Abstract
Coupling prokaryote identification with ultrastructural investigation of bacterial communities has proven difficult in environmental samples. Prokaryotes can be identified by using specific probes and fluorescence in situ hybridization (FISH), but resolution achieved by light microscopes does not allow ultrastructural investigation. In the case of symbioses involving bacteria associated with metazoan tissues, FISH-based studies often indicate the co-occurrence of several bacterial types within a single host species. The ultrastructure is then relevant to address host and bacterial morphology and the intra- or extracellular localization of symbionts. A simple protocol for correlative light and electron microscopy (CLEM) is presented here which allows FISH-based identification of specific 16S rRNA phylotypes and transmission electron microscopy to be performed on a same sample. Image analysis tools are provided to superimpose images obtained and generate overlays. This procedure has been applied to two symbiont-bearing metazoans, namely, aphids and deep-sea mussels. The FISH protocol was modified to take into account constraints associated with the use of electron microscopy grids, and intense and specific signals were obtained. FISH signals were successfully overlaid with bacterial morphotypes in aphids. We thus used the method to address the question of symbiont morphology and localization in a deep-sea mussel. Signals from a type I methanotroph-related phylotype were associated with morphotypes displaying the stacked internal membranes typical for this group and three-dimensional electron tomography was performed, confirming for the first time the correspondence between morphology and phylotype. CLEM is thus feasible and reliable and could emerge as a potent tool for the study of prokaryotic communities.
INTRODUCTION
In the last few decades, a broad range of symbiotic associations between bacteria and invertebrates have been identified (34). In addition to symbioses involving phloem-sucking insects and endosymbiotic Gammaproteobacteria (5, 9), many other symbioses were discovered between diverse groups of metazoans (e.g., families within Bivalvia, Gastropoda, Polychaeta, or Decapoda) and chemosynthetic bacteria (10, 16). First thought to involve only a single or two distinct bacterial types (11, 12, 14), more recent studies have described more complex symbiotic communities, including a larger number of bacterial taxa (6, 19, 20, 21, 40, 50). Diverse metabolisms can co-occur, and certain bacterial types are even proposed to act as consortia (17, 20, 31, 48).
The characterization of 16S rRNA phylotypes in host tissues and their localization via fluorescence in situ hybridization (FISH) experiments, using suitable corresponding probes (2), gives an indication of the potential role of the association. However, the low resolution of the classical light microscopes is rarely sufficient to ascertain precise cellular localization or morphology, whereas transmission electron microscopy (TEM), with its great higher resolution, is the suitable microscopy to address the issue of extra- or intracellular localization and ultrastructural morphology. The morphology can be determined by classical TEM observations or by tomographic investigation, allowing to access the three-dimensional (3D) structural information (4, 27, 29). The ultrastructural investigation of bacterial symbioses can also provide valuable information on the organization of the hosting-specialized cells (usually named bacteriocytes) and eventually of the hosting-specialized organ, yielding clues about the cellular processes underlying symbiosis (8, 22, 35). Furthermore, the morphology of bacteria can support metabolism information inferred by phylogenetic analyses of 16S rRNA phylotypes, such as type I methanotrophic bacteria displaying typical internal stacked membranes that play a role in methane metabolism (11, 24).
Finally, the observation of physical interactions between different types of bacteria in the host tissue, not visible in fluorescence microscopy, could support the existence of potential consortia between symbionts. However, discriminating between several bacterial types using TEM alone remains a challenging task, because of the difficulty of multiple labeling in TEM and the lack of distinctive morphological features in most groups; this problem is more tangible since the number of symbiont phylotypes in the host tissue section is greater. Symbiosis studies would thus greatly benefit from techniques allowing the combination of light microscopy, in which symbionts can be reliably identified, and electron microscopy in which structures appear in high resolution.
Correlative light and electron microscopy (CLEM) involves observing the same biological sample under both light and electron microscopy. Many protocols of CLEM were developed to investigate protein localization and organization and intercellular trafficking processes (32, 36, 39, 41, 45, 46). In the field of microbiology, CLEM was used recently to visualize matrix-embedded bacteria in a multilayered endodontic biofilm under a scanning electron microscope (42). Another approach was proposed by Wrede et al. in which the authors prepared their sections by alternating between semithin and ultrathin sections to be used in polysaccharide fluorescent labeling and electron microscopy studies, respectively (47). However, due to the relatively small sizes of the structures studied, the morphology can change from one section to another, and the problem of registration between images remains.
We present here a simple protocol, from sample preparation and labeling up to image analysis, in order to study 2D and 3D ultrastructural information for symbionts detected by FISH on a single section. This protocol is based on several routine techniques used in microbiology and microscopy. We also developed two in silico procedures to superimpose FISH and TEM information, which were validated by using fluorescent bead preparations. We tested the sample preparation on two organisms documented to harbor bacterial symbionts in their tissues, namely, aphids (Aphidoidea and Macrosiphon rosae) and deep-sea mussels (Idas sp. strain Med). The latter harbors a single bacterial symbiont related to type I methanotrophic Gammaproteobacteria (19), among six distinct bacterial 16S rRNA phylotypes associated with its gill epithelial cells (19). An Idas sp. was thus used to test FISH specificity and whether detection of methanotroph-specific FISH signal would superimpose with bacterial morphotypes displaying stacked internal membranes. Indeed, this morphology is described as typical of type I methanotrophs and is thus considered evidence for the presence of methanotrophic symbionts in mussels (11), but its actual correspondence with identified methanotroph-related 16S rRNA phylotypes has never been proven. This question is even more critical since a one study (19) suggested that these symbiotic bacteria were extracellular in Idas spp., whereas all other described associations involving methanotrophs and marine invertebrates were intracellular symbioses.
MATERIALS AND METHODS
Biological sample collection.
Aphids were chosen as test organisms because of their wide availability and their well-documented symbiosis occurring in their easily recognizable paired specialized organs called bacteriomes (15, 33). The specimens were collected from a Dog Rose (Rosa canina) shrub tree at the Université Pierre et Marie Curie. Aphids were dissected, and the posterior parts of the abdomen were fixed using 1% glutaraldehyde and 2% formaldehyde in sodium cacodylate 0.1 M solution. For mussel samples, four Idas specimens were collected with the ROV Quest during the BIONIL cruise to the Eastern Mediterranean (2006; chief scientist, A. Boetius). The samples were collected from the outer surface of the chitinous tube of the siboglinid Lamellibrachia sp. from the Amon Mud Volcano (N32°22 E31°42, 1,157-m depth). Upon recovery, mussels were dissected, and gills were fixed using 1% glutaraldehyde–2% formaldehyde in filtered seawater (0.22-μm pore size) over 3 h. For both sample types, fixation was followed by progressive dehydration in increasing ethanol series, and tissues were stored in 100% ethanol.
Sample preparation.
Fixed aphid tissues and mussel gills were directly transferred into pure LR-White Medium grade resin (London Resin Company, Reading, England) for eight successive baths of 30 min each. The samples were then placed into gelatin capsules with renewed resin and incubated at 50°C for 48 h for polymerization (as described in reference 35). Semithin sections of 200 nm for aphids and 350 nm for gills were obtained by using a Leica Ultracut R ultramicrotome (Leica Microsytems, Vienna, Austria) and a Histo diamond knife (Diatome AG, Biele, Switzerland). Sections were laid on 200-mesh-coated carbon nickel grids (EMS, Hatfield, PA). In order to validate the registration procedures between fluorescence and TEM, we also prepared a solution of fluorescent beads at 1/500 (TetraSpek 0.2-μm beads; Invitrogen, Carlsbad, CA) deposited on a 200-mesh copper grids (EMS).
FISH.
Loaded grids were deposited onto 30-μl hybridization solution drops in glass slides. Aphid sections were hybridized for 3 h at 46°C with 100 ng of the general bacterial probe Eub338 (5′-GCTGCCTCCCGTAGGAGT-3′) (3) labeled with Cy3 in a specific buffer (2× phosphate-buffered saline [PBS], 0.01% sodium dodecyl sulfate [SDS], and 30% formamide). Mussel gill sections were hybridized the same way using the Imed-M probe (5′-ACCATGTTGTCCCCCACTAA-3′) (19). The Idas methanotrophic symbiont-specific 16S rRNA probe (Imed-M) was labeled with the Cy3 fluorochrome. After hybridization, grids were washed using a solution containing 2× PBS and 0.001% SDS for 15 min at 48°C and then rinsed in a water drop and air dried. The grids were then mounted in SlowFade medium (Invitrogen) with a coverslip.
Fluorescence microscopy observations.
Hybridized sections were observed by using a BX61 microscope (Olympus Optical Co., Tokyo, Japan). For the aphid sections, images of nucleic acid-specific image (corresponding to the DAPI [4′,6′-diamidino-2-phenylindole] wavelength emission) and eubacterium-specific signal (corresponding to the Cy3 wavelength emission) were acquired by using a ×10 objective lens in order to reconstruct a single mosaic image of the section. Transmitted light and fluorescence images of the square of interest (i.e., where the labeled bacteria were detected) were obtained by using a ×60 objective lens. Mosaic images of sections and squares were reconstructed by using MosaicJ (43), a plugin for the free image analysis software ImageJ (1, 38).
TEM preparation.
After fluorescence microscope observations, glass slides were briefly plunged in liquid nitrogen, and the coverslips were removed. Grids were rinsed using a drop of ultrapure water and dried at room temperature. After a new rinsing and drying step, the sections were contrasted with uranyl-acetate and lead citrate according to classical methods. For tomographic acquisitions Idas gill sections were nonspecifically immunolabeled using a generic anti-protein A antibody coupled to 10-nm-diameter gold particles (10 min for each side with a 1/40-diluted solution in physiological serum [EMS]). Antibodies were fixed using a fixation step of 5 min in 1% glutaraldehyde solution. After a new rinsing and drying step, the sections were contrasted with uranyl-acetate and lead citrate according to classical methods.
TEM acquisitions and tomographic reconstruction.
Grid squares of interest were observed at nominal magnification (×5,000) with a Zeiss omega TEM microscope at 80 kV for aphid sections and fluorescent bead deposits. Images were acquired with a SlowScan 1,024 by 1,024 charge-coupled device (CCD) camera (Gatan, Pleasanton, CA). For the coordinate-based procedure 1, real coordinates of feature points (preferentially the corners of squares of interest) were noted. For procedure 2, whole-grid squares were acquired in mosaic mode. The mosaic TEM image was reconstructed using the 2D fast stitching ImageJ plugin (37).
TEM observations of Idas gill sections were made using a JEOL 200-kV microscope. Grids were transferred at room temperature into the special tomographic holder (JEOL). Images were acquired with a 2,048 by 2,048 UltraScan CCD camera (Gatan) at nominal magnifications of ×6,000 for acquisitions used for mosaic reconstruction and ×30,000 for tilt-series acquisitions of bacteria. Tilt series consisted of images acquired between −60° and +60° with a 2° increment step, using the acquisition software digital micrograph (Gatan). Tilt-series image alignment, determination of tilt axis, and weighted back projection algorithm volume reconstructions were performed using IMOD software (28, 30).
Image analysis for correlative microscopy.
Two procedures were developed for superimposition of fluorescence and TEM images. They were implemented as plugins or macro scripts in ImageJ. The programs are available at http://www.snv.jussieu.fr/∼wboudier/softs/correlativej.html. The first procedure (procedure 1) consisted of transferring detected positions from fluorescence to TEM image by using feature points to act as system coordinates. The procedure works as follows: (i) the fluorescence image is opened and scaled for subpixel resolution (×2 up to ×10); (ii) the user clicks to define the center Oo and the two axes Ox and Oy on the image; (iii) the user selects the point(s) of interest (Pi); (iv) the program computes the relative coordinates of the Pi into the system (Oo, Ox, and Oy); (v) when observed using TEM, real coordinates of points corresponding to Oo, Ox, and Oy are entered by the user (either directly on the microscope or via an acquired image); and (vi) the program computes the absolute coordinates of the Pi into the system that was defined in step v, i.e., the microscope coordinates or new image coordinates. This method can be used both to find the square of interest from the grid center and the points of interest within a square.
The second procedure (procedure 2) consists of directly registering, using the grid squares as reference objects, the images observed in transmitted light microscopy and TEM in order to subsequently superimpose fluorescence and TEM images to create an overlay image fluorescence-TEM image. First, the fluorescence images are scaled to match the TEM resolution. The images are then cropped to the same size before registration. The squares of the grid are automatically extracted by using classical thresholding methods (Isodata thresholding). They are then automatically registered using the TurboReg plugin (43). The overall algorithm works as follows: (i) the resolution is adjusted (scale-up bright-field and fluorescence images to match TEM resolution); (ii) squares are extracted from the images (using automatic thresholding and cropping); (iii) binarized images (squares from the bright-field image and the TEM image) are registered; (iv) transformation to fluorescence images to match TEM images is applied; and (v) the fluorescence and TEM images are overlaid.
RESULTS
Image analysis protocol.
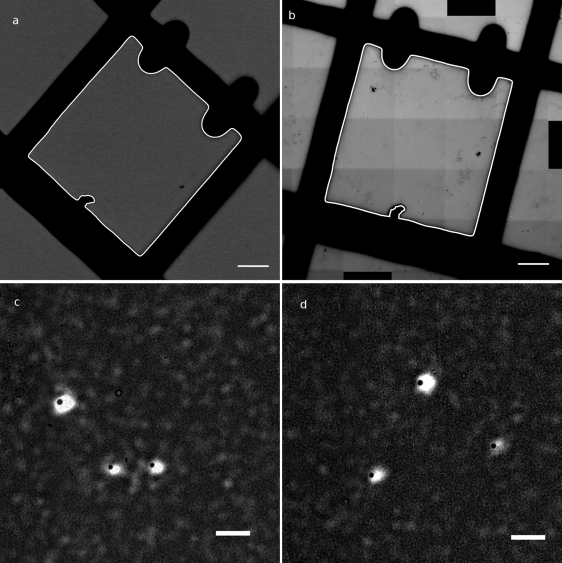
For our study we developed tools to superimpose fluorescence and TEM information based solely on the image information. Registration procedure 1, based on a coordinate system of coordinates, was tested with corners of squares as feature points and fluorescent latex beads as interest points. The procedure gave a satisfactory result, with an average distance between computed positions from fluorescence and positions observed in TEM of 0.5 μm or less depending on the accuracy of the localization of reference points. The accuracy was, as expected, better with square registration using procedure 2 (Fig. 1). The average distance between registered centers of beads observed in fluorescence and TEM was 0.375 μm (standard deviation = 0.109, n = 21). In most cases, the beads observed in TEM were inside their fluorescence spot; only two to three beads were outside. Compared to fluorescence resolution, the mean distance of 375 nm corresponds to 3 to 4 pixels in fluorescence images. The procedures are available as open-source programs to be used with a common tool for image analysis, ImageJ.
Fig. 1.
Superimposition of fluorescence and TEM images on test beads. (a) Transmitted light image of the square of interest, with automatically extracted square in white (bar, 20 μm). (b) Reconstructed mosaic TEM image of the square of interest, with automatically extracted square in white (bar, 20 μm). (c and d) Overlay of observed beads in fluorescence (white spots) after registration and in TEM (black dots) (bar, 2 μm).
FISH protocol on aphids.
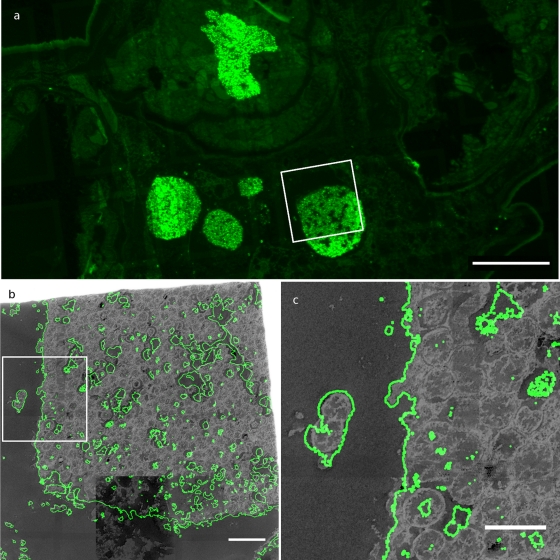
Successful hybridization of the Eub338 probe was obtained from the bacteriomes of aphid samples (Fig. 2 a). Signals were not ambiguous, as can be seen from the contrast between labeled and nonlabeled areas. Fluorescence and TEM images could then be successfully superimposed by using procedure 2 (image based) as seen in Fig. 2b. The FISH signal overlapped with typical bacterium-shaped structures that are identical to the Buchnera aphidicola large morphotypes described in aphids (33). The high correlation between TEM and FISH images is illustrated by the overlap observed in isolated bacterial morphotypes (Fig. 2c). Despite the use of a “simplified” FISH protocol (without Tris-HCl buffer), good hybridizations were obtained. The thin sections used, which were less than the diameter of a single bacterium, possibly helped by limiting the need for permeabilization.
Fig. 2.
FISH-CLEM imaging on aphid tissues. (a) Mosaic image of fluorescent signals in aphid tissues (bar, 100 μm). (b) Superimposition using the procedure illustrated in Fig. 1, of fluorescent signal (in green, contours shown) and TEM signal (in gray) (bar, 10 μm). (c) Zoom on an isolated bacterium showing very good superimposition of fluorescence and TEM signals (bar, 5 μm).
Bacteria identification on mussel gills.
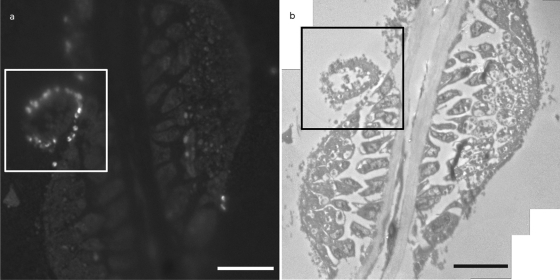
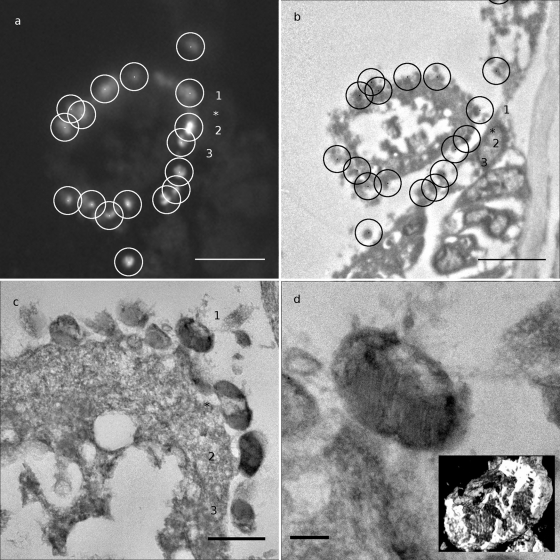
The same FISH protocol successfully labeled specific bacteria, namely, type I methanotrophic bacteria (Fig. 3). Labeled bacteria were successfully identified in TEM by using Procedure 1 (coordinate-based) (Fig. 4). These bacteria were observed mostly outside of the host cells (Fig. 4c). The methanotroph-specific probe Imed-M indeed hybridized only with bacteria displaying the expected stacked internal membranes of type I methanotrophs under TEM (Fig. 4c and d). Typical stacked internal membranes were observed using both standard TEM and 3D tomography (Fig. 4d and see the supplemental material). Idas gills displayed an unusual morphology compared to gills of well-studied related species, such as the vent mussel Bathymodiolus azoricus, whose gill epithelium consists of a single layer of cells shared between large bacteriocytes containing numerous symbiotic bacteria, and small symbiont-free intercalary cells (22). The gills observed here indeed rather resemble the gills observed in small mussels from sunken wood and bones, with small epithelial cells displaying irregular shapes (23, 18).
Fig. 3.
FISH-CLEM imaging on gills showing methanotrophic bacteria. (a) FISH image of a transverse section of a gill filament of Idas sp. strain Med. Bright signal corresponds to hybridized methanotrophs (bar, 10 μm). (b) Mosaic image of the same region observed in TEM (bar, 10 μm).
Fig. 4.
Superimposition of FISH and TEM information for detection of bacteria. (a) Higher magnification of the framed area from Fig. 3a. White dots and circled areas correspond to selected bacteria (radius, 1 μm; bar, 5 μm). (b) Higher magnification of the framed area from Fig. 3b. Circles are centered on positions computed from positions in panel a) (radius, 1 μm). Note that bacteria detected in this TEM image are inside circles (bar, 5 μm). (c) Zoom image at higher resolution (mosaic image). Note the extracellular localization of bacteria. Numbers refer to bacteria detected in panels a and b. Note that a bacterium (*) was not detected by fluorescence (bar, 1 μm). (d) Enlarged view of the bacterium 1 displaying stacked internal membranes typical of type I methanotrophic Gammaproteobacteria (bar, 200 nm). The inset shows 3D visualization of the reconstructed bacterium from tomogram (see the supplemental material).
DISCUSSION
We demonstrate here that it is possible to perform both FISH and TEM observation on the same sample, allowing the detection with an electron microscope of bacteria whose phylotype was previously identified only by using FISH and permitting the investigation of their ultrastructure, including the use of tomography.
FISH protocol.
CLEM by itself is not a new technique, but its use in microbiology required some specific adaptations. The methacrylate resin (LR-White) was chosen because it provides a good compromise between preservation of the ultrastructure and FISH labeling. Indeed, FISH can be performed on thinner sections than are usually obtained with paraffin or Steedman's wax, and sections can be visualized using TEM. Moreover, this resin, in contrast to epoxy resin, is suitable for immunolabeling. The FISH protocol has been adapted in order to eliminate some classical components of buffers such as Tris (2), which possess a strong oxidative power and lead to oxidation of the grids. For this reason, nickel grids were preferred to copper grids, which react with the ionic components of FISH buffer solutions. Probes, fluorochromes, formamide, and hybridization conditions, on the other hand, were kept standard. The method has a few drawbacks. First, the chemical fixation used to perform FISH at 46°C can result in some artifacts under electron microscopy. However, the use of formaldehyde is important in order to reduce the proportion of glutaraldehyde in the mixture, which, though suitable and often used for electron microscopy, is known to cause autofluorescence. Furthermore, this mixture is thought to have complementary cross-linking effects and thus could provide better fixation (25). Second, osmium cannot be used in the sample preparation, and thus the images obtained display a weaker contrast than would be obtained otherwise. The latter effect is partly compensated for by the use of tomography, which allows an increased signal-to-noise ratio (44).
Microscopic observations.
The fixation and preparation protocol presented here is a compromise that allowed the observation of bacteria under both a fluorescence and electron microscope. FISH images were of sufficiently good quality for the reliable identification of phylotype-specific signal using both a generalistic eubacterial probe and a methanotroph phylotype-specific probe. Although the TEM images were not as detailed as they would be if the samples were specifically fixed for TEM ultrastructure studies, their resolution was sufficient to observe the typical stacked intracytoplasmic membranes of methanotrophic mussel symbionts, as well as of host cell membranes (Fig. 2 and 4) (11).
Registration procedures.
Two procedures were developed for the superimposition of the FISH signals and TEM images. Both procedures have pros and cons. Procedure 1 is less accurate but is more generic and can be used directly under the TEM to find bacteria localized beforehand in fluorescence. The key point is the subpixel accuracy obtained by scaling up the fluorescence image in order to be less sensitive to the position of the selected points. Feature points can be chosen arbitrarily as any structural features recognizable from bright-field image to TEM image or directly under TEM, the squares of the grid being the simplest. Procedure 2, although automated, can be more difficult to set up since mosaic images of the complete square of observation must be obtained, often leading to large images (up to 10,000 by 10,000 pixels for the final reconstructed mosaic image). However, the accuracy of the registration is almost perfect, leading to a <0.4-μm error between fluorescence and TEM images. This small error may be due to several factors, including the accuracy of the segmentation of the grid squares based on thresholding, the accuracy of the registration algorithms, and the change in the resin structure under the TEM.
Potential new insights into the structure of symbiotic associations using CLEM.
The FISH-CLEM methodology, coupled with image analysis tools, presented here represents the first protocol, to our knowledge, that allows ultrastructural investigation of a phylotypically characterized bacterium. Here, the use of CLEM led for the first time to the 3D observation of a methanotrophic symbiont whose identity was formally ascertained using a specific FISH probe. In Idas species, methanotrophs were clearly located outside of host gill bacteriocytes; meanwhile, bacteria have been described as intracellular in all other documented methanotroph-associated mussels (13). This result is intriguing since symbiont localization is considered an important aspect of associations with consequences on its transmission, functioning and evolution (see reference 7 for a review). More work on additional specimens, as well as on the other symbionts present in Idas species, is needed to ascertain localization of each symbiont type and whether the preliminary observations from the present study are representative. Nevertheless, CLEM already allowed this new observation. In addition to symbiont ultrastructure and localization, the image analysis tools developed here are ideally adapted to identifying any cell-associated structure, such as bacteria, and could also be easily implemented for the identification of cellular components such as nuclei, centrosomes, Golgi bodies, or synapses.
Our approach was developed according to standard protocols so as to be easily reproduced by other teams for their own investigations. However, other techniques may provide potential relevant improvements to this method. In particularly, cryofixation followed by freeze substitution is known to carry out a better preservation of biological tissues than chemical fixation and to reduce fixation artifacts (26). Quantum dots also appear to be an attractive option in the choice of fluorescent dyes, because of their fluorescent properties and electron density (49). Despite the challenges associated with their implementation, these techniques offer interesting avenues for future research.
CLEM-FISH has many potential applications. In the case of symbiosis, intra- or extracellular localization of any type of symbiotic prokaryote can be investigated even if several symbionts without distinct morphologies occur, such as in an Idas sp. The CLEM-FISH approach can also be used to track peculiar morphological features in uncultivated prokaryotes occurring in complex communities, both within cells (inclusions, membranes, etc.) and among cells (attachment structures, extracellular matrices, etc.), and to link ultrastructural information with FISH-based identification.
Supplementary Material
ACKNOWLEDGMENTS
We thank P.-E. Gleizes and the TEM facility of IBCG, Toulouse, France, for access to their new JEOL 200 kV microscope. We also thank the SME and imaging facility at IFR83 (Université Pierre et Marie Curie) for help with image acquisition. In addition, we thank the ROV Quest team, the crew of the Meteor, and the chief scientist of the BIONIL cruise, A. Boetius, for their help with the Idas collection.
Financial support for this research was provided by IFR83, the French ANR Deep Oases (lab work), the EU network of excellence REX-3DEM (CT 502828), and the EU program HERMIONE (S.H.).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 22 April 2011.
REFERENCES
- 1. Abramoff M. D., Magelhaes P. J., Ram S. J. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 2. Amann R. I., Ludwig W., Schleifer K. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amann R. I., et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson L., et al. 2008. Tomography of bacteria-mineral associations within the deep-sea hydrothermal vent shrimp Rimicaris exoculata. C. R. Chimie 11:268–280 [Google Scholar]
- 5. Baumann P., Moran N., Baumann L. L. 2000. Bacteriocyte-associated endosymbionts of insects, p. 403–438 In Dworkin M. (ed.), The prokaryotes: a handbook on the biology of bacteria—ecophysiology, isolation, identification, applications. Springer-Verlag, New York, NY [Google Scholar]
- 6. Blazejak A., Erséus C., Amann R., Dubilier N. 2005. Coexistence of bacterial sulfide oxidizers, sulfate reducers, and spirochetes in a gutless worm (Oligochaeta) from the Peru margin. Appl. Environ. Microbiol. 71:1553–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bright M., Bugheresi S. 2010. A complex journey: transmission of microbial symbionts. Nat. Rev. Microbiol. 8:218–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bright M., Keckeis H., Fisher C. R. 2000. An autoradiographic examination of carbon fixation, transfer and utilization in the Riftia pachyptila symbiosis. Mar. Biol. 136:621–632 [Google Scholar]
- 9. Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 10. Cavanaugh C. M., McKiness Z. P., Newton I. L. G., Stewart F. J. 2006. Marine chemosynthetic symbioses, p. 475–507 In Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (ed.), The prokaryotes—a handbook on the biology of bacteria: symbiotic associations, biotechnology, applied microbiology, 3rd ed., vol. 1. Springer, New York, NY [Google Scholar]
- 11. Cavanaugh C. M., Levering P. R., Maki J. S., Mitchell R., Lidstrom M. E. 1987. Symbiosis of methylotrophic bacteria and deep-sea mussels. Nature 325:346–347 [Google Scholar]
- 12. Childress J. J., et al. 1986. A methanotrophic marine molluscan (Bivalvia, Mytilidae) symbiosis: mussels fueled by gas. Science 233:1306–1308 [DOI] [PubMed] [Google Scholar]
- 13. DeChaine E. G., Cavanaugh C. M. 2005. Symbioses of methanotrophs and deep-sea mussels (Mytilidae: Bathymodiolinae), p. 227–249 In Overmann J. (ed.), Molecular basis of symbiosis. Springer-Verlag, New York, NY: [DOI] [PubMed] [Google Scholar]
- 14. Distel D. L., Lee H. K., Cavanaugh C. M. 1995. Intracellular coexistence of methano- and thioautotrophic bacteria in a hydrothermal vent mussel. Proc. Natl. Acad. Sci. U. S. A. 92:9598–9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Douglas A. E. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17–37 [DOI] [PubMed] [Google Scholar]
- 16. Dubilier N., Bergin C., Lott C. 2008. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6:725–740 [DOI] [PubMed] [Google Scholar]
- 17. Dubilier N., et al. 2001. Endosymbiotic sulfate-reducing and sulphide-oxidizing in an oligochaete worm. Nature 411:298–302 [DOI] [PubMed] [Google Scholar]
- 18. Duperron S., Laurent M. C. Z., Gaill F., Gros O. 2008. Sulphur-oxidizing extracellular bacteria in the gills of Mytilidae associated with wood falls. FEMS Microbiol. Ecol. 63:338–349 [DOI] [PubMed] [Google Scholar]
- 19. Duperron S., Halary S., Lorion J., Sibuet M., Gaill F. 2008. Unexpected co-occurrence of six bacterial symbionts in the gills of the cold seep mussel Idas sp. (Bivalvia: Mytilidae). Environ. Microbiol. 10:433–445 [DOI] [PubMed] [Google Scholar]
- 20. Duperron S., et al. 2007. Diversity, relative abundance, and metabolic potential of bacterial endosymbionts in three Bathymodiolus mussels (Bivalvia: Mytilidae) from cold seeps in the Gulf of Mexico. Environ. Microbiol. 9:1423–1438 [DOI] [PubMed] [Google Scholar]
- 21. Durand L., et al. 2010. Microbial diversity associated with the hydrothermal shrimp Rimicaris exoculata gut and occurrence of a resident microbial community. FEMS Microbiol. Ecol. 71:291–303 [DOI] [PubMed] [Google Scholar]
- 22. Fiala-Médioni A., et al. 2002. Ultrastructural, biochemical and immunological characterisation of two populations of the mytilid mussel Bathymodiolus azoricus from the Mid Atlantic Ridge: evidence for a dual symbiosis. Mar. Biol. 141:1035–1043 [Google Scholar]
- 23. Gros O., Gaill F. 2007. Extracellular bacterial association in gills of “wood mussels. ” Cahiers Biol. Mar. 48:103–109 [Google Scholar]
- 24. Hanson R., Hanson T. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hayat M. A. 1989. Principles and techniques of electron microscopy: biological applications, 3rd ed. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 26. Hohn K., et al. 2010. Preparation of cryofixed cells for improved 3D ultrastructure with scanning transmission electron tomography. Histochem. Cell Biol. 135:1–9 [DOI] [PubMed] [Google Scholar]
- 27. Koster A. J., et al. 1997. Perspectives of molecular and cellular electron tomography. J. Struct. Biol. 120:276–308 [DOI] [PubMed] [Google Scholar]
- 28. Kremer J. R., Mastronarde D. N., McIntosh J. R. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 29. Lucić V, et al. 2007. Multiscale imaging of neurons grown in culture: from light microscopy to cryo-electron tomography. J. Struct. Biol. 160:146–156 [DOI] [PubMed] [Google Scholar]
- 30. Mastronade D. N. 1997. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120:343–352 [DOI] [PubMed] [Google Scholar]
- 31. McCutcheon J. P., Moran N. A. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. U. S. A. 104:19392–19397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mironov A. A., Beznoussenko G. V. 2009. Correlative microscopy: a potent tool for the study of rare or unique cellular and tissue events. J. Microsc. 235:308–321 [DOI] [PubMed] [Google Scholar]
- 33. Moran N. A., Russell J. A., Koga R., Fukatsu T. 2005. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71:3302–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moya A., Peretó J., Gil R., Latorre A. 2008. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 9:218–229 [DOI] [PubMed] [Google Scholar]
- 35. Nussbaumer A. D., Fisher C. R., Bright M. 2006. Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature 441:345–348 [DOI] [PubMed] [Google Scholar]
- 36. Polishchuk R. S., Mironov A. A. 2001. Correlative video light/electron microscopy. Chapter 4, unit 4.8. Curr. Protoc. Cell Biol. doi:10.1002/0471143030.cb0408s11 [DOI] [PubMed] [Google Scholar]
- 37. Preibisch S., Saalfeld S., Tomancak P. 2009. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25:1463–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rasband W. S. 1997–2008. ImageJ. National Institutes of Health, Bethesda, MD: http://rsb.info.nih.gov/ij/ [Google Scholar]
- 39. Razi M., Tooze S. A. 2009. Correlative light and electron microscopy. Methods Enzymol. 452:261–275 [DOI] [PubMed] [Google Scholar]
- 40. Ruehland C., et al. 2008. Multiple bacterial symbionts in two species of co-occurring gutless oligochaete worms from Mediterranean sea grass sediments. Environ. Microbiol. 10:3404–3416 [DOI] [PubMed] [Google Scholar]
- 41. Sartori A., et al. 2007. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J. Struct. Biol. 160:135–145 [DOI] [PubMed] [Google Scholar]
- 42. Schaudinn C., et al. 2009. Imaging of endodontic biofilms by combined microscopy (FISH/cLSM-SEM). J. Microsc. 235:124–127 [DOI] [PubMed] [Google Scholar]
- 43. Thévenaz P., Ruttimann U. E., Unser M. 1998. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Proc. 7:27–41 [DOI] [PubMed] [Google Scholar]
- 44. van Driel L. F., Knoops K., Koster A. J., Valentijn J. A. 2007. Fluorescent labeling of resin-embedded sections for correlative electron microscopy using tomography-based contrast enhancement. J. Struct. Biol. 161:372–383 [DOI] [PubMed] [Google Scholar]
- 45. Vicidomini G., et al. 2010. A novel approach for correlative light electron microscopy analysis. Microsc. Res. Tech. 73:215–224 [DOI] [PubMed] [Google Scholar]
- 46. Vicidomini G., et al. 2008. High data output and automated 3D correlative light-electron microscopy method. Traffic 9:1828–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wrede C., Heller C., Reitner J., Hoppert M. 2008. Correlative light/electron microscopy for the investigation of microbial mats from Black Sea cold seeps. J. Microbiol. Methods 73:85–91 [DOI] [PubMed] [Google Scholar]
- 48. Wu D., et al. 2006. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 4:e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu S.-M., et al. 2006. Quantum-dot-labeled DNA probes for fluorescence in situ hybridization (FISH) in the microorganism Escherichia coli. Chem. Phys. Chem. 7:1062–1067 [DOI] [PubMed] [Google Scholar]
- 50. Zbinden M., et al. 2008. New insights on the metabolic diversity among the epibiotic microbial community of the hydrothermal shrimp Rimicaris exoculata. J. Exp. Mar. Biol. Ecol. 359:131–140 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.