a waterborne outbreak in north west Wales. J. Water Health 8:311–325 [DOI] [PubMed] [Google Scholar]
- 7. Chandler D. P., Fredrickson J. K., Brockman F. J. 1997. Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol. Ecol. 6:475–482 [DOI] [PubMed] [Google Scholar]
- 8.Drinking Water Inspectorate. 2000. The water supply (water quality) regulations 2000. Drinking Water Inspectorate, London, United Kingdom. http://dwi.defra.gov.uk/stakeholders/legislation/ws_wqregs2000.pdf.
- 9. Eckert K. A., Kunkel T. A. 1991. DNA polymerase fidelity and the polymerase chain reaction. PCR Methods Appl. 1:17–24 [DOI] [PubMed] [Google Scholar]
- 10. Feng Y., et al. 2007. Cryptosporidium genotypes in wildlife from a New York watershed. Appl. Environ. Microbiol. 73:6475–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferguson C., et al. 2006. Application of genotyping methods to assess risks from Cryptosporidium in watersheds. Environ. Health Perspect. 114:430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jansen R., Ledley F. D. 1990. Disruption of phase during PCR amplification and cloning of heterozygous target sequences. Nucleic Acids Res. 18:5153–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jellison K. L., Hemond H. F., Schauer D. B. 2002. Sources and species of Cryptosporidium oocysts in the Wachusett Reservoir watershed. Appl. Environ. Microbiol. 68:569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jellison K. L., Lynch A. E., Ziemann J. M. 2009. Source tracking identifies deer and geese as vectors of human-infectious Cryptosporidium genotypes in an urban/suburban watershed. Environ. Sci. Technol. 43:4267–4272 [DOI] [PubMed] [Google Scholar]
- 15. Jiang J., Alderisio K. A., Xiao L. 2005. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 71:4446–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Judo M. S., Wedel A. B., Wilson C. 1998. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res. 26:1819–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kanagawa T. 2003. Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 96:317–323 [DOI] [PubMed] [Google Scholar]
- 18. Kim K., Gooze L., Petersen C., Gut J., Nelson R. G. 1992. Isolation, sequence and molecular karyotype analysis of the actin gene of Cryptosporidium parvum. Mol. Biochem. Parasitol. 50:105–113 [DOI] [PubMed] [Google Scholar]
- 19. Langhans M. T., Palladino M. J. 2009. Cleavage of mispaired heteroduplex DNA substrates by numerous restriction enzymes. Curr. Issues Mol. Biol. 11:1–12 [PMC free article] [PubMed] [Google Scholar]
- 20. Le Blancq S. M., Khramtsov N. V., Zamani F., Upton S. J., Wu T. W. 1997. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol. Biochem. Parasitol. 90:463–478 [DOI] [PubMed] [Google Scholar]
- 21. Leetz A. S., Sotiriadou I., Ongerth J., Karanis P. 2007. An evaluation of primers amplifying DNA targets for the detection of Cryptosporidium spp. using C. parvum HNJ-1 Japanese isolate in water samples. Parasitol. Res. 101:951–962 [DOI] [PubMed] [Google Scholar]
- 22. Lueders T., Friedrich M. W. 2003. Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Modrich P. 1987. DNA mismatch correction. Annu. Rev. Biochem. 56:435–466 [DOI] [PubMed] [Google Scholar]
- 24. Mutter G. L., Boynton K. A. 1995. PCR bias in amplification of androgen receptor alleles, a trinucleotide repeat marker used in clonality studies. Nucleic Acids Res. 23:1411–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nichols R. A., Campbell B. M., Smith H. V. 2006. Molecular fingerprinting of Cryptosporidium oocysts isolated during water monitoring. Appl. Environ. Microbiol. 72:5428–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nichols R. A., Connelly L., Sullivan C. B., Smith H. V. 2010. Identification of Cryptosporidium species and genotypes in Scottish raw and drinking waters during a one-year monitoring period. Appl. Environ. Microbiol. 76:5977–5986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Polz M. F., Cavanaugh C. M. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rochelle P. A., De Leon R., Stewart M. H., Wolfe R. L. 1997. Comparison of primers and optimization of PCR conditions for detection of Cryptosporidium parvum and Giardia lamblia in water. Appl. Environ. Microbiol. 63:106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ruecker N. J., et al. 2005. Molecular forensic profiling of Cryptosporidium species and genotypes in raw water. Appl. Environ. Microbiol. 71:8991–8994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ruecker N. J., et al. 2007. Tracking host sources of Cryptosporidium spp. in raw water for improved health risk assessment. Appl. Environ. Microbiol. 73:3945–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarkar G., Cassady J., Bottema C. D., Sommer S. S. 1990. Characterization of polymerase chain reaction amplification of specific alleles. Anal. Biochem. 186:64–68 [DOI] [PubMed] [Google Scholar]
- 32. Shuldiner A. R., Nirula A., Roth J. 1989. Hybrid DNA artifact from PCR of closely related target sequences. Nucleic Acids Res. 17:4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Speksnijder A. G., et al. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suzuki M. T., Giovannoni S. J. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thompson J. R., Marcelino L. A., Polz M. F. 2002. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 30:2083–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.U.S. Environmental Protection Agency. 2005. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. 815-R-05-002. Office of Water, EPA, Washington, DC.
- 38. von Wintzingerode F., Gobel U. B., Stackebrandt E. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213–229 [DOI] [PubMed] [Google Scholar]
- 39. Walsh P. S., Erlich H. A., Higuchi R. 1992. Preferential PCR amplification of alleles: mechanisms and solutions. PCR Methods Appl. 1:241–250 [DOI] [PubMed] [Google Scholar]
- 40. Wang G. C., Wang Y. 1996. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 142 (Pt. 5):1107–1114 [DOI] [PubMed] [Google Scholar]
- 41. Xiao L., Alderisio K., Limor J., Royer M., Lal A. A. 2000. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 66:5492–5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xiao L., Alderisio K. A., Jiang J. 2006. Detection of Cryptosporidium oocysts in water: effect of the number of samples and analytic replicates on test results. Appl. Environ. Microbiol. 72:5942–5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiao L., et al. 1999. Presence of heterogeneous copies of the small subunit rRNA gene in Cryptosporidium parvum human and marsupial genotypes and Cryptosporidium felis. J. Eukaryot. Microbiol. 46:44S–45S [PubMed] [Google Scholar]
- 44. Xiao L., Ryan U. M. 2008. Molecular epidemiology, p. 119–163 In Fayer R., Xiao L. (ed.), Cryptosporidium and cryptosporidiosis CRC Press, Boca Raton, FL [Google Scholar]
- 45. Xiao L., et al. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiao L., et al. 2006. Development and standardization of a Cryptosporidium genotyping tool for water samples. IWA Publishing, Denver, CO [Google Scholar]
- 47. Yang W., et al. 2008. Cryptosporidium source tracking in the Potomac River watershed. Appl. Environ. Microbiol. 74:6495–6504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou L., Yang C., Xiao L. 2003. PCR-mediated recombination between Cryptosporidium spp. of lizards and snakes. J. Eukaryot. Microbiol. 50(Suppl.):563–565 [DOI] [PubMed] [Google Scholar]