Abstract
Paenibacillus curdlanolyticus B-6 Xyn10D is a xylanase containing a family 3 carbohydrate-binding module (CBM3). Biochemical analyses using recombinant proteins derived from Xyn10D suggested that the CBM3 polypeptide has an affinity for cellulose and xylan and that CBM3 in Xyn10D is important for hydrolysis of insoluble arabinoxylan and natural biomass.
INTRODUCTION
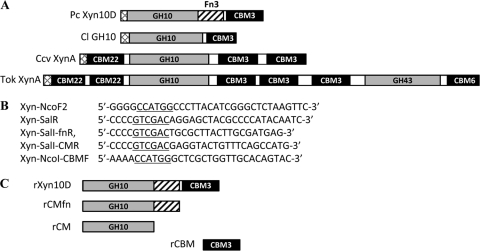
Most catalytic modules of xylanases (endo-1,4-β-xylanase; EC 3.2.1.8) are classified in family 10 or 11 of the glycoside hydrolases, based on the amino acid sequence (http://www.cazy.org/). Many xylanases consist of catalytic and ancillary modules, such as the carbohydrate-binding module (CBM) and the fibronectin type-3 homology (Fn3) module. CBMs are also classified into families based on amino acid sequence similarities. The family 3 CBMs (CBM3s) are often associated with family 9 and 48 catalytic modules or scaffolding proteins that are responsible for cellulosome (cellulolytic complex) assembly in anaerobic bacteria (3). CBM3s are sometimes found in complex modular xylanases, including Caldibacillus cellulovorans XynA (21), Caldicellulosiruptor sp. strain Rt69B.1 XynC (15), and Caldicellulosiruptor strain Tok7B.1 XynA (5) (Fig. 1 A), which contain CBM22s in addition to CBM3s. The xylan-binding activity of CBM22 in C. cellulovorans XynA has been reported (21), but the function of the CBM3s was not investigated. The CBM3 of Clostridium lentocellum putative xylanase (GenBank accession no. ADZ82311) has a simple modular structure (Fig. 1A) but has not been biochemically characterized.
Fig. 1.
Schematic of P. curdlanolyticus Xyn10D and some related xylanases (A), PCR primers for construction of rXyn10D (B), and the truncated derivatives (C). Pc, P. curdlanolyticus; Cl GH10, C. lentocellum putative family 10 glycoside hydrolase; Ccv, C. cellulovorans; Tok, Caldicellulosiruptor strain Tok7B.1; SP, signal peptide; doc, dockerin. NcoI and SalI sites in PCR primers are underlined. rXyn10D, rCMfn, rCM, and rCBM contain Xyn10D residues 32 to 577, 32 to 410, 32 to 317, and 429 to 577, respectively.
(All protein sequences cited in this paper are from the GenBank database.)
Paenibacillus curdlanolyticus strain B-6 is unique in that it produces extracellular xylanolytic-cellulolytic multienzyme complexes, mainly comprising xylanases, under aerobic conditions (17, 18, 22, 23), although the mechanism of multienzyme complex formation is unclear. Four xylanase genes, xyn10A (24), xyn10B (20), xyn10C (unpublished results), and xyn11A (17), were cloned from P. curdlanolyticus strain B-6, and the translated products were characterized. In this study, we cloned xyn10D, encoding a CBM3-containing xylanase (Xyn10D), from this bacterium and characterized the recombinant enzyme and its truncated derivatives.
A genomic library of P. curdlanolyticus strain B-6 constructed using a CopyControl fosmid library production kit (Epicenter, Madison, WI) has been described previously (20). We detected and excluded recombinant bacteria carrying the known xylanase genes by PCR using recombinant fosmid DNA as templates and specific primers. We identified xyn10D (DDBJ/EMBL/GenBank accession number AB600191), which consisted of 1,734 nucleotides (nt) and encoded 577 amino acid residues with a calculated molecular weight of 61,811, including a possible N-terminal signal peptide, as predicted by the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/). Searches using the Pfam database (http://pfam.sanger.ac.uk/) demonstrated that Xyn10D is a modular enzyme consisting of a signal peptide, a family 10 catalytic module of the glycoside hydrolases, an Fn3 module, and a CBM3, in that order, from the N terminus (Fig. 1A). CBM3 of Xyn10D showed high sequence identities to CBMs found in some characterized or hypothetical cellulolytic enzymes of Paenibacillus species, including the family 5 endoglucanase of Paenibacillus sp. KSM-N115 (64%; accession no. BAF62084) (16), Paenibacillus lautus CelA (60%; accession no. AAA22303) (8), and a putative cellobiohydrolase (69%; accession no. EFM08880) of P. curdlanolyticus YK9. It exhibited moderate sequence identities to some CBM3s present in xylanases and in scaffolding proteins that are involved in cellulosome formation, including C. cellulovorans XynA (45%; accession no. AAF61649) (21), Caldicellulosiruptor sp. strain Rt69B.1 XynC (44%; accession no. AAB95326) (15), Caldicellulosiruptor strain Tok7B.1 XynA (42%; accession no. AAD30363) (5), Clostridium thermocellum CipA (42%; accession no. CAA48312) and Clostridium cellulolyticum CipC (44%; accession no. AAC28899). There was a low identity with CBM3 of the putative C. lentocellum xylanase (27%; accession no. ADZ82311). CBM3s have been grouped into three subfamilies, 3a, 3b, and 3c (4, 9). Although the family 3a and 3b CBMs, both of which bind strongly to crystalline cellulose, are closely similar in their primary structures, family 3b CBMs lack a short region. The CBM of Xyn10D is a member of family 3b (see Fig. S1 in the supplemental material).
To investigate the function of CBM3 of Xyn10D, Xyn10D and its truncated derivatives were produced as recombinant proteins (rXyn10D, rCMfn, rCM, and rCBM) (Fig. 1C) by Escherichia coli. Plasmids used for the expression of these proteins were constructed by ligating a PCR fragment amplified using an appropriate combination of primers (Fig. 1B) into pET-28a(+) (Novagen, Madison, WI). E. coli BL21-CodonPlus (DE3)-RIPL (Novagen) cells carrying the recombinant plasmid were cultivated in Super broth (BD, Franklin Lakes, NJ) supplemented with chloramphenicol (50 μg/ml) and kanamycin (25 μg/ml) at 37°C. Cell extracts prepared by sonication were used for purification by a HisTrap HP column (GE Healthcare Japan, Tokyo, Japan). All purified proteins resulted in a single band following SDS-polyacrylamide gel electrophoresis (PAGE) (data not shown) (11).
Hydrolytic activities of the recombinant enzymes were measured with soluble xylans of different origins, such as rye arabinoxylan (Megazyme, Wicklow, Ireland), birchwood xylan (Sigma-Aldrich Japan, Tokyo, Japan), and 4-O-methyl-d-glucrono-d-xylan (Sigma-Aldrich Japan) by incubating for 10 min at 35°C in 50 mM sodium phosphate buffer (pH 7.0 for rCM, pH 6.0 for rXyn10B and rCMfn), followed by measuring reducing sugars released from the substrates by the 3,6-dinitrophthalic acid reagent (12). Enzyme assays were done in triplicate, and results were expressed as micromoles of reducing sugars/min/micromole of protein. rXyn10D was most active toward rye arabinoxylan and moderately active toward birchwood and oat spelt xylans (Sigma-Aldrich Japan, Tokyo, Japan) (Table 1). Derivatives of Xyn10D devoid of CBM, rCMfn, and rCM showed the same tendency in catalytic activities toward different xylans. They were most active toward rye arabinoxylan. These results suggest that Xyn10D and its derivatives prefer highly arabinosylated xylan to less substituted xylans since rye arabinoxylan is more arabinosylated than oat spelt xylan (19). However, since rXyn10D hydrolyzed xylo-oligosaccharides (data not shown), it does not necessarily require arabinose residues for substrate recognition. Specific activities of these enzymes were similar (Table 1). These results suggest that the presence of CBM3 in rXyn10D does not affect the catalytic activity toward soluble substrates. In contrast, when insoluble wheat arabinoxylan was used as a substrate, rXyn10D showed higher activity toward the insoluble substrate than the truncated enzymes (Table 1). It should be noted, furthermore, that rXyn10D hydrolyzed agricultural waste, such as rice straw, rice husk, and rice bran, more efficiently than the truncated enzymes (Table 2). These results strongly suggest that CBM3 in rXyn10D plays an important role in hydrolysis of insoluble xylan and native biomass materials. Although Kataeva et al. (10) found that the Fn3 repeat from the C. thermocellum cellobiohydrolase CbhA promoted hydrolysis of cellulose by modifying its surface, the function of the Fn3 module in Xyn10D is unclear, because activities of rCMfn and rCM were similar toward xylans from different origins (Table 1).
Table 1.
Activity of rCM, rCMfn, and rXyn10D toward xylans from different origins
| Enzyme | Activity (μmol/min/μmol protein ± SD) toward: |
||||
|---|---|---|---|---|---|
| Rye arabinoxylana | Birchwood xylana | 4-O-Methyl-d-glucrono-d-xylana | Oat spelt xylana | Wheat arabinoxylan (insoluble)b | |
| rXyn10D | 26,500 ± 411 | 7,470 ± 98.0 | 9,110 ± 247 | 6,880 ± 0.00 | 2,470 ± 145 |
| rCMfn | 29,600 ± 408 | 8,330 ± 312 | 10,500 ± 478 | 6,040 ± 255 | 1,760 ± 100 |
| rCM | 31,200 ± 283 | 8,190 ± 170 | 10,400 ± 94.3 | 7,090 ± 233 | 1,770 ± 37.4 |
Substrate concentration, 0.5%.
Substrate concentration, 1.0%.
Table 2.
Activity of rXyn10D, rCMfn, and rCM toward agricultural wastes
| Enzyme | Activity (μmol/min/μmol protein ± SD) towardc: |
||
|---|---|---|---|
| Rice strawa | Rice huskb | Rice branb | |
| rXyn10D | 32 ± 1.5 | 20 ± 0.5 | 91 ± 1.5 |
| rCMfn | 10 ± 7.5 | 2.0 ± 0.0 | ND |
| rCM | ND | ND | 26 ± 6.5 |
Substrate concentration, 2.5%.
Substrate concentration, 5.0%.
ND, not detectable.
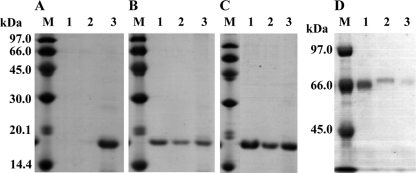
The binding of rCBM to Avicel and insoluble fractions of oat spelt xylan and wheat arabinoxylan was determined by mixing rCBM (88 mg) and insoluble polysaccharides (100 mg) in 50 mM sodium phosphate buffer (pH 7.0) and then incubating on ice for 1 h with occasional stirring. After centrifugation, the supernatant fraction was recovered, and the precipitate was resuspended in sodium phosphate buffer. The suspension was centrifuged to separate wash and precipitate fractions, which were all analyzed by SDS-PAGE. Almost all protein coprecipitated with insoluble cellulose (Avicel), and very little protein was detected in the supernatant and wash fractions (Fig. 2 A), indicating that rCBM was tightly bound. The rCBM moderately bound to the insoluble fraction of oat spelt xylan (Fig. 2B) and wheat arabinoxylan (Fig. 2C). The control protein (bovine serum albumin [BSA]) only slightly bound to xylan (Fig. 2D).
Fig. 2.
Adsorption of rCBM to Avicel and insoluble xylan preparations. rCBM was incubated with Avicel (A), an insoluble fraction of oat spelt xylan (B), and insoluble wheat arabinoxylan (C). After centrifugation, proteins in the supernatant (lane 1), wash (lane 2), and precipitate (lane 3) fractions were analyzed by SDS-PAGE. In panel D, BSA as a control protein was incubated with an insoluble fraction of oat spelt xylan, and each fraction was subjected to SDS-PAGE. In panels A to C, 15% polyacrylamide gels were used. In panel D, a 7.5% polyacrylamide gel was used.
The affinity of rCBM for rye arabinoxylan, birchwood xylan, and oat spelt xylan was examined by native affinity PAGE as described previously (2). Protein samples (2.4 μg) were loaded onto gels, and BSA was used as a control. Migration of the proteins was significantly retarded by inclusion of rye arabinoxylan and birchwood xylan in gels and less affected by oat spelt xylan (Fig. 3). Affinity of rCBM for xylans was in contrast to the observations for CBMs from C. thermocellum CipB (13) and Clostridium cellulovorans CbpA (7); that is, the latter CBMs showed no affinity for xylan.
Fig. 3.
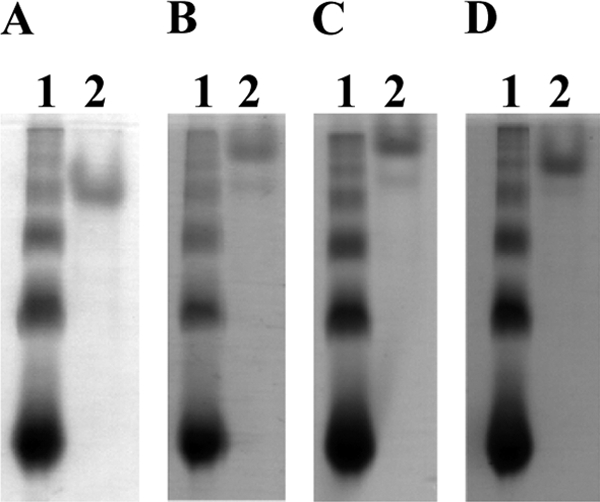
Adsorption of rCBM to soluble xylans of different origins. Affinities of rCBM (lane 2) for rye arabinoxylan (B), birchwood xylan (C), and oat spelt xylan (D) were analyzed by native affinity gel electrophoresis. Lane 1 contains BSA as a control protein. (A) A gel without a polysaccharide served as a reference.
Importance of CBMs in hydrolysis of xylan and complex substrates has been sometimes found in modular enzymes and enzyme complex (1, 6, 14). It is interesting that CBM3 has an affinity for xylan and plays an important role in hydrolysis in soluble substrates. Structural analysis of the CBM of Xyn10D should be carried out to determine substrate recognition by CBM3.
Supplementary Material
Acknowledgments
This study was supported in part by the joint program in the field of biotechnology under Japan Society for the Promotion of Science (JSPS), National Research Council of Thailand (NRCT), and National Science and Technology Development Agency of Thailand (NSTDA).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Ali M. K., et al. 2001. Importance of the carbohydrate-binding module of Clostridium stercorarium Xyn10b to xylan hydrolysis. Biosci. Biotechnol. Biochem. 65:41–47 [DOI] [PubMed] [Google Scholar]
- 2. Arai T., et al. 2003. Characterization of a cellulase containing a family 30 carbohydrate-binding module (CBM) derived from Clostridium thermocellum CelJ: importance of the CBM to cellulose hydrolysis. J. Bacteriol. 185:504–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bayer E. A., Lamed R., White B. A., Flint H. J. 2008. From cellulosome to cellulosomics. Chem. Rec. 8:364–367 [DOI] [PubMed] [Google Scholar]
- 4. Bayer E. A., Morag E., Lamed R., Yaron S., Shoham Y. 1998. Cellulosome structure: four-pronged attack using biochemistry, molecular biology, crystallography and bioinformatics, p. 39–65 In Claeyssens M., Nerinckx W., Piens K. (ed.), Carbohydrases from Trichoderma reesei and other microorganisms. Royal Society of Chemistry, London, United Kingdom [Google Scholar]
- 5. Gibbs M. D., et al. 2000. Multidomain and multifunctional glycosyl hydrolases from the extreme thermophile Caldicellulosiruptor isolate Tok7B.1. Curr. Microbiol. 40:333–340 [DOI] [PubMed] [Google Scholar]
- 6. Gill J., et al. 1999. The type II and X cellulose-binding domains of Pseudomonas xylanase A potentiate catalytic activity against complex substrates by a common mechanisms. Biochem. J. 342:473–480 [PMC free article] [PubMed] [Google Scholar]
- 7. Goldstein M. A., et al. 1993. Characterization of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A. J. Bacteriol. 175:5762–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hansen C. K., Diderichsen B., Jørgensen P. L. 1992. celA from Bacillus lautus PL236 encodes a novel cellulose-binding endo-beta-1,4-glucanase. J. Bacteriol. 174:3522–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jindou S., et al. 2006. Novel architecture of family-9 glycoside hydrolases identified in cellulosomal enzymes of Acetivibrio cellulolyticus and Clostridium thermocellum. FEMS Microbiol. Lett. 254:308–316 [DOI] [PubMed] [Google Scholar]
- 10. Kataeva I. A., et al. 2002. The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl. Environ. Microbiol. 68:4292–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 12. Miller G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426–428 [Google Scholar]
- 13. Morag E., et al. 1995. Expression, purification, and characterization of the cellulose-binding domain of the scaffoldin subunit from the cellulosome of Clostridium thermocellum. Appl. Environ. Microbiol. 61:1980–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moraïs S., et al. 2010. Contribution of a xylan-binding module to the degradation of a complex cellulosic substrate by designer cellulosomes. Appl. Environ. Microbiol. 76:3787–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morris D. D., Gibbs M. D., Ford M., Thomas J., Bergquist P. L. 1999. Family 10 and 11 xylanase genes from Caldicellulosiruptor sp. strain Rt69B.1. Extremophiles 3:103–111 [DOI] [PubMed] [Google Scholar]
- 16. Ogawa A., et al. 2007. Endoglucanases from Paenibacillus spp. form a new clan in glycoside hydrolase family 5. J. Biotechnol. 129:406–414 [DOI] [PubMed] [Google Scholar]
- 17. Pason P., et al. 2010. Purification and characterization of a multienzyme complex produced by Paenibacillus curdlanolyticus B-6. Appl. Microbiol. Biotechnol. 85:573–580 [DOI] [PubMed] [Google Scholar]
- 18. Pason P., Kyu K. L., Ratanakhanokchai K. 2006. Paenibacillus curdlanolyticus strain B-6 xylanolytic-cellulolytic enzyme system that degrades insoluble polysaccharides. Appl. Environ. Microbiol. 72:2483–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pastell H., Tuomainen P., Virkki L., Tenkanen M. 2008. Step-wise enzymatic preparation and structural characterization of singly and doubly substituted arabinoxylo-oligosaccharides with non-reducing end terminal branches. Carbohydr. Res. 343:3049–3057 [DOI] [PubMed] [Google Scholar]
- 20. Sudo M., Sakka M., Kimura T., Ratanakhanokchai K., Sakka K. 2010. Characterization of Paenibacillus curdlanolyticus intracellular xylanase Xyn10B encoded by the xyn10B gene. Biosci. Biotechnol. Biochem. 74:2358–2360 [DOI] [PubMed] [Google Scholar]
- 21. Sunna A., Gibbs M. D., Bergquist P. L. 2000. A novel thermostable multidomain 1,4-beta-xylanase from ‘Caldibacillus cellulovorans’ and effect of its xylan-binding domain on enzyme activity. Microbiology 146:2947–2955 [DOI] [PubMed] [Google Scholar]
- 22. Waeonukul R., Kyu K. L., Sakka K., Ratanakhanokchai K. 2008. Effect of carbon sources on the induction of xylanolytic-cellulolytic multienzyme complexes in Paenibacillus curdlanolyticus strain B-6. Biosci. Biotechnol. Biochem. 72:21–328 [DOI] [PubMed] [Google Scholar]
- 23. Waeonukul R., Kyu K. L., Sakka K., Ratanakhanokchai K. 2009. Isolation and characterization of a multienzyme complex (cellulosome) of the Paenibacillus curdlanolyticus B-6 grown on Avicel under aerobic conditions. J. Biosci. Bioeng. 107:610–614 [DOI] [PubMed] [Google Scholar]
- 24. Waeonukul R., et al. 2009. Cloning, sequencing, and expression of the gene encoding a multidomain endo-β-1,4-xylanase from Paenibacillus curdlanolyticus B-6, and characterization of the recombinant enzyme. J. Microbiol. Biotechnol. 19:277–285 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.