Abstract
Currently, 2,610 different Salmonella serovars have been described according to the White-Kauffmann-Le Minor scheme. They are routinely differentiated by serotyping, which is based on the antigenic variability at lipopolysaccharide moieties (O antigens), flagellar proteins (H1 and H2 antigens), and capsular polysaccharides (Vi antigens). The aim of this study was to evaluate the potential of matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry for rapid screening and identification of epidemiologically important Salmonella enterica subsp. enterica serovars based on specific sets of serovar-identifying biomarker ions. By analyzing 913 Salmonella enterica subsp. enterica strains representing 89 different serovars using MALDI-TOF mass spectrometry, several potentially serovar-identifying biomarker ions were selected. Based on a combination of genus-, species-, subspecies-, and serovar-identifying biomarker ions, a decision tree classification algorithm was derived for the rapid identification of the five most frequently isolated Salmonella enterica serovars, Enteritidis, Typhimurium/4,[5],12:i:-, Virchow, Infantis, and Hadar. Additionally, sets of potentially serovar-identifying biomarker ions were detected for other epidemiologically interesting serovars, such as Choleraesuis, Heidelberg, and Gallinarum. Furthermore, by using a bioinformatic approach, sequence variations corresponding to single or multiple amino acid exchanges in several biomarker proteins were tentatively assigned. The inclusivity and exclusivity of the specific sets of serovar-identifying biomarker ions for the top 5 serovars were almost 100%. This study shows that whole-cell MALDI-TOF mass spectrometry can be a rapid method for prescreening S. enterica subsp. enterica isolates to identify epidemiologically important serovars and to reduce sample numbers that have to be subsequently analyzed using conventional serotyping by slide agglutination techniques.
INTRODUCTION
Salmonella is a major zoonotic food-borne pathogen, causing outbreaks and sporadic cases of gastroenteritis in humans worldwide (24). Two species are currently recognized in the genus Salmonella, Salmonella enterica and Salmonella bongori. S. enterica has been further subdivided into six subspecies (48), of which Salmonella enterica subsp. enterica is the most frequently isolated and is associated with gastrointestinal disease in a wide range of mammalian hosts. Traditionally, a number of biochemical reactions differentiate Salmonella species and subspecies (16). The Salmonella serotyping scheme according to White-Kauffmann-Le Minor is accepted worldwide as a gold standard for the differentiation of salmonellae below the subspecies level (19). It is based on a combination of biochemical reactions and serotyping of the somatic O, flagellar H, and capsular Vi antigens. However, serotyping, which is usually performed by slide agglutination, is a laborious, time-intensive, and expensive method requiring more than 250 different antisera.
Whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) based on intact protein profiling, a technique also known as direct bacterial profiling, has increasingly been studied for bacterial species identification in recent years (1, 8, 12, 13, 13, 29, 34, 35, 45). Reproducibility issues had been a major concern, but it was shown that the technique provides reliable results despite some experimental and biological variabilities that are inherent to such a phenotypic approach (14, 41, 49, 51, 54–56). A sufficient number of stable mass signals of major housekeeping proteins, mainly ribosomal proteins, can reproducibly be detected and used for bacterial species identification by using simple mass pattern-matching approaches or more sophisticated algorithms to compare and estimate the similarities between spectra (3, 9, 22, 23, 26, 27, 29, 37, 43). These approaches don't rely on actual identification of the biomarker ion peaks in an MS spectrum but on the characteristic mass profile generated by a set of ion peaks that constitute a bacterial “fingerprint.” While the majority of these studies described the usage of the technique for species identification, bacterial characterization below the species level, e.g., for discrimination of serovars, was rarely approached (4, 28, 33, 43, 46). Recently, we optimized and evaluated the use of whole-cell MALDI-TOF mass spectrometry for Salmonella identification and differentiation on the subspecies level (13). Here, we extended the analysis for Salmonella enterica subsp. enterica (subspecies I) in order to study the suitability of the technique for differentiation of salmonellae at the serovar level. Generally, only a few serovars represent the majority of isolates from humans and animals. For example, in Europe the 10 most frequently isolated Salmonella serovars in humans include serovars Enteritidis, Typhimurium, Virchow, Infantis, and Hadar and accounted for more than 85% of the confirmed salmonellosis cases in humans during the years 2006 and 2007. Serovar Enteritidis was by far the predominant serovar (67.8%), followed by serovar Typhimurium (15.6%) (2).
The aim of this study was to establish and validate a rapid MALDI-TOF MS-based method that allows Salmonella identification at the species, subspecies, and serovar levels at once. As a proof of principle, the study focused on the recognition and validation of serovar-identifying biomarker ions for the frequently isolated Salmonella subsp. enterica serovars Typhimurium, Enteritidis, Virchow, Infantis, and Hadar, enabling rapid screening and identification.
MATERIALS AND METHODS
Bacterial strains.
Altogether, 913 Salmonella enterica subsp. enterica strains were selected for this study. A strain list is shown in Table 1. All strains were biochemically differentiated on the subspecies level (30–32) and serotyped by slide agglutination with O antigen-specific and H antigen-specific sera (Sifin Diagnostics, Germany, Berlin) according to the White-Kauffmann-Le Minor scheme (19). The strain collection represents epidemiologically unlinked strains originating from different regions and herds in Germany that were received at the National Reference Laboratory for Salmonella, Berlin, Germany, on a routine basis for serotyping. The strains selected were frequently isolated serovars in humans and animals and included host-restricted and host-adapted serovars, as well as more rare serovars displaying similar antigenic formulas.
Table 1.
Salmonella enterica subsp. enterica strains (n = 913) used in this study
| Serovar | No. of strains | Source(s) | Yr(s) isolated |
|---|---|---|---|
| Enteritidis | 53 | Poultry, bovine, swine, food, environmental | 2003–2008, 2010 |
| Typhimurium/4,[5],12:i:- | 101/27 | Bovine, poultry, swine, food, others | 2000–2010 |
| Infantis | 51 | Bovine, poultry, swine, human, food, environmental | 2000–2010 |
| Virchow | 32 | Poultry, swine, human, cat, food, environmental | 2000–2001, 2003–2009 |
| Hadar | 52 | Bovine, poultry, human, cat, dog, bird, food | 2001–2009 |
| Bovismorbificans | 14 | Poultry, swine, bovine, food | 2002–2008 |
| Derby | 74 | Poultry, swine, human, dog, environmental, food | 2001–2010 |
| Anatum | 17 | Poultry, swine, bovine, food | 2002–2008 |
| Newport | 48 | Poultry, bovine, human, dog, fish, food, environmental | 2000, 2002–2010 |
| Goldcoast | 24 | Swine, bovine, rabbit, environmental, food | 2001–2006, 2008–2009 |
| 4,12:d:- | 13 | Poultry, swine, bovine | 2003–2004, 2008 |
| Abony | 6 | Bovine, horse | 2007–2008 |
| Agama | 1 | Swine | 2007 |
| Agona | 7 | Swine, dog, poultry, food, environmental | 2005, 2008 |
| Albany | 1 | Fertilizer | 2006 |
| Amersfoort | 1 | Swine | 2005 |
| Augustenborg | 1 | Poultry | 2002 |
| Bareilly | 2 | Environmental, food | 2006 |
| Blockley | 11 | Poultry, bovine, swine | 2000, 2005–2009 |
| Braenderup | 4 | Swine, food, mussel | 2005–2007 |
| Brandenburg | 9 | Swine, poultry, bovine, human, fertilizer | 2002, 2005–2007, 2010 |
| Bredeney | 10 | Poultry, environmental, dog, bird, food | 2005, 2008, 2010 |
| Chandans | 1 | Food | 2010 |
| Cerro | 1 | Chicken | 2008 |
| Choleraesuis | 5 | Swine, game, cat | 2002, 2004–2006, 2009 |
| Coeln | 2 | Chicken | 2008 |
| Colindale | 1 | Food | 2003 |
| Corvallis | 2 | Animal food | 2006 |
| Cubana | 6 | Poultry, animal food, fertilizer | 2006, 2008–2009 |
| Dublin | 11 | Swine, bovine, poultry, dog, food | 2005–2008 |
| Ealing | 5 | Dog, animal food, environmental | 2002–2004, 2006 |
| Eastbourne | 4 | Food, poultry | 2005, 2009 |
| Emek | 1 | Sludge | 2007 |
| Falkensee | 2 | Animal food | 2004–2005 |
| Ferruch | 1 | Poultry | 2002 |
| Gallinarum/Gallinarum var. Pullorum | 4/10 | Poultry | 2004–2010 |
| Give | 4 | Swine, poultry, food | 2006, 2008 |
| Glostrup | 4 | Swine, environmental, food | 2000, 2006, 2008 |
| Haardt | 3 | Poultry | 2000–2001 |
| Haifa | 5 | Swine, human, fertilizer, food | 2005–2006, 2008 |
| Heidelberg | 8 | Poultry, rabbit | 2003, 2005, 2007–2008 |
| Indiana | 8 | Poultry, environmental | 2006, 2008 |
| Isangi | 4 | Poultry | 2006, 2008–2009 |
| Israel | 1 | Deer | 1998 |
| Javiana | 4 | Food, dog, swine, environmental | 2004, 2006–2008 |
| Jerusalem | 1 | Poultry | 2008 |
| Kentucky | 3 | Swine, human, food | 2005, 2007–2008 |
| Kiambu | 11 | Cat, poultry, dog, environmental | 2000, 2004–2006, 2008–2009 |
| Kottbus | 16 | Poultry, bird, bovine, swine, environmental, sludge | 2000, 2002–2006, 2008 |
| Lagos | 1 | Not known | 1983 |
| Lexington | 5 | Swine, poultry, food | 2006–2008 |
| Litchfield | 5 | Swine, poultry, environmental | 2000–2001, 2005–2006 |
| Livingstone | 17 | Poultry, swine, food, dog | 2006–2008, 2010 |
| London | 16 | Swine, bovine, poultry, food | 2000–2009 |
| Manchester | 2 | Poultry | 2002, 2005 |
| Manhattan | 12 | Poultry, bovine, swine | 2000–2001, 2004–2009 |
| Mbandaka | 9 | Food, poultry, swine, environmental | 2001, 2006–2010 |
| Meleagridis | 1 | Human | 2009 |
| Minnesota | 1 | Not known | 2010 |
| Molade | 1 | Food | 2009 |
| Montevideo | 4 | Poultry | 2005, 2007, 2010 |
| Muenchen | 5 | Bovine, dog, game | 2004, 2006, 2008–2009 |
| Muenster | 3 | Poultry, swine, food | 2005, 2010 |
| Ohio | 4 | Poultry, food | 2006, 2008 |
| Oranienburg | 2 | Sludge, environmental | 2006, 2010 |
| Orion | 12 | Poultry, bovine, fish, environmental, food, sludge | 2000–2001, 2005–2007, 2009 |
| Pakistan | 1 | Poultry | 2000 |
| Panama | 12 | Swine, poultry, food | 2000, 2003–2004, 2006–2008, 2010 |
| Paratyphi A | 1 | Human | 1999 |
| Paratyphi B | 12 | Poultry, bovine | 2003, 2004, 2008, 2010 |
| Poona | 1 | Food | 2008 |
| Reading | 6 | Food, poultry, dog | 2001, 2003–2005, 2007 |
| Rissen | 5 | Poultry, bovine, swine, food | 2004, 2008 |
| Saintpaul | 20 | Poultry, swine | 2003, 2006, 2008, 2010 |
| Sandiego | 7 | Dog, mouse, bird, fertilizer, food | 2005–2007, 2009–2010 |
| Schleissheim | 7 | Dog, bird, game, environmental | 2002, 2006, 2007 |
| Schwarzengrund | 4 | Swine, poultry, food, fertilizer | 2006, 2008 |
| Senftenberg | 13 | Poultry, swine, food, environmental | 2001, 2003, 2005, 2008–2010 |
| Stanley | 8 | Swine, dog, food | 2004–2005, 2007–2008 |
| Stanleyville | 8 | Poultry, bovine, food wastewater | 1998, 2001, 2007, 2009 |
| Stourbridge | 1 | Badger | 2010 |
| Teddington | 1 | Food | 2010 |
| Tennessee | 3 | Food, fertilizer | 2006, 2008 |
| Thompson | 4 | Swine, poultry, bovine | 2005, 2007–2008 |
| Weltevreden | 4 | Food, bovine, seafood | 2005, 2008 |
| Wien | 3 | Dog, environmental, food | 2004, 2006–2007 |
| Wilhelmsburg | 1 | Duck | 1998 |
| Zanzibar | 4 | Fish, wild pig | 2000, 2006–2007 |
Cell culturing and preparation of samples for whole-cell MALDI-TOF MS.
The strains were grown at 37°C for 24 h ± 1 h on Mueller-Hinton agar (Oxoid, Greve, Denmark). Bacteria were removed from agar plates by using a sterile pipette tip and applied directly as a thin film onto a 384-position MALDI sample target (Bruker Daltonics, Bremen, Germany). The samples were immediately mixed with 1 μl matrix solution (25 mg/ml 3,5-dimethoxy-4-hydroxycinnamic acid [sinapinic acid; Bruker Daltonics, Bremen, Germany]) in 50% (vol/vol) acetonitrile (Sigma-Aldrich) supplemented with 0.6% (vol/vol) trifluoroacetic acid (Roth, Germany). The matrix sample spots were crystallized by air drying.
Whole-cell MALDI-TOF MS parameters.
The MALDI-TOF MS analysis has been described previously (13). Briefly, mass spectra were acquired with an Ultraflex II MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) equipped with an all-solid-state Smartbeam Nd:YAG laser and operated at 100 Hz in the positive linear mode (delay, 100 ns; voltage, 25 kV; molecular mass range, 2.2 to 40 kDa) under the control of Flexcontrol software (version 3.0; Bruker Daltonics). Each spectrum was obtained by averaging up to 7,000 laser shots acquired at the minimum laser power necessary for ionization of the samples. Automated spectrum acquisition was performed using the Auto Execute software integrated within Flexcontrol.
Data evaluation.
Mass data files were transferred to the Flexanalysis software (version 3.0; Bruker Daltonics) and processed with baseline correction, Gaussian smoothing, and peak finding. Average mass values were determined. Spectra were internally calibrated using a set of ribosomal biomarker proteins common to most Salmonella enterica subsp. enterica strains, including ribosomal proteins RL36 (m/z 4,365.3), RL32 (m/z 6,316.2), RL30 (m/z 6,383.6), RS15 (m/z 10,067.5), RS19 (m/z 10,286.1), RL25 (m/z 10,542.2), RS14 (m/z 11,478.3), RS10 (m/z 11,767.6), and DNA-binding protein H-NS (m/z 15,412.5) (10). A maximum of 300 peaks with a signal-to-noise ratio minimum of 2 were selected in the range of 2,000 to 25,000 Da. For species and subspecies identifications, peak lists were imported into the SARAMIS software (Spectral Archiving and Microbial Identification System, release 3.4; Anagnostec GmbH, Germany). In the first step, a consensus spectrum, called the superspectrum, was calculated based on duplicate spectra of 221 S. enterica subsp. enterica strains belonging to different serovars. This superspectrum was used for identification of S. enterica subsp. enterica at the subspecies level and consisted of sets of biomarker ions that were present in >90% of the strains and displayed specificities at the genus, species, and subspecies levels. Clinprotools software (version 2.1; Bruker Daltonics) was used for peak definition (signal to noise ratio of >1), integration (endpoint level), mass recalibration (maximal peak shift of 200 ppm), area normalization (against total ion count). Receiver operating characteristic (ROC) analysis was performed in order to prescreen for serovar-identifying biomarker ions for the most prevalent serovars. ROC curves were generated for each peak as two subgroups (a set of spectra from multiple strains of the “target” serovar and a second subset of spectra from all other serovars) and were compared by using a graphical display of the false-positive rate (horizontal axis) and the true-positive rate (vertical axis). The optimal ROC curve, meaning the highest true-positive rate for a given false-positive rate, corresponds to an area under the ROC curve (AUC) of 1. Potential biomarker candidates were further analyzed by visual inspection of the mass spectra by Flexanalysis (overlaid view). As a general rule, mass signals were preferably selected as potentially serovar specific if they were present in all sets of data obtained for a given serovar and were absent in all other serovars, resulting in an AUC value of near 1. For identification of Salmonella strains below species level, weighted pattern-matching approaches were conducted. By using the SARAMIS software, weighted superspectra were created, and highly discriminative biomarkers were upweighted (given an overemphasized value) in the identification routine, while nonspecific, variable, and low-intensity peaks were downweighted or ignored in the identification process. Automated computer-aided identification was performed by comparing peak lists for individual samples with the established reference database of superspectra, generating a ranked list of matching spectra based on the point value system. The established procedures were then used to screen an additional 692 S. enterica subsp. enterica strains comprising 89 serovars for the presence of serovar-identifying biomarker ions.
Identification of biomarker proteins based on database searches.
The m/z peaks obtained were subjected to an online TagIdent protein database search (18). An unrestricted search with pI values was performed. Theoretical masses of protein sequences were calculated using the PeptideMass tool (18). The BLAST servers at www.sanger.ac.uk and www.expasy.ch were used for protein-versus-translated DNA BLAST searches in 19 Salmonella genome completed sequences and 20 Salmonella whole-genome available shotgun sequences.
RESULTS
Salmonella enterica subsp. enterica identification.
Whole-cell MALDI-TOF MS for S. enterica subsp. enterica (subspecies) identification, which is a prerequisite for serovar identification, was performed with 221 Salmonella strains essentially as described in reference 13. As a major Salmonella enterica subsp. enterica-specific biomarker ion, a mass peak at m/z 14,395, corresponding to the ribosomal protein L17, was used and given an overemphasized value in the superspectrum. This peak was present in almost all 89 different S. enterica subsp. enterica serovars tested (except serovars Gallinarum var. Pullorum and Schleissheim) but absent in all other S. enterica subspecies and S. bongori. Non-S. enterica subsp. enterica and S. bongori strains, on the other hand, were characterized by a mass peak at m/z 14,453, corresponding to ribosomal protein L17 with an additional serine residue and a threonine-to-alanine mutation.
Identification of potential serovar-identifying biomarker ions.
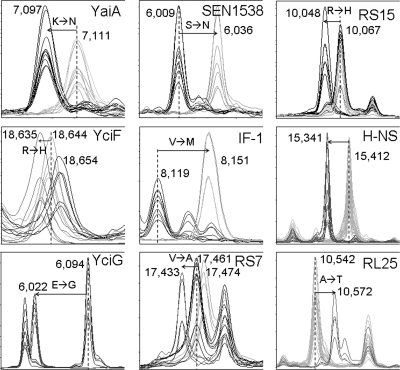
Discrimination ability at the serovar level was determined by analyzing data subsets comprised of multiple MALDI-TOF MS spectra obtained from different serovars. Typically, when two different serovars were compared, not more than 15 potentially serovar-discriminating peaks per spectrum were observed, of which usually several were present in more than one serovar. Therefore, the spectra were carefully inspected for reproducible serovar-identifying biomarkers and combinations thereof. Promising markers displaying high AUC values (near to 1) were selected accordingly and evaluated for their usage to discriminate either single serovars or groups of serovars (Table 2). Table 3 shows additional mass peaks displaying characteristic mass shifts in subsets of strains belonging to different serovars that assisted serovar identification based on major serovar-identifying signals. Based on sequence data from completed genome sequencing or whole-shotgun genome sequencing of more than 24 Salmonella strains belonging to different serovars, the detected mass signals that could be assigned to known proteins and possible point mutations explaining the detected mass shifts were identified. While mass spectrometric species identification mainly relies on sets of ribosomal proteins, many of the amino acid polymorphisms that resulted in potentially serovar-identifying mass variations were found in other protein classes, including many proteins with unknown functions. For example, a mass signal at m/z 6,036 was uniquely found in serovar Enteritidis and therefore useful for identifying this serovar. On the other hand, the presence of a mass signal of the same protein at m/z 6,009 indicated another serovar. Based on genome database searches, the corresponding mass difference of 27 Da could be explained by an S→N exchange in a putative uncharacterized protein (Fig. 1). Similarily, Leuschner et al. (33) found a mass signal at m/z 6,030 as one peak that was present in all of the five serovar Enteritidis strains they analyzed. A mass signal at m/z 7,097 of a putative uncharacterized protein, YaiA, was selected to differentiate serovar Typhimurium or its related monophasic variant (15, 57) from almost all other serovars, which mostly displayed a peak of the corresponding protein at 7,111 (K→N). The same peak was also found in serovar Virchow and one subtype of serovar Newport (Newport-II), but those could be differentiated from serovar Typhimurium by using additional markers. Serovar Virchow could be clearly identified by the presence of a mass peak at m/z 10,048 instead of m/z 10,067, corresponding to an R→H substitution in ribosomal protein S15. Serovar Infantis (6,7,14:r:1,5) and the related but rarely isolated serovars Augustenborg (6,7,14:i:1,2) and Colindale (6,7:r:1,7) showed a characteristic mass shift in the putative uncharacterized protein YciF at m/z 18,635 (Fig. 1). Serovars Augustenborg and Colindale in turn could be differentiated from serovar Infantis, e.g., by using the protein Gns as an assisting biomarker at m/z 6,484, which displayed a mass signal at m/z 6,512 in serovar Infantis (Table 2). Serovar Hadar was characterized by the presence of a mass peak at m/z 10,927 (protein YbgS), which was also found in strains of serovars Blockley, Ealing, Ferruch, Haardt, Kiambu, Kottbus, Orion, and 4,12:d:-. By using assisting peaks at m/z 6,484 (Gns), 6,771 (YhfG), 8,699 (putative inner membrane protein), 9,404 (unidentified), 10,542 (RL25), 17,461 (RS7), and 18,655 (YciF), serovar Hadar could be differentiated from all other serovars tested. Notably, host-restricted or host-adapted serovars, like serovar Choleraesuis (swine), serovar Paratyphi A (human), and serovar Gallinarum/Pullorum (chicken), showed more mass variations than other serovars and could be readily identified by several serovar-identifying biomarkers, even allowing subtyping between Salmonella enterica serovar Gallinarum biotype Gallinarum and biotype Pullorum. These nonmotile strains cannot be distinguished by serological methods and have to be subtyped on the basis of biochemical characteristics. Using MALDI-TOF MS, biotype Pullorum could readily be differentiated from biotype Gallinarum on the basis of at least 9 mass variations caused by amino acid polymorphisms in several proteins (Table 4). Moreover, biotype Pullorum was the only serovar that displayed a serovar-specific deficiency in posttranslational modification of, e.g., ribosomal proteins RS11 (m/z 13,701 instead of 13,715; methylation), RS12 (m/z 13,607 instead of 13,653; β-methylthiolation), RL33 (m/z 6241 instead of 6255; methylation), and RL16 (m/z 15,194 instead of 15,225; unknown modification). Similarly, potential serovar-specific mass signals were detectable for serovars Manhattan, Heidelberg, London, Tennessee, Schleissheim, and 4,12:d:- (Table 2).
Table 2.
Selected biomarkers for serovar-level identification of single serovars or groups of serovars (major serovar-identifying peaks)
| Exptl mass (avg m/z) | Tentative protein identity | Predicted mass (Da) | Serovar(s) in which biomarker found | Antigenic formula | Mass in other serovars (avg m/z) | Predicted mass (Da) | Amino acid exchange |
|---|---|---|---|---|---|---|---|
| 5,966 | Probably putative uncharacterized protein | NAa | Newport-I | 6,8:e,h:1,2 | 6,009 | 6,009.3b | NA |
| 6,022 | Putative uncharacterized protein YciG | 6,022.3a | Choleraesuis | 6,7:c:1,5 | 6,094 | 6,094.4 | E→G |
| 6,036 | Putative uncharacterized protein | 6,036.3b | Enteritidis | 1,9,12:g,m:- | 6,009 | 6,009.3b | S→N |
| 6,512 | Gns | 6,511.6 | Newport-I | 6,8:e,h:1,2 | 6,484 | 6,483.5 | A→V |
| Infantis | 6,7,14:r:1,5 | ||||||
| Manhattan | 6,8:d:1,5 | ||||||
| Mbandaka | 6,7,14:z10:e,n,z15 | ||||||
| 4,12:d:- | 4,12:d:- | ||||||
| Paratyphi A | 1,2,12:a:- | ||||||
| Stanleyville | 1,4,12:z4, z23:(1,2) | ||||||
| Zanzibar | 3,(10),(15):k:1,5 | ||||||
| Lexington | 3,10:z10:1,5 | ||||||
| 6,587 | Ribosome modulation factor | 6,586.6 | Paratyphi A | 1,2,12:a:- | 6,572 | 6,572.5 | D→E |
| 7,097 | Putative uncharacterized protein YaiA | 7,096.8b | Typhimurium/4,5,12:i:- | 4,5,12:i:(1,2) | 7,111 | 7,110.9b | K→N |
| Virchow | 6,7,14:r:1,2 | ||||||
| Newport-II | 6,8:e,h:1,2 | ||||||
| 7,139 | Putative uncharacterized protein YaiA | 7,138.9b | Heidelberg | 1,4,[5],12:r:1,2 | 7,111 | 7,110.9b | A→V |
| 7,983 | Uncharacterized protein YibT | NA | Derby-I | 1,4,[5],12:f,g:[1,2] | 7,993 | 7,993.1 | NA |
| 8,023 | Uncharacterized protein YibT | 8,023.1b | Heidelberg | 1,4,[5],12:r:1,2 | 7,993 | 7,993.1 | G→S |
| NA | Thompson | 6,7,14:k:1,5 | NA | ||||
| Blockley | 6,8:k:1,5 | ||||||
| Haardt | 8:k:1,5 | ||||||
| 8,027 | Uncharacterized protein YibT | 8,027.1a | Tennessee | 6,7,14:z29:[1,2,7] | 7,993 | 7,993.1 | L→F |
| 8,061 | Probably putative cytoplasmic protein YecF | NA | 4,12:d:- | 4,12:d:- | 8,051 | 8,051.2b | NA |
| 8,151 | RNA binding translation initiation factor IF-1 | 8,151.5b | Paratyphi A | 1,2,12:a:- | 8,119 | 8,119.4 | V→M |
| 8,342 | Putative stress response protein YjbJ | 8,342.3b | Tennessee | 6,7,14:z29:[1,2,7] | 8,329 | 8,329.3b | T→N |
| 9,548 | Probably DNA-binding protein HU-α | NA | Manhattan | 6,8:d:1,5 | 9,523 | 9,521.9 | NA |
| 10,048 | Ribosomal protein S15 | 10,048.5b | Virchow | 6,7,14:r:1,2 | 10,067 | 10,067.5b | R→H |
| 10,381 | Probably ribosomal protein L17 | NA | Schleissheim | 4,12,27:b:- | 14,395 | 14,395.6 | NAc |
| Gallinarum var. Pullorum | 1,9,12:-:- | ||||||
| 10,528 | Ribosomal protein L25 | 10,528.2 | Manhattan | 6,8:d:1,5 | 10,542 | 10,542.2 | I→V |
| Braenderup | 6,7:e,h:enz15 | ||||||
| Newport-III | 6,8:e,h:1,2 | ||||||
| Haifa | 1,4,[5],12:z10:1,2 | ||||||
| Stanley | 1,4,[5],12:z10:1,2 | ||||||
| 4,5,12,27:d:1,2 | |||||||
| 10,569 | Probably RL25 | NA | Schleissheim | 4,12,27:b:- | 10,542 | 10,542.2 | NA |
| 10,872 | Probable σ54 modulation protein | 10,872.4 | Heidelberg | 1,4,[5],12:r:1,2 | 10,858 | 10,858.4 | V→I |
| 10,886 | Probable σ54 modulation protein | 10,886.4 | Cholerasuis | 6,7:c:1,5 | 10,957 | 10,958.5c | K→R |
| 10,899 | Uncharacterized protein YbgS | 10,900.5c | Manhattan | 6,8:d:1,5 | 10,957 | 10,958.5c | D→G |
| Choleraesuis | 6,7:c:1,5 | ||||||
| Muenchen | 6,8:d:1,2 | ||||||
| Haifa | 1,4,[5],12:z10:1,2 | ||||||
| 10,927 | Uncharacterized protein YbgS | 10,928.5c | Hadar | 6,8:z10:e,n,x | 10,957 | 10,958.5c | T→A |
| Kottbus | 6,8:e,h:1,5 | ||||||
| Haardt | 8:k:1,5 | ||||||
| Blockley | 6,8:k:1,5 | ||||||
| Ferruch | 8:e,h:1,5 | ||||||
| Kiambu | 1,4,12:z:1,5 | ||||||
| 4,12:d:- | 4,12:d:- | ||||||
| Orion | 3,10:y:1,5 | ||||||
| Ealing-I | 35:g,m,s:- | ||||||
| 10,947 | Probably uncharacterized protein YbgS | NA | Brandenburg | 4,[5],12:l, v:enz15 | 10,957 | 10,958.5c | NA |
| Panama | 1,9,12:l,v:1,5 | ||||||
| Israel | 9,12:e,h:enz15 | ||||||
| Reading | 4,12:e,h:1,5 | ||||||
| Sandiego | 1,4,[5],12:e,h:enz15 | ||||||
| Eastbourne | 1,9,12:e,h:1,5 | ||||||
| 10,969 | Probably uncharacterized protein YbgS | NA | Aba | 6,8:i:enz15 | 10,957 | 10,958.5c | NA |
| Sandiego | 1,4,[5],12:e,h:enz15 | ||||||
| 10,988 | Uncharacterized protein YbgS | 10,988.5c | Heidelberg | 1,4,[5],12:r:1,2 | 10,957 | 10,958.5c | G→S |
| 11,591 | Ribosomal protein L21 | 11,591.8 | Paratyphi A | 1,2,12:a:- | 11,579 | 11,578.4 | V→I |
| 14,381 | Ribosomal protein RL17 | NA | Schleissheim | 4,12,27:b:- | NA | 14,395.6 | NA |
| 14,981 | Ribosomal protein RL15 | NA | London | 3,(10)(15):l,v:1,6 | 14,967 | 14,967.4 | NA |
| 15,342 | Probably DNA binding protein H-NS | NA | Paratyphi B-I | 1,4,[5],12:b:1,2 | 15,412 | 15,412.5b | NA |
| 15,471 | DNA binding protein H-NS | 15,470.5b | Paratyphi A | 1,2,12:a:- | 15,412 | 15,412.5b | G→D |
| 17,432 | Ribosomal protein RS7 | 17,432.1b | Choleraesuis | 6,7:c:1,5 | 17,461/474 | V→A | |
| 18,635 | Putative uncharacterized protein YciF | 18,634.2 | Infantis | 6,7,14:r:1,5 | 18,644/655 | 18,643.7 | R→H/P→S |
| Augustenborg | 6,7,14:i:1,2 | ||||||
| Colindale | 6,7:r:1,7 |
NA, no sequence data available.
Methionine loss.
Signal peptide cleavage.
Table 3.
Mass peaks frequently displaying mass shifts detected by whole-cell MALDI-TOF MS of Salmonella enterica subsp. enterica serovarsa
| Tentative protein identity | Observed mass (Da) | Predicted mass (Da) | Amino acid substitution(s) compared to serovar Typhimurium sequence | Example(s) of prevalent serovars containing signal |
|---|---|---|---|---|
| Uncharacterized protein YhfG | 6,799 | 6,799.8 (M+H)+ | Typhimurium, Virchow | |
| 6,771 | 6,771.8 (M+H)+ | D→S | Enteritidis, Hadar | |
| 6,718 | 6,718.7 (M+H)+ | R→C/V→A | Infantis | |
| 6,757 | NA | NA | Goldcoast | |
| NA | 8,418 | NA | NA | Typhimurium, Virchow, Infantis, |
| 8,445 | NA | NA | Hadar | |
| Putative inner membrane protein STM0348 | 8,686 | 8,687.3 (M+H)+ | Typhimurium, Enteritidis | |
| 8,699 | 8,699.3 (M+H)+ | M→L/A→T | Virchow, Infantis, Hadar | |
| NA | 9,404 | Typhimurium, Virchow, Hadar | ||
| 9,390 | Enteritidis | |||
| 9,374 | Infantis | |||
| Ribosomal protein RL21 | 11,565 | 11,565.4 (M+H)+ | I→V | |
| 11,579 | 11,578.4 (M+H)+ | Typhimurium, Enteritidis, | ||
| Virchow, Hadar, Infantis | ||||
| Putative inner membrane protein YgaM | 11,848 | 11,848.4 (M+H)+ | Typhimurium, Virchow, Hadar | |
| 11,857 | 11,857.5 (M+H)+ | Q→H | Enteritidis, Infantis | |
| Ribosomal protein RS7 | 17,461 | 17,460.2 (M-M+H)+ | Typhimurium, Virchow, Infantis, Hadar | |
| 17,474 | 17,474.2 (M-M+H)+ | D→E | ||
| Putative uncharacterized protein YciF | 18,654 | 18,653.2 (M+H)+ | Typhimurium, Virchow, Hadar | |
| 18,644 | 18,643.2 (M+H)+ | P→S | Enteritidis | |
| 18,635 | 18,634.2 (M+H)+ | R→H | Infantis | |
| Superoxide dismutase (Mn) | 22,949 | 22,948.9 (M-M+H)+ | Typhimurium | |
| 23,012 | 23,010.9 (M-M+H)+ | S→D/L→F | Enteritidis | |
| 29,979 | 22,976.9 (M-M+H)+ | S→D/N | Infantis, Virchow, Hadar |
NA, not assigned.
Fig. 1.
Examples of serovar-identifying biomarker ions found in MALDI-TOF MS spectra of S. enterica subsp. enterica. For specificities and details, see Tables 2 and 3.
Table 4.
Differentiation of serovars Gallinarum and Gallinarum biotype Pullorum based on specific sets of serovar-identifying biomarker ions
| Tentative protein identity | Observed mass (Da) |
Amino acid substitution or posttranslational modification | |
|---|---|---|---|
| Gallinarum biotype Pullorum | Gallinarum | ||
| RL33 | 6,241 | 6,255 | + Methyl |
| YibT | 7,993 | 7,965 | K→T |
| YecF | 8,051 | 8,038 | T→S |
| RS20 | 9,523 | 9,553 | A→V |
| RL25 | 10,542 | 10,572 | A→T |
| RP5 M | 10,830 (only subtype) | 10,858 | NAa |
| RL7/RL12 | NDb | 12,170 | NA |
| Unknown | 12,211 | 12,211 | NA |
| RL18 | 12,771 | 12,776 | NA |
| RS12 | 13,606 | 13,652 | + β-Methylthiolation |
| RS11 | 13,701 | 13,715 | + Methyl |
| RL17 | 14,382 | 14,395 | NA |
| RL16 | 15,194 | 15,225 | Unknown |
| Unknown | 16,306 | 16,306 | NA |
| Superoxide dismutase | 23,011 | 23,045 | L→F |
NA, no sequence data available.
ND, not determined.
Some serovars showed a notable degree of intraserovar variability in their MALDI-TOF profiles. For example, serovar Derby showed at least two MALDI-TOF subtypes, designated Derby-I and Derby-II, of which one could be identified using peaks at m/z 6,302 (instead of 6,316 for ribosomal protein L32) and m/z 7,983 (instead of 7,993 for protein YibT). Serovar Newport could be also divided in different subtypes, one of which was characterized by mass signals at m/z 5,966 and 17,474 (RS7) (mainly avian strains), whereas another contained corresponding signals at m/z 6,009 and 17,461 (Newport-I and Newport-III). Whole-cell MALDI-TOF MS subtypes have been also identified in serovar Paratyphi B d-tartrate positive and Ealing. Serovars Newport, Paratyphi B, and Derby have been already described to be polyphyletic according to both multilocus enzyme electrophoresis (MLEE) and multilocus sequence typing (MLST) studies, while most other Salmonella serovars represent monophyletic lineages (6, 42, 47). Apparently, MALDI-TOF MS subtyping, similar to MLST, reflects different evolutionary lineages for polyphyletic serovars, in contrast to serotyping.
Validation for identification of five frequently isolated Salmonella serovars.
To validate the approach for the identification of the five frequently isolated serovars Enteritidis, Typhimurium, Virchow, Hadar, and Infantis in humans in Europe, 692 various Salmonella strains comprising 89 serovars were analyzed by automated MALDI-TOF MS analyses. The results were compared with traditional serotyping results. The selectivity for the top 5 serovars of the newly developed method is shown in Table 5, including the major discriminative serovar-identifying biomarker ions. Exclusivity was 100% for all five serovars, meaning that none of the nontarget samples was false positively identified. Inclusivity was 100% for serovars Typhimurium, Virchow, and Infantis. In cases of serovars Enteritidis and Hadar, a reduced inclusivity was observed (86.5% and 85.7%), resulting in a reduced selectivity of the method. While 32 strains were correctly identified as serovar Enteritidis (true positives), 5 strains lacked the diagnostic marker ion for serovar Enteritidis at m/z 6,036. Those strains lacked a detectable expression of a set of about 10 to 15 proteins, including some of the diagnostic marker peaks used in this study. This set of proteins included, besides several uncharacterized proteins with unknown functions, the putative homeobox protein YbgS and the hydrophilins YciG, YciF, and YciE and YjbJ, which are known to be highly abundant under certain stress conditions (Table 6). Expression levels of proteins listed in Table 6 varied strongly according to peak intensities observed in whole-cell MALDI-TOF MS. However, when fresh isolates from routine diagnostics were analyzed, absence of these peaks was rarely observed, keeping the number of false negatives in routine diagnostics to a minimum. In addition, it was reproducibly observed that expression of this set of proteins was not detected or barely detectable in some strains when bacterial biomass was taken from single colonies instead of bacterial smears taken from the same agar plate. When bacteria were grown as biofilm-like smears, these proteins usually were more highly expressed, and therefore both sample preparation procedures were routinely performed for MALDI-TOF MS-based Salmonella serovar screening.
Table 5.
Major serovar-identifying peaks used for MALDI-TOF screening of the top 5 serovars and results of selectivity validationa
| Parameter | Enteritidis | Typhimurium | Virchow | Infantis | Hadar |
|---|---|---|---|---|---|
| Antigenic formula | 1,9,12:g,m:- | 1,4,[5],12:i:1,2/1,4,[5],12:i:- | 6,7,14:r:1,2 | 6,7,14:r:1,5 | 6,8:z10:e,n,x |
| Selectivity (%) | 99.3 | 100 | 100 | 100 | 99.1 |
| Exclusivity (%) | 100 | 100 | 100 | 100 | 100 |
| Inclusivity (%) | 86.5 | 100 | 100 | 100 | 85.7 |
| m/z | |||||
| Putative uncharacterized protein | 6,036 | 6,009 | 6,008 | 6,009 | 6,009 |
| Protein Gns | 6,484 | 6,484 | 6,484 | 6,512 | 6,484 |
| Uncharacterized protein YhfG | 6,771 | 6,799 | 6,799 | 6,718 | 6,771 |
| Putative uncharacterized protein YaiA | 7,111 | 7,097 | 7,097 | 7,111 | 7,111 |
| Putative inner membrane protein STM0348 | 8,686 | 8,686 | 8,699 | 8,699 | 8,699 |
| NAa | 9,390 | 9,404 | 9,404 | 9,374 | 9,404 |
| Ribosomal protein RS15 | 10,067 | 10,067 | 10,048 | 10,067 | 10,067 |
| Uncharacterized protein YbgS | 10,958 | 10,958 | 10,958 | 10,958 | 10,927 |
| Putative inner membrane protein YgaM | 11,857 | 11,848 | 11,848 | 11,857 | 11,848 |
| Ribosomal protein RS7 | 17,474 | 17,461 | 17,461 | 17,461 | 17,461 |
| Putative uncharacterized protein YciF | 18,644 | 18,655 | 18,655 | 18,635 | 18,654 |
| Superoxide dismutase SodM (Mn) | 23,012 | 22,949 | 22,979 | 22,979 | 22,979 |
Highly specific biomarker combinations are indicated in boldface.
Table 6.
Major highly upregulated (differentially expressed) proteins (potentially rpoS dependent)
| Tentative protein identity | Observed mass(es) (Da) | Function/regulation | Reference(s)a |
|---|---|---|---|
| Putative cytoplasmic protein (STM1513) | 6,009, 6,036 | YciG paralogue, upregulated in 2-day-old minimal medium, hydrophilin | 40 |
| Putative uncharacterized protein YciG (Q7CQG0_SALTY) | 6,094 | Cotranscribed with YciF, induced in stationary phase, hydrophilin | 40, 53 |
| Putative uncharacterized protein YhfG (YHFG_SALTY) | 6,799, 6,771, 6,718 | Unknown/stationary-phase protein, biofilm-specific pattern of expression | 53 |
| Putative uncharacterized protein YaiA (Q8ZRE9_SALTY) | 7,097, 7,111 | Peroxide induction | 53 |
| Putative periplasmic or secreted protein YahO (Q7CR49_SALTY) | 7,661 | Unknown | 25, 38 |
| Uncharacterized protein YibT (YIBT_SALTY) | 7,993 | Unknown | |
| Putative uncharacterized protein YjbJ (YJBJ_SALTY) | 8,329 | Highly abundant, very high RpoS dependence in stationary phase, induced by osmotic stress, dispensable in E. coli grown in rich or minimal medium, hydrophilin | 50, 53 |
| Putative inner membrane protein (Q8ZRH2_SALTY) | 8,686, 8,699 | Putative inner membrane protein | |
| Putative homeobox/exported protein YbgS (YBGS_SALTY) | 10,927, 10,956 | Function unknown, upregulated in 2-day-old minimal medium, upregulated in biofilm | 38, 53 |
| Inner membrane protein YgaM (Q8ZML4_SALTY) | 11,848, 11,857 | Stress induced, upregulated in biofilm | 53 |
| Putative LysM domain protein YgaU (Q8ZML9_SALTY) | 15,991 | Cell wall catabolic process | 25, 50, 53 |
| Putative uncharacterized protein YciF (Q7CQF9_SALTY) | 18,644, 18,655 | Upregulated in 2-day-old minimal medium, induced by osmotic stress imposed by NaCl, unique to biofilm, putative structural protein, osmotically induced, belongs to the σS regulon | 25, 40, 53 |
| Putative cytoplasmic protein YciE (Q7CQF8_SALTY) | 18,974 | Induced by acid shock | 40 |
Reference(s) describing RpoS dependence under one or more environmental conditions in E. coli or Salmonella for the given protein.
To assess the repeatability of the method, selected strains of each top 5 serovar were tested in series of eight replicates. Furthermore, five strains of each top 5 serovar were reanalyzed after a period of 2 years under the same conditions. The presence and absence of the serovar-identifying biomarker ions have been confirmed in all spectra, and all strains have been reidentified on the serovar level (data not shown).
DISCUSSION
The aim of this study was primarily to evaluate whole-cell MALDI-TOF mass spectrometry as a prescreening tool for rapid identification of epidemiologically important serovars, with emphasis on five frequently isolated Salmonella subsp. enterica serovars in Europe, namely, Enteritidis, Typhimurium, Infantis, Virchow, and Hadar, thereby reducing numbers of samples that have to be analyzed by traditional serotyping in combination with biochemical test reactions.
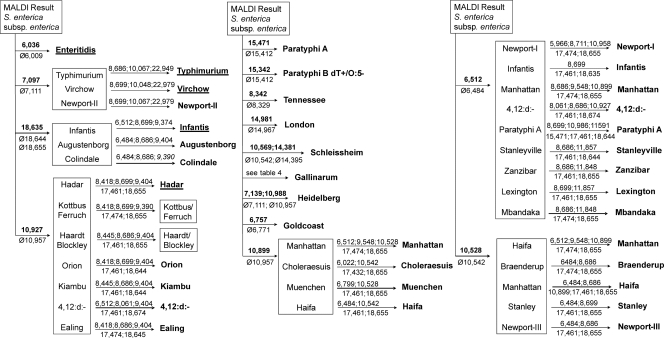
For identification of specific serovars, a hierarchical decision tree network-like approach was developed and initially validated (Fig. 2). It consists of a sequential scanning of mass spectra for the presence or absence of peaks displaying specificities at different taxonomic levels, namely, sets of genus-, species-, subspecies-, or serovar-identifying biomarkers. A future computer-aided identification scheme could be based on such a hierarchical approach, applying suitable algorithms for different taxonomic levels in a sequential mode. Generally, when higher taxonomic resolution is required, as with serovar level identification, a pattern recognition approach would be of limited use, because of the complexity of the peak patterns. Weighted pattern matching approaches could be established using average spectra (superspectra, main spectra) that contain a limited number of selected highly serovar-discriminative peaks, which are given high discriminative values (upweighted peaks) in the identification routines. Variable expression rates of biomarker proteins due to differential regulation, especially of nonhousekeeping proteins, might be observed, possibly leading to false negatives when the biomarker concentration is below the detection limit. As in the case of serovar Enteritidis, some strains reproducibly lacked expression of a subset of proteins under different conditions. Consequently, the obligatory presence of those biomarkers can be regarded as internal control mass peaks. It is interesting that those affected proteins are very likely to be controlled by the global regulator RpoS, which is known to control expression of proteins involved in stress response and virulence (17, 52). Under certain conditions, mutations within rpoS appear to result in a growth advantage in Escherichia coli and Salmonella, leading to a selection of rpoS mutants in bacterial populations (36). Since RpoS frequently is a target for point mutations modifying bacterial fitness, it can be assumed that some of the strains acquired point mutations during passage and storage that led to inhibition of RpoS-regulated proteins. Such strains typically displayed wrinkled colony morphology when grown on agar plates. It has already been reported that cultures of virulent S. enterica serovar Enteritidis can convert from the smooth to a new wrinkled (lacy) colonial phenotype without loss of antigen after storage in nutrient agar stabs for longer periods (20). RpoS mutants were also frequently found from highly passaged laboratory strains of Salmonella (39).
Fig. 2.
Decision tree classification for identification of selected Salmonella enterica subsp. enterica serovars using MALDI-TOF MS. Starting from a MALDI-TOF MS-based identification result (here, at the subspecies level), spectra are screened for the absence/presence of a limited number of serovar-identifying mass peaks. Major serovar-identifying biomarker ions are indicated in bold. Ions absent from respective serovars are indicated by Ø followed by the m/z value. Selected ions useful for further discrimination within groups of serovars (framed) are shown above and below the right set of arrows. Underlined serovars were validated with an extended reference strain collection (Table 5). For detailed information, see Tables 2 and 3.
This method could be a valuable tool when integrated into microbiological routine diagnostics, because prevalent serovars could easily be identified at low costs, while the number of isolates that would have to be subsequently identified by traditional and more laborious methods would be significantly reduced. Several studies have calculated the cost of MALDI-TOF mass spectrometry when used for typing of organisms (7, 10, 11, 44). Generally, they concluded that the technique is rather cost-efficient and rapid compared to other typing methods. We estimated that the average cost for serovar identification using MALDI-TOF MS of a strain is between $6.00 and $10.00, including consumables and personnel costs. In our calculation, this price is at least 3-fold lower than the one for serotyping and biochemical test reactions. However, laboratories need to purchase a MALDI-TOF MS instrument, but the expense of such an instrument is meanwhile comparable to other common laboratory equipment needed for molecular bacterial typing (e.g., sequencers or apparatus for pulsed-field gel electrophoresis [PFGE]). Furthermore, the higher the throughput rate of samples in a laboratory, the lower the costs of the analysis per strain. Sample preparation and duration of analysis times are rather rapid. On average, with the strain cultured as a single colony on an agar plate, MALDI analysis requires only a few minutes, compared to hours or days for other molecular typing methods, such as PFGE.
Whole-cell MALDI-TOF MS generates numerical data (mass peaks). Those data are easy to exchange between laboratories, compared to molecular fingerprinting methods, such as PFGE, which is currently accepted as the gold standard method for bacterial typing (5). Therefore, it can be compared with DNA sequence data handling used, for example, in MLST. However, whole-cell MALDI-TOF MS assesses the allelic variation in multiple genes (mainly housekeeping genes) in a strain by determining variations on protein level, in this case, mass variations. Silent mutations are therefore not assessed and, due to the fact that such variations accumulate very slowly in housekeeping genes, the discriminative power might be generally lower than for MLST.
Nevertheless, apparently MALDI-TOF MS subtyping, as described here, similar to MLST reflects different evolutionary lineages for polyphyletic serovars such as Newport, Paratyphi B, or Derby, in contrast to serotyping, while most other Salmonella serovars represent monophyletic lineages (21, 42, 47). In a future study the potential of bacterial MALDI-TOF MS subtyping compared to DNA-based typing methods needs to be elucidated in more detail.
Since identification is not based on the direct detection of antigenic determinants but on surrogate marker proteins, this opens the possibility to assign rough strains to their respective serotypes by using the MALDI-TOF MS approach. Preliminary experiments in our laboratory indicated the usefulness of the method to allocate rough strains derived from serovars Enteritidis and Typhimurium. Suggesting that approximately 3% of Salmonella enterica subsp. enterica strains received for routine diagnosis in our laboratory are serotyped as rough, MALDI-TOF MS provides a promising advantage in sensitivity compared to serotyping.
In conclusion, the mass spectrometric approach presented here may complement traditional approaches, e.g., as a tool for rapid prescreening of isolates, but the subsequent identification of the majority of serovars will still rely on traditional serotyping. MALDI-TOF MS can function as a prescreening method for Salmonella serovar identification. The identification relies rather on certain specific biomarker identification than on pattern recognition. High selectivity was exemplarily shown for five important serovars. Further studies should focus on other frequently isolated serovars.
ACKNOWLEDGMENTS
We kindly thank the serology team of the NRL-Salmonella for serotyping of Salmonella isolates. We are thankful to Reiner Helmuth for valuable advice and critical reading of the manuscript.
This work was supported by PRO INNO II, grant KF 0350101 MD6, of the Federal Ministry of Economics and Technology, Germany.
Footnotes
Published ahead of print on 22 April 2011.
REFERENCES
- 1. Alispahic M., et al. 2010. Species-specific identification and differentiation of Arcobacter, Helicobacter and Campylobacter by full-spectral matrix-associated laser desorption/ ionization time of flight mass spectrometry analysis. J. Med. Microbiol. 59:295–301 [DOI] [PubMed] [Google Scholar]
- 2. Anonymous. 2009. The Community Summary Report on Trends and Sources of Zoonoses and Zoonotic Agents in the European Union in 2007. EFSA J. 9:2090 [Google Scholar]
- 3. Arnold R. J., Reilly J. P. 1998. Fingerprint matching of E. coli strains with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of whole cells using a modified correlation approach. Rapid Commun. Mass Spectrom. 12:630–636 [DOI] [PubMed] [Google Scholar]
- 4. Barbuddhe S. B., et al. 2008. Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74:5402–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barrett T. J., Gerner-Smidt P., Swaminathan B. 2006. Interpretation of pulsed-field gel electrophoresis patterns in foodborne disease investigations and surveillance. Foodborne Pathog. Dis. 3:20–31 [DOI] [PubMed] [Google Scholar]
- 6. Beltran P., et al. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. U. S. A. 85:7753–7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bizzini A., Durussel C., Bille J., Greub G., Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 48:1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carbonnelle E., et al. 2007. Rapid identification of staphylococci isolated in clinical microbiology laboratories by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 45:2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen P., Lu Y., Harrington P. B. 2008. Application of linear and nonlinear discrete wavelet transforms to MALDI-MS measurements of bacteria for classification. Anal. Chem. 80:7218–7225 [DOI] [PubMed] [Google Scholar]
- 10. Cherkaoui A., et al. 2010. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dhiman N., Hall L., Wohlfiel S. L., Buckwalter S. P., Wengenack N. L. 2011. Performance and cost analysis of MALDI-TOF mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49:1614–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dieckmann R., Alter T., Strauch E. 2010. Rapid identification and characterization of Vibrio species using whole-cell MALDI-TOF mass spectrometry. J. Appl. Microbiol. 109:199–211 [DOI] [PubMed] [Google Scholar]
- 13. Dieckmann R., Helmuth R., Erhard M., Malorny B. 2008. Rapid classification and identification of salmonellae at the species and subspecies levels by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74:7767–7778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Domin M. A., Welham K. J., Ashton D. S. 1999. The effect of solvent and matrix combinations on the analysis of bacteria by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 13:222–226 [DOI] [PubMed] [Google Scholar]
- 15. Echeita M. A., Herrera S., Usera M. A. 2001. Atypical, fljb-negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:i:- appears to be a monophasic variant of serovar typhimurium. J. Clin. Microbiol. 39:2981–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farmer J. J., III 2003. Enterobacteriaceae: introduction and identification, p. 636–653 In Murray P. R., Baron E. J., Jorgensen J. H., Pfaller M. A., Yolken R. H. (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washinghton, DC [Google Scholar]
- 17. Garay-Arroyo A., Colmenero-Flores J. M., Garciarrubio A., Covarrubias A. A. 2000. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem. 275:5668–5674 [DOI] [PubMed] [Google Scholar]
- 18. Gasteiger E., et al. 2005. Protein identification and analysis tools on the ExPASy server, p. 571–607 In Walker J. M. (ed.), The proteomics protocol handbook. Humana Press, Totowa, NJ [Google Scholar]
- 19. Grimont P. A. D., Weill F.-X. 2007. Antigenic formulae of the Salmonella serovars. WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France [Google Scholar]
- 20. Guard-Petter J., Keller L. H., Mahbubur Rahman M., Carlson R. W., Silvers S. 1996. A novel relationship between O-antigen variation, matrix formation, and invasiveness of Salmonella enteritidis. Epidemiol. Infect. 117:219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hauser E., et al. 2010. Pork contaminated with Salmonella enterica serovar 4,[5],12:i:-, an emerging health risk for humans. Appl. Environ. Microbiol. 76:4601–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hayek C. S., Pineda F. J., Doss O. W., III, Lin J. S. 1999. Computer-assisted interpretation of mass spectra. Johns Hopkins APL Tech. Dig. 20:363–370 [Google Scholar]
- 23. Hettick J. M., et al. 2006. Discrimination of intact mycobacteria at the strain level: a combined MALDI-TOF MS and biostatistical analysis. Proteomics 6:6416–6425 [DOI] [PubMed] [Google Scholar]
- 24. Humphrey T. 2000. Public-health aspects of Salmonella infection, p. 245–263 In Way C., Way A. (ed.), Salmonella in domestic animals. CABI Publishing, Oxon, United Kingdom [Google Scholar]
- 25. Ibanez-Ruiz M., Robbe-Saule V., Hermant D., Labrude S., Norel F. 2000. Identification of RpoS (σs)-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:5749–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jarman K. H., et al. 2000. An algorithm for automated bacterial identification using matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 72:1217–1223 [DOI] [PubMed] [Google Scholar]
- 27. Jarman K. H., Wahl K. L. 2005. Development of spectral pattern-matching approaches to matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for bacterial identification. Chem. Anal. 169:153–160 [Google Scholar]
- 28. Karger A., et al. 2011. Determination of serotypes of shiga toxin-producing Escherichia coli isolates by intact cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 77:896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lasch P., et al. 2009. Identification of Bacillus anthracis by using matrix-assisted laser desorption ionization-time of flight mass spectrometry and artificial neural networks. Appl. Environ. Microbiol. 75:7229–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Le Minor L. 1984. Genus III. Salmonella, p. 427–458 In Krieg N. R., Bergey D. H., Holt J. G. (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, MD [Google Scholar]
- 31. Le Minor L., Popoff M. Y., Laurent B., Hermant D. 1986. Characterization of a 7th subspecies of Salmonella: S. choleraesuis subsp. indica subsp. nov. Ann. Inst. Pasteur Microbiol. 137B:211–217 [PubMed] [Google Scholar]
- 32. Le Minor L., Veron M., Popoff M. 1982. The taxonomy of Salmonella. Ann. Microbiol. (Paris) 133:223–243 [PubMed] [Google Scholar]
- 33. Leuschner R. G. K., Beresford-Jones N., Robinson C. 2004. Difference and consensus of whole cell Salmonella enterica subsp. enterica serovars matrix-assisted laser desorption/ionization time-of-flight mass spectrometry spectra. Lett. Appl. Microbiol. 38:24–31 [DOI] [PubMed] [Google Scholar]
- 34. Mandrell R. E., et al. 2005. Speciation of Campylobacter coli, C. jejuni, C. helveticus, C. lari, C. sputorum, and C. upsaliensis by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 71:6292–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mazzeo M. F., et al. 2006. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the discrimination of food-borne microorganisms. Appl. Environ. Microbiol. 72:1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Notley-McRobb L., King T., Ferenci T. 2002. RpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184:806–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parisi D., et al. 2008. Analysis and classification of bacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and a chemometric approach. Anal. Bioanal. Chem. 391:2127–2134 [DOI] [PubMed] [Google Scholar]
- 38. Patten C. L., Kirchhof M. G., Schertzberg M. R., Morton R. A., Schellhorn H. E. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272:580–591 [DOI] [PubMed] [Google Scholar]
- 39. Robbe-Saule V., Algorta G., Rouilhac I., Norel F. 2003. Characterization of the RpoS status of clinical isolates of Salmonella enterica. Appl. Environ. Microbiol. 69:4352–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robbe-Saule V., Coynault C., Ibanez-Ruiz M., Hermant D., Norel F. 2001. Identification of a non-haem catalase in Salmonella and its regulation by RpoS. Mol. Microbiol. 39:1533–1545 [DOI] [PubMed] [Google Scholar]
- 41. Saenz A. J., et al. 1999. Reproducibility of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for replicate bacterial culture analysis. Rapid Commun. Mass Spectrom. 13:1580–1585 [DOI] [PubMed] [Google Scholar]
- 42. Sangal V., et al. 2010. Evolution and population structure of Salmonella enterica serovar Newport. J. Bacteriol. 192:6465–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sauer S., et al. 2008. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS One 3:e2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Seng P., et al. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 45. Siegrist T. J., et al. 2007. Discrimination and characterization of environmental strains of Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). J. Microbiol. Methods 68:554–562 [DOI] [PubMed] [Google Scholar]
- 46. Stackebrandt E., Päuker O., Erhard M. 2005. Grouping myxococci (Corallococcus) strains by matrix-assisted laser desorption ionization time-of-flight (MALDI TOF) mass spectrometry: comparison with gene sequence phylogenies. Curr. Microbiol. 50:71–77 [DOI] [PubMed] [Google Scholar]
- 47. Sukhnanand S., et al. 2005. DNA sequence-based subtyping and evolutionary analysis of selected Salmonella enterica serotypes. J. Clin. Microbiol. 43:3688–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tindall B. J., Grimont P. A. D., Garrity G. M., Euzeby J. P. 2005. Nomenclature and taxonomy of the genus Salmonella. Int. J. Syst. Evol. Microbiol. 55:521–524 [DOI] [PubMed] [Google Scholar]
- 49. Valentine N., Wunschel S., Wunschel D., Petersen C., Wahl K. 2005. Effect of culture conditions on microorganism identification by matrix-assisted laser desorption ionization mass spectrometry. Appl. Environ. Microbiol. 71:58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vijayakumar S. R. V., Kirchhof M. G., Patten C. L., Schellhorn H. E. 2004. RpoS-regulated genes of Escherichia coli identified by random lacZ fusion mutagenesis. J. Bacteriol. 186:8499–8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Z., Russon L., Li L., Roser D. C., Long S. R. 1998. Investigation of spectral reproducibility in direct analysis of bacteria proteins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 12:456–464 [DOI] [PubMed] [Google Scholar]
- 52. Weber A., Kögl S. A., Jung K. 2006. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J. Bacteriol. 188:7165–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weber H., Polen T., Heuveling J., Wendisch V. F., Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Welham K. J., Domin M. A., Ashton D. S. 1999. Solvent and matrix combinations in the quality of matrix-assisted laser desorption/ionization-time of flight mass spectra obtained from microorganisms. Pharm. Pharmacol. Commun. 5:73–78 [Google Scholar]
- 55. Williams T. L., Andrzejewski D., Lay J., Musser S. M. 2003. Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J. Am. Soc. Mass Spectrom. 14:342–351 [DOI] [PubMed] [Google Scholar]
- 56. Wunschel S., et al. 2002. Interlab comparison study of bacterial analysis by MALDI mass spectrometry, p. 581–582 Proc. 50th ASMS Conf. Mass Spectrom. Allied Topics. American Society for Mass Spectrometry, Santa Fe, NM [Google Scholar]
- 57. Zamperini K., et al. 2007. Molecular characterization reveals Salmonella enterica serovar 4,[5],12:i:- from poultry is a variant Typhimurium serovar. Avian Dis. 51:958–964 [DOI] [PubMed] [Google Scholar]